SUMMARY
There has never been a wholesale way of identifying neurons that are monosynaptically connected either to some other cell group or, especially, to a single cell. The best available tools, transsynaptic tracers, are unable to distinguish weak direct connections from strong indirect ones. Furthermore, no tracer has proven potent enough to label any connected neurons whatsoever when starting from a single cell. Here we present a transsynaptic tracer that crosses only one synaptic step, unambiguously identifying cells directly presynaptic to the starting population. Based on rabies virus, it is genetically targetable, allows high-level expression of any gene of interest in the synaptically coupled neurons, and robustly labels connections made to single cells. This technology should enable a far more detailed understanding of neural connectivity than has previously been possible.
INTRODUCTION
Recent advances in our knowledge of the complexity and specificity of neural circuits suggest that understanding how neural circuits generate perception and behavior will be nearly impossible with presently available techniques. Because different neuron types involved in distinct subcircuits are intermingled, and even neighboring neurons of the same type differ in their connectivity and function (DeAngelis et al., 1999; Ohki et al., 2005; Song et al., 2005; Yoshimura et al., 2005), methods are required that can reveal the connections both of specific cell types and of single neurons (Crick, 1979). Existing techniques (Callaway and Katz, 1993; Douglas and Martin, 2004; Gilbert, 1983; Gray, 1959; Mercer et al., 2005; Shepherd and Svoboda, 2005; Timofeeva et al., 2005; Zarrinpar and Callaway, 2006) have been extremely valuable but all are limited in various ways. None has been capable of identifying en masse the cells that are directly connected to either a cell type of interest or a single cell.
Transsynaptic tracers, out of all the available techniques, might appear to offer a solution to this problem. By introducing a tracer into a particular cell or cell type, synaptically connected cells should be labeled by the tracer and therefore be identifiable as those in synaptic contact with the starting cell(s) in question. Several approaches to selectively introducing either conventional or viral tracers into particular genetically identified populations have been taken (Braz et al., 2002; DeFalco et al., 2001; Maskos et al., 2002; Zou et al., 2001).
However, due to their dependence on cellular machinery for transport to and across synapses (Vercelli et al., 2000), transsynaptic tracers cross different synapses at different rates: the more hardware servicing a given connection, the more efficiently it will be traversed by the tracer. Tracer that accrues in transsynaptically labeled cells will begin spreading in turn to the cells that are connected to them and in fact can label the most strongly connected of these even before weakly connected synaptic partners of the starting population are labeled (Ugolini, 1995a; Ugolini et al., 1987). The result is an inescapable ambiguity in the number of synapses crossed: transsynaptic tracers are incapable of distinguishing strong indirect connections from weak direct ones. They therefore cannot be used in their current form to determine monosynaptic connections in any kind of comprehensive way.
We have overcome this obstacle by constructing a viral tracer that crosses only one synaptic step to cells directly connected to the starting population. Monosynaptically connected cells are therefore labeled unambiguously. In addition, the system is genetically targetable, and, unlike all previous methods, it is powerful enough to label neurons that connect to a single starting cell.
Background and Theory
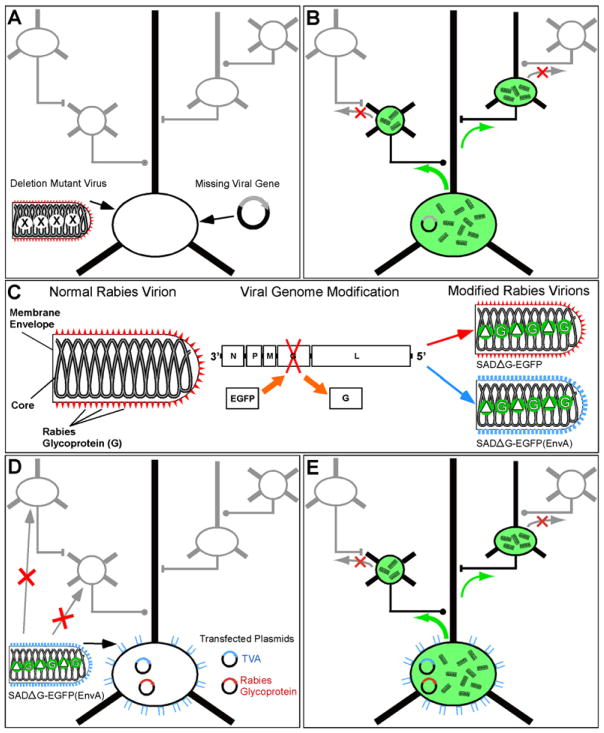
The core idea of the system is to infect the cell population of interest with a deletion-mutant tracing virus that is missing one or more genes that are required for transsynaptic spread, and to complement this deletion by providing the missing viral genes in trans in the initially infected neurons only (Figure 1A). With all the viral genes present in the starting cells, the virus can spread from them to cells in monosynaptic contact (Figure 1B). Because those genes are not in the secondarily infected cells, however, the virus can’t spread beyond them.
Figure 1. Transcomplemented Transsynaptic Tracing.
(A) A deletion mutant tracing virus missing one or more genes required for transsynaptic spread, as well as the separate missing viral gene(s), are introduced into a cell or cell type of interest. Both the initial infection and the complementing viral genes must be restricted to the neuronal population of interest.
(B) Because all viral genes are present in the initially infected population, the virus can spread transsynaptically to cells in direct synaptic contact with them. Since the missing viral genes are not present in these cells, however, the virus cannot spread further.
(C) The rabies virion. The viral core consists of the RNA genome and associated proteins and is surrounded by a host-cell-derived membrane in which is embedded the rabies virus glycoprotein (G). The EGFP gene was substituted for that of the glycoprotein within the viral genome (center). Versions of this virus can be made that incorporate either its native glycoprotein (designated SADΔG-EGFP, right, top) or the glycoprotein of some other virus. In this study the rabies virus was pseudotyped with the glycoprotein from ASLV-A, termed EnvA, and this virus is designated SADΔG-EGFP(EnvA) (right, bottom).
