Abstract
BACKGROUND
The chitinase-like protein YKL-40 is involved in inflammation and tissue remodeling. We recently showed that serum YKL-40 levels were elevated in patients with asthma and were correlated with severity, thickening of the subepithelial basement membrane, and pulmonary function. We hypothesized that single-nucleotide polymorphisms (SNPs) that affect YKL-40 levels also influence asthma status and lung function.
METHODS
We carried out a genomewide association study of serum YKL-40 levels in a founder population of European descent, the Hutterites, and then tested for an association between an implicated SNP and asthma and lung function. One associated variant was genotyped in a birth cohort at high risk for asthma, in which YKL-40 levels were measured from birth through 5 years of age, and in two populations of unrelated case patients of European descent with asthma and controls.
RESULTS
A promoter SNP (−131C→G) in CHI3L1, the chitinase 3–like 1 gene encoding YKL-40, was associated with elevated serum YKL-40 levels (P = 1.1×10−13), asthma (P = 0.047), bronchial hyperresponsiveness (P = 0.002), and measures of pulmonary function (P = 0.046 to 0.002) in the Hutterites. The same SNP could be used to predict the presence of asthma in the two case–control populations (combined P = 1.2×10−5) and serum YKL-40 levels at birth (in cord-blood specimens) through 5 years of age in the birth cohort (P = 8.9×10−3 to 2.5×10−4).
CONCLUSIONS
CHI3L1 is a susceptibility gene for asthma, bronchial hyperresponsiveness, and reduced lung function, and elevated circulating YKL-40 levels are a biomarker for asthma and decline in lung function.
Chitinases are evolutionarily conserved proteins that mediate airway inflammation in mouse models of asthma.1 The chitinase-like protein YKL-40 lacks chitinase activity but binds ubiquitously expressed chitin and has been implicated in inflammation and tissue remodeling.2–6 We recently demonstrated that serum YKL-40 levels are elevated in patients with asthma and that circulating YKL-40 levels are correlated with asthma severity, thickness of the subepithelial basement membrane, and pulmonary function,7 suggesting that circulating YKL-40 levels are a biomarker for asthma. The YKL-40 protein is encoded by the chitinase 3–like 1 gene CHI3L1, and single-nucleotide polymorphisms (SNPs) in the CHI3L1 promoter have been associated with elevated serum YKL-40 levels,8,9 differential gene expression9 and transcript levels,10 and a higher risk of schizophrenia.9 We conducted a genomewide association study to identify genes that affect serum YKL-40 levels and hypothesized that variation in associated genes influences the risk of asthma and bronchial hyperresponsiveness and is associated with reduced lung function.
METHODS
Written informed consent was provided by all adult participants; the children who participated provided oral assent and their parents provided written informed consent. The study was approved by the institutional review boards at the participating centers.
THE HUTTERITES
To minimize the confounding effects of genetic and environmental heterogeneity, we focused our genetic studies of common diseases in the Hutterites. 11,12 The 753 Hutterites we studied live on communal farms in South Dakota and are related to each other through multiple lines of descent in a 3028-person, 13-generation pedigree with 62 founders.13,14 The small number of founding genomes reduces genetic heterogeneity, and the communal lifestyle of the Hutterites ensures that nongenetic factors are remarkably uniform among persons. Smoking is prohibited (and rare) in this community, minimizing exposure to firsthand or secondhand smoke.
Asthma was assessed in 652 Hutterites by obtaining a history of symptoms (cough, wheeze, shortness of breath), bronchial hyperresponsiveness to methacholine inhalation or airway reversibility, and a doctor’s diagnosis, according to previously published protocols.12,15 A total of 76 (11.7%) met the criteria for asthma; 80 others (12.3%) had bronchial hyperresponsiveness only, and 423 (64.9%) did not have bronchial hyperresponsiveness and were not symptomatic.12
Persons were considered to have atopy if they had a positive skin-prick test for at least 1 of 14 airborne allergens12; 311 of 702 Hutterites (44.3%) had atopy.
YKL-40 levels were measured in frozen serum specimens from 632 Hutterites who were 6 years of age or older.7 The clinical characteristics of these 632 Hutterites are shown in Table 1.
Table 1.
