Abstract
Escherichia coli expressing the Dr family of adhesins adhere to epithelial cells by binding to decay-accelerating factor (DAF) and carcinoembryonic antigen (CEA)-related cell surface proteins. The attachment of bacteria expressing Dr adhesins to DAF induces clustering of DAF around bacterial cells, and also recruitment of CEA - related cell adhesion molecules. (CEA, CEACAM1, CEACAM3 and CEACAM6) have been shown to serve as receptors for some Dr adhesins (DraE, DaaE, and AfaE-III). We demonstrate that DraE, DaaE, AfaE-V and AfaE-I adhesins bind to the N-domain of CEA. We utilized binding analysis of naturally occuring Dr variants and constructed mutants, an nuclear magnetic resonance (NMR) analysis to identify the amino acids of DraE and CEA involved in binding interactions. We identified distinct binding sites for DAF and CEA on opposite faces of the adhesin. These findings imply that the adhesin could be bound simultaneously by both receptors on the epithelial cell surface. The occurrence at high frequency of point mutations in the amino acids of Dr adhesins involved in CEA binding indicate that interaction with these receptors play an important role in niche adaptation of E. coli expressing Dr adhesins.
The Dr family of adhesins of Escherichia coli is associated with diarrhea and urinary tract infections (UTI), in particular, gestational pyelonephritis and recurring cystitis [1,2,3]. This family includes Dr hemagglutinin (DraE), Dr-II, DaaE, AfaE-I, AfaE-II, AfaE-III, AfaE-V, and NfaE-I [4]. Dr adhesins recognize as receptors decay-accelerating factor (DAF) and carcinoembryonic antigen (CEA) - related cell adhesion molecules (CEACAM) [5,6]. DAF is a complement regulatory protein present on a variety of epithelial surfaces, including gastrointestinal mucosa, exocrine glands, renal pelvis, ureter, bladder, cervix and uterine mucosa [7].
The CEACAM family is a group of highly glycosylated homotypic/heterotypic cell surface intracellular adhesion molecules which includes CEA, CEACAM1, CEACAM3, CEACAM4, CEACAM6, CEACAM7 and CEACAM8 [8]. It has been reported recently that E.coli expressing some Dr adhesins, DraE, DaaE, and AfaE-III, adhered to CHO cells expressing CEACAM1, CEA or CEACAM6 [9]. These adhesins also elicit the recruitment of CEACAM1, CEA, CEACAM6 and CEACAM3 to the sites of adherent bacteria [9]. Recognition of CEA and CEACAM6 but not CEACAM1 is accompanied by tight attachment of the bacteria to elongated cell surface microvillus-like extensions. This cellular response results from activation of Rho GTPase Cdc42 and phosphorylation of ezrin/radixin/moesin (ERM) [9].
The CEA family is a member of the immunoglobulin (Ig) superfamily [8,10]. Each CEA family member consists of an N-terminal Ig variable (IgV)-like domain. At the amino acid level, the N-terminal domain exhibits greater than 90% identity with other members of the CEA subgroup. The N-terminal domain may be followed by up to six IgC2 domains (A1, B1, A2, B2, A3, B3) [8], which are all present in CEA. CEACAM1, CEACAM3, CEACAM4 are inserted into cellular membrane via a carboxy-terminal transmembrane and cytoplasmic domain, while CEA, CEACAM6, CEACAM7, CEACAM8 are anchored to the membrane via GPI. These molecules are expressed on numerous cells including epithelial, endothelial and myeloid cells [11]. Within the family, CEACAM1, a signaling receptor, is the most widely expressed in distinct human tissues, being present in granulocytes, monocytes and epithelial cells in different organs including colonic and respiratory epithelia [8,12,13].
It has been reported that E.coli and Salmonella enterica bind CEACAM molecules via their mannosyl residues [14,15,16]. Several microorganisms including Neisseria meningitidis, N. gonorrhoea, Haemophilus influenzae, and Moraxella catarrhalis target members of the CEACAM family via the proteinaceous component of the N-terminal domain [17,18,19,20,21,22]. Neisseria spp. bind CEACAM molecules via the structurally related Opa proteins, whilst in the case of H. influenzae and M.cattarrhalis the ligands appear to be distinct from this family [22,23]. Targeting of CEACAM molecules by Neisseria leads to cellular invasion and passage across polarized monolayers [24].
In this study we provide evidence that Dr adhesins, including DraE, DaaE, AfaE-V and AfaE-I bind to the N-domain of CEA. Using a combined nuclear magnetic resonance (NMR) and mutagenesis approach we identified amino acids of Dr ahdesins and CEA involved in the interactions. We demonstrate that Dr/CEA interaction is sensitive to Cm inhibition due to direct disruption of the CEA-binding surface of the adhesin. Using NMR we also show that CEA and DAF binding sites do not overlap and that DAF does not inhibit binding to CEA.
EXPERIMENTAL PROCEDURES
Bacterial strains
Bacterial strains were grown in Luria-Bertani (LB) or Super Broth (SB) medium at 37°C. Derivatives of pUC-Cm were grown in the presence of 25 μg/ml chloramphenicol (Cm). Derivatives of pET-21d and pCC90-D54stop were grown in 100 μg/ml ampicillin or carbenicillin. E. coli DH5α (Life Technologies, Inc., Rockville, Md.) and BL21 (DE3) (Novagen, San Diego, CA) were host for the plasmids. The purification of E.coli chromosomal DNA, plasmid isolation, E. coli transformation, restriction enzyme digestion or ligation were carried out as described [25]. Enzymes were purchased from New England Biolabs (Beverly, Ma) and used as recommended by the manufacturer.
The Chinese hamster ovary (CHO) cell transfectant clones that express human CEA or the vector alone were used. Cells were cultured in Ham’s F12 supplemented with 10% fetal bovine serum (FBS) and 400 μg/ml of hygromicin B, and cultured according to standard tissue culture techniques.
All constructs were confirmed by sequencing using Big Dye Terminator method and ABI sequencing (PE Applied Biosystems, Foster City, CA).
Cloning, expression and purification of N-domain of CEA, CEACAM1, CEACAM6, CEACAM8 and CEA-CEACAM8 chimera
The sequences corresponding to the mature amino acid sequence of N-domain of CEA, CEACAM1, CEACAM6, CEACAM8 were PCR amplified using cDNA clones (image ID:5184800, 4540619, 3640231, 4618311; ATCC) as templates and inserted into pET-21d (Novagen). The CEA-CEACAM8 chimera was synthesized using the PCR-based splicing by an overlap extension approach using CEA and CEACAM8 cDNA clones as templates and the following oligonucleotide pairs: CEAf, 5′-ccatggccaagctcactattgaatc-3′; CEACAM8-CEAr, 5′-gattgctgtatgcaggccctggggtagcttgttgagttc-3′, and CEACAM8r, 5′-gaagcttttaagtctccggatgtacgctgaac-3′; CEA-CEACAM8f, 5′-gaactcaacaagctaccccagggcctgcatacagcaatc-3′. The chimera was then amplified by adding CEAf and CEACAM8r primers to the PCR mix. The hybrid molecule was cloned into pET-21d (Novagen). The proteins were expressed in E. coli BL21 (DE3) and purified from inclusion bodies. The inclusion bodies were dissolved overnight in buffer containing 30 mM Tris/HCl (pH 8.5), 150 mM NaCl, 1 mM EDTA and 8 M urea. One volume of the protein sample was added slowly to 20 volumes of buffer containing 50 mM CHES (pH 9.2) and 500 mM arginine, and the sample was left overnight at 4 °C. The refolded protein was concentrated by ultrafiltration and purified by gel-filtration using a Superdex 75 column (Amersham Corporation, Piscataway, N.J) in 30 mM Tris/HCl (pH 8.5), 150 mM NaCl. Calibration of the column was performed using gel filtration markers (Amersham).