(D) Targeting infection. ASLV-A-pseudotyped rabies virus [SADΔG-EGFP(EnvA)] can’t infect mammalian neurons unless the gene for ASLV-A’s receptor, TVA, has been introduced into them. This causes the receptor to be expressed on the cells’ surface, allowing infection by the pseudotyped virus. Therefore, the basic requirements of the system are as follows. First insert two genes into the cell or cell type of interest: the gene for TVA, so the virus can enter, and the gene for the rabies virus glycoprotein, so the virus can spread to synaptically coupled cells. Then apply the ASLV-A-pseudotyped virus.
(E) Following these steps, there is specific infection of the TVA-expressing cell; complementation with the rabies virus glycoprotein allows the virus to spread to directly presynaptic neurons. These cells all express the EGFP encoded in the viral genome, but the virus cannot spread beyond these directly connected cells because they do not express the viral glycoprotein.
We have implemented this idea using rabies virus (Figure 1C, left) because of its significantly lower cytopathicity and far greater infection efficiency than the other widely used family of tracing viruses, the α-herpesviruses (Card et al., 1999; Ito et al., 2001; Lafay et al., 1991; Norgren and Lehman, 1998; Ugolini, 1995a; Ugolini et al., 1987). It has been used with great success in its intact form as a transsynaptic tracer, crossing synapses exclusively in the retrograde direction (Hoshi et al., 2005; Kelly and Strick, 2000; Nassi et al., 2006; Ugolini, 1995a; Ugolini et al., 1989). Furthermore, the transsynaptic spread of rabies virus has been observed to be specific to connected neurons, and not to adjoining neurons that are not connected (Ugolini, 1995b). The system presented here is therefore a means of identifying cells presynaptic to a cell or cell type of interest.
Because intact rabies virus will infect nonspecifically and replicate and spread across multiple synapses, our strategy required the use of modified rabies virus. Two key changes were required to implement our strategy. The first was to modify the viral genome by deletion of a gene required for the production of infectious viral particles. This would allow viral spread to be monosynaptically restricted using the transcomplementation approach described above. The second modification was to alter the tropism of the virus so that it could only infect a genetically specified neuronal population.
We implemented the transcomplementation strategy by deletion of the rabies virus glycoprotein gene. The glycoprotein, embedded in the membrane that surrounds the viral core (Figure 1C, left), is not required for transcription of the viral genes or for replication of the genome within infected cells, but is required for transsynaptic spread (Etessami et al., 2000; Mebatsion et al., 1996; Wickersham et al., 2007). We have recently described a recombinant rabies virus with its glycoprotein gene replaced with the coding sequence for enhanced green fluorescent protein (EGFP) (termed SADΔG-EGFP, Figure 1C, middle and top right) that cannot spread beyond initially infected neurons but, because the deletion of its glycoprotein gene leaves intact its ability to replicate the viral core, produces levels of EGFP sufficient to brightly label even fine dendritic and axonal details (Wickersham et al., 2007). We reasoned that this virus would be ideal for our transcomplementation approach because viral spread would be monosynaptically restricted, yet the virus would still be able to replicate and express abundant EGFP in infected cells. This would have the potential to greatly amplify the signal from the small number of viral particles that might spread across a small number of synapses.
To target the initial SADΔG-EGFP rabies virus infection to a genetically defined target neuronal cell population, we have taken advantage of the exquisite specificity of subgroup A avian sarcoma and leukosis virus (ASLV-A)-receptor interactions. The envelope protein of this virus (EnvA) can direct virus infection specifically into cells that express the cognate TVA viral receptor, a protein which is found in birds but not mammals (Barnard et al., 2006; Bates et al., 1993; Federspiel et al., 1994; Young et al., 1993). Thus, by pseudotyping the rabies virus with EnvA (Figure 1C, right, bottom), we can restrict infection of the resulting virus—termed SADΔG-EGFP(EnvA)—to only a small subpopulation of neuronal cells engineered to express TVA (Figure 1D).
By also supplying the rabies virus glycoprotein gene in trans within these initially infected cells, we permit rabies virus assembly and spread to monosynaptically connected cells. Because the viral glycoprotein gene is not present in the transsynaptically infected cells, the virus cannot spread beyond them (Figure 1E). Monosynaptically connected cells will therefore be identified unambiguously.
Here we demonstrate the validity of our inferences. We show the following. (1) Our strategy for targeting the initial infection with rabies virus works, with SADΔG-EGFP-(EnvA) specifically infecting mammalian neurons that express TVA. (2) Transcomplementation with rabies virus glycoprotein is necessary and sufficient for the spread of SADΔG-EGFP beyond the initially infected neuron(s). When the glycoprotein is not expressed, infection is restricted to TVA-expressing neurons, but when the glycoprotein is expressed, the infection spreads transsynaptically. (3) These same data also demonstrate that the spread of rabies virus is monosynaptically restricted. Since glycoprotein expression is required for viral spread, it cannot spread transsynaptically from secondarily infected cells that do not express the glycoprotein. (4) This system is sufficiently sensitive to label neurons that are presynaptic to a single starting cell.
RESULTS
We have tested the system in cultured slices of neonatal rat brain, using the “gene gun” (Bio-Rad, Hercules, CA) to transfect small numbers of relatively isolated neurons within each slice with plasmid DNA encoding TVA, rabies virus glycoprotein, and DsRed2 to label the transfected cell population. A day after transfection, SADΔG-EGFP-(EnvA) was added to the culture wells, and virus infection was subsequently monitored using fluorescence microscopy to score EGFP expression.