Baseline Characteristics of the Hutterites with Measured YKL-40 Levels.*
| Characteristic | Males (N = 280) | Females (N = 352) | Total (N = 632) |
|---|---|---|---|
| Age — yr | |||
| Mean | 32.7 | 33.8 | 33.3 |
| Range | 6–92 | 6–88 | 6–92 |
| YKL-40 — ng/ml | 96.7±4.7 | 88.6±3.5 | 92.2±2.9 |
| Asthma — no./no. tested (%) | 36/251 (14.3) | 27/295 (9.2) | 63/546 (11.5) |
| Bronchial hyperresponsiveness — no./no. tested (%) | 63/251 (25.1) | 58/295 (19.7) | 121/546 (22.2) |
| Atopy — no./no. tested (%) | 124/263 (47.1) | 116/320 (36.3) | 240/583 (41.2) |
| Serum IgE — IU | 151.2±21.7 | 49.8±5.7 | 94.9±10.3 |
| FEV1 — % of predicted value | 100.2±1.0 | 101.2±0.8 | 100.8±0.6 |
| FVC — % of predicted value | 105.5±0.9 | 106.5±0.8 | 106.0±0.6 |
| FEV1:FVC — % of predicted value | 79.6±0.5 | 82.2±0.5 | 81.0±0.4 |
| FEF25–75 — % of predicted value | 3.7±0.1 | 3.1±0.1 | 3.3±0.1 |
Plus–minus values are means ±SE. Data for serum IgE were available for 610 Hutterites (271 males and 339 females). Data for forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), the FEV1:FVC ratio, and the forced expiratory flow between 25% and 75% of the FVC (FEF25–75) were available for 599 Hutterites (272 males and 327 females).
For genetic studies, we used a natural-log transformation of the serum YKL-40 level to fulfill the distributional requirements of our methods, and we included age and sex as covariates. The heritability of YKL-40 levels was estimated with the use of variance-component methods.13,14 We used as a test of association the general two-allele model for quantitative measures (YKL-40 level, pulmonary-function measures, and total serum IgE level).16 (For more details of the variance component estimate of heritability, the general two-allele model test of association, and an estimate of the effect size of associated alleles on the trait variance, see the Supplementary Appendix, available with the full text of this article at www.nejm.org.)
Associations with binary phenotypes (asthma, bronchial hyperresponsiveness, and atopy) were assessed using the case–control quasi-likelihood test, which takes into account the relatedness between persons with the phenotypes and controls. 17
THE CHILDHOOD ORIGINS OF ASTHMA COHORT
The Childhood Origins of Asthma (COAST) cohort consists of 206 children of European descent (56.8% of whom are boys) who participated in genetic studies in a birth-cohort study of the origins of asthma,18 with asthma diagnosed at 6 years of age. Serum levels of YKL-40 were measured in 125 of these children at birth (in cord-blood specimens) and at 1 and 3 years of age and in 105 of these children at 5 years of age.
YKL-40 levels were measured in frozen serum specimens, according to the same protocols used for studies of the Hutterites.7 At 6 years of age, the children in the COAST cohort received a diagnosis of asthma if they met at least one of the following criteria: doctor-diagnosed asthma, use of doctor-prescribed albuterol for episodes of coughing or wheezing more than once between 60 and 72 months of age, daily use of controller medication, implementation of a step-up plan as prescribed by a doctor (including the use of albuterol or the short-term use of inhaled corticosteroids during illness), or use of doctor-prescribed prednisone for the treatment of an asthma exacerbation.
The difference in YKL-40 levels between children with asthma and those without asthma were tested using a Wilcoxon rank-sum test. Associations between CHI3L1 SNPs and YKL-40 levels were examined with the use of log-transformed YKL-40 levels at birth and at 1, 3, and 5 years of age in a linear-regression model, with sex included as a covariate.
ASTHMA CASE PATIENTS AND CONTROLS
Two populations of European descent were used to replicate the associations with asthma. The Freiburg population consists of 344 children with asthma and 294 control children without asthma, recruited from clinics at the Children’s University Hospital in Freiburg, Germany. Asthma was defined by the presence of self-reported symptoms (cough, wheeze, or shortness of breath), current use of asthma medications, a doctor’s diagnosis, and bronchial hyperresponsiveness (i.e., a 15% decrease in the baseline value of the forced expiratory volume in 1 second [FEV1] after either inhalation of ≤8 mg per deciliter of histamine or ≤6 minutes of exercise). The controls did not have a history of asthma, recurrent wheezing, or atopy. A total of 64.7% of the case patients were male, with a mean age of 10.1 years (range, 6 to 16) at the time of evaluation; 59.4% of the controls were male, with a mean age of 7.9 years (range, 4 to 16) at the time of enrollment.
The Chicago population consisted of 99 case patients recruited through the adult and pediatric asthma clinics at the University of Chicago Medical Center and 197 controls recruited from the same medical center. Diagnosis of asthma in the case patients was based on fulfillment of all of the following criteria: age of 6 or more years, presence of at least two of three symptoms (cough, wheeze, and shortness of breath), a physician’s diagnosis of asthma (with no conflicting pulmonary diagnosis), either bronchial hyperresponsiveness (defined as a ≥20% decrease in FEV1 after inhalation of ≤25 mg of methacholine per milliliter) or an increase by 15% or more in FEV1 after treatment with a short-acting bronchodilator or treatment with inhaled corticosteroids, and less than 3 pack-years of cigarette smoking. 15 The controls were adults recruited from the University of Chicago Medical Center who did not have a history of asthma (either personally or among first-degree relatives). In all, 32.5% of the case patients were male, with a mean age of 24.4 years (range, 7 to 74) at the time of evaluation; 52.5% of the controls were male, with a mean age of 31.6 years (range, 18 to 69) at the time of evaluation.
