Abstract
In this study, a reverse-engineering strategy was used to infer and analyze the structure and function of an aging and glucose repressed gene regulatory network in the budding yeast Saccharomyces cerevisiae. The method uses transcriptional perturbations to model the functional interactions between genes as a system of first-order ordinary differential equations. The resulting network model correctly identified the known interactions of key regulators in a 10-gene network from the Snf1 signaling pathway, which is required for expression of glucose-repressed genes upon calorie restriction. The majority of interactions predicted by the network model were confirmed using promoter-reporter gene fusions in gene-deletion mutants and chromatin immunoprecipitation experiments, revealing a more complex network architecture than previously appreciated. The reverse-engineered network model also predicted an unexpected role for transcriptional regulation of the SNF1 gene by hexose kinase enzyme/transcriptional repressor Hxk2, Mediator subunit Med8, and transcriptional repressor Mig1. These interactions were validated experimentally and used to design new experiments demonstrating Snf1 and its transcriptional regulators Hxk2 and Mig1 as modulators of chronological lifespan. This work demonstrates the value of using network inference methods to identify and characterize the regulators of complex phenotypes, such as aging.
Keywords: chronological aging, gene networks, Snf1 pathway, systems biology
Characterizing biomolecular interaction networks can shed new insight into biological processes, including the complex mechanisms regulating longevity and aging. Organisms like the budding yeast Saccharomyces cerevisiae have become valuable model systems to discover genes modulating longevity and to identify their associated interaction networks, many of which are conserved in metazoans (1, 2). Replicative lifespan (RLS), the number of daughter cells an individual mother can produce before senescence, and chronological lifespan (CLS), the length of time cells from stationary phase cultures can remain viable in a quiescent state, are two definitions of yeast age that have achieved common acceptance (1, 3). RLS and CLS have been proposed as models for mitotically active and postmitotic metazoan cells, respectively (1, 3).
Longevity extension in response to calorie restriction has been observed in organisms ranging from yeasts to mammals (1, 2, 4) and can be observed in S. cerevisiae by limiting the glucose concentration in the growth medium (2, 5). Consequently, many longevity genes have been identified by their role in relevant cellular processes, such as glucose signaling (5–9). Identifying these genes and growth conditions is a first step in understanding the mechanisms linking aging and calorie restriction. Defining the network of regulatory interactions between these genes could further our understanding of the processes that underlie aging.
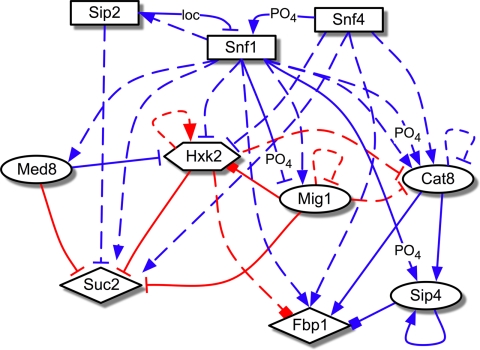
The development of methods to characterize biomolecular networks has been an active area of research (10–18). Recently, we developed an integrated experimental/computational reverse-engineering strategy, network identification by regression (NIR) (19), for the elucidation of gene regulatory networks. In the present study, we applied this method to 10 genes selected from the glucose-responsive Snf1 pathway (Fig. 1 and Table S1). Snf1, the homolog of human AMP kinase, is essential for release of glucose repression (20). It is the catalytic subunit of the heterotrimeric SNF1 complex, which also contains the coactivating γ subunit Snf4 and 1 of 3 ß subunits (Sip2 in our network) that influence subcellular localization of the complex (21). Many genes in the Snf1 network are known to affect RLS when perturbed, including Snf1, Sip2, Snf4 (22), Mig1 (23), and Hxk2 (5, 8). In a glucose-rich environment, the transcription factor (TF) Mig1 represses alternative carbon source metabolism and gluconeogenesis gene expression, including enzyme SUC2 and TF CAT8 (7, 9, 21). Hxk2, the predominant glucose kinase in the first step of glycolysis, shows glucose-dependent nuclear localization and associates with DNA-binding factors Mig1 and Mediator subunit Med8 at the promoter of SUC2 to repress its expression (24, 25). Glucose exhaustion activates SNF1, which translocates from the cytoplasm to the nucleus and deactivates Mig1 by phosphorylation. This triggers the translocation of Mig1 and Hxk2 to the cytoplasm, resulting in increased expression of SUC2, as well as CAT8, TF SIP4, and their downstream target FBP1 (7, 25). Expression changes in response to calorie restriction (Fig. S1) are thus consistent with the known interactions between these genes, as illustrated by the condensed representation of the Snf1 gene regulatory network architecture shown in Fig. 1.
