Abstract
Kainate receptors are allosterically regulated by sodium ions. Removal of Na+ from the extracellular solution, or replacement of Na+ by larger monovalent cations, inhibits kainate receptor activity. Sodium binds at a negatively charged cavity in the extracellular neurotransmitter binding domain that is capped by a small hydrophobic residue. Prior work revealed that introduction of aspartic acid at this site strongly reduces GluR6 sensitivity to monovalent cations of different size. We found that the GluR6 M739D mutant is also insensitive to substitution of Na+ by the large organic cations Tris and NMDG. Because these are excluded from the Na+ binding site by steric hindrance, we investigated the possibility that divalent cations can substitute for Na+. We show that in Na+ free solutions with low concentrations of Ca2+ and Mg2+ the GluR6 M739D mutant is inhibited by EDTA; that divalent ions in the micromolar concentration range act as positive allosteric modulators; and that the chemistry of the mutant cation binding site is typical of Ca2+ and Mg2+ binding sites found in protein crystal structures. Hence, the apparent insensitivity of the M739D mutant to monovalent cations is due to the adventitious allosteric effects of divalent ions at physiological concentrations and below.
Keywords: Glutamate Receptor, Cation binding site, Calcium, Magnesium
1. Introduction
Glutamate receptor ion channels mediate excitatory neurotransmission at fast chemical synapses in the brain. The subtypes are named for their selective agonists, NMDA, AMPA and kainate (Watkins and Jane, 2006), and form a family with diverse functions, which is however encoded by genes for proteins with similar structures (Hollmann and Heinemann, 1994). NMDA and AMPA receptors have well defined roles, underpinning the slow and fast components of the classical excitatory synaptic current. Kainate receptors are less well understood, being found at both pre- and post-synaptic sites (Chittajallu et al., 1996), and give rise to synaptic currents that can be as brief as those of AMPA receptors (DeVries, 2000), or as long lived as those due to NMDA receptors (Kidd and Isaac, 1999).
Glutamate receptor ion channels are tetrameric proteins, the extracellular domains of which assemble as two pairs of dimers. Each subunit contains an extracellular ligand binding core shaped like a clamshell, with two domains, D1 and D2, forming the jaws (Armstrong et al., 1998). When glutamate binds, the clamshell closes through rotation of D2. A critical inter-subunit interface is formed between D1 in the resting and open state of the receptor (Sun et al., 2002) and permits clamshell closure to exert a lateral force on the membrane ion channel and open it. In the AMPA subtype of glutamate receptor that undergoes profound desensitization, the D1 dimer interface is ruptured in the desensitized state (Armstrong et al., 2006), and it is likely that this also occurs in kainate receptors. The resulting relaxed conformation of the dimer binds agonist but cannot do work on the pore. As a consequence, the stability of the D1 interface determines the mean lifetime of receptor activation in the presence of glutamate (Horning and Mayer, 2004; Sun et al., 2002). The D1 dimer interface is also a hotspot for alternative splicing and RNA editing in AMPA receptors (Greger et al., 2006; Lomeli et al., 1994; Mosbacher et al., 1994) and the binding site for allosteric modulators like cyclothiazide, aniracetam and CX-614 (Jin et al., 2005; Sun et al., 2002).
Like many other ion channels, glutamate receptor activity is modified by numerous endogenous small molecules and ions. Magnesium binds within the pore of NMDA receptors, and the resultant voltage-dependent block (Mayer et al., 1984; Nowak et al., 1984) allows Hebbian coincidence detection critical to synaptic plasticity. Intracellular polyamines bind in the pore of unedited non-NMDA receptors (Bowie and Mayer, 1995; Kamboj et al., 1995) and confer rapid, activity-dependent changes of AMPA receptor activity at synapses (Rozov et al., 1998). Modulatory mechanisms for ions that act directly or indirectly through the ligand binding core dimer were discovered more recently. In kainate receptors a chloride ion is buried in the D1 dimer interface in the active conformation (Plested and Mayer, 2007), and is flanked by two sodium binding sites (Plested et al., 2008), which in combination permit allosteric control of receptor activity by these ions. The glutamate receptor delta-2 subtype is modulated by calcium (Wollmuth et al., 2000), and Ca2+ ions bind to the dimer interface in a similar location to allosteric monovalent cations in kainate receptors (Naur et al., 2007). Zinc binds to the amino terminal domain of NMDA-receptors, but controls the activity of the NMDA-receptor pore at a distance by altering the configuration of the ligand binding core dimer (Gielen et al., 2008).
In the course of our experiments on kainate receptor cation binding sites we examined the GluR6 M739D mutant that lacks the strong sensitivity to replacement of Na+ by K+ or Cs+ observed for wild-type channels (Wong et al., 2007). We aimed to understand this apparent insensitivity in terms of the chemistry of the binding site. We examined a range of probable causes and present evidence that the mutation converts the monovalent cation-preferring site of wild-type receptors into one that prefers divalent ions.