Purification of CEA N-A3 domain
CEA containing N and A3 domains and an oligohistidine tag at the N-terminal end (CEA N-A3) was purified from recombinant Pichia pastoris which was generously provided by Dr. John E. Shively (Beckman Research Institute City of Hope, Duarte, CA). CEA N-A3 was expressed in P. pastoris and purified from the supernatant of induced cultures by Ni-NTA chromatography as described previously [26,27]. For SPR experiments, the protein was then purified by gel filtration chromatography using a Superdex 200 column (Amersham) in HBS-E buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA).
Purification of Dr family fimbriae
Genes encoding Dr adhesins were amplified by using the following primers: for DraE and AfaE-III: DraE-BamHI, ggatccgaaggagatatacatatgaaaaaattagcgatcatggcc, and DraE-PstI, cacgcacgtcctgcagtcattttgcccagtaacc; for AfaE-V: DraE-BamHI primer and AfaE-V-PstI, cacgcacgctgcagtcaactcacccagtagccccagt; for NfaE: NfaE-BamHI, cgaggatccgaaggagatatacatatgaaaataaaatatacgatg, and NfaE- PstI, cacgcacgctgcagttattggctgtacactgcggc. Products were digested with BamHI and PstI and inserted into BamHI and PstI restriction sites of pUC-Cm. Gene encoding DaaE adhesin was amplified by using the following primers: DaaE-BamHI, cgatccgaacaggtaatcaatatgaaaaaattagcgataatg and DaaE-EcoRI, gaattcttagttcgtccagtaacccc. Product was digested with BamHI and EcoRI and inserted into BamHI and EcoRI restriction sites of pUC-Cm. Gene encoding AfaE-I adhesin was amplified by using the following primers: AfaE-I-EcoR1, gaattcgaaggagatatacatatgaaaaaattagcgatcatag, and AfaE-I-PstI, cacgcacgctgcagttattttgtccagaacccgccttcg. Product was digested with EcoRI and PstI and inserted into EcoRI and PstI restriction sites of pUC-Cm. The resulting plasmids were transformed into E. coli 191A (pCC90-D54stop). This strain contains the necessary genes of the dra operon for fimbrial expression, with a premature stop codon at codon 54 within draE. Dr fimbriae were purified from recombinant strains as described previously [28]. For SPR analysis, fimbriae were purified by gel-filtration chromatography using a Superdex 200 column (Amersham) in HBS-E buffer.
Purification of DAF
DAF234 contains short consensus repeats 2, 3, and 4 and an oligohistidine tag at the C-terminal end and was purified from recombinant P. pastoris kindly provided by Susan Lea, Oxford University, as previously described [29].
Surface plasmon resonance studies
SPR measurements were carried out in HBS-EP buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% P-20 surfactant [BIAcore AB, Uppsala, Sweden]) using a BIAcore 2000 system (BIAcore AB). To analyze the interaction between Dr fimbriae and N-domains of CEACAM receptors, native fimbriae were immobilized on a CM5 research-grade sensor chip (BIAcore AB) by amine coupling chemistry using the manufacturer’s protocols. Immobilization of 500 response units (RU) resulted in optimal responses. The N-domain of each CEACAM was dissolved in running buffer and analyzed using a ∼102 dilution series, i.e., (3-300 μM). Analyte was injected over the surface at a flow rate of 20 μl/min for 2 min. For competition experiments, DAF234 (30 μM final concentration), or Cm (1 mM final concentration) were added to the running buffer. The affinities of the interactions were studied under steady-state conditions. Average equilibrium responses were measured for six to seven concentrations of CEACAM receptor. Raw sensorgrams were corrected using the double-subtraction protocol [30] and by subtracting both the reference flow cell response and the average of eight buffer injections. The resulting data were analyzed with BIAevaluation 3.0 software (BIAcore AB) to globally fit the data and derive equilibrium constants describing the intermolecular interactions. The reported KD values are the average of at least three independent experiments. Error estimates were propagated from the standard error (SE) of the association constant (KA).
Chemical shift mapping for the AfaE-dsc and N-CEA interaction
The recombinant strain expressing AfaE-III-dsc described previously [31]. An 15N-labelled sample of AfaE-III-dsc was produced in minimal media, containing 0.07% 15NH4Cl and 0.2% C-glucose, supplemented with 50 μg/ml ampicillin. AfaE-III-dsc was purified under denaturing conditions using the binding of the N-terminal hexahistidine tag to Nickel-bound agarose beads as described previously. Purified protein was refolded by dialysis into 50 mM sodium acetate buffer, pH 5.0 and concentrated to approximately 0.2 mM for NMR. N-CEA in the same buffer was introduced at several molar ratios until saturation was achieved and 2D 15N-1H HSQC experiments were recorded at each stage under identical experimental conditions. 1H-15N resonances shift changes and line widths were monitored by analysis of the 2D 1H-15N HSQC spectra.
PCR mutagenesis of DraE
DraE was subjected to PCR mutagenesis using the GeneMorph II Random Mutagenesis Kit as directed by the manufacturer (Stratagene, La Jolla, CA) The pCC90 vector containing draE [32] was used as a template with following primers: 5ranmut, 5′-cccgcccgccgattcggggtaagacagc-3′ and 3ranmut, 5′-cccgcccgccctgcagtcattttgcccagtaacc-3′. Transformants containing mutant derivatives of draE were constructed as described previously [28].
Selection for CEA-binding deficient mutants
CEA N-A3 protein was used to coat seven 6-well microtiter plates at a concentration of 5 μg/ml in bicarbonate buffer at 37 °C for 1 h. Anti-Dr rabbit antisera [32] was dissolved in bicarbonate buffer at a 1:100 dilution and used to coat 6-well microtiter plates at 37 °C for 1 h. The wells were blocked with 0.01 M phosphate-buffered saline pH 7.2 (PBS) containing 1% BSA for 15 min at 37 °C. Transformants containing the mutant draE derivatives were resuspended in 12 ml of PBS to an OD540 of 0.025. 2 ml of cell suspension were added to each well coated with CEA and allowed to bind for 45 min at 37 °C. The supernatant containing unbound bacteria was collected and added to a fresh plate coated with CEA. These steps were repeated successively for a total 6 times. The final supernatant was added to a microtiter plate coated with anti-Dr antisera and allowed to bind for 45 min at 37 °C. The unbound bacteria were removed by 7-9 washes with PBS and 2 ml of SB medium were added to each well. The plate was incubated with shaking at 37°C for 1 h. The resulting bacterial suspension was plated on LB medium containing Amp, Cm and X-gal. White colonies were selected and examined for erythrocyte binding by a mannose-resistant hemagglutination assay (MRHA, see below). Those transformants that were MRHA+ were analyzed for CEA binding.
Mannose resistant hemagglutination assay (MRHA)
MRHA with human erythrocytes of blood group O was performed as previously described [28].
CEA binding growth assay
Those transformants that were MRHA+ were examined for the ability to bind CEA. CEA N-A3 was used to coat 96-well microtiter plates as described above. 100 μl of an overnight culture of each transformant, OD540 = 4.0, were added to individual wells and allowed to bind for 45 min at 37 °C. The unbound bacteria were removed with PBS then 150 μl of SB medium were added to each well. The plate was incubated with shaking at 37 °C for 2 h. The resulting growth was then measured at OD540. Those transformants that exhibited little or no growth were further analyzed to confirm DraE expression.
Surface expression assay and sequencing analysis of mutants
To confirm that the transformants express the various mutant adhesin variants at equivalent levels, they were examined for the ability to bind polyclonal antisera prepared against DraE [32]. This assay was performed in the same manner as the CEA binding growth assay with following modification. Plates were coated with anti-Dr antisera at a 1:200 dilution for 2 hours. Those transformants that exhibited growth were selected for sequencing.