Fig. 1.
Known interactions in the Snf1 network. Diagram of transcriptional regulatory influences previously described in the literature (see Table S1) by regulator proteins (source of arrows) on the expression of target genes (arrowheads) in glucose repressing (2% glucose, red edges) vs. low glucose growth conditions (including nonfermentable carbon sources) (blue edges). Solid lines denote known physical TF binding to regulatory sequences of target genes; dashed lines denote functional interactions in which the target's expression is regulated through possible intermediaries. Arrows denote activation, bars denote repression, and boxes denote physical associations established but not completely characterized. Nontranscriptional regulatory interactions are included where relevant: “PO4” denotes phosphorylation and “loc” denotes regulation through subcellular localization. Shapes of nodes indicate known functional attributes of proteins: rectangle, kinase or kinase-associated protein; circle, TF; diamond, enzyme; hexagon, dual enzyme/TF.
The NIR method uses mRNA expression changes that arise in response to network gene perturbations (applied here as small, second copy over-expressions) to formulate a first-order network model, which provides a quantitative, directed, and unsupervised description of functional transcriptional interactions. We previously applied the NIR method to a nine-gene subnetwork of the SOS DNA damage response pathway in Escherichia coli (19). Here, we investigate the utility of this approach in a eukaryotic organism using the known interactions of the Snf1 gene regulatory network as an initial benchmark.
Results
mRNA Expression Profiling and Network Inference.
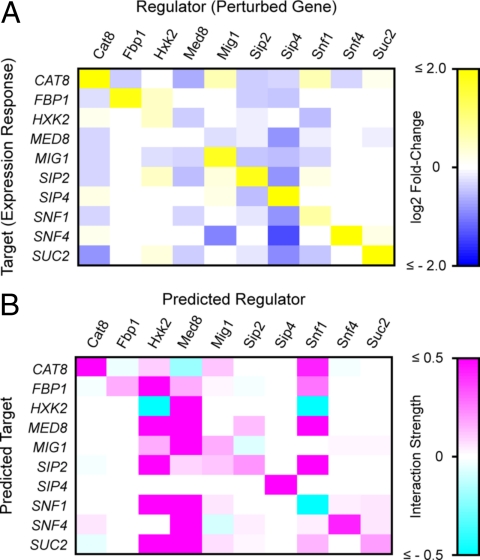
Expression changes in response to ≈2- to 4-fold over-expression of each network gene were profiled using strains containing an integrated second copy of each gene under the control of a doxycycline-inducible promoter (26). After induction, cultures were grown overnight in 2% glucose synthetic media to OD600 ≤ 0.5, which maintained cells in log phase at transcriptional steady-state conditions. Real-time quantitative RT-PCR (qRT-PCR) was used to assay mRNA expression changes relative to an isogenic control strain expressing GFP (Tables S2–S4). GFP mRNA levels in control cultures were measured as an indicator of perturbation size. Expression changes in response to these perturbations were nearly all less than 2-fold relative to control (Fig. 2A), and included responses from regulators normally active in low-glucose conditions, such as Snf1 (21), Cat8, and Sip4 (7). Thus, exogenous cis-regulatory elements in the doxycycline-inducible promoter construct (26) allowed forced expression of genes that are normally glucose repressed. Notable among these was the autotranscriptional activation of SIP4 (7): a 2-fold level of SIP4 overexpression from the second copy of the gene resulted in a 26-fold overexpression of total SIP4 mRNA.
Fig. 2.
(A) mRNA expression profiling of network genes in response to systematic perturbations. Color intensities represent the magnitude of mRNA expression changes for each gene (Rows) in response to 2- to 4-fold over-expression of each other network gene (Columns), relative to an isogenic control strain expressing GFP. Values are log2-transformed ratios measured by quantitative RT-PCR, normalized by internal standard genes ACT1 and RDN18–1. Only significant expression changes (greater than the propagated standard error) were used for network model inference and are displayed here. (B) NIR-predicted transcriptional interactions in the Snf1 gene network. This matrix is a quantitative model of predicted regulatory influences. Color intensities denote the relative strength of regulators (Column vectors) upon mRNA expression of predicted target genes (Row vectors).