2. Results
2.1 The M739D mutant is insensitive to monovalent cation exchange
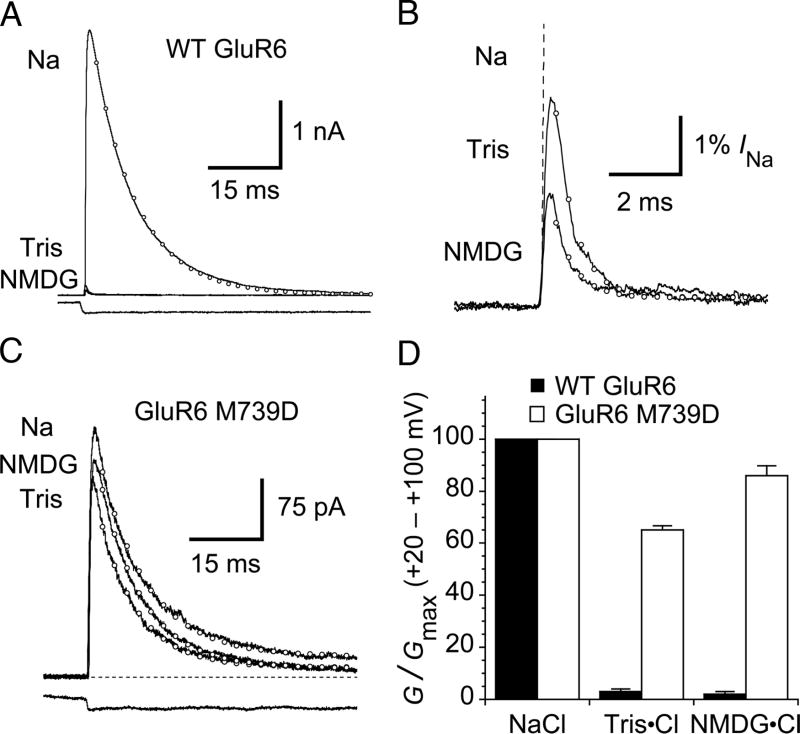
With few exceptions, mutation of amino acids that form the binding sites for allosteric anions and cations in kainate receptors speeds desensitization (Paternain et al., 2003; Plested and Mayer, 2007; Wong et al., 2007; Plested et al., 2008). In contrast, the GluR6 M739D mutant shows minimal difference from wild type, with time constants for desensitization in the presence of 150 mM NaCl of 5.8 and 5.5 ms, respectively (Wong et al., 2007). Surprisingly, in contrast to the extreme sensitivity of WT GluR6, responses to glutamate for the M739D mutant showed minimal change in amplitude or desensitization kinetics when extracellular Na+ was replaced by Tris or NMDG (Figure 1). At + 60 mV, with 150 mM Na+, wild-type GluR6 currents desensitize with rate of 120 ± 9 s−1. Under these conditions, the rate of desensitization of the GluR6 M739D mutant is about 1.3-fold slower (89 ± 7 s−1, n = 11), identical to the slow desensitization asymptote of 91 s−1 for wild-type GluR6 in 600 mM NaCl, which we showed previously was a sufficient concentration to saturate the Na+ binding site (Plested et al., 2008). When Na+ was replaced by the organic cations Tris or NMDG, the slope conductance for wild type GluR6 at positive potentials decreased to 3 ± 1 % and 2 ± 1% of that in 150 mM NaCl (n = 5), and the rate of desensitization approached the fast asymptote of ~1800 s−1 measured when sucrose was used to replace Na+ (Plested et al., 2008). In contrast, for the GluR6 M739D mutant, responses recorded with Tris or NMDG as the major external cation, showed only small changes in amplitude (Figure 1). These measurements, by necessity, were performed at +ve membrane potentials because Tris and NMDG act as weakly permeant channel blockers for GluR6 (Burnashev et al., 1996; Bähring et al, 1997).
Figure 1. GluR6 M739D mutant is relatively insensitive to NMDG and Tris.
(A) In normal saline (Na), outward currents at + 60 mV due to wild-type GluR6 receptors activate rapidly in response to a 100 ms pulse of 10 mM glutamate and then desensitize (open circles represent a monoexponential fit, kdes = 110 s−1). In the same patch, when external sodium is replaced by either Tris or N-methyl-D-glucamine (NMDG), outward currents are strongly inhibited. The lower trace shows the solution exchange into glutamate, measured after the experiment.
(B) Expanded view of the current response in NMDG and Tris at +60 mV. The response in Na is shown as a dashed line. Monoexponential fits are shown with open circles. The rate of desensitization is ~ 15-fold faster (in Tris, 1600 ± 400 s−1 and in NMDG, 1600 ± 100 s−1 n = 5) than in sodium-containing solutions.
(C) The desensitization and peak current of the GluR6 M739D mutant is much less sensitive to substitution of sodium with NMDG or Tris. Monoexponential fits are shown as open circles (in this patch, Na kdes = 99 s−1; NMDG kdes = 110 s−1; Tris kdes = 124 s−1). The lower trace is the solution exchange measured after the experiment.