CEA binding assay with radiolabeled bacteria
This assay was performed in the same manner as the CEA binding growth assay with the following modifications. Plates were coated with soluble CEA (Xema, Moscow, Russia) or CEA N-A3 at a concentration of 15, 10, 5 and 3 μg/ml in PBS. The bacterial strains were grown overnight in SB supplemented with 10 μCi/ml of 3H-thymidine at 37 °C. The cells were then pelleted and resuspended in PBS and adjusted to OD540 = 4.0. 50 μl of each of the bacterial strain suspensions were added to a microtiter plate well together with 50 μl of PBS containing 0.2% BSA and allowed to bind for 1 hour at 37 °C. To test the effect of competitors (DAF234, N-domain of CEA, and Cm) on bacterial binding to CEA, 20 μM of N-domain of CEA, 80 μM DAF, or 10 mM Cm were added to PBS containing 0.2% BSA. Unbound bacteria were removed by washing with PBS and the wells were dried at 60 °C. The wells were placed in scintillation fluid and bound bacteria quantified by scintillation counting.
Site-directed mutagenesis of CEA gene
Mutations were introduced into the CEA (N-domain) gene on plasmid pET-21d (this study) by site-directed mutagenesis using the Quick Change Kit as directed by the manufacturer (Stratagene). Constructs containing mutations were identified by sequence analysis.
CHO cell-binding assay
CHO cells were split into 24-well plates with glass coverslips and grown to confluence. Before the assay, cells were washed twice with Hanks’ balanced saline solution (HBSS) and incubated with fresh medium without antibiotics and without FBS for 1 h. The bacterial strains were grown overnight on LB medium and harvested and resuspended in PBS to OD540 = 0.6. The bacterial cells were pelleted and resuspended in the tissue culture medium. Then 0.1 ml of each bacterial strain were added to each well. The adherence assay was performed as described previously [28] and repeated in triplicate.
RESULTS
Adhesins of the Dr family, including DraE, DaaE, AfaE-V and AfaE-I, bind soluble CEA, CEA N-A3 and CHO cells expressing CEA
Previously it has been shown that Dr adhesins including DraE, AfaE-III and DaaE bind and elicit recruitment of CEA, CEACAM1 and CEACAM6 [9]. To determine if other members of Dr family could bind CEA, recombinant E.coli expressing different Dr adhesins were constructed. Dr adhesins were expressed in a background strain containing a mutant dra operon expressing genes necessary for assembly of the Dr fimbriae, but not the adhesive subunit. Transfected CHO cell lines expressing human recombinant CEA or containing the expression vector alone were used to study the recognition of CEA by bacteria expressing Dr adhesins. None of the tested strains showed binding to CHO cells containing the vector alone. Strains expressing DraE/AfaE-III alleles, DaaE, and AfaE-V were able to bind 60-80% of CHO-CEA cells, whereas the parent strain with a premature stop codon in DraE did not bind the cells. The recombinants expressing AfaE-I demonstrated poor binding to CHO-CEA cells, while E. coli expressing NfaE did not mediate adherence to this cell line.
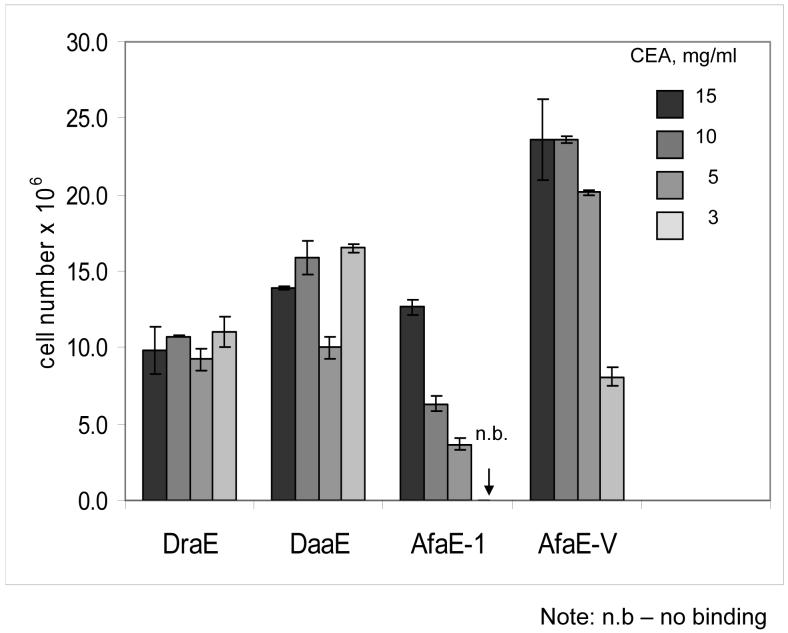
To further investigate the CEA-binding phenotype of Dr adhesins, we coated plates with soluble CEA (15, 10, 5, and 3 mg/ml) and incubated them with radiolabelled E.coli expressing the adhesins. Strains expressing DraE, DaaE, and AfaE-V adhered strongly to plates coated with CEA at all tested concentrations (Fig.1). Moreover, these strains also bound to a CEA derivative construct consisting of only the N and A3 domains (data not shown).
Fig. 1. CEA binding assay with radiolabeled bacteria expressing Dr family adhesins.
Bacterial cells were labeled with 3H-thymidine and incubated in microtiter wells coated with soluble CEA at concentrations of 15, 10, 5, or 3 mg/ml. Bound bacteria remaining in the wells were quantified by scintillation counting. Results shown are the average of three independent experiments with duplicate wells of each sample. Error bars indicate standard deviation.
Previously observed phenotypes of AfaE-I and NfaE correlated well with the ability of the recombinants to bind to CEA-coated plates (Fig. 1). NfaE did not mediate bacterial binding to CEA and CEA N-A3 plates. When bacteria expressing AfaE-I were incubated with the plates coated with 15 and 10 mg of CEA, this adhesin demonstrated a level of adherence comparable to DraE, DaaE and AfaE-V. However, AfaE-I mediated a reduced level of binding to plates coated with CEA at a concentration of 5 mg/ml and failed to bind CEA at 3 μg/ml. These data indicate that the affinity of AfaE-I for CEA is significantly lower than DraE, DaaE or AfaE-V, but as multiple subunits of adhesion complex contribute an avidity effect to the interaction, the binding of the adhesins could be compared only at low concentrations of CEA. Taken together, these results demonstrate that recognition of CEA is expressed by most adhesins of Dr family and N-terminal and A3 domains are sufficient for these interactions.