To infer the Snf1 gene regulatory network, expression changes less than the propagated standard error were first filtered from the dataset. This significance threshold was chosen based on the 12 replicate RT-PCR reactions for each gene in each perturbation and control strain and the small magnitude of most expression changes. The optimal model recovered from this analysis (Fig. 2B and Tables S5–S9) correctly identified Hxk2, Med8, and Snf1 as the major transcriptional regulators in this network, as evidenced by the proportion of predicted targets and comparative magnitude of regression coefficients (indicative of interactive strength) for these regulators. Snf1 and Hxk2 are well-established key regulators of glucose repression (7, 9, 21, 27), and the Mediator complex, of which Med8 is a DNA-binding subunit, is considered a general TF required for RNA polymerase II transcription (28). In contrast, network genes with no previous known roles as transcriptional regulators, such as enzymes Fbp1 and Suc2, were predicted to have distinctly fewer low-strength regulatory influences or had regression coefficients statistically not different from zero.
We compared evidence from the literature (summarized in Table S10) with our network model to determine the sensitivity (the percentage of known interactions the NIR model successfully identified) and precision (the percentage of predicted interactions that are consistent with known interactions) of the NIR algorithm, which were found to be 65% and 22%, respectively. Correctly identified known interactions between Snf1 and its targets contributed largely to the sensitivity measure. However, the NIR model also predicted many interactions with no previous literature evidence. The highest proportion of these were observed as input predictions for SNF1 complex subunits SNF1, SNF4, and SIP2, whose regulatory roles have been well-characterized (21) but whose transcriptional regulation has not been previously detailed. We therefore performed additional experiments to test and validate these predicted interactions, as described below.
Experimental Verification of Network Model Predictions.
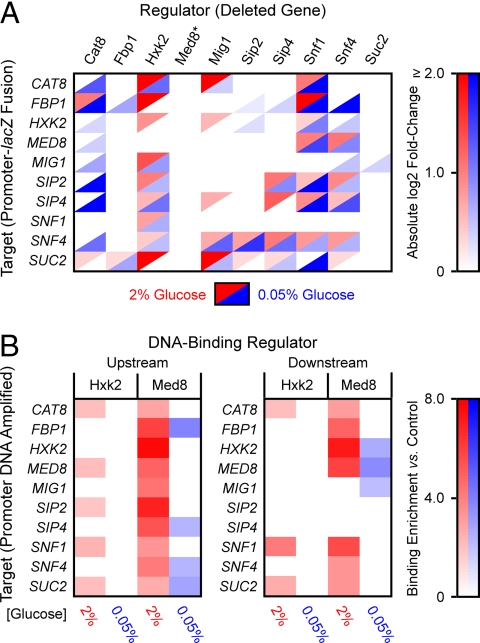
In predicted functional interactions, a putative regulator affects the expression of its target through one or more intermediaries. These were tested using deletion strains for each network gene (except MED8, whose deletion mutant is inviable), transformed with plasmid shuttle vectors containing the promoter region from each potential target cloned in-frame with the lacZ reporter gene. ß-galactosidase (ß-gal) activity for each possible target promoter-lacZ plasmid/regulator gene deletion pair was compared to activity from the same plasmid construct in the isogenic wild-type strain. These experiments were performed with cultures grown in both 2% and 0.05% glucose, as some NIR model predictions were observed for regulators previously characterized to be active either during calorie restriction or in both conditions.
Fig. 3A summarizes experimental results for statistically significant (P ≤ 0.05) ratios of ß-gal activity in gene deletion strains relative to control strains (see also Tables S11–S14). The functional importance of Hxk2 and Snf1 as key regulators, based on the number of targets and the relative magnitude of expression changes, was confirmed by these data. Fbp1 and Suc2 deletions caused relatively few low-magnitude changes in target-gene promoter activity, also consistent with the NIR model. Only one network regulator (Snf4) caused significant changes in target promoter activity not predicted in the NIR model (potential false negatives). These results may be because of Snf4's role as coactivator of Snf1 (see Fig. 1), so that over-expression of Snf4 does not lead to appreciable changes in activity of Snf1, while deletion of Snf4 does.
Fig. 3.