Associations with asthma were tested with the use of Fisher’s exact test for differences in the genotypes and allele frequencies between case patients and controls. The 95% confidence intervals were obtained from the hypergeometric distribution of the entries, conditional on fixed margins. The analysis for the two case–control populations combined was done using the Cochran–Mantel–Haenszel method.
GENOTYPING AND QUALITY CHECKS
We first carried out a genomewide association study to identify SNPs associated with serum YKL-40 levels (see the Supplementary Appendix). We then selected SNPs to tag all common haplotypes in CHI3L1 and within the 15 kb upstream of its transcriptional start site. We included in the analysis the validated nonsynonymous SNP rs880633, the functional promoter SNP rs4950928,9 and a SNP (rs946263) previously shown to be associated with levels of expression of CHI3L1.10 The tag SNPs rs4950928 (−131C→G), rs880633 (Arg145→Gly), rs10399805, rs1538372, and rs2275352 were genotyped using TaqMan Assay-on-Demand (ABI). An additional five SNPs in the 15-kb upstream region (including one tag SNP) were genotyped in specimens from the Hutterites, with the use of the Affymetrix GeneChip Mapping 500K Array; genotypes were determined by means of the BRLMM algorithm.19 Some redundant SNPs were included because they were present on the Affymetrix chip. The 10 SNPs were successfully genotyped in more than 95% of the persons studied, were in Hardy–Weinberg equilibrium (P>0.20), and in the Hutterites, had no mendelian errors (see the Supplementary Appendix). Allele frequencies and Hardy–Weinberg calculations for the Hutterites were adjusted for relatedness. 20,21
RESULTS
CHI3L1, YKL-40, AND ASTHMA PHENOTYPES IN THE HUTTERITES
Serum YKL-40 levels increased significantly with increasing age in the Hutterites (Pearson’s r = 0.21, P<0.001) but did not differ between males and females (t = 0.52, P = 0.61). Mean YKL-40 levels were increased among Hutterites with asthma (102.7 ng per milliliter) or bronchial hyperresponsiveness (96.5 ng per milliliter), as compared with controls (87.2 ng per milliliter) (P = 0.005 and P = 0.002, respectively), but as in our previous study7 the levels did not differ between subjects with atopy (99.4 ng per milliliter) and those without atopy (85.1 ng per milliliter) (P = 0.68). Among the Hutterites, serum YKL-40 levels were significantly inversely correlated with FEV1 (P = 0.02) but not with forced vital capacity (FVC) (P = 0.16), the FEV1:FVC ratio (P = 0.98), or forced expiratory flow between 25% and 75% of the FVC (FEF25–75) (P = 0.41).
To assess the relative contribution of genes to the variance in YKL-40 levels among subjects, we first estimated the heritability of the YKL-40 level. The narrow heritability (h2) of this trait in the Hutterites (±SE) is 0.51±0.10 and the broad heritability (H2) is 1.0±0.16. The high estimate for broad heritability indicates that differences in serum YKL-40 levels among individual Hutterites are due nearly entirely to genetic differences between individual persons. The comparatively large broad heritability indicates the presence of autosomal loci with significant nonadditive (e.g., dominant) effects on YKL-40 levels.
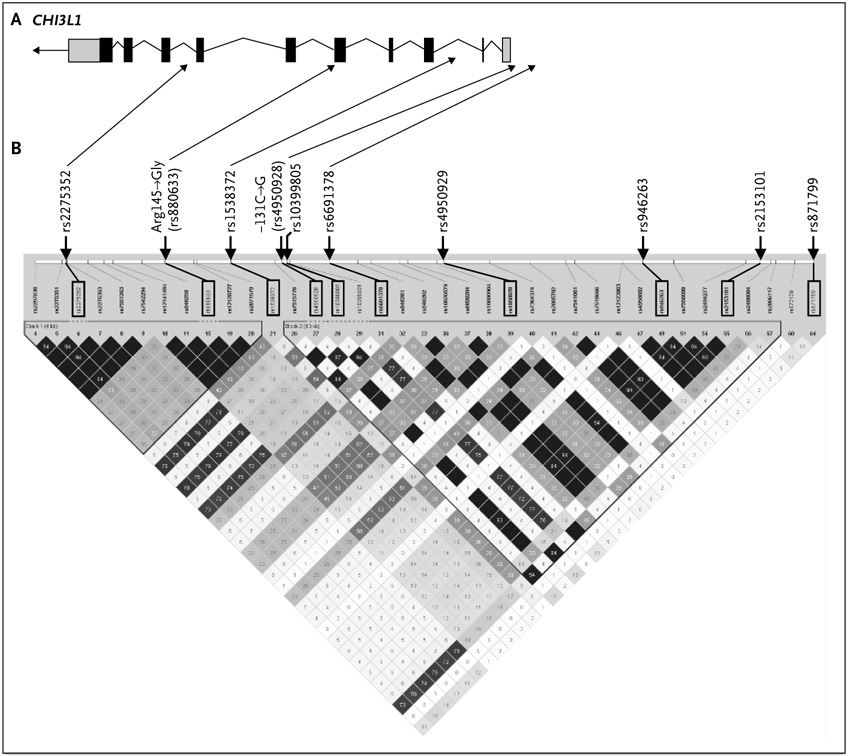
The most significant associations in the genomewide association study were found between the YKL-40 level and SNPs upstream of the gene encoding YKL-40, CHI3L1 (Fig. S1 and S2 in the Supplementary Appendix). The P values for all tested SNPs calculated with the use of the general two-allele model can be obtained from the National Institutes of Health Genotype and Phenotype database, dbGaP (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gap). To further evaluate the specific contribution of the CHI3L1 locus to the variance in YKL-40 levels, five additional SNPs were genotyped in the Hutterites; the location of these SNPs and the linkage-disequilibrium structure of the gene in this population are shown in Figure 1. Three SNPs on the Affymetrix chip (rs4950929, rs946263, and rs2153101) are in perfect linkage disequilibrium with the functional promoter SNP −131C→G (rs4950928) (r2 = 1.0) (Fig. 1B, and Fig. S3 in the Supplementary Appendix).