The N-terminal domain of CEA mediates binding to DraE, AfaE-III, AfaE-V and AfaE-I
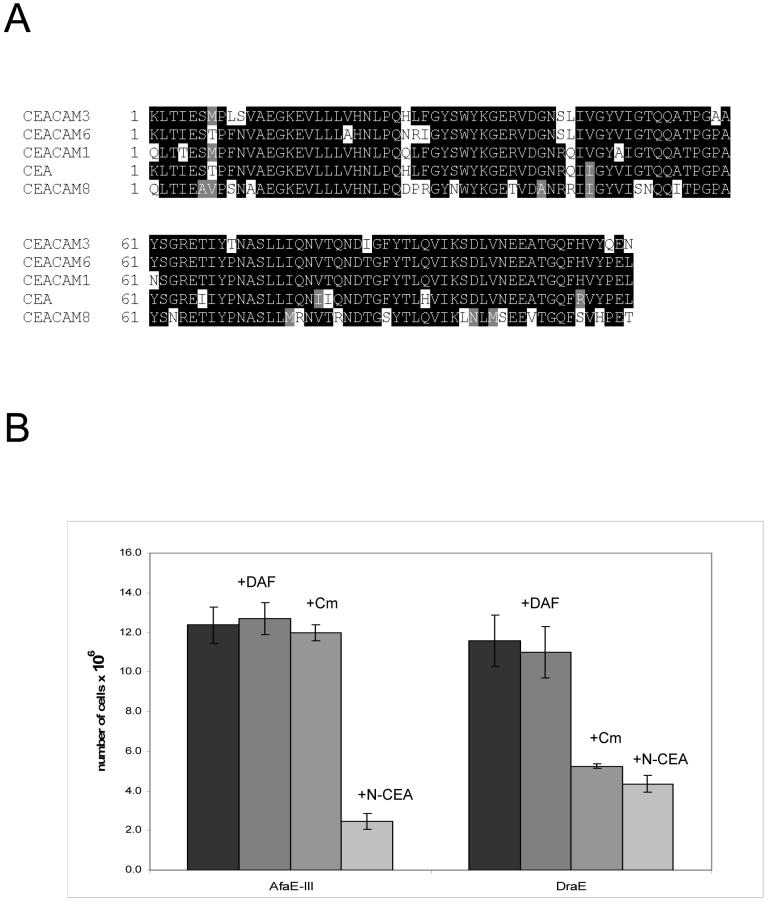
Dr adhesins recognize as receptors CEA, CEACAM1 and CEACAM6 and also elicit the recruitment of CEACAM3 around adhering bacteria [9]. Since the N-terminal and A3 domains of CEA are sufficient for bacterial adherence and the N-terminal domains of CEACAMs have high homology (Fig. 2,A), we hypothesized that the N-domain of CEA, CEACAM1 and CEACAM6 is the target for Dr adhesins. To test our hypothesis we constructed E.coli recombinants expressing the N-domain of CEA (N-CEA), and isolated the protein from inclusion bodies. In a similar manner, the N-domain of CEACAM8 (N-CEACAM8) was cloned, expressed and purified. When N-CEA was tested as an inhibitor in the CEA binding assay with radiolabeled strains expressing DraE and AfaE-III adhesins, bacterial adherence was inhibited significantly, suggesting that the adhesins recognize the N-domain on CEA (Fig.2,B).
Fig. 2. The N-domain of CEACAMs is important for Dr adhesin binding.
A) Sequence alignments for N domains of CEACAMs. B) CEA binding assay with E.coli expressing DraE and AfaE-III. Bacterial cells were labeled with 3H-thymidine and incubated with or without the competitor in microtiter wells coated with soluble CEA at a concentration of 5 mg/ml. Competitors were added at final concentrations of 20 μM N-CEA, 80 μM DAF, and 10 mM Cm. Bound bacteria remaining in the wells were quantified by scintillation counting. Results shown are the average of three independent experiments with duplicate wells of each sample. Error bars indicate standard deviation.
We examined the affinity of Dr adhesins (DraE, AfaE-III, DaaE, AfaE-V, AfaE-I and NfaE) to N-CEA and N-CEACAM8 by surface plasmon resonance (SPR) analysis. N-CEACAM8 has a high degree of amino acid homology to N-CEA (Fig. 2,A) but CHO cells expressing CEACAM8 are not recognized by Dr adhesins [9]. The affinities of the interactions were studied by immobilizing purified adhesin to the sensor surface and flowing CEACAMs over the surface. No change in the resonance signal was detectable when N-CEACAM8 was injected to flow over adhesin-immobilized sensor surface, indicating the absence of detectable binding. No binding was observed when NfaE was immobilized to the sensor surface and N-CEA was used as analyte.
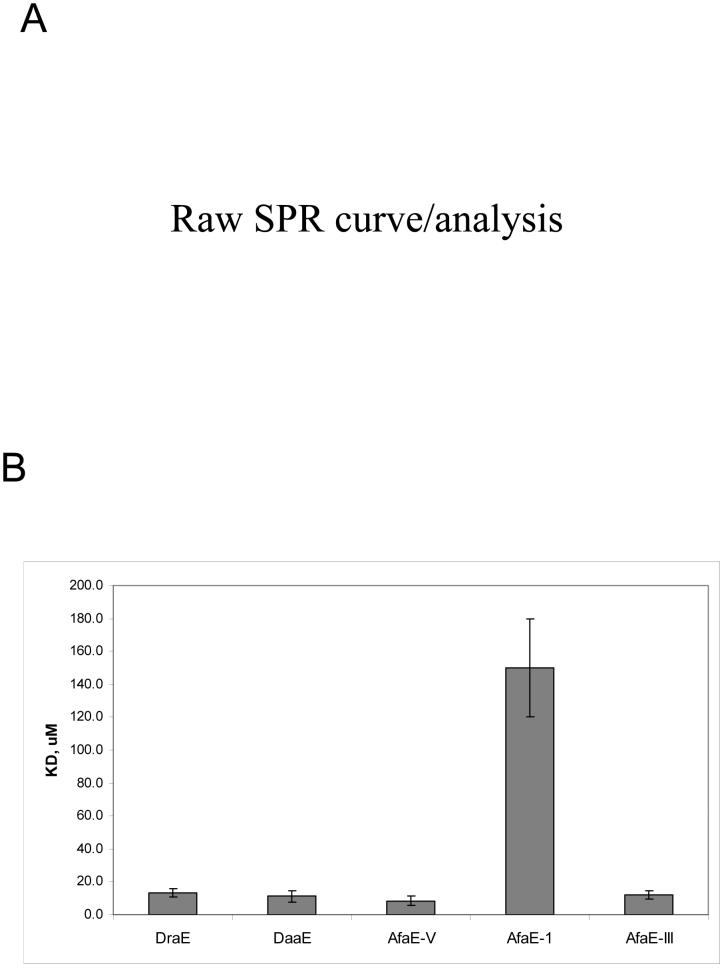
SPR studies indicated that the interactions between DraE, AfaE-III, DaaE, AfaE-V and AfaE-I adhesins and N-CEA are characterized by very fast on and off rates, therefore steady-state conditions were used to calculate affinity (Fig. 3A). Interestingly, AfaE-V, which does not mediate MRHA and displays very low affinity to DAF (unpublished observations), was the strongest binding variant among the tested adhesins. AfaE-I demonstrated the lowest affinity to N-CEA, consistent with previously observed phenotype (Fig. 3B).
Fig. 3. SPR measurement of N-CEA affinity (KD) to Dr adhesins.
A) SPR binding curves of the interaction between N-CEA and DraE fimbriae in steady state. The resulting data were analyzed with BIAevaluation 3.0 software to globally fit data and derive the dissociation constants (KD). A fit of these data is shown. RU, relative units. RE, response difference. B) Bar chart showing a comparison of dissociation constants (KD) for different Dr adhesins.
Effect of competitors (Cm and DAF) on DraE and AfaE-III binding to CEA
It has been shown that the binding of DAF to DraE is inhibited by chloramphenicol (Cm) [3]. However, adherence of DAF to AfaE-III, differing from DraE by three non-synonymous nucleotide changes (D52N, T88M, I111T), is resistant to Cm [3]. X-ray studies of DraE in the complex with Cm indicated that Cm interacts with P40, G42, P43, I111, G113, I114 and Y115 of DraE, and Cm sensitivity of DraE is caused by direct disruption of DAF-binding surface of the adhesin [33]. To investigate the effect of Cm on DraE/CEA interactions, bacterial cells expressing DraE or AfaE-III were mixed with 15 mM Cm and incubated with CEA-coated plates (Fig. 2,B). Binding of DraE-expressing bacteria was inhibited by Cm, while AfaE-III-expressing E.coli were resistant to Cm inhibition (Fig. 2,B). These data suggest that DraE binding to Cm disrupts the CEA-binding surface of the adhesin.
Amino acids involved in DAF/DraE and DAF/AfaE-III interactions were implicated by mutagenesis and NMR studies [28,31]. NMR analysis has suggested that the DAF binding region of AfaE-III involves a large surface comprising domains A1, A2, B, C2, E, F and Gd [31]. To determine if binding of DraE adhesins to DAF disrupts the CEA-binding surface of the adhesin, DAF234 was tested as a competitor in the CEA-binding assay with radiolabeled bacteria. DAF had no effect on bacterial adherence to the CEA-coated plates, implying that the binding domains for DAF and CEA are distinct (Fig. 2B).