(A) Experimental confirmation of NIR-predicted functional interactions. Color intensities represent the absolute magnitude of ß-gal activity in deletion strains (Columns) expressing lacZ target-promoter fusions (Rows) relative to the same construct in the isogenic wild-type strain (BY4742). The essential Med8 protein (asterisk) could not be tested with this method. For clarity of presentation, absolute values of statistically significant (P ≤ 0.05) expression ratios are displayed. (B) In vivo ChIP-qPCR experimental results for regulators Hxk2 and Med8. Upstream denotes detection of Hxk2 or Med8 binding (Columns) to promoter DNA for each network gene (Rows); downstream indicates binding detected within the target coding region. Color intensities represent the ratio of enrichment of target DNA captured by immunoprecipitated Hxk2 or Med8 relative to control (isogenic wild-type strain BY4742), determined by real-time qPCR. Only statistically significant (P ≤ 0.10) expression changes are displayed.
In vivo TF-promoter binding for the repressor Mig1 (10) and the gluconeogenic activators Cat8 (29) and Sip4 (10) has been observed by chromatin immunoprecipitation/DNA microarrays (ChIP-chip) and other experimental methods (see Table S1). Hxk2 and Med8 were previously identified with Mig1 as physical repressors of SUC2 expression (24, 25), and Med8 as a repressor of HXK2, binding cis-regulatory sequences within its coding region (24). We used ChIP with detection by real-time quantitative PCR (ChIP-qPCR) to test for Hxk2 and Med8 binding to the promoters [≈350 base pairs (bp) before the ATG start codon] and downstream regions (≈300 bp after the start codon) of all network genes for cells grown in both 2% and 0.05% glucose. These experiments enabled us to test the NIR network model predictions for the essential Med8 protein, and to ascertain whether Hxk2 acts as a TF for network genes other than SUC2.
ChIP-qPCR results revealed that Med8 and Hxk2 bind in varying combinations to regulatory regions of all network genes in a statistically significant manner (Fig. 3B, Tables S15–S18). A P value of ≤0.10 was used reflecting the lower sample numbers, diminished template quality, and consequent measurement noise associated with the ChIP assay. Med8 results were consistent with the NIR network model (see Fig. 2B) and expectations that the Mediator complex acts as a general RNA Pol II transcription factor (28). Interestingly, both promoter and downstream chromatin fragments of the same gene immunoprecipitated with Med8 in many instances, which may be indicative of DNA secondary structures spanning both regions. Evidence for this type of Mediator regulation has been reported for the HXK2 gene (30), the GAL1 gene (31), and in genome-wide ChIP-chip studies of Mediator in the fission yeast Schizosaccharomyces pombe (31). Dual binding was not observed in reactions for the control actin gene ACT1 (see Tables S15–S18), ruling out artifacts of inefficient chromatin shearing. Data for Hxk2 were consistent with evidence for binding to the SUC2 gene (25). We also saw enrichment of Hxk2-bound chromatin from the CAT8 and SNF1 promoters and downstream regions, and from the promoter regions of MED8 and SIP2 (see Fig. 3B). No significant enrichment of Hxk2-bound chromatin was observed in cells grown in 0.05% glucose, consistent with the cytoplasmic localization of Hxk2 in the absence of glucose (25).
Compared to our experimental data (in both 2% and 0.05% glucose culture growth) and literature evidence (Table S19), the NIR network model showed 62% sensitivity and 69% precision. False-positive interactions contributing to the precision result may be a result of posttranscriptional influences predicted in the NIR model that are not identified by measuring promoter-lacZ fusion expression. This type of regulation has been reported for other glucose-repressed genes (7, 32). Of the interactions predicted by NIR, 24% had the opposite sign; that is, a repressing influence was predicted when an activating influence was indicated by experiments, or vice versa. The majority (9 of 16) of these were TF Hxk2 and Mig1 interactions. As an additional means of assessing performance, we performed a permutation test with models generated from 10,000 iterations of NIR using randomized expression data. The true NIR model had higher measures of sensitivity and precision than 88% and 98% of the randomized models, respectively (Fig. S2). Thus, our results show that the NIR algorithm infers the Snf1 gene regulatory network with a high degree of sensitivity, precision and significance.
Regulation of SNF1 Gene Expression and its Effect on Chronological Lifespan.
