Abstract
Recent evidence suggests that the microbial community in the human intestine may play an important role in the pathogenesis of obesity. We examined 184,094 sequences of microbial 16S rRNA genes from PCR amplicons by using the 454 pyrosequencing technology to compare the microbial community structures of 9 individuals, 3 in each of the categories of normal weight, morbidly obese, and post-gastric-bypass surgery. Phylogenetic analysis demonstrated that although the Bacteria in the human intestinal community were highly diverse, they fell mainly into 6 bacterial divisions that had distinct differences in the 3 study groups. Specifically, Firmicutes were dominant in normal-weight and obese individuals but significantly decreased in post-gastric-bypass individuals, who had a proportional increase of Gammaproteobacteria. Numbers of the H2-producing Prevotellaceae were highly enriched in the obese individuals. Unlike the highly diverse Bacteria, the Archaea comprised mainly members of the order Methanobacteriales, which are H2-oxidizing methanogens. Using real-time PCR, we detected significantly higher numbers of H2-utilizing methanogenic Archaea in obese individuals than in normal-weight or post-gastric-bypass individuals. The coexistence of H2-producing bacteria with relatively high numbers of H2-utilizing methanogenic Archaea in the gastrointestinal tract of obese individuals leads to the hypothesis that interspecies H2 transfer between bacterial and archaeal species is an important mechanism for increasing energy uptake by the human large intestine in obese persons. The large bacterial population shift seen in the post-gastric-bypass individuals may reflect the double impact of the gut alteration caused by the surgical procedure and the consequent changes in food ingestion and digestion.
Keywords: methanogen, microbial community, syntrophy, pyrosequencing, microbiome
Obesity is an enormous public health problem, arising as a consequence of alterations in eating behavior and how the body regulates energy intake, expenditure, and storage. Although an increased intake of energy-dense foods, especially when combined with reduced physical activity, surely contributes to the high prevalence of obesity, the existence of complex systems that regulate energy balance requires that this paradigm be considered in a larger context (1). In particular, recent evidence suggests that the gut microbiota may play a role in obesity by increasing the host's energy-harvesting efficiency (2–4). A mouse model has shown that Methanobrevibacter smithii, the predominant archaeon in the human gut, enhanced short-chain fatty acid (SCFA) production by fermentative bacteria by removing H2 and formate (3). Gut microbial diversity surveys have demonstrated a lower percentage of Bacteroidetes and proportionally more Firmicutes in obese mice compared with their lean counterparts (5). Similar to these mice experiments, Ley et al. (6) have shown that the relative proportion of Bacteroidetes increased while Firmicutes decreased in humans on a weight-loss program. But with Firmicutes containing at least 250 genera and Bacteroidetes containing more than 20 genera, the observed differences at the higher division level have yet not pinpointed the specific bacteria exclusively associated with obesity (7).
The treatment of obesity is challenging. Bariatric surgery is currently the only available treatment for morbid obesity that consistently achieves and sustains substantial weight loss (8). Various surgical procedures designed to interfere with the ingestion and/or absorption of foods have been developed over the last 50–60 years. The Roux-en-Y gastric bypass (RYGB), currently the most commonly performed bariatric operation, involves creating a small (about 15–30 mL) gastric pouch from the fundus of the stomach. The distal stomach and proximal small intestine are bypassed by attaching the distal end of the mid-jejunum to the proximal gastric pouch (creating the Roux limb), and then reattaching the biliary and pancreatic limb at a specific location along the Roux limb. This surgery leads to changes in acid exposure to the gastric remnant and proximal small bowel, restricts the amount and types of food that can be comfortably ingested, promotes a modest degree of nutrient malabsorption by shortening the length of the small bowel, and may result in intestinal dysmotility, all of which might be expected to alter the gut microbiota. Presently, very little is known about the changes in the gut microbiota that occur after RYGB (9), and, to the best of our knowledge, no information has been published on changes in microbial diversity after RYGB in humans.