Figure 1. Single-Nucleotide Polymorphisms (SNPs) in CHI3L1 and Its Upstream Region on Chromosome 1q32.1.
Part A shows the structure of the CHI3L1 gene. Coding exons are indicated by black rectangles, and untranslated regions are shown as gray rectangles; the arrow indicates the direction of transcription. The gene spans from 201,414,557 bp to 201,422,504 bp. Part B shows the linkage disequilibrium (r2) among SNPs in HapMap CEPH samples (of persons of European ancestry collected by the Centre d’Etude du Polymorphisme Humain) from 201,416,807 bp to 201,436,499 bp (Haploview). SNPs typed in the Hutterites and the SNP typed in the case and control populations (−131C→G) are indicated by black rectangles. SNPs in the linkage-disequilibrium plot are equally spaced across the region (and thus are not to physical scale). The linkage-disequilibrium pattern in the Hutterites and in the European CEPH sample is shown in Figure S3 in the Supplementary Appendix.
These four SNPs showed the strongest association with serum YKL-40 levels of all the SNPs tested in the Hutterites (P≤1.3×10−12 for all four comparisons) (Table 2), and remained statistically significant after correction for the number of SNPs present on the Affymetrix chip (see the Supplementary Appendix). None of the 10 SNPs (Table 2) had significant sex-specific effects on serum YKL-40 levels. The nonsynonymous SNP in exon 5 (Arg145→Gly) was not significantly associated with YKL-40 levels (P = 0.67) or any other phenotypic characteristic (Table 2). The major (most common) allele at each of the associated SNPs in CHI3L1 is the ancestral allele, according to the sequence of the orthologous gene in the chimpanzee. The −131C→G SNP rs4950928 accounts for 9.4% of the variance in YKL-40 levels in the Hutterites, with the minor G allele having an additive (negative) effect on YKL-40 levels (Fig. 2A).
Table 2.
Results of Association Studies of Single-Nucleotide Polymorphisms (SNPs) in the CHI3L1 Gene and Its Upstream Region on Chromosome 1q32.1 among the Hutterites.*
| SNP | Location Relative to Translational Start Site |
Distance from p Terminus | Minor-Allele Frequency |
P Values for Association | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serum YKL-40 Level |
Percent of Predicted FEV1 |
Percent of Predicted FVC |
FEV1:FVC | FEF25–75 | Total Serum IgE |
Asthma | Bronchial Hyperresponsiveness |
Atopy | ||||
| base pairs | ||||||||||||
| rs871799 | −14,120 | 201,436,494 | 0.18 | 0.03 | 0.29 | 0.23 | 0.99 | 0.83 | 0.68 | 0.86 | 0.68 | 0.93 |
| rs2153101 | −12723 | 201,435,097 | 0.21 | 9.7×10−13 | 0.02 | 0.10 | 6.7×10−4 | 0.03 | 0.40 | 0.008 | 5.9×10−4 | 0.24 |
| rs946263 | −9630 | 201,432,004 | 0.21 | 9.7×10−13 | 0.02 | 0.10 | 6.7×10−4 | 0.03 | 0.40 | 0.008 | 5.9×10−4 | 0.24 |
| rs4950929 | −4375 | 201,426,749 | 0.21 | 1.3×10−12 | 0.02 | 0.11 | 7.3×10−4 | 0.03 | 0.39 | 0.008 | 5.9×10−4 | 0.23 |