(D) Bar plot indicating inhibition of the slope conductance at positive potentials for wild-type and M739D receptors. The limited inhibition of the M739D mutant by NMDG and Tris probably derives in part from channel block. Data are mean ± SEM for five patches.
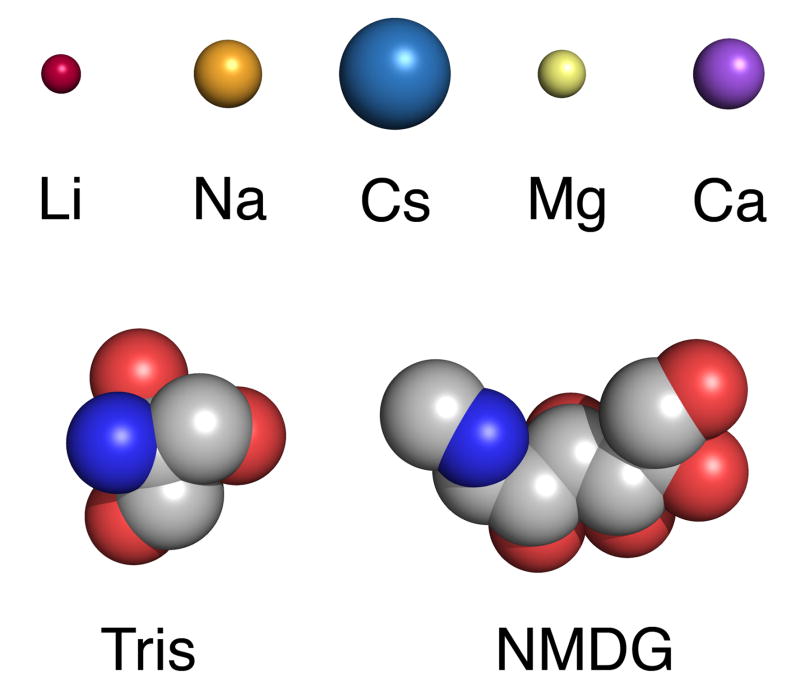
The GluR6 M739D mutation also greatly reduces the increase in rate of desensitization which occurs when external Na+ is replaced by K+ or Cs+ (Wong et al., 2007). Crystal structures of GluR5 solved for different ion complexes reveal that all of these ions can bind to wild-type kainate receptors, but with different affinities (Plested et al., 2008). Thus it is possible that the M739D mutation increases the binding affinity of these ions (Na, K and Cs), but this would not explain why the M739D mutant is insensitive to replacement of Na+ by Tris or NMDG. It is very unlikely that Tris or NMDG can bind to the site for allosteric cations since they are much too large to enter the funnel-shaped cavity which binds Na+ (Figure 2). This leaves open the possibility that divalent cations act as positive allosteric modulators for GluR6 M739D mutant receptors, even though in our experiments Ca2+ and Mg2+ were each present at a concentration of only 100 μM in the NMDG and Tris substituted solutions, which are nearly as effective as 150 mM Na+.
Figure 2. Organic cations and monovalent and divalent metal cations used in this study.
Alkali earth metal ions that bind in the kainate receptor cation binding site, Magnesium and Calcium are shown as spheres with radii according to Shannon (Li, 0.59Å; Na, 1.02 Å; Cs, 1.67 Å; Mg, 0.72 Å; Ca, 1.06 Å). Tris and N-methyl-D-glucamine are drawn in CPK representation (see Methods).
2.2 The M739D mutant creates a binding site for divalent cations
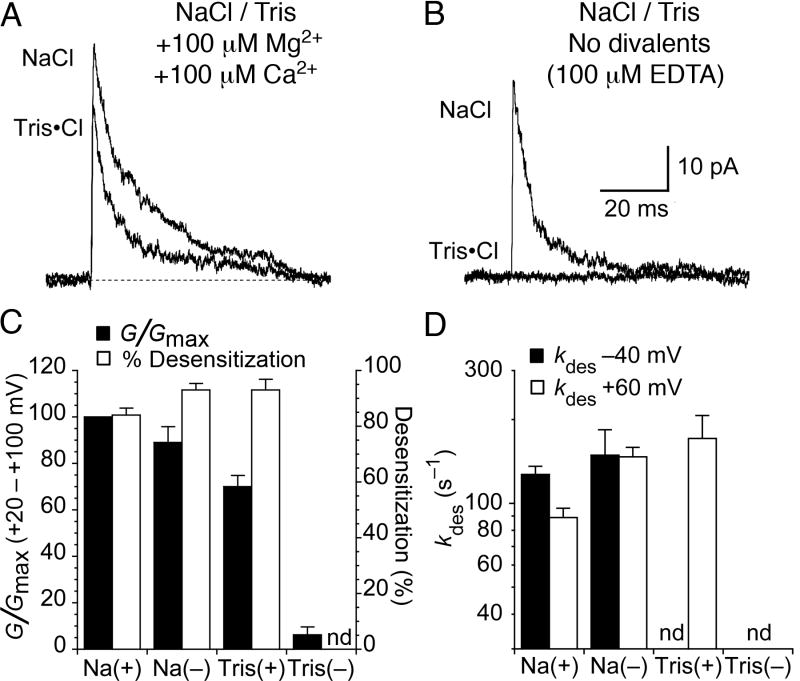
Because our prior structural studies excluded the possibility that NMDG and Tris can bind to the allosteric cation site for Na+, we considered the possibility that divalent cations present in the extracellular solution were responsible for maintaining function of the GluR6 M739D mutant. Our standard recording conditions, as well as those of Wong et al., (2007), contained 100 μM each Ca2+ and Mg2+ to facilitate stable patch recordings. To test whether these low concentrations of divalent cations can substitute for Na+ in the binding site of the GluR6 M739D mutant we recorded currents from the same patch in the presence of either 150 mM external Na+ or 150 mM Tris, with either 100 μM each of Ca2+ or Mg2+, or no added divalent ions and 100 μM EDTA to chelate contaminant divalent cations (Figure 3). We found that with 150 mM Na+ the rate of desensitization of the M739D mutant was mildly voltage-sensitive in the presence of 100 μM each Ca2+ and Mg2+, kdes 130 ± 10 s−1 at −40 mV, and 89 ± 7 s−1 at +60 mV (n = 5-11 patches). The faster onset of desensitization at -40 mV most likely is a result of the channel block by divalent ions at negative potentials. Glutamate-evoked responses in 150 mM Na+ for the GluR6 M739D mutant were not greatly changed when divalent cations were chelated by EDTA, suggesting that at 150 mM the cation binding site of the M739D mutant is occupied