In six slices quantitatively examined 6 days postinfection, 242 cells were identified that expressed DsRed2, 62 of which also expressed EGFP, indicating infection with the rabies virus. Spectacularly, the double-labeled cells were surrounded by large clusters of virus-infected neurons—totaling 5424—expressing only EGFP (Figures 2E–2H and Figure 3). None of the green cells, upon closer examination, had the appearance of glia. Qualitatively similar results were obtained with dozens of other brain slices tested. These observations suggested that the red TVA-expressing cells at the centers of these clusters were initially infected by the EnvA-pseudotyped rabies virus and that the additional thousands of green cells were connected directly to the initially infected ones. To evaluate the validity of these inferences, we further tested the specificity of infection with EnvA-pseudotyped virus, the requirement of G expression for viral spread, and the existence of functional connections between labeled neurons.
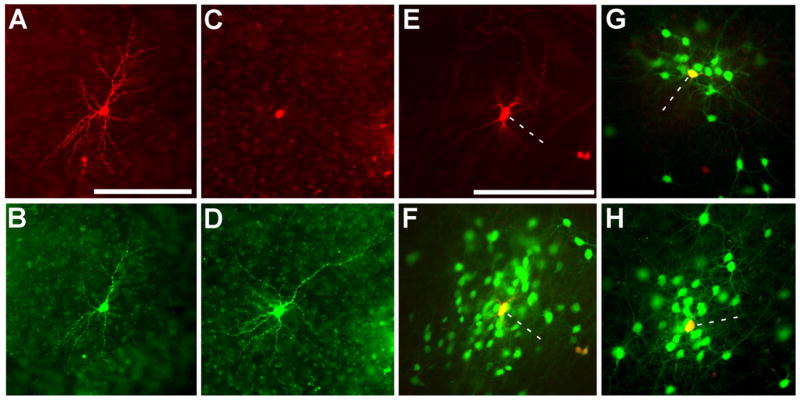
Figure 2. Selective Infection and In Situ Complementation in Slice Culture.
(A–D) Initial infection is restricted to cells expressing the ASLV-A receptor, TVA. In these control experiments to test infection selectivity, isolated neurons in cultured brain slices were transfected using the gene gun with two genes, one encoding TVA, and the other, DsRed2. ASLV-A-pseudotyped rabies virus [SADΔG-EGFP(EnvA)] was applied the next day and images were taken 6 days following. (A and C) DsRed2 expression marking transfection with TVA and (B and D) EGFP expression indicating subsequent selective infection of the same TVA-expressing cells with pseudotyped virus. (E–H) In situ complementation permits transsynaptic spread from a single initially infected cell to a cluster of monosynaptically connected cells. Isolated neurons were transfected with three genes encoding TVA to permit viral infection, DsRed2 to mark transfected cells, and the rabies virus glycoprotein gene to complement the deletion in the viral genome. (E) DsRed2 fluorescence indicating a transfected cell, marked with a dotted line, at the center of the cluster shown in (F). (G and H) Two more examples of clusters surrounding a single transfected cell. The initially infected cells expressing both EGFP and DsRed2 appear yellow. Scale bars: 200 μm.
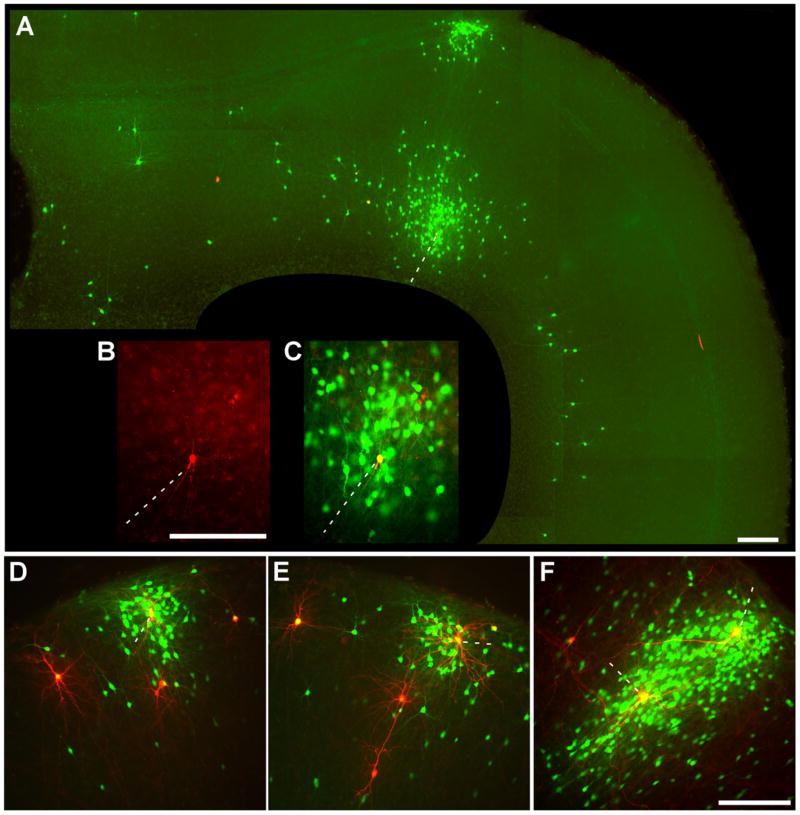
Figure 3. More Examples of Transcomplemented Tracing.