| rs6691378 | −1371 | 201,423,745 | 0.21 | 3.8×10−5 | 0.03 | 0.02 | 0.59 | 0.59 | 0.68 | 0.82 | 0.46 | 0.54 |
| rs10399805 (tag SNP) | −247 | 201,422,621 | 0.24 | 5.8×10−5 | 0.10 | 0.05 | 0.30 | 0.74 | 0.94 | 0.83 | 0.68 | 0.97 |
| rs4950928 (tag SNP) | −131 | 201,422,505 | 0.22 | 1.1×10−13 | 0.046 | 0.50 | 0.002 | 0.03 | 0.37 | 0.047 | 0.002 | 0.20 |
| rs1538372 (tag SNP) | +1220 (intron 2) | 201,421,155 | 0.38 | 3.1×10−3 | 0.89 | 0.67 | 0.05 | 0.37 | 0.69 | 0.33 | 0.43 | 0.92 |
| rs880633 (tag SNP)† | +2951 (exon 5) | 201,419,424 | 0.41 | 0.67 | 0.27 | 0.79 | 0.20 | 0.45 | 0.07 | 0.33 | 0.16 | 0.78 |
| rs2275352 (tag SNP) | +5573 (intron 7) | 201,416,802 | 0.24 | 1.8×10−4 | 0.11 | 0.047 | 0.58 | 0.97 | 0.57 | 0.49 | 0.24 | 0.53 |
Associations were evaluated with use of the general two-allele model test for quantitative phenotypes16 and the case–control quasi-likelihood test17 for binary phenotypes. Allele frequencies are corrected for relatedness.21 Distances are based on build 126 of the National Center for Biotechnology Information’s SNP database (dbSNP). FEF25–75 denotes forced expiratory flow between 25% and 75% of the forced vital capacity (FVC), and FEV1 forced expiratory volume in 1 second.
This SNP is the validated nonsynonymous SNP Arg145→Gly.
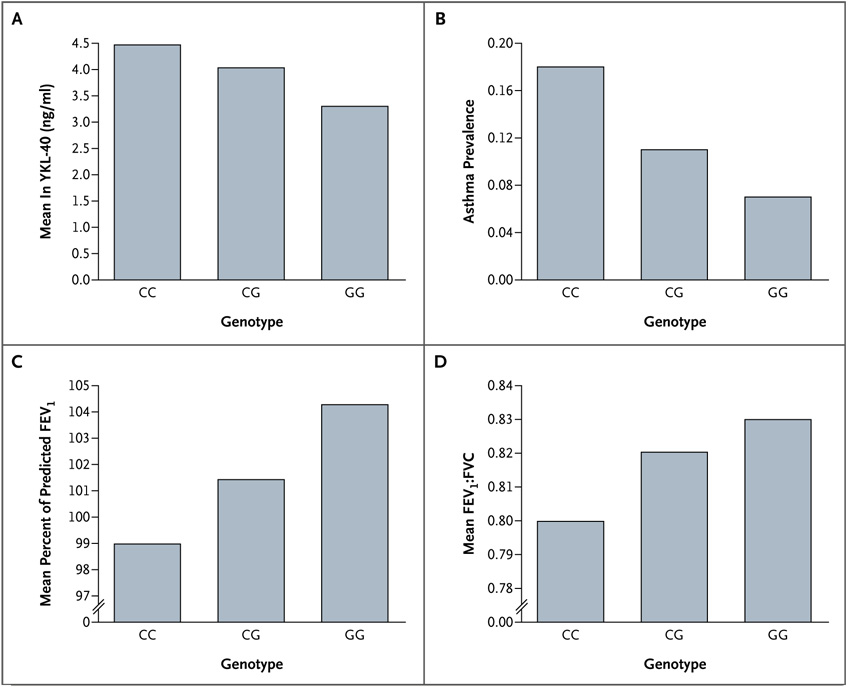
Figure 2. Serum YKL-40 Level, Asthma Prevalence, and Lung-Function Measures in Hutterites, According to −131C→G Genotype (rs4950928).
All measures differed significantly among the three genotypes. Panel A shows the mean natural-log–transformed serum YKL-40 levels (P = 1.1×10−13 by the general two-allele model). Panel B shows asthma prevalences among 554 Hutterites (P = 0.047 by the case–control quasi-likelihood test). Panel C shows the mean percent of the predicted forced expiratory volume in 1 second (FEV1) (P = 0.046 by the general two-allele model). Panel D shows the mean ratio of FEV1 to forced vital capacity (FVC) (P = 0.002 by the general two-allele model).
In the Hutterites, the frequency of the rs4950928 C allele was 0.84 in persons with asthma, 0.83 in persons with bronchial hyperresponsiveness, and 0.79 in controls; the allele was significantly associated with the asthma phenotype (P = 0.047 by the case–control quasi-likelihood test) and the bronchial hyperresponsiveness phenotype (P = 0.002 by the case–control quasi-likelihood test) (Table 2 and Fig. 2B). This SNP was not significantly associated with atopy (P = 0.20 by the case–control quasi-likelihood test) or total serum IgE level (P = 0.37 by the general two-allele model). The rs4950928 C allele was also a significant predictor of decreased FEV1 (P = 0.046 by the general two-allele model), decreased FEV1:FVC (P = 0.002 by the general two-allele model), and decreased FEF25–75 (P = 0.03 by the general two-allele model) in the Hutterites (Table 2 and Fig. 2C and 2D).