In order to confirm with the effect of DAF or Cm on binding of N-domain of CEA to fimbrial preparations, we utilized SPR analysis. DAF234 (30 μM) or Cm (1 mM) was added to buffer containing N-CEA, and flowed over DraE fimbriae immobilized on the sensor surface. Results as clearly consistent with the CEA binding assay with DraE-expressing bacteria described above. DAF had no effect on the affinity of DraE for N-CEA (KD=13.1±1.5 μM). However when DAF was replaced with Cm (1 mM), N-CEA failed to bind the DraE immobilized surface.
Analysis of CEA binding phenotypes of DraE/AfaE-III clinical isolates
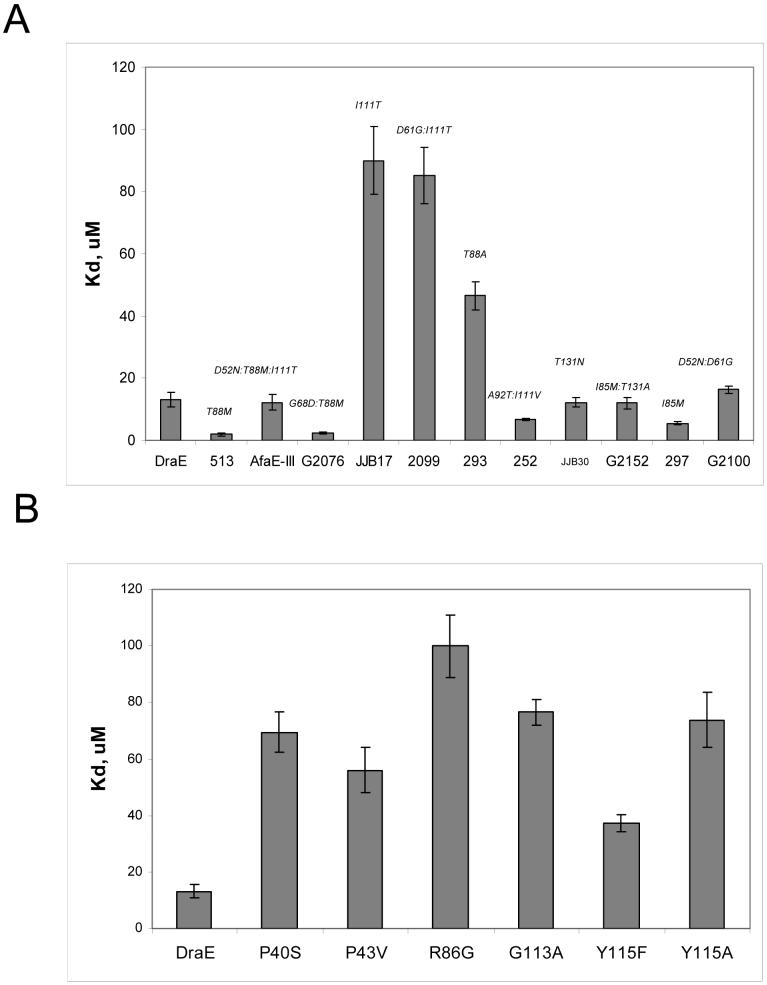
We have demonstrated that the Dr family of adhesins in general, and the DraE/AfaE-III family in particular, are under strong selective pressure for functional variation. E.coli expressing DraE/AfaE-III adhesins are adapting to a pathogenic life style by selection for mutations in the adhesin that enhance binding to DAF and decrease sensitivity to Cm (Korotkova et al., manuscript in preparation). To investigate the functional variability of DraE/AfaE-III alleles with regard to CEA binding, we purified naturally occuring DraE-related fimbrial variants and utilized SPR to determine the affinity to N-CEA as described above. We found that three variants, JJB17, 2099 and 293 had low affinity to N-CEA (Fig. 4,A). JJB17 and 293 each differ from DraE by a single non-synonymous mutation, I111T and T88A, respectively. 2099 differs from JJB17 by one mutation, D61G.
Fig. 4. N-CEA affinity (KD) for DraE mutants and naturally-occuring DraE/AfaE-III adhesins.
A) DraE mutants and B) DraE/AfaE-III-related adhesins determined by SPR. Fimbriae were immobilized on a CM5 sensor chip. N-CEA was dissolved in running buffer and injected over the surface at a flow rate of 20 μl/min for 2 min. Average equilibrium responses were measured for seven concentrations of N-CEA. The resulting data were analyzed with BIAevaluation 3.0. Reported KDs are the average of at least three independent experiments. Error estimates were propagated from the SE of the KA.
Four adhesins, 513, G2076, 252 and 297 have increased affinity to N-CEA (Fig. 4,A). 513 and 297 differ from DraE by single non-synonymous mutation, T88M and I85M, respectively. G2076 differs from 513 by one mutation, G68D. 252 differs from DraE by two non-synonymous mutations, A92T and I111V. Interestingly, AfaE-III, differing from 513 by two non-synonymous nucleotide changes (D52N, I111T) demonstrated N-CEA affinity comparable to DraE. This adhesin acquired two CEA-binding mutations, T88M and I111T. It is possible that the positive effect of the T88M mutation on CEA binding compensates the negative effect of I111T mutation. Thus, our data suggest that: (i) residues I111, T88 and I85 are important for DraE/N-CEA interactions; (ii) the DraE sub-group accumulates a high number of non-synonymous substitutions leading to variation in CEA-binding phenotypes of the adhesins.
Identification of amino acids of DraE involved in CEA binding by random mutagenesis
To independently identify amino acids of the adhesin critical for recognition of CEA and provide further evidence for our NMR-derived N-CEA binding surface, we performed random mutagenesis of DraE. Several mutant strains were isolated that mediated MRHA but demonstrated very weak binding to CEA-coated plates. We chose to further characterize 6 mutants that contained single amino acid substitutions within DraE (P40S, P43V, R86G, G113A, Y115F, Y115A).
To examine the effect of the mutations in DraE upon its binding properties, fimbriae were isolated from the bacterial mutants. The affinities of DraE/CEA interactions were analyzed by SPR as described above. Figure 4,B summarizes the DraE mutations and their effects on CEA binding. All mutants mediated binding to N-CEA, but the mutations significantly weaken the interactions between DraE and CEA. The mutation at positions R86G has the largest effect, decreasing the affinity more than seven fold. The position of DraE mutations affecting CEA binding are shown in Figs. 5B, 6A and 6C.
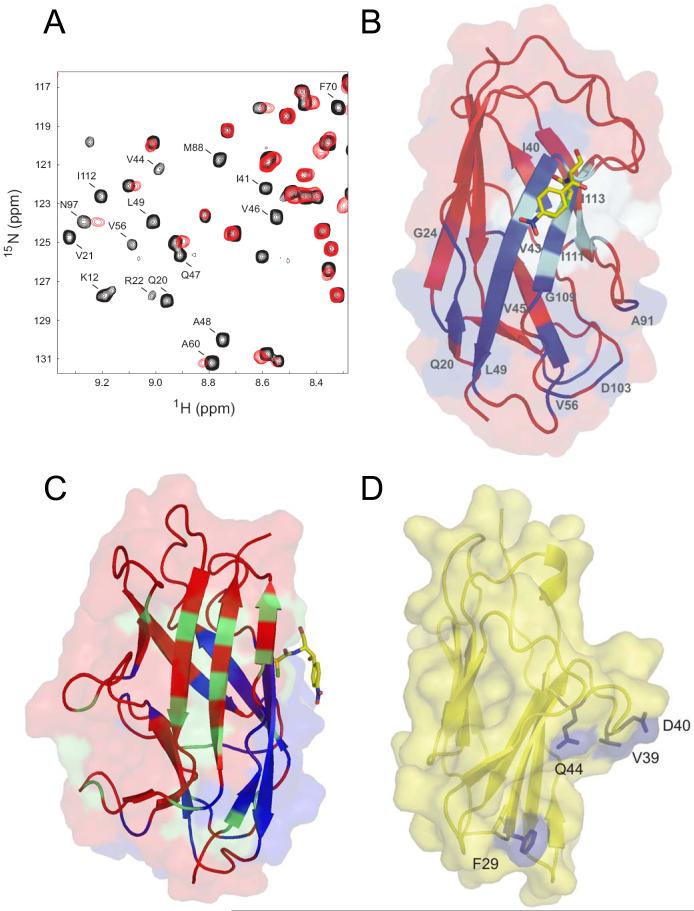
Fig. 5. Chemical shift mapping of the N-CEA/AfaE-III interaction.