(A–C) Long-range viral spread from a single initially infected cell. (A) A huge cluster of green cells surrounds a single red/green deep-layer cortical neuron (dotted line) at 8 days postinfection. Another dense cluster of cells is also infected in the superficial cortical layers immediately above it, consistent with known projections of superficial layers to deeper ones; distant deep-layer pyramidal cells are also infected, again consistent with known patterns of long-range intralaminar connectivity. To the left of the putatively initially infected cell is a second yellow (double-labeled) cell, apparently secondarily—and recently—infected because of the lack of green cells surrounding it. (B–C) Closeup of central cluster from (A). (D–F) More examples of in situ complementation: clusters of infected cells surrounding isolated putatively postsynaptic ones. Scale bars: 200 μm.
For these purposes, SADΔG-EGFP(EnvA) was also used to infect slices that either had not been transfected or were transfected with plasmid DNA encoding TVA and DsRed2, but not rabies virus glycoprotein. Consistent with the low-level tropism of ASLV-A for mammalian cells, examination of 20 independent untransfected brain slices led to the identification of only a single EGFP-expressing cell. Another set of 12 slice cultures was transfected with plasmid DNA encoding TVA and DsRed2, but not rabies virus glycoprotein. A day after transfection, SADΔG-EGFP(EnvA) was added to the culture wells, and virus infection was again monitored using fluorescence microscopy to score EGFP expression. In 12 slices, 43 cells expressed DsRed2; 23 of these red cells, and again only one untransfected cell, also expressed EGFP. Thus, in the absence of the rabies virus glycoprotein gene, virus infection was restricted almost entirely to the TVA-expressing cells and did not spread beyond the initially infected cells (Figures 2A–2D).
These experiments indicate that in situ complementation of the deletion mutant rabies virus worked extremely effectively. The initial virus infection was restricted to TVA-positive cells, transcomplementation with rabies virus glycoprotein was necessary for spread beyond these cells, and this transcomplementation was sufficient to unleash a viral infection from these cells to thousands of others surrounding them in the slices.
These results also indicated that the complemented virus was quite capable of spreading from a single starting cell to a vast number of neighboring neurons. Due to the limitations of the gene gun technique, cells are generally shot in clumps when they are shot at all, typically resulting in multiple red cells per slice; we were not able to adjust parameters so as to transfect one neuron or fewer per slice. Despite this limitation, in many cases, clusters of secondarily infected neurons were clearly centered on a single DsRed2-expressing cell, as seen in Figures 2E–2H) as well as the striking example shown in Figures 3A–3C). Cells expressing both DsRed2 and EGFP, which were at the center of EGFP-positive cell clusters, included those with morphologies of both inhibitory and excitatory neurons, indicating that the transcomplemented rabies virus can efficiently spread from either cell type (see also paired recording data below).
Testing Synaptic Specificity by Paired Recording
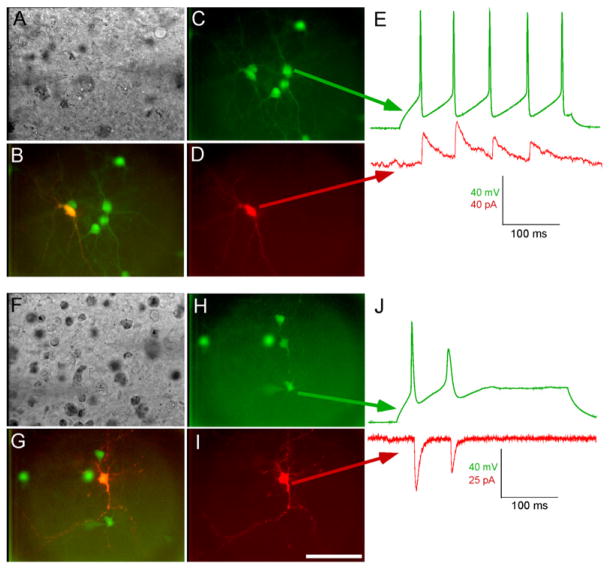
The observed clusters of green neurons with transfected neurons at their centers (Figure 2 and Figure 3) strongly suggested that virus was spreading transsynaptically from single cells to cells presynaptic to them. To test the synaptic specificity of viral spread, we conducted paired whole-cell recordings from putatively pre- and postsynaptic cells (Figure 4). Cells fluorescent in both the red channel (indicating transfection with TVA and rabies virus glycoprotein) and the green channel (indicating infection with SADΔG-EGFP(EnvA) (red/green cells) were recorded under voltage clamp while nearby green-only cells (infected by viral spread), held in current clamp, were depolarized to fire action potentials. Synaptic currents in the voltage-clamped red/green cell that were simultaneous with the green cell’s action potentials indicated a monosynaptic connection, as seen in the example traces in Figures 4E and 4J. Nonfluorescent cells at similar distances from the red/green cells were also stimulated as controls. Because the gene gun typically transfects large numbers of cells within a slice, there were no ideal cases of only a single transfected cell per slice (see above). We therefore established a criterion of only recording from red/green cells with no other red/green cells present within 250 μm. Of 11 green cells stimulated, synaptic currents were detected in the nearby red/green, putatively postsynaptic partners, in 9 cases (2 failures). Of these nine connected pairs, five elicited excitatory currents (e.g., Figure 4J), while four elicited inhibitory currents (e.g., Figure 4E). In striking contrast, nine control pairs, each consisting of a nonfluorescent neuron near a red/green cell, were recorded, and none was found to be connected. For comparison, randomly sampled adjacent neurons in acute brain slices from rat cortex are connected about 10%–20% of the time or less (Thomson et al., 2002; Yoshimura et al., 2005); there are not comparable data available for slice cultures matched to our experimental conditions.
Figure 4. Viral Spread Is Specific to Cells Presynaptic to the Initially Infected Cell.
(A) DIC image of slice and recording pipettes targeting putatively pre- and postsynaptic neurons, (B) combined fluorescent image, and (C–D) single-channel fluorescence images. (E) Inhibitory postsynaptic currents in the putatively postsynaptic cell are coincident with action potentials in a nearby infected one, demonstrating a monosynaptic connection. (F–J) Similar demonstration of spread to an excitatory pre-synaptic cell. Scale bar: 100 μm, applies to all panels.