REPLICATION STUDIES IN THE COAST COHORT
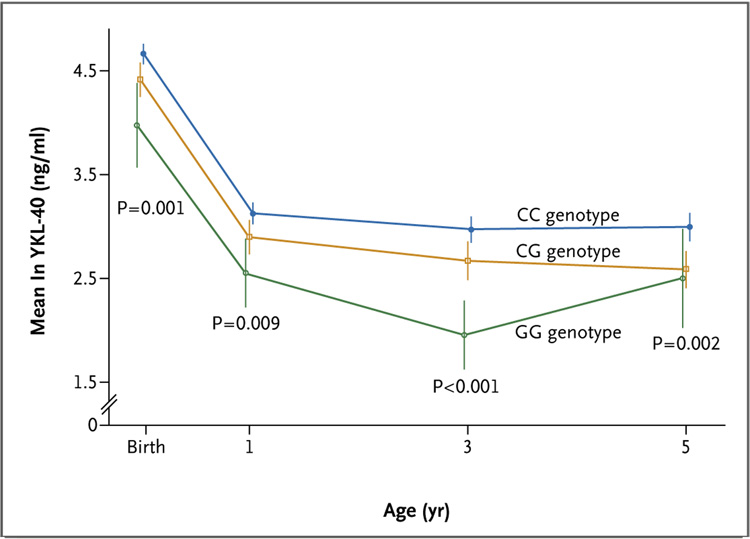
Serum YKL-40 levels were highest at birth and decreased through 3 years of age but were relatively stable between 3 and 5 years of age (Fig. 3, and Table S1 in the Supplementary Appendix). Serum YKL-40 levels at each age were not significant predictors of asthma diagnosis at 6 years of age, although the association at 3 years of age approached statistical significance (P = 0.85 for the 121 subjects at birth, P = 0.82 for the 121 subjects at 1 year of age, P = 0.08 for the 121 subjects at 3 years of age, and P = 0.29 for the 103 subjects at 5 years of age) (all P values by the Wilcoxon test).
Figure 3. Mean Serum YKL-40 Levels in the Childhood Origins of Asthma Cohort, According to Age and −131C→G Genotype (rs4950928).
P values were calculated for the differences in mean natural-log–transformed serum YKL-40 levels among the three genotype groupings by means of an analysis of variance. Vertical bars indicate standard errors.
We also genotyped the −131C→G SNP (rs4950928) in the children in the COAST cohort. The −131C allele was associated with elevated YKL-40 levels at each age (Fig. 3), indicating that genotype-specific effects on circulating YKL-40 levels are present at birth and remain throughout the first 5 years of life. The changes among ages within genotype groupings were not significant. Among the 178 children whose asthma status was evaluated at 6 years of age, 52 (29.2%) received a diagnosis of asthma. The −131C→G genotype and allele frequencies did not differ significantly between children with asthma and those without asthma at 6 years of age. This result could be due to the different criteria (based on clinical criteria) used to diagnose asthma in the children in the COAST cohort, the influence of the CHI3L1 SNPs on YKL-40 levels before the onset of asthma-related sequelae, or the SNP having an independent effect on the risk of asthma later in life (i.e., after age 6).
REPLICATION STUDIES IN THE CASE–CONTROL SAMPLES
In contrast to the COAST cohort, in the Freiburg population, the prevalence of the −131C allele was significantly greater in the case patients with asthma as compared with controls (frequency of the C allele, 0.81 among the case patients and 0.71 among the controls; P = 1.6×10−4) (Table 3). In particular, the CC genotype was more common in patients with asthma (frequency, 0.66) than in controls (frequency, 0.51); both the CG and GG genotypes were more common among controls (CG frequency, 0.41 vs. 0.29 among the case patients; GG frequency, 0.08 vs. 0.05; P = 5.6×10−4 by Fisher’s exact test, assuming a dominant model). This pattern is similar to that found among the Hutterites. The odds ratio for the presence of one or two −131G alleles (CG or GG, vs. CC) was 0.54 (95% confidence interval [CI], 0.39 to 0.75), indicating that the minor −131G allele that is associated with reduced levels of circulating YKL-40 protein confers protection against asthma.
Table 3.
The Single-Nucleotide Polymorphism −131C→G (rs4950928) in Case Patients with Asthma and Controls from Freiburg and Chicago.
| Population | Total No. of Subjects | Genotype | Frequency of G Allele | Odds Ratio for Allele (95% CI)* | ||
|---|---|---|---|---|---|---|
| CC | CG | GG | ||||
| no. of subjects (frequency) | ||||||
| Freiburg | 0.54 (0.39–0.75) | |||||
| Case patients | 344 | 227 (0.66) | 100 (0.29) | 17 (0.05) | 0.19 | |
| Controls | 294 | 150 (0.51) | 120 (0.41) | 24 (0.08) | 0.29 | |
| Chicago | 0.56 (0.32–0.95) | |||||
| Case patients | 99 | 69 (0.70) | 25 (0.25) | 5 (0.05) | 0.18 | |
| Controls | 197 | 111 (0.56) | 80 (0.41) | 7 (0.04) | 0.24 | |
The odds ratio for the two populations combined is 0.54 (95% CI, 0.41 to 0.71) (P = 1.2×10−5 by the Cochran–Mantel–Haenszel method).