by Na+ instead of Ca2+ or Mg2+ (Figure 3). The slope conductance at positive potentials was 89 ± 7 % of control in the presence of divalent cations, and desensitization was faster but no longer voltage sensitive, kdes 150 ± 40 s−1 at −40 mV, and 150 ± 10 s−1 at +60 mV (n = 5), consistent with our proposal that divalent cations act as channel blockers. However, in Tris/EDTA solutions the amplitude of the current activated by 10 mM glutamate was reduced to 6 ± 3 % of control (n=5). This strongly suggests that the M739D creates a high-affinity binding site for divalent cations, and that large organic cations such as Tris cannot bind to this site.
Figure 3. The GluR6 M739D mutant is modulated by divalent cations.
(A) Glutamate activated outward currents recorded in the same patch, in control NaCl solution, and when Na+ was replaced by Tris. In contrast to the near complete loss of response for wild type GluR6, the rate of onset of desensitization increased only 1.6-fold, and the peak amplitude decreased by only 30 ± 5 %. Both solutions contained 100 μM Ca2+ and 100 μM Mg2+.
(B) In the same patch as in (A), when Ca2+ and Mg2+ were omitted and trace divalent cations chelated with 100 μM EDTA, the removal of divalent ions had little effect on the outward current in 150 mM NaCl, but resulted in near-total abolition of responses recorded with Tris as the external cation (6 ± 3% of control, n = 5).
(C) Bar plot showing the effect of 100 μM Ca2+ and 100 μM Mg2+ (+) and EDTA (-) on slope conductance and %desensitization for responses recorded with either Na or Tris. Steady-state desensitization in Tris in the absence of divalent cations was not determined (nd) because the residual current was too small to measure. Data shown from six patches; error bars indicate SEM.
(D) Consistent with channel block by divalent cations, desensitization of the M739D mutant was slower at positive potentials for responses with 100 μM Ca2+ and 100 μM Mg2+ present (Na+). The rates of desensitization were the same at −40mV and at +60 mV when divalent cations were chelated with EDTA (Na−). We did not observe appreciable inward currents in external Tris, and the outward current in Tris without divalent cations was too small to reliably determine the desensitization rate. Bars represent the mean ± SEM for five to eleven patches.
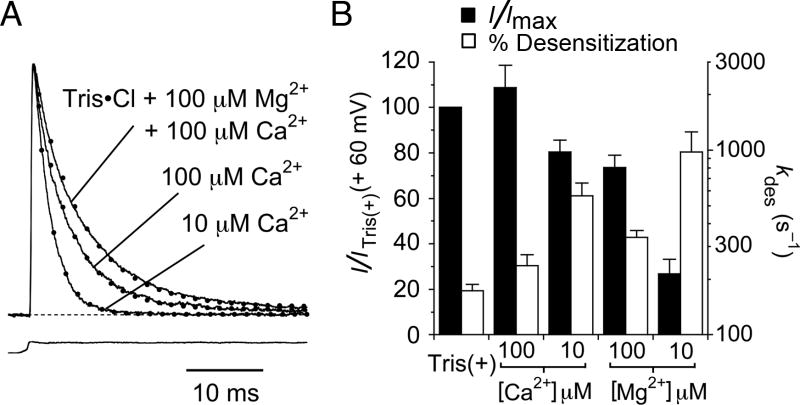
We investigated the selectivity of the novel divalent cation binding site in the GluR6 M739D mutant by measuring responses in buffered solutions containing 10 to 100 μM 7 free Ca2+ or Mg2+. We found that both divalent ion species could support kainate receptor activity. Ca2+ was more potent than Mg2+ and even 10 μM Ca2+ robustly supported function of the M739D mutant (Figure 4). Although Mg2+ was less efficacious than Ca2+ (Figure 4B) overall the M739D mutant is only weakly selective for Ca2+. Our results suggest that both Ca2+ and Mg2+ ions can bind in the engineered cation binding site and stabilize the dimer interface in the same way that sodium does in wild-type GluR6, and that binding of divalent cations to the M739D mutant is approximately three orders of magnitude tighter than the binding of Na+ to the wild-type receptor.