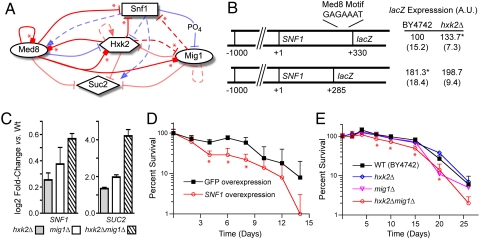
The NIR network model predicted that Med8, Hxk2, Snf1, and Mig1 have the largest effects on SNF1 gene expression. ChIP-qPCR data further confirmed Med8 and Hxk2 as direct regulators of SNF1, with higher affinity to downstream than upstream regions for both regulators. In previous studies, SNF1 gene expression was insensitive to growth on a nonfermentable carbon source (33) and increased only slightly during the diauxic shift (6, 34). However, we observed consistent reductions in log-phase growth rate in response to modest levels of SNF1 overexpression during perturbation expression profiling (a doubling time of 1.70 h−1 vs. 1.46 h−1 for the GFP overexpression control strain; P < 0.01), suggesting that small changes in SNF1 mRNA levels are physiologically important. Hxk2 functions as both a glycolytic enzyme and a transcriptional regulator; consequently, a clear definition of its role in glucose signaling has remained elusive (9, 24, 27). Because Snf1 is required for release from glucose repression, and Hxk2 was confirmed as a direct regulator of SNF1, we carried out additional experiments to clarify its role in SNF1 transcriptional control (see hypothesized schematic in Fig. 4A).
Fig. 4.
Characterization of SNF1 transcriptional regulation and its effects on chronological lifespan. (A) Schematic of hypothesis tested in subsequent experiments that SNF1 gene expression is repressed by Hxk2, Med8, and Mig1 in 2% glucose (red arrows), in a manner analogous to the previously detailed regulation of SUC2 (asterisks denote new interactions predicted by the network model and confirmed experimentally in this study). Definitions of other symbols are the same as in Fig 1. (B) A putative Med8 binding motif in the coding sequence of SNF1 represses expression of SNF1 promoter-lacZ fusions with Hxk2. Plasmids containing SNF1 promoters and variable lengths of the SNF1 coding region to include (SNF1+330) or exclude (SNF1+285), a sequence similar to the Med8 consensus motif fused in-frame to the lacZ reporter gene were transformed into hxk2Δ and isogenic wild-type strain BY4742. ß-gal activity was measured in SC media + 2% glucose. Values are normalized such that ß-gal activity in SNF1 + 330/BY4742 = 100, to compare data across multiple experiments. Asterisks indicate P < 0.05 relative to SNF1 + 330/BY4742. (C) Double deletion of HXK2 and MIG1 derepresses SNF1 expression synergistically. SNF1 and SUC2 (positive control) mRNA expression in hxk2Δ, mig1Δ, and hxk2Δmig1Δ deletion mutants relative to isogenic wild-type strain BY4742 was determined by qRT-PCR for cultures grown in SC media + 2% glucose. Error bars denote propagated standard error. (D and E) Effects of SNF1 expression on chronological longevity. Percent survival of poststationary phase cultures was determined from CFUs of batch cultures grown in SC media + 2% glucose. SNF1 and GFP overexpression strains (D) were the same as those used in perturbation experiments, constructed in the W303-derived strain BMA64–1A. Error bars denote standard deviation; data points with asterisks (*) signify statistically significant differences (P < 0.05) in CFUs relative to control at the same time point.
We first tested if putative Med8 and Mig1 cis-regulatory elements in the SNF1 coding region affected expression of SNF1-reporter gene constructs. Motifs with close similarity to the known Med8 binding sequence (24) were identified at + 247, +305, and + 377 nucleotides (nts) past the ATG start codon. Inclusion of the + 305 motif significantly decreased ß-gal activity compared to a SNF1-lacZ fusion construct truncated at + 285 nts (Fig. 4B). HXK2 deletion also derepressed ß-gal activity in constructs containing the + 305 motif, suggesting Hxk2 acts as a corepressor in this region. A putative Mig1 motif (7) at + 336 nts in similar constructs did not cause consistent, significant differences in ß-gal activity in mig1Δ vs. wild-type strains (data not shown). However, SNF1 expression measured by qRT-PCR exhibited a small but significant increase in a hxk2Δmig1Δ double-deletion mutant in 2% glucose growth, an effect that was greater than the deletion of either HXK2 or MIG1 individually (Fig. 4C). This increase in SNF1 expression in the hxk2Δmig1Δ strain was of a comparable magnitude to that reported previously in the diauxic shift (34), and in response to calorie restriction (see Fig. S1), which is consistent with the translocation of Hxk2 and Mig1 from the nucleus to the cytoplasm in response to glucose depletion (25). Additionally, a log-phase growth defect was observed in the hxk2Δmig1Δ strain relative to its wild-type parent (doubling time 1.79 h−1 vs. 1.55 h−1 for BY4742; P < 0.01), similar to that seen in response to SNF1 overexpression. These results, together with our ChIP-qPCR data, support a model for direct transcriptional regulation of SNF1 by Hxk2 and Med8 and indirect regulation by Mig1 (see Fig. 4A), revealing a mechanism by which Hxk2 affects glucose-regulated gene expression.