An ideal test of synaptic specificity in our slice culture system would involve transfection of only a single neuron per slice and unambiguous physiological testing for functional connections between large numbers of EGFP-positive, putatively presynaptic neurons. Several technical limitations prevented such a test based on available methods and resources. First, as noted above, we were not able to restrict transfection to single neurons using the gene gun. As a result, EGFP-positive neurons could not be unambiguously linked to a particular, putative post-synaptic partner. This problem was compounded by the fact that there is considerable cell death in slice cultures, leaving open the possibility that, even when there appeared to be an isolated transfected cell, there might have previously been a second postsynaptic neuron that died before observation. Finally, the fact that synaptic contacts can be formed transiently and then eliminated (De Paola et al., 2006) raises the possibility of labeling cells that are not connected at the time of the functional assay. The results described above must, therefore, be viewed as a preliminary assessment and represent a lower limit on the specificity of this system. Future studies using single-cell electroporation and high-throughput stimulation of hundreds of presynaptic neurons will likely provide a more definitive measure.
DISCUSSION
There has never before been a method of identifying cells that are monosynaptically connected to a population of interest, and there has never been a method of transsynaptically labeling cells connected—by any number of synapses—to a single starting cell. The system we have introduced here does both. We expect it to be rapidly applicable in vivo, as outlined briefly below, to answer major outstanding questions of connectivity and function and to open entirely new avenues of investigation.
These in vitro results demonstrate proof of concept: the viral spread resulting from in situ complementation is extremely effective, with the virus reliably proceeding from initially infected neurons to hundreds of their peers surrounding them in the slice. We also demonstrate that EnvA-pseudotyped rabies virus specifically infects TVA-expressing neurons so that it is possible to restrict initial infection to a genetically defined population. Furthermore, we demonstrate that it is possible to label presynaptic neurons from a single postsynaptic parent cell. Although we were not able to routinely restrict biolistic transfection to a single neuron, there are cases in which EGFP-expressing cells were found in slices with only one DsRed2-expressing neuron (e.g., Figure 3). In addition, because of the known sparse connectivity in cortex, even in cases where there are several possible postsynaptic (DsRed2- plus EGFP-expressing) cells, the great majority of presynaptic (EGFP-only) cells must have been connected to only one of the postsynaptic cells. Our paired recordings also provide important insight. They indicate, first, that the virus can spread effectively from individual cells, and second, that this spread is to cells directly presynaptic in the strong majority of cases. At this point we cannot answer the question of whether the virus would eventually label every cell connected to a given starting neuron, except to point to our limited paired recording data finding that none of the cells that were not green were connected to nearby red/green cells. The answers to this question will depend on further testing and experience with the system.
The most begrudging interpretation of the data presented here, given that 2 out of 11 recorded green cells were not found to be connected to the nearest transfected cell, might be that viral spread in this system is to directly connected presynaptic cells most of the time. However, there is reason to have more confidence in the technique than that, because of the limitations of the strategy used here for testing it. As stated above, the gene gun typically transfects large numbers of cells within a slice rather than the single cell that would be required to ideally test the system. Because, as seen in Figure 3A, a single postsynaptic cell can label cells across hundreds of microns, we suspect that the two infected cells that were not found to be connected to the recorded transfected cells were connected to some other transfected cell elsewhere in the slice. Death of some of the transfected cells is also a possibility. A better way to test the system would be to transfect only one cell per slice using microinjection or single-cell electroporation (Haas et al., 2001; Rathenberg et al., 2003). A further improvement would be to use optical stimulation of putatively presynaptic cells, as well as controls, for a much higher throughput than we have managed here. Such rigorous testing is needed and would be welcomed.
Fundamentally, though, we have no reason to believe that the spread of the transcomplemented deletion mutant virus used here would be any less synaptically specific than that of intact rabies virus, which has been found to be significantly more specific in its transsynaptic spread than the much more commonly used, but much more cytotoxic, α-herpesviruses (Ugolini, 1995b; Ugolini et al., 1987).
The system presented here uses a virus that travels retrogradely and therefore labels presynaptic neurons; since postsynaptic cells are as interesting as presynaptic ones, it would also be desirable to have a similar system based on a virus that travels anterogradely. Unfortunately, there are no known strains or mutants of rabies virus that do so. The α-herpesviruses, by contrast, travel bidirectionally in their wild forms, and the HSV-1 tracing strain H129 appears to travel solely anterogradely (Rinaman and Schwartz, 2004; Zemanick et al., 1991). Although our attempts to implement the transcomplementation idea using a widely used α-herpesvirus, pseudorabies virus, met with little success (data not shown)—a failure we attributed to the high cytopathicity and low efficiency of the herpesviruses relative to rabies virus (Card et al., 1999; Ito et al., 2001; Lafay et al., 1991; Norgren and Lehman, 1998; Ugolini, 1995a; Ugolini et al., 1987)—it may be worth revisiting these efforts.
Implementation In Vivo
The requirements of our system are simply to introduce the genes for TVA and the rabies virus glycoprotein into the cell or cells of interest, then to apply the pseudotyped virus after TVA has been expressed. There are many options for introducing the two genes into a particular cell type in vivo, such as genetically modifying mice or using a helper virus with a cell-type-specific promoter (Davis et al., 2001; Gou et al., 2004; van den Pol et al., 2004). Single-cell electroporation (Haas et al., 2001; Rathenberg et al., 2003) offers the promise of identifying the cells pre-synaptic to a single identified neuron whose activity and response properties have been characterized. In combination with brain slice preparation or in vivo optical methods, labeling of inputs to single neurons should also facilitate studies of the functional properties and interactions between connections that are presently not possible based on random sampling. For example, labeled pre-synaptic cells could be stimulated while recording intracellularly from the postsynaptic cell. This would overcome the difficulty in finding presynaptic cells in sparsely connected structures such as the cerebral cortex.