Many previous studies examining the diversity of the human gut microbiota have relied on the generation of clone libraries of the 16S rRNA gene, followed by sequencing using the Sanger method. Using this methodology, none of the largest human gut microbial diversity surveys to date has sampled more than 20,000 bacterial sequences (6, 10, 11). Nonparametric estimations and extrapolations from collector's curves predict that obtaining a much higher number of sequences can reveal as many as 500–15,000 species (10, 11), which include relatively rare members of the microbial community that collectively could have a profound impact on gut health and disease, including obesity. Pyrosequencing, a sequencing-by-synthesis method, can achieve the much higher throughput, or number of sequences, needed to reveal the full diversity of the intestinal microbial community at a lower cost than the Sanger method (12). Pyrosequencing has been used successfully to study the microbial community in animals (2), humans (13, 14), soils (15), and oceans (16).
In the current study, we used the traditional Sanger and the high-throughput 454 pyrosequencing methods to analyze the human gut microbiota in 9 individuals, 3 in each of the categories of normal weight, morbidly obese, and post-gastric bypass surgery. Our goals were to identify specific microbial lineages that may play important roles in the development of obesity and also to determine whether the presence or abundance of these microorganisms changes after successful RYGB. Using 454 pyrosequencing, we were able to analyze 184,094 16S rRNA gene sequence tags of the human intestinal bacterial community from the 9 individuals. We also quantified the abundance of Bacteria and Archaea using quantitative real-time PCR (QPCR). To the best of our knowledge, this is the first report of a molecular survey of the gut microbiota after a surgical weight-loss procedure, in this case RYGB.
Results
Subject Characteristics.
We studied stool samples from 9 subjects, 3 each in 3 groups: normal weight (nw), obese (ob), and post-gastric bypass (gb). Two of the 3 subjects in each group were female. Mean (± SD) subject age was similar in the 3 groups (nw, 36.7 ± 4.0 years; ob, 35.7 ± 4.2 years; gb, 43.3 ± 8.1 years). Mean body mass index (BMI) was 22.7 ± 2.3 kg/m2 in the nw group, 48.3 ± 7.7 kg/m2 in the ob group, and 27.7 ± 4.1 kg/m2 in the gb group. The mean preoperative BMI in the gb group was 40.6 ± 5.4 kg/m2, and the mean weight loss after RYGB was 40.7 ± 5.9 kg. Stool samples were collected between 8 and 15 months after RYGB in the 3 gb subjects; at that point, weight loss had ceased in 1 of the subjects, whereas the 2 others were continuing to lose weight.
Human Gut Microbial Community Revealed by Sanger Sequencing and High-Throughput Pyrosequencing.
We first studied the human gut microbial community by traditional Sanger sequencing by building 9 bacterial clone libraries from the test subjects. We analyzed 2,817 sequences of ca. 600 bp in length; the results are shown as a phylogenetic tree in supporting information (SI) Fig. S1. This tree clearly shows that the gut community contained sequences from 6 bacterial divisions/phyla. Most of the sequences belonged to Firmicutes and Bacteroidetes, with the rest distributed among Proteobacteria, Actinobacteria, Fusobacteria, and Verrucomicrobia. The family Prevotellaceae within the phylum Bacteroidetes and the family Erysipelotrichaceae within phylum Firmicutes harbored sequences mostly from obese individuals (indicated in red). The obesity-specific Prevotellaceae group contains carbohydrate- and protein-fermenting, acetate and H2 producers, as inferred by their cultured phylogenetic relatives, such as Prevotella ruminicola (17). Erysipelotrichaceae is peripherally related to the butyrate-producing superfamily Lachnospiraceae, including Clostridium XIVa and IV subgroups within the order Clostridiales, such as Lachnospira and Roseburia (17). Butyrate produced by the gut bacteria is a major energy source for the colonic epithelium (18). In contrast, Fusobacteria and the family Enterobacteriaceae within Proteobacteria were found only in the gb group.
To reveal the fine details of the human gut microbial community structures, we conducted massively parallel pyrosequencing on the hypervariable V6 region of the 16S rRNA gene. Used as a “tag” or “bar code,” the V6 region is part of the full-length 16S rRNA gene (16). Table 1 shows that most of the V6 tags in this study were highly similar to reference sequences. For example, more than 92% of the tags had matches in the reference database with a distance shorter than 0.05, or 95% similarity, and the remainder (8%) had matches with a distance of 0.06–0.14. During taxonomic assignments, we found that a hypervariable V6 tag can match to multiple sequences in the reference database with the same shortest distance. In the majority of cases, all of the matches had the same taxonomy. In very few cases (<3%), 2 equally short distances we seen, pointing to different taxonomic classifications; however, these differences were quite deep in the classification (meaning that the 2 references were very similar in taxonomic rankings) and did not affect family-level classification.