We next applied knowledge of the interaction network governing SNF1 expression to examine its regulators' influence on CLS. Increased Snf1 activity, through deletion of repressor subunit SIP2 or forced overexpression of the SNF1 gene, has been previously shown to decrease RLS (22). We tested whether Snf1 similarly affects CLS using overexpression strains from perturbation experiments grown in synthetic CLS media (3) containing 2% glucose. CLS was determined from CFUs from aliquots after cultures reached stationary phase. SNF1 over-expression caused a marked decrease in CLS compared to the GFP-expressing control strain (Fig. 4D), with statistically significant differences (P < 0.05) in CFU counts at several time points, despite a high level of variance among biological replicates. On the basis of the model of interactions regulating SNF1 (see Fig. 4A), we expected to observe a decrease in CLS in the hxk2Δmig1Δ strain relative to its wild-type parent strain, and this was indeed the case (Fig. 4E). No significant changes in CLS were observed in the single hxk2Δ or mig1Δ mutant strains. Detailed characterization of the interaction network thus enabled the identification of CLS mediators Hxk2 and Mig1 acting synergistically; these influences were not detectable by perturbation of the regulators individually.
Discussion
In this study we show that the NIR reverse-engineering strategy (19) can be successfully applied to infer gene regulatory networks in eukaryotic organisms. We assessed the performance of the method by comparing the NIR-inferred Snf1 network model to interactions previously described in the literature, which suggested that many interactions with no previous literature evidence were predicted by the model. The majority of these interactions were validated in experiments employing promoter-lacZ fusion constructs in gene deletion strains and ChIP-qPCR assays for physical targets of the Med8 and Hxk2 TFs. The NIR model showed good measures for sensitivity and precision, as compared with confirmation experiments and known literature interactions, and revealed a greater degree of complexity between regulators in the network than previously appreciated.
An equally important assessment of a network-identification method is the utility of the inferred model to suggest biologically meaningful, testable hypotheses for phenotype regulation. We focused on the NIR-predicted transcriptional regulation of SNF1 by Hxk2, Med8, and Mig1, and confirmed Hxk2 and Med8 as direct regulators and Mig1 as an indirect regulator of SNF1 expression in 2% glucose growth. Hxk2 and Med8 were also found to repress CAT8, a major activator of gluconeogenic genes (7, 29). These results suggest a glucose-responsive signaling mechanism for Hxk2 worthy of further study, given the previously reported challenges in clarifying its downstream targets (9, 24, 27).
We also showed that SNF1 up-regulation reduces CLS, and knowledge of the network architecture governing SNF1 expression led to the identification of Hxk2 and Mig1 as synergistic but not individual modulators of CLS. Because the HXK2 and MIG1 single-deletion mutants caused no change in CLS, it is unlikely that these two modulators of CLS would be identified without knowledge of the gene regulatory network architecture. The sequence, function, and regulatory interactions of Hxk2, Med8, and Snf1/AMP kinase are highly conserved among eukaryotes (21, 24, 28), and therefore, these results may have implications for understanding the role of AMP kinase in regulating metazoan organism lifespan (1).
Materials and Methods
Strains and Culture Growth.
Strains for network inference were constructed in the W303-dervied parent BMA64–1A (MATa ura3–1 ade2–1 leu2–3, 112 his3–11, 15 trp1Δ can1–100) using a tetracycline-inducible expression system (both were obtained from the EUROSCARF repository). Plasmid pCM252 (26) from this set was modified for chromosomal integration at the his3 auxotrophic marker. Network gene ORFs or yeast-enhanced GFP were PCR cloned into this vector, which was transformed into BMA64–1A. For ß-gal assay strains, network gene promoters (–1,000 to + 18 bp relative to the start codon, except when noted) were PCR cloned in-frame with the lacZ gene in YEp356R and YEp357 shuttle vectors (purchased from the American Type Culture Collection or ATCC), and transformed into Saccharomyces Gene Deletion Project strains and isogenic parent BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) (Invitrogen). ChIP strains were constructed in BY4742 by tagging the 3′ termini of HXK2 and MED8 genes with the 13-Myc epitope (35). The hxk2Δmig1Δ strain was constructed by replacement of HXK2 with the LEU2 gene amplified from pRS305 (ATCC) in the mig1Δ deletion strain. See SI Materials and Methods for details.