Figure 4. The GluR6 M739D mutant is preferentially binds calcium.
(A) Normalized outward currents for the M739D mutant recorded from the same patch with 100 μM each Ca2+ and Mg2+ (kdes = 154 s−1); 100 μM Ca2+ (kdes = 212 s−1); or 10 μM Ca2+ (kdes = 430 s−1) with 150 mM Tris present in all solutions. EDTA was included to chelate other trace divalent ions, and buffer free calcium for the responses measured in 10 and 100 μM Ca2+. Dotted lines represent single exponential fits.
(B) Bar plots summarizing changes in peak amplitude and desensitization rate for responses recorded with either Ca2+ or Mg2+ at concentrations of 10 and 100 μM indicate a weak preference for Ca2+. Bars show the means of 5 observations, error bars represent SEM.
3. Discussion
Sodium and chloride ions both bind to the ligand binding core dimer of kainate receptors, stabilizing the active state of the receptor and therefore slowing desensitization. Crystallographic and functional experiments have elucidated the chemistry of the ion binding sites and the mechanism by which ion binding stabilizes kainate receptor ligand binding core dimers (Plested and Mayer, 2007; Plested et al., 2008). The results provide a rationalization for the role M739 in GluR6 in the loop preceding helix J that caps the allosteric sodium binding site in kainate receptors and the substitution of Lys at this position delineates between AMPA and kainate receptors (Paternain et al., 2003; Wong et al., 2006; Wong et al., 2007; Plested et al., 2008).
In kainate receptors, this residue is an isoleucine in GluR5, a methionine in GluR6 and GluR7, a leucine in KA1 and a valine in KA2. For GluR6, replacement of this residue by arginine or lysine accelerates the rate of onset of desensitization, but abolishes cation sensitivity, suggesting that the basic amino acid side chain acts as a tethered cation (Wong et al., 2007), which however is less efficacious than Na+. Consistent with this, a similar effect is observed for wild type GluR6 when NH4+ is substituted for Na+ (Plested et al., 2008). The tethered cation hypothesis is also supported by the ability of NH4+ to bind at the same site as Na+ and Li+ (Plested et al., 2008), by the structure of the GluR5 UBP310 complex (Mayer et al., 2006) in which a lysine side chain plugs the cation binding site, and by the insensitivity of AMPA receptors, which have a lysine residue at this position, to cation exchanges (Bowie, 2002; Paternain et al., 2003). Although these observations explained the ability of basic side chains to act as surrogate cations, the apparent insensitivity of mutants with acidic substitutions, GluR6 M739D and GluR6 M739E (Wong et al., 2007) was perplexing. We have now shown that the M739D mutant is not insensitive to cations, and that it can function with either small monovalent cations, or divalent ions, provided that chloride is also present.
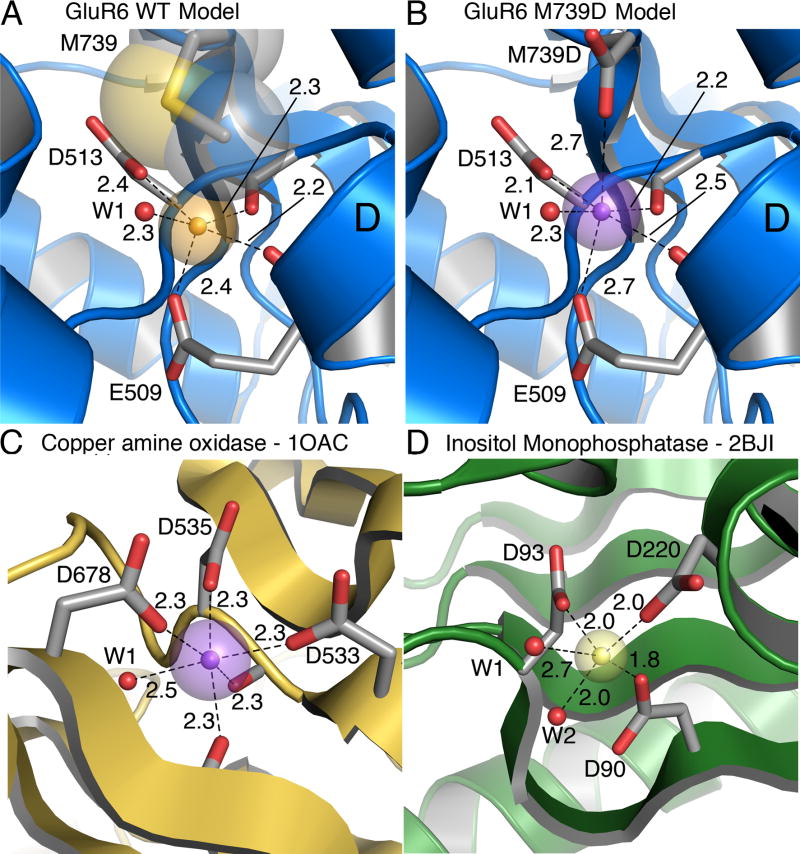
Attempts to express and crystallize the GluR5 I755D mutant protein for structural studies were not successful. To gain insight into the novel chemistry of the cation binding site we replaced Ile755 in the crystal structure for cation binding site of the GluR5 Na+ complex (PDB 3C32) with a methionine to model wild type GluR6 (Figure 5A), and for the M739D mutant replaced this with an aspartate rotamer from the crystallographic building program COOT (Figure 5B). Rotation of the χ1 angle of the aspartic acid side chain by 20° brought one of the side chain carboxylate oxygen atoms to