Table 1.
Distribution of the calculated nucleotide distances from 4 random libraries in our study
| Distance | No. of tags | Tags, % | Cumulative tags, % | Distance within |
|---|---|---|---|---|
| 0.13–0.14 | 1 | 0.001 | 100.000 | 0.14 |
| 0.12–0.13 | 71 | 0.083 | 99.999 | 0.13 |
| 0.11–0.12 | 1,332 | 1.549 | 99.916 | 0.12 |
| 0.10–0.11 | 954 | 1.110 | 98.367 | 0.11 |
| 0.09–0.10 | 583 | 0.678 | 97.257 | 0.1 |
| 0.08–0.09 | 1,012 | 1.177 | 96.579 | 0.09 |
| 0.07–0.08 | 252 | 0.293 | 95.402 | 0.08 |
| 0.06–0.07 | 1,714 | 1.994 | 95.109 | 0.07 |
| 0.05–0.06 | 823 | 0.957 | 93.115 | 0.06 |
| 0.04–0.05 | 721 | 0.839 | 92.158 | 0.05 |
| 0.03–0.04 | 3,547 | 4.126 | 91.319 | 0.04 |
| 0.02–0.03 | 122 | 0.142 | 87.194 | 0.03 |
| 0.01–0.02 | 15,328 | 17.829 | 87.052 | 0.02 |
| 0.00–0.01 | 59,513 | 69.223 | 69.223 | 0.01 |
Table 2 summarizes the number of pyrosequencing tags found for each of the 9 human subjects. The average pyrosequencing read length was 105 bp (SD = 22 bp). After applying stringent sequence trimming criteria to remove low-quality reads, we obtained a total of 184,094 high-quality trimmed reads, or 92.1% of the raw reads. The tag sequence per individual ranged from 13,963 to 31,835, with an average of 20,455. The subtotals of tags obtained within each weight group were comparable (nw, 56,600; ob, 61,916; gb, 59,016). Table 2 also shows that 419–575 phylotypes were obtained from pyrosequencing tags of the 9 subjects. Here phylotypes are defined as distinct, multiple-sequence alignment-corrected best matches to the V6 reference database V6RefDB. Our phylotype observations agree well with a previous prediction of at least 500 phylotypes in the human gut (10).
Table 2.
High-throughput pyrosequencing data summary and phylotypes
| Sample ID | Sample description | Total tags | Trimmed tags | Phylotypes |
|---|---|---|---|---|
| nw1 | Normal weight | 33,788 | 31,835 | 521 |
| nw2 | Normal weight | 18,038 | 16,260 | 423 |
| nw3 | Normal weight | 17,690 | 16,940 | 419 |
| ob1 | Obese | 14,884 | 13,963 | 424 |
| ob2 | Obese | 28,587 | 26,915 | 540 |
| ob3 | Obese | 19,478 | 17,334 | 575 |
| gb1 | Gastric bypass | 18,934 | 17,544 | 512 |
| gb2 | Gastric bypass | 26,621 | 24,794 | 434 |
| gb3 | Gastric bypass | 19,976 | 18,509 | 495 |
| Total | 197,996 | 184,094 |
Phylotypes were determined by obtaining each pyrosequencing tag's best match in the V6RefDB database and then combining all tags with the same reference.
Fig. 1 illustrates the breakdown of the bacterial taxonomy at the class level, represented by pyrosequencing tags. Despite highly diverse bacterial communities and interindividual differences, obesity and gastric bypass clearly affected the intestinal microbial community. Compared with the nw and ob groups, the gb group had a marked increase in the relative abundance of Gammaproteobacteria (P < 0.005 for both cases) and proportionally fewer Clostridia (P = 0.002 for nw and 0.029 for ob). Verrucomicrobia were generally abundant in the nw and gb groups but rare in the ob group.
Fig. 1.
Taxonomic breakdown of human intestinal bacterial V6 tags obtained by pyrosequencing in the normal-weight (nw1, nw2, and nw3), obese (ob1, ob2, and ob3), and post-gastric bypass (gb1, gb2, and gb3) subjects.