Cells were cultured at 30 °C with shaking at 300 RPM in the appropriate selective synthetic complete (SC) dropout media (Sigma), except for ChIP experiments performed in YPD (36). For network inference experiments, saturated overnight (o/n) cultures diluted ≈1/400 in fresh media were grown 8 h to OD600 ≈ 0.2 to 0.5, then diluted again in media containing doxycycline and grown 14 h to OD600 ≈ 0.5 for RNA extraction. Perturbation and GFP strains were cultured concurrently in quadruplicate for each experiment. Doxycycline concentration was varied from 0.8 to 3 μg/ml to induce over-expression from 2- to 4-fold, based upon estimated basal expression of the endogenous gene. For confirmation experiments in 2% glucose, saturated o/n cultures were diluted in fresh media to obtain OD600 ≈ 0.8 to 1.0 cultures after 5 to 6 h growth. Calorie restriction experiments used cultures pregrown in 2% glucose 4 h to OD600 ≈ 0.5, which were then washed and resuspended in 0.05% glucose media and grown 4 to 5 h to OD600 ≈ 0.8–1.0.
Network Inference Approach.
The NIR system-identification method (19) models the regulatory interactions between transcripts as a system of ordinary differential equations describing the rate of accumulation of each network species as a weighted sum of the quantity of other species in the network:
where xi is the level of transcript i, aij is the influence of transcript j on transcript i, xj is the level of transcript j, and pi is the level of an external perturbation of transcript i, applied here as additional copies of transcript i. At steady-state experimental conditions, where the quantities of transcripts remain constant over time (dxi/dt = 0), Eq. 1 reduces to:
The coefficients aij are then learned using an iterative algorithm employing multiple linear regression to return the combination of k regulatory inputs per gene fitting the expression data with the minimal least-squares error. The matrix A of coefficients aij represents the functional effects of gene j (regulator) upon transcription of i (target). The sign of each coefficient indicates an activating (aij > 0), repressing (aij < 0) or null (aij = 0) influence. For this study, the algorithm was modified to (i) consider values of k inputs from 3 to 8 [reflecting an assumption informed by previous evidence (see Fig. 1A) that no gene is completely unregulated or regulated by others], where k is selected as the maximum value for each row in which the significance of the regression model (P value of the F test) is ≤0.05, and (ii) perform a t test on individual regression coefficients to disregard those not significantly different (P ≤ 0.10) from zero. See SI Materials and Methods for details.
qRT-PCR mRNA Expression Profiling.
RNA was extracted using an acid phenol method, then treated with DNA-Free RNase-free DNase (Ambion). Reverse transcription of normalized total RNA and qPCR were performed using TaqMan and SYBR Green reagents (Applied Biosystems) according to the manufacturer's instructions. See SI Materials and Methods for details.
ß-Galactosidase Assays.
All experiments used 3 to 5 cultures grown from fresh transformations of promoter-lacZ fusion plasmids into the gene-deletion strain of interest and control strain BY4742. Control and experimental strains were grown concurrently using identical media. β-gal activity was determined with the Yeast β-Galactosidase Assay Kit (Pierce Biotechnology), according to the manufacturer's instructions.
ChIP-qPCR Analysis.
Three biological replicates from epitope-tagged Hxk2 and Med8 and isogenic wild-type strain BY4742 were grown and processed in parallel. ChIP was performed as previously described (10), with minor alterations detailed in SI Materials and Methods. Following final DNA extraction and purification, qPCR was used to detect significant enrichment (P ≤ 0.10) of network gene promoter and coding region DNA in immunoprecipitates from tagged strains compared to wild-type (See SI Materials and Methods).
Chronological Lifespan Assays.
CLS experiments were performed in standard recipe SC media (36) supplemented with a 4-fold excess of each parent strains' auxotrophic nutrients (Uracil, Adenine, Leu, His and Trp for BMA64–1A; Uracil, Leu, His and Lys for BY4742) (3). Triplicate CLS cultures were grown from 60-μl saturated o/n cultures inoculated into 6.0 ml of the appropriate SC media maintained at 30 °C, 300 RPM in 14-ml tubes. SNF1 and GFP over-expression strains were induced with 1.0-μg/ml doxycycline. We then removed 100-μl aliquots of CLS cultures at the indicated timepoints in Fig. 4, which were serially diluted to 1,000 to 3,000 cells/ml. Of these, 100 μl were spread on YPD plates, and colonies were counted manually after 48 h growth at 30 °C. The starting number of CFUs (“Day 0” in Fig. 4) was sampled at 72 h growth, after which no appreciable OD600 changes were observed.