its furthest excursion into the cation binding site. Solvent exposure of the mutant carboxyl group reduces its influence on surface potential compared to that generated by Glu509 and Asp513, both which are located deeper in the cation binding pocket (Plested et al., 2008), and thus it is unlikely that the increased affinity for divalent cations results primarily from electrostatic effects. Instead, introduction of an additional aspartate group at the entrance of the cation binding creates an environment typical of that found in high resolution crystal structures of proteins with Ca2+ and Mg2+ binding sites. When a Ca2+ ion is placed equidistant between the side chain carboxylate oxygen atoms of E509 and I755D, the short metal-oxygen distances, mean, 2.4 Å, range, 2.1 - 2.7 Å, to the five protein ligands and a water found in the Na+ complex, are typical of those expected for bound Ca2+ ions (Harding, 2006).
Figure 5. Model divalent ion binding sites.
(A) Structural model of the GluR6 wild-type cation binding site based on the Na-GluR5 complex (3c32), with sodium as a gold sphere. Transparent spheres represent the Shannon radius. The Methionine residue that caps the binding site is shown in CPK representation.
(B) Structural model of the GluR6 M739D mutant with calcium bound (violet sphere). The model is derived from the Na-GluR5 complex (3c32) with the ‘capping’ isoleucine replaced by an aspartic acid. The bound sodium ion was replaced by a calcium ion (violet sphere) placed equidistant between the proximal carboxyl groups of E509 and M739D.
(C) Ca (violet sphere) bound to copper amine oxidase (1OAC) with five protein ligands. The Ca ion is buried but the crown of three aspartic acid side chains that coordinate it is partly solvent exposed. Two backbone carbonyl oxygen atoms and a water molecule (W1) complete the six-fold coordination.
(D) Mg (yellow sphere) bound in the active site of inositol monophosphatase in a substrate-free crystal form (2BJI). The coordination sphere has three carboxylates and two waters (red spheres, W1 and W2). A site is available for third water molecule to give octahedral coordination, although it was not modeled.
To further confirm the plausibility of our model divalent ion binding sites in GluR5 and GluR6, we searched the PDB for high-resolution structures of proteins where multiple carboxylate groups coordinate Ca2+ and Mg2+ ions. Calcium is one of the most abundant elements on earth and calcium ions are widely used in biological functions. An example of a Ca2+ binding site in the copper amine oxidase of E.coli (1OAC; Parsons et al., 1995) is shown in figure 5C. The calcium ion is buried in tandem with a water molecule, and, like the putative Ca2+ binding site in the M739D mutant, it is coordinated by carboxylate oxygen atoms from three acidic amino acid side chains. Two carbonyl oxygen atoms complete the coordination sphere. One of the best-known examples of a calcium binding motif is the ‘EF-hand’, one of the most abundant of all protein domains (Rubin et al., 2000), which has 7-fold coordination of calcium. Analyses of a diverse set of calcium binding structures, from which EF hands were deliberately excluded to reduce bias, suggest that the coordination chemistry of the site in 1OAC, and hence the mutant site in GluR6 M739D, is typical of a generic structure which has six fold coordination, with mean contribution of 2.5 oxygens from carboxyl groups, 2.1 from carbonyl groups and 1.4 from waters (Pidcock and Moore, 2001).