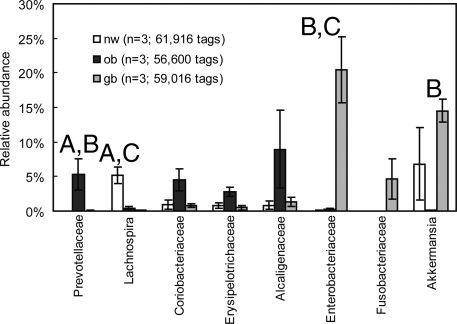
Fig. 2 shows the relative abundance of pyrosequencing tags at the family level. Prevotellaceae within the class Bacteroidetes were significantly enriched in the ob group compared with the nw group (P = 0.040), even though the difference in the relative abundance of Bacteroidetes between the 2 groups was insignificant (P = 0.061). Three other families—Coriobacteriaceae (phylum Actinobacteria), Erysipelotrichaceae (phylum Firmicutes), and Alcaligenaceae (phylum Proteobacteria)—also were somewhat enriched in the ob group (Fig. 2). Higher numbers of Coriobacteriaceae have been reported in formula-fed infants compared with breast-fed infants (19). The family Alcaligenaceae, within the order Burkholderiales of Betaproteobacteria, also has been reported in the human gut (20). In contrast, Enterobacteriaceae and Fusobacteriaceae were markedly increased in the gb group. The relative abundance of Enterobacteriaceae was higher in the gb group than in the the nw and ob groups (P = 0.006 in both cases). The relative abundance of sequences affiliated with Lachnospira within the order Clostridiales was depleted in the ob and gb groups compared with the nm group (P = 0.007 and 0.006, respectively). The beneficial Lachnospira are known pectin degraders (21) and play important roles in the colonic fermentation of dietary fibers. Interestingly, Verrucomicrobia phylotypes related to an uncultured Akkermansia sp. (GenBank accession: AJ400275) were prominent in 2 of the 3 subjects in the nw group and all 3 subjects in the gb group (3,813/61,916 and 8,597/59,016 sequences, respectively) but were poorly represented in the ob group (76/56,000 sequences).
Fig. 2.
Bacterial families enriched in the ob and gb groups. (A) Significant (P < 0.05) difference between the nw and ob groups. (B) Significant difference between the ob and gb groups. (C) Significant difference between the gb and nw groups. Error bars represent the standard error of the mean (n = 3). The P values are based on the 2-sample t-test assuming equal variances.
Fig. 3 compares taxonomic assignments by Sanger sequences and pyrosequencing tags. In general, these 2 methods were in good agreement for the more abundant taxa (Sanger reads > 10; pyrosequencing tags > 100). Data points lying along the x-axis are rare taxa missed by Sanger sequencing but captured by the higher-throughput pyrosequencing.
Fig. 3.
Correlation of Sanger sequence- and pyrosequencing tag- predicted taxonomic assignments. The number of sequences within a taxon by full-length sequences is plotted against the number of tags from the same taxon using pyrosequencing.
Archaea and Bacteria QPCR.
We used QPCR to enumerate total Bacteria and Archaea. We also quantified the order Methanobacteriales, which harbors hydrogenotrophic methanogens, including the predominant human intestinal archaeon Methanobrevibacter smithii. As shown in Fig. 4, Bacteria counts in the test subjects ranged from 1.5 × 1010 to 1.1 × 1011 copies of 16S rRNA gene per gram of wet stool. Intriguingly, we found higher numbers of Archaea in the ob group than in the other 2 groups. On average, the subjects in the ob group had 5.5 × 106 copies (n = 3) of 16S rRNA genes per gram of stool, compared with nondetectable levels in the nw group and 7.5 × 103 copies/g from only 1 of the 3 subjects (gb1) in the gb group. The abundance of Methanobacteriales in the test subjects matched very well with that of total Archaea, confirming the dominance of hydrogen-consuming methanogens in the human intestinal archaeal population. In samples harboring Archaea (from subjects ob1, ob2, and ob3 and gb1), QPCR demonstrated no amplification with primers targeting methanogen subgroups other than Methanobacteriales, and PCR-DGGE with archaeal primers detected only a single band (data not shown). Thus, our results suggest a low archaeal diversity in our samples.
Fig. 4.
The numbers of Bacteria, Archaea, and Methanobacteriales quantified by real-time QPCR. Error bars represent the standard error of the mean (n = 3).