Numerics.
NIR algorithm computations and data analysis were performed using MATLAB v7.4 (The Mathworks). Statistical analyses and outlier determination for data are detailed in SI Materials and Methods. All P values were calculated using Student's t test (unpaired, heteroscedastic), unless otherwise noted.
Supplementary Material
Acknowledgments.
We thank Gábor Balázsi, William J. Blake, and Michael J. Thompson for helpful discussions with experimental design and analysis, and Joseph F. Ryan for comments on the manuscript. This work was supported by the Ellison Medical Foundation, the National Institutes of Health through the National Institutes of Health Director's Pioneer Award Program, Grant DP1 OD003644, and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812551106/DCSupplemental.
References
- 1.Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 2.Kaeberlein M, Burtner C, Kennedy B. Recent developments in yeast aging. PLoS Genet. 2007;3:e84. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 4.Bitterman KJ, Medvedik O, Sinclair D. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 6.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 7.Schüller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- 8.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Devt. 2005;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 11.Yeung MK, Tegnér J, Collins JJ. Reverse engineering gene networks using singular value decomposition and robust regression. Proc Natl Acad Sci USA. 2002;99:6163–6168. doi: 10.1073/pnas.092576199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tegner J, Yeung MK, Hasty J, Collins JJ. Reverse engineering gene networks: integrating genetic perturbations with dynamical modeling. Proc Natl Acad Sci USA. 2003;100:5944–5949. doi: 10.1073/pnas.0933416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 14.di Bernardo D, et al. Chemogenomic profiling on a genome-wide scale using reverse-engineered gene networks. Nat Biotechnol. 2005;23:377–383. doi: 10.1038/nbt1075. [DOI] [PubMed] [Google Scholar]
- 15.Bonneau R, et al. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006;7:R36. doi: 10.1186/gb-2006-7-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Workman CT, et al. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faith J, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. Plos Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee I, Li Z, Marcotte E. An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS ONE. 2007;2:e988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 20.Celenza JL, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 21.Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashrafi K, Lin SS, Manchester JK, Gordon JI. Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes Dev. 2000;14:1872–1885. [PMC free article] [PubMed] [Google Scholar]
- 23.Harkness TA, Shea KA, Legrand C, Brahmania M, Davies GF. A functional analysis reveals dependence on the anaphase-promoting complex for prolonged life span in yeast. Genetics. 2004;168:759–774. doi: 10.1534/genetics.104.027771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno F, Herrero P. The hexokinase 2-dependent glucose signal transduction pathway of Saccharomyces cerevisiae. FEMS Microbiol Rev. 2002;26:83–90. doi: 10.1111/j.1574-6976.2002.tb00600.x. [DOI] [PubMed] [Google Scholar]
- 25.Moreno F, Ahuatzi D, Riera A, Palomino CA, Herrero P. Glucose sensing through the Hxk2-dependent signalling pathway. Biochem Soc Trans. 2005;33:265–268. doi: 10.1042/BST0330265. [DOI] [PubMed] [Google Scholar]
- 26.Bellí G, Garí E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisson LF, Kunathigan V. On the trail of an elusive flux sensor. Res Microbiol. 2003;154:603–610. doi: 10.1016/S0923-2508(03)00170-0. [DOI] [PubMed] [Google Scholar]
- 28.Casamassimi A, Napoli C. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie. 2007;89:1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Tachibana C, et al. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25:2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palomino A, Herrero P, Moreno F. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 2006;34:1427–1438. doi: 10.1093/nar/gkl028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu X, et al. Genome-wide occupancy profile of mediator and the Srb8–11 module reveals interactions with coding regions. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 32.Bennett MR, et al. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celenza JL, Carlson M. Structure and expression of the SNF1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:54–60. doi: 10.1128/mcb.4.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang YW, et al. Roles of cis- and trans-changes in the regulatory evolution of genes in the gluconeogenic pathway in yeast. Mol Biol Evol. 2008;25:1863–1875. doi: 10.1093/molbev/msn138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.