Finally, the recombinant rabies virus could of course contain genes for any proteins of interest besides EGFP, raising an endless variety of possibilities. A genetically encoded sensor of neural activity (Chanda et al., 2005; Reiff et al., 2005), for example, combined with two-photon imaging in vivo (Helmchen and Denk, 2005), should allow identification of the functional properties of cells directly connected to an originally identified, functionally characterized, and electroporated postsynaptic neuron. Conversely, inclusion of a gene encoding a photosensitive ion channel (Boyden et al., 2005; Li et al., 2005) should allow patterned stimulation of presynaptic cells while recording from the cell they converge on. A transactivator or recombinase gene could be used to further direct gene expression in synaptically coupled neurons, and indeed, since there is ample room in the rabies virus genome for additional genes (McGettigan et al., 2003), any of these approaches could be combined. In our view this technology and those that derive from it are likely to have a significant impact on neuroscience.
EXPERIMENTAL PROCEDURES
Production of Packaging Cell Line
The extracellular and transmembrane domains of the ASLV-A envelope protein from the plasmid pAB6 (Boerger et al., 1999) and the cytoplasmic domain region of the SAD B19 glycoprotein gene from pHCMV-RabiesG (Sena-Esteves et al., 2004) were combined by PCR and cloned into the murine leukemia virus (MLV) transfer vector pCMMP-IRES-GFP (Melikyan et al., 2004). Vesicular stomatitis virus (VSV)-pseudotyped MLV was then produced as described (Melikyan et al., 2004) and applied to BHK-21 cells (ATCC) at a multiplicity of infection (MOI) of ~4 overnight. After four passages, cells were sorted for high EGFP fluorescence with a FACSDiva (BD Biosciences, San Jose, CA). The resulting cell line was termed BHK-EnvARGCD.
Production of Pseudotyped Rabies Virus
BHK-EnvARGCD cells were plated in 12-well plates at 2E5 cells/well. The following day, the glycoprotein-deleted rabies virus SADΔG-EGFP (Wickersham et al., 2007) was added at an MOI of 1.5. One day later, the cells in each well were trypsinized and replated into a 10 cm plate. Virus-containing supernatants were harvested 2 days later, filter sterilized, and frozen at −80°C in 1 ml aliquots. Virus titers were determined by serial dilution and overnight infection of 293T-TVA800 cells (Narayan et al., 2003) and 293T cells followed by determination of fluorescent fraction on a FACscan (BD Biosciences) 3 days later.
Biolistics and Virus Application
Brain slices were prepared from the cortex of 3- to 7-day-old rats as described previously for ferrets (Dantzker and Callaway, 1998; McAllister et al., 1995). One day following, slices were transfected using the Helios Gene Gun (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. The following plasmids were used per 12.5 mg of gold microcarriers. Controls: pCAG-DsRed2, 5 μg; pCMMP-TVA800 (Narayan et al., 2003), 30 μg. Experimental: pCAG-DsRed2, 5 μg; pCMMP-TVA800, 30 μg, pHCMV-RabiesG (Sena-Esteves et al., 2004), 15 μg. All transgenes were expressed under the control of the human cytomegalovirus (CMV) immediate-early promoter except for DsRed2, which was driven by the CAG hybrid promoter (Borrell et al., 2005; Niwa et al., 1991). One day following transfection, 20 μl of virus stock solution (7.8E4 pfu/ml) was applied to the surface of each slice.
Electrophysiological Recordings
Three to nine days following application of virus, slices were transferred to recording chambers perfused with room temperature artificial cerebral spinal fluid (ACSF), composed of 124 mM NaCl, 5 mM KCl, 1.25 mM KH2PO4, 1.3 mM MgSO4, 3.2 mM CaCl2, 26 mM NaHCO3, and 10 mM glucose. Glass recording electrodes (7–10 MΩ resistance), filled with an intracellular solution consisting of 130 mM potassium gluconate, 6 mM KCl, 2 mM MgCl2, 0.2 mM EGTA, 10 mM HEPES, 2.5 mM Na2ATP, 0.5 mM Na2GTP, 10 mM potassium phosphocreatine, and 0.3% biocytin, adjusted to 7.25 pH with KOH, were used for whole-cell current-clamp recordings. Cells were targeted using fluorescence and DIC optics.
During paired recordings used to test functional connectivity between cell pairs, the potential presynaptic cell was recorded in current clamp and the potential postsynaptic cell in voltage clamp. Action potentials were generated in the presynaptic cell by current injection. The postsynaptic cell was routinely tested from both negative holding potentials to detect excitatory postsynaptic currents and depolarized potentials to detect inhibitory currents and/or “silent” synapses. When postsynaptic currents were not detected following generation of single spikes in the presynaptic cell, trains were generated using current injected for longer durations.
Acknowledgments
We thank Dr. Cristina Garcia-Frigola for technical assistance, Dr. Lynn Enquist for pseudorabies virus stocks, and Dr. Miguel Sena-Esteves for pHCMV-RabiesG. This work was supported by NIH grant numbers MH63912, EY10742, and CA70810 and by Deutsche Forschungsge-meinschaft grant numbers SFB455 and SPP1175.