A) Region of the 2D 1H-15N HSQC spectrum of AfaEIII-dsc in the absence (black) and presence (red) of excess N-CEA. Peaks undergoing significant shift changes and/or broadening are labelled. B) Combined surface and ribbon representation of AfaE-III-dsc with perturbed amide resonances upon N-CEA addition colored in dark blue, Cm shown in stick representation and position of mutants with altered CEA binding in cyan. C) Surface and ribbon representation of AfaE-III-dsc with NMR-derived binding sites for CEA and DAF colored blue and green respectively with Cm as a yellow stick representation [31,33]. Orientation is chosen is approximately 90y° rotation relative to Figure 5,B D) Surface and ribbon representation (yellow) of a homology model of human N-CEA derived from the crystal structure of murine N-CEACAM1 [34]. Solvent exposed mutant resides that display altered DraE binding are shown in blue.
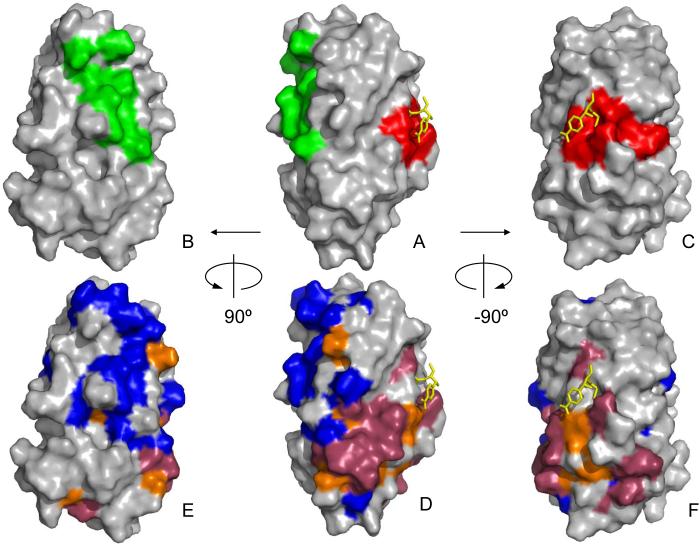
Fig. 6. DraE/AfaE-III adhesin binding-associated surfaces.
Surface representation of AfaE-III-dsc with Cm as a yellow stick representation [31,33]. Orientation of A-C the same as D-F, respectively. A-C) DAF [28] and CEA binding sites derived from DraE mutagenesis are shown in green and red, respectively. D-F) DAF-specific interactions derived from chemical shift mapping analysis of AfaE-III-dsc shown in blue [31]. CEA-specific interactions derived from chemical shift analysis of AfaE-III-dsc is shown in dark red. Chemical shift mapping of interactions common to DAF and CEA shown in orange [31].
Identification of binding site for CEA in Dr adhesin by NMR chemical shift mapping
Our earlier work on the structure and assembly of the Dr adhesins provides further opportunity to perform NMR titration experiments to directly investigate the binding site for CEA [31]. An analysis of amide line-width and chemical shift changes for AfaE-III-dsc in the presence of N-CEA was carried out (Fig. 5,A). The CEA binding region of AfaE-III-dsc may be delineated, and lies entirely on one side of the molecule, forming surface comprising side-chains from principal strands A1, B and E and adjacent loop regions (Fig. 5B,C, Fig 6E-F). Key amides perturbed in the presence of N-CEA include G6, K12, Q20-G24, I41, G42, V44-L49, R54-V56, A60, G68, F70, M88, S91, A92, N97, D104, G106, W108, G110-I112, I114, and A138.
Identification of amino acids of CEA involved in Dr adhesin binding
To investigate the affinity of DraE to other members of CEACAM family we constructed E.coli recombinant expressing N-domains of CEACAM1 (N-CEACAM1) and CEACAM6 (N-CEACAM6). N-CEACAM1 affinity for DraE was comparable to N-CEA (Table 1). However, N-CEACAM6 showed weak binding to the adhesin. DraE does not interact with N-CEACAM8, as discussed above. Sequence alignment of the N-domains of CEACAM-related proteins, including CEACAM1, CEACAM3, CEA, CEACAM6, and CEACAM8 revealed exceptional homology (Fig.2,A). In order to identify amino acids of CEA N domains involved in CEA/Dr interactions we constructed a CEA-CEACAM8 chimera consisting of the residues 1-58 of the N domain of CEA fused to residues (59-110) of the CEACAM8 N-domain and analyzed the ability of the chimeric protein to bind DraE by SPR. We found that the affinities obtained for wild-type and the mutant were the same (Table 1) implying that the N-terminal 58 amino acids of CEA in CEA/DraE interactions.
Table. 1. Affinity (KD) of DraE adhesin to N-CEACAMs and N-CEA mutants.
| N-domains of CEACAMs and N-CEA mutants | KD,μM |
|---|---|
| N-CEA | 13.1±2.5 |
| N-CEACAM1 | 15.3±3.8 |
| N-CEACAM6 | 30.1±2.8 |
| CEA-CEACAM8 chimera | 14.4±2.1 |
| F9A | 18.4±1.5 |
| V11A | 24.9±1.2 |
| L28P | 6.6±0.5 |
| F29R | >200 |
| S32N | 12.8±1.3 |
| V39A | 9.35±1.3 |
| D40A | 48.9±3.1 |
| Q44R | >200 |
| G51D | 12.0±1.4 |
| T52N | 11.0±2.1 |
Using homologue scanning mutagenesis of the N-terminal 58 amino acids of CEA, we replaced single residues of CEA with corresponding residues of CEACAM8 and determined the affinity of the mutants to DraE. As is shown in Table 1, mutations in F9, V11, L28, S32, V39, G51 and T52 had no significant effect on DraE/N-CEA interactions. Mutation D40A caused an almost four fold drop in the DraE/N-CEA affinity and two mutations, F29R and Q44R, abolished the interaction.
DISCUSSION
Diffusely adhering Escherichia coli (DAEC) strains express adhesins of Dr family and cause symptomatic urinary tract or intestinal infections [4]. The Dr family of adhesins is comprised of a number of related adhesins demonstrating extensive structural diversity. The structural variability of Dr adheins is emerging under strong positive selection resulting in generation of clades within the family, and the accumulation of point mutations within the clades (Korotkova et al., manuscript in preparation). Variability in the Dr family of adhesins results in functional diversity with regard to receptor specificity [9,32].
E.coli expressing Dr adheins adhere to human cells by recognition of the brush border-associated DAF [4]. The attachment of bacteria expressing DraE, AfaE-III, DaaE, AfaE-I to DAF induces clustering of DAF around bacterial cells, and also recruitment of the brush border proteins, CEA, CEACAM1, CEACAM3 and CEACAM6 [6,9]. It has been shown that Dr adhesins including DraE, AfaE-III and DaaE are involved in adherence to CEA, CEACAM1 and CEACAM6, thus indicating that these molecules might be important for DAEC colonization [9].