At the time of writing almost one in ten of the 50160 structures in the PDB contain either bound magnesium or manganese ions. The essential role of magnesium as a cofactor in phosphate transfer reactions at catalytic centers almost certainly underlies the enrichment of the PDB with structures containing Mg2+. Indeed, bound magnesium ions are often coordinated by phosphate groups, either from nucleic acids, ATP, sugars or other enzyme substrates. Because the chemistry of coordination by phosphates is not relevant to the coordination of Mg2+ in the GluR6 M739D mutant, where it is likely that all the ligands are oxygen atoms from protein or water molecules, we searched the PDB for high-resolution structures where phosphate groups did not coordinate the magnesium ion. One of the few examples is shown in Figure 5D (2BJI; Gill et al., 2005). The magnesium ion shown is one of three in the active site of inositol monophosphatase. This structure was solved in the absence of substrate, and water molecules that hydrate the Mg2+- ion mimic the coordination that is provided by the substrate during the catalytic reaction. Three carboxylate groups and two water molecules coordinate the Mg2+ with a mean metal-oxygen distance of 2.1 Å. Octahedral coordination is very strongly favored for magnesium (Katz et al., 1996) and in the GluR6 M739D mutant water molecules must supply the missing ligands which the protein lacks; this likely contributes to the lower affinity for Ca2+ versus Mg2+ observed in functional experiments.
In summary, we show that the GluR6 M739D mutant binds divalent cations with micromolar affinity, and that in the presence of the low concentrations of Ca2+ and Mg2+ typically used in patch clamp recording experiments, this accounts for the apparent lack of requirement for allosteric monovalent cations when Na+ is replaced by Tris or NMDG. Although the GluR6 M739D mutant has no counterpart in nature, it represents an interesting example of a divalent cation binding site created by protein engineering.
4. Methods
Wild-type and mutant glutamate receptors were expressed in HEK-293 cells for outside-out patch recording with rapid solution exchange, as described previously (Plested and Mayer, 2007). Mutations were introduced by overlap PCR and the entire open reading frame of the amplified region confirmed by sequencing. Brief pulses of glutamate (10 mM) were applied to outside-out patches using a piezo-driven perfusion system under computer control. Solution exchange was complete in ~200 μs, and patches which had exchange times slower than 400 μs (as assessed by junction currents recorded after the end of the experiment) were discarded. Data were filtered at 2-12 kHz and sampled at 10-50 kHz. Except were noted, the external solution contained 0.1 mM MgCl2 and 0.1 mM CaCl2 to increase patch stability. Solutions also contained 150 mM Na, Tris or NMDG. The counter anion was chloride. Tris base and NMDG were titrated directly with HCl to pH 7.3; this added 133 and 149 mM Cl− respectively, as a result of pKa values of 8.1 for Tris and 9.5 for NMDG. The NaCl control solution was buffered with 5 mM HEPES that was titrated to pH 7.3 with NaOH. We titrated the glutamate in the test solution to pH 7.3 with Tris base, in order to avoid adding sodium to the Tris and NMDG solutions. The internal solution contained (in mM): 115 NaCl, 10 NaF, 0.5 CaCl2, 1 MgCl2, 5 Na4BAPTA, 5 HEPES and 10 Na2ATP, pH 7.3. For experiments on the M739D mutant where we used Mg2+ and Ca2+ as surrogate allosteric cations in the presence of Tris, we chelated other trace divalent cations, and buffered Mg2+ and Ca2+ with EDTA (7 - 50 μM) to give the appropriate free concentration, according calculations made using MaxChel (www.maxchelator.stanford.edu). We observed no loss of patch stability in 10 μM free divalents, but always obtained seals in 100 μM Mg and Ca. Because we regularly used weakly permeant blockers in the extracellular solution at high concentrations, we generally estimated the proportion of active receptors from the slope conductance of peak responses at positive potentials, where the measured response is the efflux of pipette sodium. Data were fitted and plotted in Kaleidagraph (Synergy Software). All results are mean ± standard error of the mean.
The Tris molecule structure is taken from a high-resolution crystal structure of Arsenate Reductase (1jf8, 1.12A resolution) that had unambiguous density for all ligand atoms. No three- dimensional structure of NMDG is available from the ligands of proteins in the databank. The structure shown is the lowest energy conformer, generated by MarvinView (ChemAxon) via the ChemDB website (cdb.ics.uci.edu). Images in Figures 2 and 5 were generated using PyMOL (DeLano, 2002).