A similar pattern of association was present in the smaller Chicago population, in which the −131G allele was overrepresented in the controls as compared to the case patients (P = 0.11 by the Fisher’s exact test; P = 0.03 by Fisher’s exact test, assuming a dominant model; odds ratio for the G allele, 0.56; 95% CI, 0.32 to 0.95) (Table 3). The odds ratio for the G allele (CG or GG, vs. CC) in the two populations combined was 0.54 (95% CI, 0.41 to 0.71) (P = 1.2×10−5 by the Cochran–Mantel–Haenszel method).
DISCUSSION
Our study shows that serum YKL-40 level is a highly heritable, quantitative trait in humans and confirms that YKL-40 level is a significant biomarker for asthma susceptibility and reduced lung function. Moreover, genetic variation in CHI3L1 influences serum YKL-40 levels and is associated with the risk of asthma, bronchial hyperresponsiveness, and reduced lung function. Although we cannot statistically distinguish between the implicated SNPs that are in perfect linkage disequilibrium, −131C→G SNP (rs4950928) seems likely to be the causal SNP; it is in the core promoter of CHI3L1, within a binding site for the MYC and MAX transcription factors. The minor allele (−131G on the forward strand) disrupts binding and was reported to be associated with reduced transcription in a luciferase reporter assay, lower messenger RNA levels in peripheral-blood cells, and reduced levels of circulating YKL-40 protein.9 Furthermore, a SNP in strong linkage disequilibrium with −131C→G (rs946263) was found to influence CHI3L1 transcript levels in a genomewide study of gene expression in cells from children with asthma.10 Our results are consistent with these findings and indicate that the −131G allele is protective against asthma and decline in lung function, that this effect is independent of allergic (atopic) pathways, and that the effect of this SNP on circulating levels of YKL-40 is present at birth.
Does the −131C→G SNP, or another SNP in strong linkage disequilibrium with −131C→G, directly influence the risk of asthma and decline in lung function, or are these phenotypic effects mediated through its effect on circulating levels of YKL-40? Our study and previous studies showing an association between serum YKL-40 levels and a number of inflammatory conditions,2,8 including asthma,7 or between SNPs in CHI3L1 and serum YKL-40 levels8,9 and gene expression,9,10 suggest that YKL-40 is an intermediate phenotype for asthma susceptibility. However, our results in the Hutterites and the COAST cohort do not allow us to reach this conclusion. For example, in the Hutterites, YKL-40 levels are not associated with the lung-function measures of FEV1:FVC and FEF25–75, yet the −131C→G SNP is a significant predictor of both (Table 2). In the COAST cohort, the −131C→G SNP is associated with YKL-40 levels at birth through 5 years of age but not with asthma at 6 years of age (Fig. 3). Thus, the possibility remains that variation in CHI3L1 exerts effects on the risk of asthma and on lung function that are independent of circulating levels of YKL-40. Formal proof will require studying a larger population, with determinations of both serum YKL-40 levels and asthma status.
In summary, we identified an asthma susceptibility locus, CHI3L1, and showed that studying the genetics of quantitative traits (serum biomarkers) associated with asthma can identify asthma susceptibility loci. In the Hutterites, the CHI3L1 locus explains 9.4% of the variance in serum YKL-40 levels, suggesting that additional loci influence YKL-40 levels. Identifying the remaining loci that contribute to differences in serum YKL-40 levels and related proteins could identify additional genes with a significant effect on the risk of asthma and on lung function.
Supplementary Material
Acknowledgments
Supported by grants from the National Heart, Lung, and Blood Institute (HL85197, HL72414, HL56399, HL66533, and HL49596, to Dr. Ober; HL61879 and HL70831, to Dr. Lemanske; HL81638 and HL56389, to Dr. Elias; and M01 RR00055, to the University of Chicago Clinical Research Center).
Dr. Elias reports receiving grant support from MedImmune, a company exploring the regulation of YKL-40 as a therapy for asthma, and consulting fees from Eli Lilly, Pfizer, Promedior, MedImmune, Roche, and Millennium. A U.S. patent (no. 7, 214, 373) entitled “Methods, compositions and kits relating to chitinases and chitinase-like molecules and inflammatory disease” was issued on May 8, 2007, and is exclusively licensed to MedImmune (now owned by AstraZeneca); Dr. Elias is one of the inventors. Dr. Chupp reports receiving speakers’ fees from GlaxoSmith-Kline, Merck, AstraZeneca, and Novartis and grant support from Wyeth, Centocor, Pfizer, and Novartis. No other potential conflict of interest relevant to this article was reported.