In this study we have demonstrated that the bacteria expressing DraE/AfaE-III alleles, DaaE, AfaE-V and AfaE-I recognized the N-domain of CEA. Thus, these studies add to the list of bacterial ligands that target the N-domain of CEA. Adherence to this receptor by apparently structurally distinct proteins of several bacterial genera points to the importance of CEA for the colonization of pathogens.
We found that the DraE adhesin also recognizes the N-domains of CEACAM1 and CEACAM6, demonstrating low affinity to CEACAM6 and high affinity to CEACAM1. Homology scanning mutagenesis of N-CEA revealed that F29 and Q44 (and to a lesser extent D40) of CEA are required for maximal DraE binding affinity. These residues are located in the exposed loops of the GFCC’C” face of the CEA N-domain, which is not sheltered by carbohydrate as revealed by crystal structure of murine CEACAM1 [34,35] and would be accessible for pathogen binding (Fig 5,D). The solvent exposed area around F29 in CEA is hydrophobic and might be important for contacts with Dr adhesins. It has been shown that other pathogenic bacteria also target this exposed protein face of the N-terminal IgV-like domain of CEACAM receptors [18,19,20,36].
CEACAM family members can exist as dimers in the plasma membrane of eukaryotic cells, and recombinant N domains of CEA have been shown to form oligomers in solution [37,38,39]. It has also been demonstrated that residues on the GFCC’C” face of the CEA, are directly engaged in homophilic cell adhesion [34,40]. Amino acid residues V39A, D40 and Q44 in CEACAM1 have been reported to play an important role in homophilic cell adhesion of CEACAM1 [41,42]. We do not know if Dr adhesins interact with CEA oligomer, CEA monomer, or both. It is possible that CEA mutations affecting DraE binding are involved in receptor oligomerization rather than in the direct contact with the adhesin, and that Dr adhesins have higher affinity to a dimer than to a monomer. If this is the case, then the disruption of homophilic adhesion would decrease the binding to CEA. Structural studies of the CEA-DraE complex needed to provide detailed information about the mechanism of DraE recognition of the receptor.
NMR chemical shift mapping and analysis of recently published NMR and X-ray structures of AfaE/DraE (structures 1RXL and IUSQ; [31,33]) identify the CEA-N binding on the face of the DraE, implying that these residues form a part of binding surface for CEA (Fig. 6E-F, dark red and orange surfaces). Key residues for a contiguous patch included residues from strands A1, B and E and neighboring loop regions. Random mutagenesis together with functional analysis of the DraE/AfaE-III alleles identified residues P40, P43, R86, I85, T88, I111, G113, and Y115 that are important for binding to CEA. Seven of these are solvent accessible, but only two (88,111) overlap the surface identified by NMR. However the mutation-derived and NMR-derived surfaces are adjacent on the same face of the molecule in the proximity of the Cm binding pocket. Indeed, these results are consistent with the discovery that Cm inhibits CEA binding by DraE. One residue, I85, maps to a site where the side-chains point toward the center of the protein. However, this amino acid is located near the cluster of surface residues involved in CEA binding. Therefore, the mutation might influence the overall stability of that cluster and thus affect the CEA affinity.
Previously it has been shown that DraE mutations at positions T88 and I111 affected type IV collagen binding of the adhesin [32]. According to our unpublished observations, the amino acids P40, P43, I114, Y115 are also important for DraE/collagen interactions. DraE mutations P40A, P43V, I114A and Y115A resulted in a complete loss of recognition of collagen by DraE. Therefore, the hydrophobic surface on DraE that included these residues is involved in the binding of three molecules, CEA, Cm and the 7S domain of type IV collagen. DAF/DraE interactions are also sensitive to the presence of Cm, and NMR analysis indicates an overlap between DAF and CEA binding surfaces near the Cm binding site (Fig 6D,E, orange surface). Therefore one might expect that DraE binding to CEA would be inhibited by DAF. However, our data demonstrate that DAF has no inhibitory effect on CEA/DraE interactions, implying that the adhesin could be bound simultaneously by both receptors on host tissue. Figure 6 illustrates that the mutation-derived interaction sites for DAF and CEA are mutually exclusive consistent with the inability of DAF to inhibit CEA binding. The NMR-mapped DAF interaction sites near the Cm binding pocket are not contiguous with the cluster of NMR-derived DAF binding amino acids overlapping the mutation-site on the opposite face of the molecule. These observations suggest that Cm inhibition of DAF binding may be allosteric rather than competitive. However, the precise mechanism by which a weakly interacting, small molecule can potently inhibit two independent protein-protein interactions remains to be understood.
Dr adhesin-expressing E. coli penetrate into epithelial cells utilizing a zipper-like mechanism of internalization [43], although neither the mechanism of internalization nor the roles of specific binding activities of Dr adhesins in internalization have been defined in detail. It is likely that interactions of Dr adhesins, and the Dr invasin (afaD-related proteins) contribute to the process of internalization [44,45,46,47]. DAF is concentrated in lipid rafts [43], cell surface invaginations that are believed to be important for signal-transduction [48]. We hypothesize that the role for DAF in Dr-mediated infection is to promote initial binding to the cell membrane. This hypothesis is supported by the observation that all adhesins of Dr family bind DAF [4], and that DraE-family alleles are adapting to the uropathogenic niche by strong positive selection for mutations in the adhesin that enhance binding to DAF (Korotkova et al, manuscript in preparation). Bacterial binding to DAF is followed by sequential recruitment of adjacent GPI-anchored receptors, such as DAF and CEACAMs, and interactions with surface structures such as β1 integrins that participate in internalization and/or signalling events [6,43].
The role of CEACAMs in E.coli infection mediated by Dr adhesins is not clear yet. However, the occurrence of point mutations in Dr adhesins which affect CEACAM binding, under positive selection, and at high frequency, suggests that CEACAMs receptors play an important role in niche adaptation of E. coli. It has been shown that CEACAMs receptors are involved in N.gonorrhoeae internalization by epithelial cells [49]. Moreover, CEACAM5 and CEACAM6 mediate zipper-like internalization of N.gonorrhoeae [49]. We suggest that these receptors might have similar function during E.coli colonization, providing tight association between bacteria and cell membrane which results in the envelopment of the bacterial body by the cell membrane and sequential bacterial uptake through a zipper-like process.
The ability of DraE to interact simultaneously with two receptors is a very intriguing observation. Two distinct receptor binding sites on the adhesin could be the result of two independent evolution processes directed toward preserving both activities. Genetic adaptation to one environment is often associated with loss of fitness in other environments. If two binding sites for two receptors overlap, positive selection associated with the adaptation to the first receptor might lead to the loss of binding to the second receptor. Independent receptor binding sites may be important for Dr-mediated E.coli persistence in the host environment because they would contribute to the maintenance of lipid raft integrity and provide multiple high affinity interactions with cell membrane that could resulting bacterial internalization.
ACKNOWLEDGEMENTS
This work was supported by grant DK-064229 from the National Institute of Diabetes Digestive and Kidney Diseases and The Wellcome Trust. We are grateful to Evgeni Sokurenko, Veronika Chesnokova and Konstantin Korotkov for helpful criticism of the manuscript, Anh-Linh Bui and Diane Capps for technical assistance. We thank Dr. John E. Shively (Beckman Research Institute City of Hope, Duarte, CA) for generously providing the construct for CEA N-A3 expression. We also thank David J. Evans (Institute of Virology, University of Glasgow) for kindly providing the constructs for DAF expression.