Microbial Community Comparison by UniFrac.
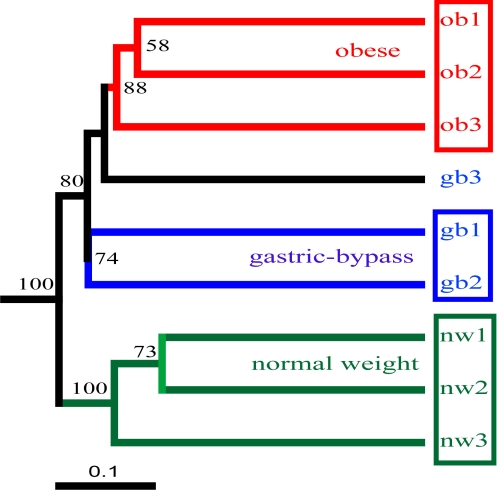
Fig. 5 shows the clusters based on UniFrac metrics. The most important finding is that the 3 subjects in the nw group formed a cluster very distinct from the ob group, indicating that they harbored different microbial communities. Two of the 3 subjects in the gb group formed their own cluster.
Fig. 5.
Clustering of human intestinal microbial communities in the 3 test groups (nw, ob, and gb), based on the unweighted UniFrac analysis of the phylogenetic tree shown in Fig. S1. The branch length represents the distance between environments in UniFrac units, as indicated by the scale bar. Jackknife counts are based on 100 replicates, and only values > 50 are shown.
Species Richness Revealed by 16S rDNA V6 Pyrosequencing.
Table S1 shows the great species richness in the human intestinal microbial communities. The nonparametric estimators Chao1 and ACE (abundance-based coverage estimator) (22, 23) project a total of 1,206–2,217 bacterial operational taxonomic units (OTUs). Additional sampling would be needed to capture the difference between the observed number of OTUs and the Chao1 or ACE estimates, as shown by rarefaction curves (Fig. S2) based on the best matches in the V6refDB. Because different pyrosequencing tags can match to the same reference sequence in V6refDB, rarefaction curves based on best-match groupings could underestimate species richness. One way to overcome this is to cluster sequence tags into OTUs of defined distances (e.g., 0.03, 0.06, 0.09). Fig. S3 shows the species accumulation curve of sample nw1 using the DOTUR program. Clearly, even after sampling 31,835 sequences, the number of OTUs continued to increase at 3% (species level) or 6% (genus level). This observation warrants additional sampling to determine the true microbial diversity in the microbial community of the human intestine.
Discussion
In this study, we interrogated the genetic diversity of the microbial community of the human intestine in relation to a disease (obesity) and a surgical weight-loss procedure (RYGB) using culture-independent molecular phylogenetic and ecological statistic methods. Our results are notable for the following: (i) Obese individuals have distinctly different intestinal communities than normal-weight individuals, and (ii) RYGB alters the intestinal microbial community in a unique way.
Our results confirm an association between methanogenic Archaea and obesity. The normal-weight individuals had no methanogens, and only 1 post-gastric bypass individual had a small number of methanogens (Fig. 4). This corroborates findings from a recent metagenomic study that detected more Archaea-derived gene fragments in genetically obese ob+/ob+ mice than in their lean ob+/− or ob−/− relatives (2). The mechanism connecting obesity to methanogens may be, at least in part, the transfer of hydrogen gas (H2) from a H2-producing bacterium to a H2-using methanogen. Microbiologists and environmental engineers have long recognized interspecies H2 transfer in rumen and engineered reactor systems (24, 25). Recent animal studies have shown that methanogens increase host energy extraction from indigestible polysaccharides (3). The authors suggested that methanogens remove fermentation intermediates, such as H2 or formate, thus relieving thermodynamic limitations and allowing greater production of SCFAs that are available to be absorbed across the intestinal epithelium. If interspecies H2 transfer is indeed active in the human model, then the obesity-specific Archaea we found in this study (which are H2 consumers) are likely to form a syntrophic partnership with H2-producing bacteria. Possible H2-producing bacterial partners could be members of Prevotellaceae (phylum Bacteroidetes), which we also found associated with obese individuals. Thus, our QPCR results for methanogens and bacterial diversity results from clone libraries and pyrosequencing support the hypothesis that Bacteria–Archaea syntrophy may be a novel biomarker of susceptibility to obesity.