We thank the Hutterites and the subjects in Chicago, Freiburg, and Madison for their participation in these studies and Kathy Roberg, Christopher Tisler, and Rebecca Anderson for technical support.
Footnotes
FULL TEXT OF ALL JOURNAL ARTICLES ON THE WORLD WIDE WEB
Access to the complete text of the Journal on the Internet is free to all subscribers. To use this Web site, subscribers should go to the Journal’s home page (www.nejm.org) and register by entering their names and subscriber numbers as they appear on their mailing labels. After this one-time registration, subscribers can use their passwords to log on for electronic access to the entire Journal from any computer that is connected to the Internet. Features include a library of all issues since January 1993 and abstracts since January 1975, a full-text search capacity, and a personal archive for saving articles and search results of interest. All articles can be printed in a format that is virtually identical to that of the typeset pages. Beginning 6 months after publication, the full text of all Original Articles and Special Articles is available free to nonsubscribers who have completed a brief registration.
REFERENCES
- Zhu Z, Zheng T, Homer RJ, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- Johansen JS. Studies on serum YKL-40 as a biomarker in diseases with inflammation, tissue remodelling, fibroses and cancer. Dan Med Bull. 2006;53:172–209. [PubMed] [Google Scholar]
- Johansen JS, Jensen HS, Price PA. A new biochemical marker for joint injury: analysis of YKL-40 in serum and synovial fluid. Br J Rheumatol. 1993;32:949–955. doi: 10.1093/rheumatology/32.11.949. [DOI] [PubMed] [Google Scholar]
- Johansen JS, Williamson MK, Rice JS, Price PA. Identification of proteins secreted by human osteoblastic cells in culture. J Bone Miner Res. 1992;7:501–512. doi: 10.1002/jbmr.5650070506. [DOI] [PubMed] [Google Scholar]
- Hakala BE, White C, Recklies AD. Human cartilage gp-39, a major secretory product of articular chondrocytes and synovial cells, is a mammalian member of a chitinase protein family. J Biol Chem. 1993;268:25803–25810. [PubMed] [Google Scholar]
- Kelleher TB, Mehta SH, Bhaskar R, et al. Prediction of hepatic fibrosis in HIV/HCV co-infected patients using serum fibrosis markers: the SHASTA index. J Hepatol. 2005;43:78–84. doi: 10.1016/j.jhep.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Chupp GL, Lee CG, Jarjour N, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–2027. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- Kruit A, Grutters JC, Ruven HJ, van Moorsel CC, van den Bosch JMA. CHI3L1 gene polymorphism is associated with serum levels of YKL-40, a novel sarcoidosis marker. Respir Med. 2007;101:1563–1571. doi: 10.1016/j.rmed.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Zhao X, Tang R, Gao B, et al. Functional variants in the promoter region of chitinase 3-like 1 (CHI3L1) and susceptibility to schizophrenia. Am J Hum Genet. 2007;80:12–18. doi: 10.1086/510438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Ober C, Abney M, McPeek MS. The genetic dissection of complex traits in a founder population. Am J Hum Genet. 2001;69:1068–1079. doi: 10.1086/324025. [Erratum, Am J Hum Genet 2002;70:284.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. 2000;67:1154–1162. doi: 10.1016/s0002-9297(07)62946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abney M, McPeek MS, Ober C. Broad and narrow heritabilities of quantitative traits in a founder population. Am J Hum Genet. 2001;68:1302–1307. doi: 10.1086/320112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Ober C, Abney M. Heritability estimation of sex-specific effects on human quantitative traits. Genet Epidemiol. 2007;31:338–347. doi: 10.1002/gepi.20214. [DOI] [PubMed] [Google Scholar]
- Lester LA, Rich SS, Blumenthal MN, et al. Ethnic differences in asthma and associated phenotypes: Collaborative Study on the Genetics of Asthma. J Allergy Clin Immunol. 2001;108:357–362. doi: 10.1067/mai.2001.117796. [DOI] [PubMed] [Google Scholar]
- Abney M, Ober C, McPeek MS. Quantitative trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites. Am J Hum Genet. 2002;70:920–934. doi: 10.1086/339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgain C, Hoffjan S, Nicolae R, et al. Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am J Hum Genet. 2003;73:612–626. doi: 10.1086/378208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanske RF., Jr The Childhood Origins of Asthma (COAST) study. Pediatr Allergy Immunol. 2002;13 Suppl 15:38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22:7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- Bourgain C, Abney M, Schneider D, Ober C, McPeek MS. Testing for Hardy-Weinberg equilibrium in samples with related individuals. Genetics. 2004;168:2349–2361. doi: 10.1534/genetics.104.031617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Wu X, Ober C. Best linear unbiased allele-frequency estimation in complex pedigrees. Biometrics. 2004;60:359–367. doi: 10.1111/j.0006-341X.2004.00180.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.