Acknowledgments
We thank Carla Glasser for preparing cDNAs; the NINDS DNA sequencing facility for support and Drs. S. Heinemann and P. Seeburg for the gift of wild type cDNAs. This work was supported by the intramural research program of NICHD, NIH, DHHS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- Bowie D. External anions and cations distinguish between AMPA and kainate receptor gating mechanisms. J Physiol. 2002;539:725–733. doi: 10.1113/jphysiol.2001.013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D, Mayer ML. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15:453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- Bähring R, Bowie D, Benveniste M, Mayer ML. Permeation and block of rat GluR6 glutamate receptor channels by internal and external polyamines. J Physiol (Lond) 1997;502:575–589. doi: 10.1111/j.1469-7793.1997.575bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N, Villarroel A, Sakmann B. Dimensions and ion selectivity of recombinant AMPA and kainate receptor channels and their dependence on Q/R site residues. J Physiol. 1996;496(Pt 1):165–173. doi: 10.1113/jphysiol.1996.sp021674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittajallu R, Vignes M, Dev KK, Barnes JM, Collingridge GL, Henley JM. Regulation of glutamate release by presynaptic kainate receptors in the hippocampus. Nature. 1996;379:78–81. doi: 10.1038/379078a0. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- DeVries SH. Bipolar cells use kainate and AMPA receptors to filter visual information into separate channels. Neuron. 2000;28:847–856. doi: 10.1016/s0896-6273(00)00158-6. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R, Mohammed F, Badyal R, Coates L, Erskine P, Thompson D, Cooper J, Gore M, Wood S. High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy. Acta Crystallographica Section D-Biological Crystallography. 2005;61:545–555. doi: 10.1107/S0907444905004038. [DOI] [PubMed] [Google Scholar]
- Greger IH, Akamine P, Khatri L, Ziff EB. Developmentally regulated, combinatorial RNA processing modulates AMPA receptor biogenesis. Neuron. 2006;51:85–97. doi: 10.1016/j.neuron.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallographica Section D-Biological Crystallography. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Horning MS, Mayer ML. Regulation of AMPA receptor gating by ligand binding core dimers. Neuron. 2004;41:379–388. doi: 10.1016/s0896-6273(04)00018-2. [DOI] [PubMed] [Google Scholar]
- Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005;25:9027–9036. doi: 10.1523/JNEUROSCI.2567-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj SK, Swanson GT, Cull-Candy SG. Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J Physiol. 1995;486(Pt 2):297–303. doi: 10.1113/jphysiol.1995.sp020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz AK, Glusker JP, Beebe SA, Bock CW. Calcium ion coordination: A comparison with that of beryllium, magnesium, and zinc. Journal of the American Chemical Society. 1996;118:5752–5763. [Google Scholar]
- Kidd FL, Isaac JT. Developmental and activity-dependent regulation of kainate receptors at thalamocortical synapses. Nature. 1999;400:569–573. doi: 10.1038/23040. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP. A molecular determinant for submillisecond desensitization in glutamate receptors. Science. 1994;266:1059–1062. doi: 10.1126/science.7973663. [DOI] [PubMed] [Google Scholar]
- Naur P, Hansen KB, Kristensen AS, Dravid SM, Pickering DS, Olsen L, Vestergaard B, Egebjerg J, Gajhede M, Traynelis SF, Kastrup JS. Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci U S A. 2007;104:14116–14121. doi: 10.1073/pnas.0703718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium Gates Glutamate-Activated Channels in Mouse Central Neurons. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- Parsons MR, Convery MA, Wilmot CM, Yadav KDS, Blakely V, Corner AS, Phillips SEV, Mcpherson MJ, Knowles PF. Crystal-Structure of a Quinoenzyme - Copper Amine Oxidase of Escherichia-Coli at 2-Angstrom Resolution. Structure. 1995;3:1171–1184. doi: 10.1016/s0969-2126(01)00253-2. [DOI] [PubMed] [Google Scholar]
- Paternain AV, Cohen A, Stern-Bach Y, Lerma J. A role for extracellular Na+ in the channel gating of native and recombinant kainate receptors. J Neurosci. 2003;23:8641–8648. doi: 10.1523/JNEUROSCI.23-25-08641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidcock E, Moore GR. Structural characteristics of protein binding sites for calcium and lanthanide ions. J Biol Inorg Chem. 2001;6:479–489. doi: 10.1007/s007750100214. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Vijayan R, Biggin PC, Mayer ML. Molecular Basis of Kainate Receptor Modulation by Sodium. Neuron. 2008;58:720–735. doi: 10.1016/j.neuron.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Zilberter Y, Wollmuth LP, Burnashev N. Facilitation of currents through rat Ca2+-permeable AMPA receptor channels by activity-dependent relief from polyamine block. J Physiol. 1998;511(Pt 2):361–377. doi: 10.1111/j.1469-7793.1998.361bh.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LS, Gong F, Guan P, Harris NL, Hay BA, Hoskins RA, Li J, Li Z, Hynes RO, Jones SJ, Kuehl PM, Lemaitre B, Littleton JT, Morrison DK, Mungall C, O’Farrell PH, Pickeral OK, Shue C, Vosshall LB, Zhang J, Zhao Q, Zheng XH, Lewis S. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Watkins JC, Jane DE. The glutamate story. Br J Pharmacol. 2006;147(Suppl 1):S100–108. doi: 10.1038/sj.bjp.0706444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Jatzke C, Seeburg PH, Heintz N, Zuo J. The lurcher mutation identifies delta 2 as an AMPA/kainate receptor-like channel that is potentiated by Ca2+ Journal of Neuroscience. 2000;20:5973–5980. doi: 10.1523/JNEUROSCI.20-16-05973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AY, Fay AM, Bowie D. External ions are coactivators of kainate receptors. J Neurosci. 2006;26:5750–5755. doi: 10.1523/JNEUROSCI.0301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AY, MacLean DM, Bowie D. Na+/Cl- dipole couples agonist binding to kainate receptor activation. J Neurosci. 2007;27:6800–6809. doi: 10.1523/JNEUROSCI.0284-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]