References
- Barnard RJ, Elleder D, Young JA. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane fusion. Virology. 2006;344:25–29. doi: 10.1016/j.virol.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Bates P, Young JA, Varmus HE. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- Boerger AL, Snitkovsky S, Young JA. Retroviral vectors preloaded with a viral receptor-ligand bridge protein are targeted to specific cell types. Proc Natl Acad Sci USA. 1999;96:9867–9872. doi: 10.1073/pnas.96.17.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, Yoshimura Y, Callaway EM. Targeted gene delivery to telencephalic inhibitory neurons by directional in utero electroporation. J Neurosci Methods. 2005;143:151–158. doi: 10.1016/j.jneumeth.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Braz JM, Rico B, Basbaum AI. Transneuronal tracing of diverse CNS circuits by Cre-mediated induction of wheat germ agglutinin in transgenic mice. Proc Natl Acad Sci USA. 2002;99:15148–15153. doi: 10.1073/pnas.222546999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM, Katz LC. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc Natl Acad Sci USA. 1993;90:7661–7665. doi: 10.1073/pnas.90.16.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Enquist LW, Moore RY. Neuroinvasiveness of pseudorabies virus injected intracerebrally is dependent on viral concentration and terminal field density. J Comp Neurol. 1999;407:438–452. doi: 10.1002/(sici)1096-9861(19990510)407:3<438::aid-cne11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Chanda B, Blunck R, Faria LC, Schweizer FE, Mody I, Bezanilla F. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat Neurosci. 2005;8:1619–1626. doi: 10.1038/nn1558. [DOI] [PubMed] [Google Scholar]
- Crick FH. Thinking about the brain. Sci Am. 1979;241:219–232. doi: 10.1038/scientificamerican0979-219. [DOI] [PubMed] [Google Scholar]
- Dantzker JL, Callaway EM. The development of local, layer-specific visual cortical axons in the absence of extrinsic influences and intrinsic activity. J Neurosci. 1998;18:4145–4154. doi: 10.1523/JNEUROSCI.18-11-04145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JR, McVerry J, Lincoln GA, Windeatt S, Lowenstein PR, Castro MG, McNeilly AS. Cell type-specific adenoviral transgene expression in the intact ovine pituitary gland after stereotaxic delivery: an in vivo system for long-term multiple parameter evaluation of human pituitary gene therapy. Endocrinology. 2001;142:795–801. doi: 10.1210/endo.142.2.7963. [DOI] [PubMed] [Google Scholar]
- De Paola V, Holtmaat A, Knott G, Song S, Wilbrecht L, Caroni P, Svoboda K. Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 2006;49:861–875. doi: 10.1016/j.neuron.2006.02.017. [DOI] [PubMed] [Google Scholar]
- DeAngelis GC, Ghose GM, Ohzawa I, Freeman RD. Functional micro-organization of primary visual cortex: receptive field analysis of nearby neurons. J Neurosci. 1999;19:4046–4064. doi: 10.1523/JNEUROSCI.19-10-04046.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, Friedman JM. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- Douglas RJ, Martin KA. Neuronal circuits of the neocortex. Annu Rev Neurosci. 2004;27:419–451. doi: 10.1146/annurev.neuro.27.070203.144152. [DOI] [PubMed] [Google Scholar]
- Etessami R, Conzelmann KK, Fadai-Ghotbi B, Natelson B, Tsiang H, Ceccaldi PE. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol. 2000;81:2147–2153. doi: 10.1099/0022-1317-81-9-2147. [DOI] [PubMed] [Google Scholar]
- Federspiel MJ, Bates P, Young JA, Varmus HE, Hughes SH. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc Natl Acad Sci USA. 1994;91:11241–11245. doi: 10.1073/pnas.91.23.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert CD. Microcircuitry of the visual cortex. Annu Rev Neurosci. 1983;6:217–247. doi: 10.1146/annurev.ne.06.030183.001245. [DOI] [PubMed] [Google Scholar]
- Gou D, Narasaraju T, Chintagari NR, Jin N, Wang P, Liu L. Gene silencing in alveolar type II cells using cell-specific promoter in vitro and in vivo. Nucleic Acids Res. 2004;32:e134. doi: 10.1093/nar/gnh129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EG. Electron microscopy of synaptic contacts on dendrite spines of the cerebral cortex. Nature. 1959;183:1592–1593. doi: 10.1038/1831592a0. [DOI] [PubMed] [Google Scholar]
- Haas K, Sin WC, Javaherian A, Li Z, Cline HT. Single-cell electroporation for gene transfer in vivo. Neuron. 2001;29:583–591. doi: 10.1016/s0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Hoshi E, Tremblay L, Feger J, Carras PL, Strick PL. The cerebellum communicates with the basal ganglia. Nat Neurosci. 2005;8:1491–1493. doi: 10.1038/nn1544. [DOI] [PubMed] [Google Scholar]
- Ito N, Takayama M, Yamada K, Sugiyama M, Minamoto N. Rescue of rabies virus from cloned cDNA and identification of the pathogenicity-related gene: glycoprotein gene is associated with virulence for adult mice. J Virol. 2001;75:9121–9128. doi: 10.1128/JVI.75.19.9121-9128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/s0165-0270(00)00296-x. [DOI] [PubMed] [Google Scholar]