A previous human weight-loss study of 12 obese subjects found an increase in the fraction of Bacteroidetes relative to each subject's baseline level (6). We did not study the longitudinal effect on the gut microbiota associated with weight loss by individuals. We found somewhat more Bacteroidetes in the obese individuals than in normal-weight individuals, but the difference was not significant. But our results clearly show that Prevotellaceae, a subgroup of Bacteroidetes, was significantly enriched in the obese individuals. Ley et al. (6) did not report a change in subgroups within the Bacteroidetes. It is possible that weight loss affects these subgroups differently, causing one subgroup to increase while another decreases. Diet also may help explain the apparently different results. The test subjects in the study of Ley et al. (6) were on either a fat-restricted or carbohydrate-restricted diet, whereas we did not limit dietary components in our test subjects. A recent study found no difference between the fraction of Bacteroidetes in obese and nonobese individuals (26), in agreement with our results. The same study also reported no significant change in the relative abundance of Bacteroidetes in obese individuals on a weight-loss diet.
Although the connection between the relative abundance of Bacteroidetes and diet-induced weight loss of obese humans remains under debate (6, 26), our results clearly show that an effective surgical treatment for morbid obesity, RYGB, markedly altered the stool microbial community structure toward a large increase in Gammaproteobacteria (96.2% of which were members of the family Enterobacteriaceae), a proportional decrease in Firmicutes, and a loss of methanogens. Although the reasons for this large shift are not yet clear, we propose a 4-pronged explanation. First, the anatomical and physiological changes caused by RYGB, particularly relating to changes in acid exposure to the gastric remnant and Roux limb and the shortened small intestinal length, likely favor the fast-growing facultative anaerobes of Gammaproteobacteria over such obligate anaerobes as Clostridia within Firmicutes. Second, facultative anaerobes are favored by the input of dissolved oxygen due to the shortened small bowel; normally, oxygen is completely removed before the beginning of the colon. Third, the surgical procedure, which bypasses the upper small intestine, also might relocate some of the typical small intestine microbiota, such as Enterobacteriaceae, to the large intestine, given the shorter length of the small bowel, possibly leading to more rapid transit of ingested materials to the colon. Fourth, because it drastically alters the anatomy of the gastrointestinal tract, gastric bypass surgery may affect food ingestion and digestion due to the reduced stomach and the shortened small intestine. Therefore, our results on the large microbial population shift in gastric bypass individuals may reflect the double impact of the gut alteration caused by the surgical procedure and the consequent changes in food ingestion and digestion.
To minimize the effect of antibiotics on microbial composition changes, we purposely chose gastric bypass patients who had not taken any antibiotics within 3 months before stool sample collection. Previous studies have shown that the dominant human microbiota is resilient to antibiotic treatment, and the microbial community composition generally returns to a predrug state after 30 days (27). Another recent study found that the microbial composition in 3 individuals who had received a 5-day course of ciprofloxacin closely resembled that found before treatment by 4 weeks after the drug challenge (14). Although our findings reflect the community shift for a single time point after surgery, the time point coincides with dramatic weight loss. More investigation involving more study subjects and temporal samplings before and after surgery is needed to gain more insight into the mechanisms involved and the cause-and-effect relationship among obesity, weight loss, the intestinal microbiota, and the influence of dietary changes after gastric bypass surgery.
Our approach of using V6 pyrosequencing sampled a large number of sequences in the human large intestine. The good agreement between taxonomic assignments done by Sanger sequences and pyrosequencing tags indicates that pyrosequencing tags described microbial community structures largely equivalent to those by the longer Sanger sequences at the phylum, class, order, and genus levels. At the same time, pyrosequencing revealed more rare taxa than Sanger sequencing. Our findings agree with a recent in-depth study that found equivalent taxonomic assignments by Sanger and tag pyrosequencing of V3 and V6 regions (28). But species-level diversity generally requires longer 16S rRNA gene PCR amplicons, which can be obtained with additional primers. We are aware of the limitations of using the sequence similarity of the 16S rRNA gene to define phylotypes. Multiple rRNA genes in a bacterial genome can be 3% divergent (29); thus, distance-based prediction may overestimate species richness. In this study, we used a lower-end 97% cutoff for V6 tag phylotypes in rarefaction curves, which allowed us to more readily compare our bacterial diversity results with those of other studies. As with other molecular diversity surveys, primer coverage is a factor that may underestimate the true microbial diversity in a sample; however, this should not affect our comparisons across samples, given that primer coverage is not sample-dependent. Consequently, despite the implications of primer selection, our comparative conclusions still hold.