- Lafay F, Coulon P, Astic L, Saucier D, Riche D, Holley A, Flamand A. Spread of the CVS strain of rabies virus and of the avirulent mutant AvO1 along the olfactory pathways of the mouse after intranasal inoculation. Virology. 1991;183:320–330. doi: 10.1016/0042-6822(91)90145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskos U, Kissa K, St Cloment C, Brulet P. Retrograde trans-synaptic transfer of green fluorescent protein allows the genetic mapping of neuronal circuits in transgenic mice. Proc Natl Acad Sci USA. 2002;99:10120–10125. doi: 10.1073/pnas.152266799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- McGettigan JP, Naper K, Orenstein J, Koser M, McKenna PM, Schnell MJ. Functional human immunodeficiency virus type 1 (HIV-1) Gag-Pol or HIV-1 Gag-Pol and env expressed from a single rhabdovirus-based vaccine vector genome. J Virol. 2003;77:10889–10899. doi: 10.1128/JVI.77.20.10889-10899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T, Konig M, Conzelmann KK. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- Melikyan GB, Barnard RJ, Markosyan RM, Young JA, Cohen FS. Low pH is required for avian sarcoma and leukosis virus Env-induced hemifusion and fusion pore formation but not for pore growth. J Virol. 2004;78:3753–3762. doi: 10.1128/JVI.78.7.3753-3762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer A, West DC, Morris OT, Kirchhecker S, Kerkhoff JE, Thomson AM. Excitatory connections made by presynaptic cortico-cortical pyramidal cells in layer 6 of the neocortex. Cereb Cortex. 2005;15:1485–1496. doi: 10.1093/cercor/bhi027. [DOI] [PubMed] [Google Scholar]
- Narayan S, Barnard RJ, Young JA. Two retroviral entry pathways distinguished by lipid raft association of the viral receptor and differences in viral infectivity. J Virol. 2003;77:1977–1983. doi: 10.1128/JVI.77.3.1977-1983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Lyon DC, Callaway EM. The parvocellular LGN provides a robust disynaptic input to the visual motion area MT. Neuron. 2006;50:319–327. doi: 10.1016/j.neuron.2006.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Norgren RB, Jr, Lehman MN. Herpes simplex virus as a transneuronal tracer. Neurosci Biobehav Rev. 1998;22:695–708. doi: 10.1016/s0149-7634(98)00008-6. [DOI] [PubMed] [Google Scholar]
- Ohki K, Chung S, Ch’ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- Rathenberg J, Nevian T, Witzemann V. High-efficiency transfection of individual neurons using modified electrophysiology techniques. J Neurosci Methods. 2003;126:91–98. doi: 10.1016/s0165-0270(03)00069-4. [DOI] [PubMed] [Google Scholar]
- Reiff DF, Ihring A, Guerrero G, Isacoff EY, Joesch M, Nakai J, Borst A. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaman L, Schwartz G. Anterograde transneuronal viral tracing of central viscerosensory pathways in rats. J Neurosci. 2004;24:2782–2786. doi: 10.1523/JNEUROSCI.5329-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena-Esteves M, Tebbets JC, Steffens S, Crombleholme T, Flake AW. Optimized large-scale production of high titer lentivirus vector pseudotypes. J Virol Methods. 2004;122:131–139. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Shepherd GM, Svoboda K. Laminar and columnar organization of ascending excitatory projections to layer 2/3 pyramidal neurons in rat barrel cortex. J Neurosci. 2005;25:5670–5679. doi: 10.1523/JNEUROSCI.1173-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Sjostrom PJ, Reigl M, Nelson S, Chklovskii DB. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. 2005;3:e68. doi: 10.1371/journal.pbio.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AM, Bannister AP, Mercer A, Morris OT. Target and temporal pattern selection at neocortical synapses. Philos Trans R Soc Lond B Biol Sci. 2002;357:1781–1791. doi: 10.1098/rstb.2002.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva E, Dufresne C, Sik A, Zhang ZW, Deschenes M. Cholinergic modulation of vibrissal receptive fields in trigeminal nuclei. J Neurosci. 2005;25:9135–9143. doi: 10.1523/JNEUROSCI.3073-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini G. Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J Comp Neurol. 1995a;356:457–480. doi: 10.1002/cne.903560312. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Transneuronal tracing with alpha-herpesviruses: a review of the methodology. In: Loewy AD, Keplitt M, editors. Viral Vectors: Tools for the Study and Genetic Manipulation of the Nervous System. New York: Academic Press; 1995b. [Google Scholar]
- Ugolini G, Kuypers HG, Simmons A. Retrograde trans-neuronal transfer of herpes simplex virus type 1 (HSV 1) from motoneurones. Brain Res. 1987;422:242–256. doi: 10.1016/0006-8993(87)90931-0. [DOI] [PubMed] [Google Scholar]
- Ugolini G, Kuypers HG, Strick PL. Transneuronal transfer of herpes virus from peripheral nerves to cortex and brainstem. Science. 1989;243:89–91. doi: 10.1126/science.2536188. [DOI] [PubMed] [Google Scholar]
- van den Pol AN, Acuna-Goycolea C, Clark KR, Ghosh PK. Physiological properties of hypothalamic MCH neurons identified with selective expression of reporter gene after recombinant virus infection. Neuron. 2004;42:635–652. doi: 10.1016/s0896-6273(04)00251-x. [DOI] [PubMed] [Google Scholar]
- Vercelli A, Repici M, Garbossa D, Grimaldi A. Recent techniques for tracing pathways in the central nervous system of developing and adult mammals. Brain Res Bull. 2000;51:11–28. doi: 10.1016/s0361-9230(99)00229-4. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Dantzker JL, Callaway EM. Excitatory cortical neurons form fine-scale functional networks. Nature. 2005;433:868–873. doi: 10.1038/nature03252. [DOI] [PubMed] [Google Scholar]
- Young JA, Bates P, Varmus HE. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Callaway EM. Local connections to specific types of layer 6 neurons in the rat visual cortex. J Neurophysiol. 2006;95:1751–1761. doi: 10.1152/jn.00974.2005. [DOI] [PubMed] [Google Scholar]
- Zemanick MC, Strick PL, Dix RD. Direction of trans-neuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc Natl Acad Sci USA. 1991;88:8048–8051. doi: 10.1073/pnas.88.18.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Horowitz LF, Montmayeur JP, Snapper S, Buck LB. Genetic tracing reveals a stereotyped sensory map in the olfactory cortex. Nature. 2001;414:173–179. doi: 10.1038/35102506. [DOI] [PubMed] [Google Scholar]