Summary Hypothesis and Implications.
Our results extend the findings of others by demonstrating that obese individuals harbor unique H2-producing bacterial groups, particularly members of the Prevotellaceae family and certain groups within Firmicutes. These H2-producing bacteria coexist in the gastrointestinal tracts of obese subjects with relatively high numbers of H2-oxidizing methanogenic Archaea. Methanogens may comprise up to 10% of all anaerobes in the colon. They may be more likely to inhabit epithelium biofilms than stools because of their slower growth rates (25). On the other hand, living closer to the intestinal epithelium may expose the strictly anaerobic methanogens to a more oxidizing environment. Analysis of colon mucosal biopsy specimens is needed to address the question of whether proportions of methanogens are higher in mucosal biofilms than in stool samples. This analysis also will be useful in determining whether higher numbers of H2-producing bacteria inhabit the gut mucosal layer. Previous studies have found differing bacterial composition in mucosa and stool samples (10), and the mucosal bacteria perform beneficial functions, including enhancing nutrient absorption, inducing host immunity, and modifying host gene expression (30).
Plant polysaccharides and dietary fibers are fermented by gut bacteria with the production of SCFAs, including formate, acetate, propionate, butyrate, and lactate. An increase in H2-oxidizing methanogenesis facilitates fermentation, which produces more SCFA. Formate also can be directly used by hydrogenotrophic methanogens. Propionate, butyrate, and lactate can be fermented to acetate and H2, the latter of which is used by hydrogenotrophic methanogens. Consequently, the increase of H2-oxidizing methanogenesis should increase the conversion of plant polysaccharides to SCFAs, particularly acetate. Our findings have a clear link to energy homeostasis in humans, because the SCFAs produced by the fermentative bacteria are taken up through the epithelium of the human intestine, whereas H2 is a well-known factor for energy exchange within microbial communities (31).
Our findings lead us to the following hypothesis regarding interspecies H2 transfer between bacterial and archaeal species and how it affects energy uptake by humans. Rapid H2 uptake by methanogens accelerates the fermentation of plant polysaccharides and other carbohydrates by H2-producing fermenters such as Prevotellaceae. The accelerated fermentation stimulates the hydrolysis of usually indigestible organic matter and leads to increased production of acetate, which is absorbed by the human gut.
Materials and Methods
Human Subjects and Stool Sampling.
We studied 9 unrelated human subjects without current gastrointestinal symptoms or a history of chronic gastrointestinal problems, divided into 3 groups: 3 individuals of normal weight (subjects nw1, nw2, and nw3; BMI, 20–25 kg/m2), 3 morbidly obese individuals (subjects ob1, ob2, and ob3; BMI > 35 kg/m2), and 3 individuals who had undergone RYGB at least 6 months earlier (subjects gb1, gb2, and gb3; preoperative BMI > 35 kg/m2). None of the human subjects lived in the same household, and none had received any antibiotic, probiotic, or prebiotic agents in the 3-month period before the collection of fecal samples. After collection, stool samples were frozen at −80 °C until the molecular analyses were conducted. The Mayo Clinic's Institutional Review Board approved the study, and all subjects provided written informed content.
Clone Library Construction, Phylogenetic Analysis, and Comparison of Communities by UniFrac.
We isolated genomic DNA from human stool samples (wet weight, 0.2 g) with the QIAamp DNA Stool Kit (Qiagen), following the manufacturer's instructions. We constructed 16S rDNA clone libraries with primers 8f and 1525r (32) and sequenced the inserts with an ABI 3730xl capillary sequencer. Sequences were aligned with NAST (33), imported into an ARB database (greengenes.arb, version 23, May 2007), and added into a backbone tree (“tree_all” with 137,916 near-full-length reference sequences) with the “LanemaskPH” filter. Possible differences in microbial community structures among the 3 study groups were explored using unweighted UniFrac metrics (5). The library and tree construction is described in more detail in SI Materials and Methods.
16S rDNA V6 Pyrosequencing and Data Analysis.
We used bacterial primers 967f and 1046r (16) to amplify the V6 region of the 16S rRNA gene. Amplicon pyrosequencing was performed with standard 454/Roche GS-FLX protocols (12). For data analysis, we modified the methodology described by Sogin et al. (16) by restricting taxonomy assignments to those alignments longer than 57 bp; by doing so, we excluded very short BLAST matches that were not informative. We used the Ribosomal Database Project's Classifier 2.0 (34) to assign taxonomy. For distance-based species richness estimation, we aligned the tags using MAFFT with the PartTree algorithm (35), calculated a distance matrix with QuickDist (16), and clustered sequences into OTUs with DOTUR (36). Details of tag analysis are described in SI Materials and Methods.
Real-Time QPCR.
To quantify Bacteria, Archaea, and the archaeal subgroups, we performed 16S rRNA gene-targeted QPCR with either SYBR-green (for Bacteria) or TaqMan detection (for Archaea, Methanobacteriales, Methanomicrobiales, and Methanosaetaceae) (37, 38). Details of the standards and PCR conditions are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Julie Huber for the QuickDist code, Patrick Schloss for guidance on using the DOTUR program, Doug Fuller for cluster computing support, and Andrew Kato Marcus for his critical review of the manuscript. Funding for this research was provided by an Arizona State University–Mayo Clinic research collaboration grant.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences obtained in this study have been deposited in the GenBank database (accession nos. FJ452085–FJ454865).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812600106/DCSupplemental.
References
- 1.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 3.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiBaise JK, et al. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc. 2008;83:460–469. doi: 10.4065/83.4.460. [DOI] [PubMed] [Google Scholar]
- 5.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Cole JR, et al. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchwald H, et al. Bariatric surgery: A systematic review and meta-analysis. JAMA. 2004;292:1724–1737. doi: 10.1001/jama.292.14.1724. [DOI] [PubMed] [Google Scholar]
- 9.Bjorneklett A, Viddal KO, Midtvedt T, Nygaard K. Intestinal and gastric bypass: Changes in intestinal microecology after surgical treatment of morbid obesity in man. Scand J Gastroenterol. 1981;16:681–687. doi: 10.3109/00365528109182030. [DOI] [PubMed] [Google Scholar]
- 10.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson AF, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roesch LFW, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marounek M, Duskova D. Metabolism of pectin in rumen bacteria Butyrivibrio fibrisolvens and Prevotella ruminicola. Lett Appl Microbiol. 1999;29:429–433. [Google Scholar]
- 18.Roediger WEW. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen HJM, et al. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl Environ Microbiol. 2000;66:4523–4527. doi: 10.1128/aem.66.10.4523-4527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 21.Rode LM, Genthner BRS, Bryant MP. Syntrophic association by cocultures of the methanol-utilizing CO2-H2-utilizing species Eubacterium limosum and pectin-fermenting Lachnospira multiparus during growth in a pectin medium. Appl Environ Microbiol. 1981;42:20–22. doi: 10.1128/aem.42.1.20-22.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- 23.Chao A, Ma MC, Yang MCK. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika. 1993;80:193–201. [Google Scholar]
- 24.Bryant MP. In: Microbial Energy Conversion. Schlegel HG, Barnea J, editors. New York: Pergamon; 1977. pp. 107–117. [Google Scholar]
- 25.Rittmann BE, McCarty PL. Environmental Biotechnology: Principles and Applications. Boston: McGraw–Hill; 2001. [Google Scholar]
- 26.Duncan SH, et al. Human colonic microbiota associated with diet, obesity, and weight loss. Int J Obes. 2008 doi: 10.1038/ijo.2008.155. in press. [DOI] [PubMed] [Google Scholar]
- 27.De La Cochetiere MF, et al. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huse SM, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooper LV, Midtvedt T, Gordon JI. How host–microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 31.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane DJ. In: Modern Microbiological Methods. Stackebrandt E, Goodfellow M, editors. Chichester, UK: Wiley; 1991. pp. 115–175. [Google Scholar]
- 33.DeSantis TZ, et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008 doi: 10.1093/bib/bbn013. bbn013. [DOI] [PubMed] [Google Scholar]
- 36.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritalahti KM, et al. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microbiol. 2006;72:2765–2774. doi: 10.1128/AEM.72.4.2765-2774.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng. 2005;89:670–679. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.