The abbreviations used are
- DAF
decay accelerating factor
- UTI
urinary tract infection
- SPR
surface plasmon resonance
- MRHA
mannose resistant hemagglutination assay
- CEA
carcinoembrionic antigen
- CEACAM
(CEA) - related cell adhesion molecule
- CEA N-A3
CEA containing N and A3 domains
- N-CEA
CEA containing N domain
- NMR
nuclear magnetic resonance
- DAEC
diffusely adhering Escherichia coli
- GPI
glycosylphosphatidylinositol
- PBS
phosphate-buffered saline
- Cm
chloramphenicol
REFERENCES
- 1.Garcia MI, Gounon P, Courcoux P, Labigne A, Le Bouguenec C. The afimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol Microbiol. 1996;19:683–693. doi: 10.1046/j.1365-2958.1996.394935.x. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B, Zhang L, Tallman P, Palin K, Rode C, et al. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J Infect Dis. 1995;172:1536–1541. doi: 10.1093/infdis/172.6.1536. [DOI] [PubMed] [Google Scholar]
- 3.Nowicki B, Labigne A, Moseley S, Hull R, Hull S, et al. The Dr hemagglutinin, afimbrial adhesins AFA-I and AFA-III, and F1845 fimbriae of uropathogenic and diarrhea-associated Escherichia coli belong to a family of hemagglutinins with Dr receptor recognition. Infect Immun. 1990;58:279–281. doi: 10.1128/iai.58.1.279-281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Servin AL. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev. 2005;18:264–292. doi: 10.1128/CMR.18.2.264-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowicki B, Moulds J, Hull R, Hull S. A hemagglutinin of uropathogenic Escherichia coli recognizes the Dr blood group antigen. Infect Immun. 1988;56:1057–1060. doi: 10.1128/iai.56.5.1057-1060.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guignot J, Peiffer I, Bernet-Camard MF, Lublin DM, Carnoy C, et al. Recruitment of CD55 and CD66e brush border-associated glycosylphosphatidylinositol-anchored proteins by members of the Afa/Dr diffusely adhering family of Escherichia coli that infect the human polarized intestinal Caco-2/TC7 cells. Infect Immun. 2000;68:3554–3563. doi: 10.1128/iai.68.6.3554-3563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medof ME, Walter EI, Rutgers JL, Knowles DM, Nussenzweig V. Identification of the complement decay-accelerating factor (DAF) on epithelium and glandular cells and in body fluids. J Exp Med. 1987;165:848–864. doi: 10.1084/jem.165.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- 9.Berger CN, Billker O, Meyer TF, Servin AL, Kansau I. Differential recognition of members of the carcinoembryonic antigen family by Afa/Dr adhesins of diffusely adhering Escherichia coli (Afa/Dr DAEC) Mol Microbiol. 2004;52:963–983. doi: 10.1111/j.1365-2958.2004.04033.x. [DOI] [PubMed] [Google Scholar]
- 10.Obrink B. CEA adhesion molecules: multifunctional proteins with signal-regulatory properties. Curr Opin Cell Biol. 1997;9:616–626. doi: 10.1016/S0955-0674(97)80114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prall F, Nollau P, Neumaier M, Haubeck HD, Drzeniek Z, et al. CD66a (BGP), an adhesion molecule of the carcinoembryonic antigen family, is expressed in epithelium, endothelium, and myeloid cells in a wide range of normal human tissues. J Histochem Cytochem. 1996;44:35–41. doi: 10.1177/44.1.8543780. [DOI] [PubMed] [Google Scholar]
- 12.Kodera Y, Isobe K, Yamauchi M, Satta T, Hasegawa T, et al. Expression of carcinoembryonic antigen (CEA) and nonspecific crossreacting antigen (NCA) in gastrointestinal cancer; the correlation with degree of differentiation. Br J Cancer. 1993;68:130–136. doi: 10.1038/bjc.1993.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metze D, Bhardwaj R, Amann U, Eades-Perner AM, Neumaier M, et al. Glycoproteins of the carcinoembryonic antigen (CEA) family are expressed in sweat and sebaceous glands of human fetal and adult skin. J Invest Dermatol. 1996;106:64–69. doi: 10.1111/1523-1747.ep12327258. [DOI] [PubMed] [Google Scholar]
- 14.Leusch HG, Drzeniek Z, Markos-Pusztai Z, Wagener C. Binding of Escherichia coli and Salmonella strains to members of the carcinoembryonic antigen family: differential binding inhibition by aromatic alpha-glycosides of mannose. Infect Immun. 1991;59:2051–2057. doi: 10.1128/iai.59.6.2051-2057.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauter SL, Rutherfurd SM, Wagener C, Shively JE, Hefta SA. Binding of nonspecific cross-reacting antigen, a granulocyte membrane glycoprotein, to Escherichia coli expressing type 1 fimbriae. Infect Immun. 1991;59:2485–2493. doi: 10.1128/iai.59.7.2485-2493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauter SL, Rutherfurd SM, Wagener C, Shively JE, Hefta SA. Identification of the specific oligosaccharide sites recognized by type 1 fimbriae from Escherichia coli on nonspecific cross-reacting antigen, a CD66 cluster granulocyte glycoprotein. J Biol Chem. 1993;268:15510–15516. [PubMed] [Google Scholar]
- 17.Virji M, Watt SM, Barker S, Makepeace K, Doyonnas R. The N-domain of the human CD66a adhesion molecule is a target for Opa proteins of Neisseria meningitidis and Neisseria gonorrhoeae. Mol Microbiol. 1996;22:929–939. doi: 10.1046/j.1365-2958.1996.01548.x. [DOI] [PubMed] [Google Scholar]
- 18.Virji M, Evans D, Hadfield A, Grunert F, Teixeira AM, et al. Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N-domain of CD66 molecules. Mol Microbiol. 1999;34:538–551. doi: 10.1046/j.1365-2958.1999.01620.x. [DOI] [PubMed] [Google Scholar]
- 19.Virji M, Evans D, Griffith J, Hill D, Serino L, et al. Carcinoembryonic antigens are targeted by diverse strains of typable and non-typable Haemophilus influenzae. Mol Microbiol. 2000;36:784–795. doi: 10.1046/j.1365-2958.2000.01885.x. [DOI] [PubMed] [Google Scholar]
- 20.Bos MP, Kuroki M, Krop-Watorek A, Hogan D, Belland RJ. CD66 receptor specificity exhibited by neisserial Opa variants is controlled by protein determinants in CD66 N-domains. Proc Natl Acad Sci U S A. 1998;95:9584–9589. doi: 10.1073/pnas.95.16.9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toleman M, Aho E, Virji M. Expression of pathogen-like Opa adhesins in commensal Neisseria: genetic and functional analysis. Cell Microbiol. 2001;3:33–44. doi: 10.1046/j.1462-5822.2001.00089.x. [DOI] [PubMed] [Google Scholar]
- 22.Hill DJ, Virji M. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol Microbiol. 2003;48:117–129. doi: 10.1046/j.1365-2958.2003.03433.x. [DOI] [PubMed] [Google Scholar]
- 23.Hill DJ, Toleman MA, Evans DJ, Villullas S, Van Alphen L, et al. The variable P5 proteins of typeable and non-typeable Haemophilus influenzae target human CEACAM1. Mol Microbiol. 2001;39:850–862. doi: 10.1046/j.1365-2958.2001.02233.x. [DOI] [PubMed] [Google Scholar]
- 24.Gray-Owen SD. Neisserial Opa proteins: impact on colonization, dissemination and immunity. Scand J Infect Dis. 2003;35:614–618. doi: 10.1080/00365540310016042. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1982. p. x.p. 545. [Google Scholar]
- 26.You YH, Hefta LJ, Yazaki PJ, Wu AM, Shively JE. Expression, purification, and characterization of a two domain carcinoembryonic antigen minigene (N-A3) in pichia pastoris. The essential role of the N-domain. Anticancer Res. 1998;18:3193–3201. [PubMed] [Google Scholar]
- 27.Hellwig S, Robin F, Drossard J, Raven NP, Vaquero-Martin C, et al. Production of carcinoembryonic antigen (CEA) N-A3 domain in Pichia pastoris by fermentation. Biotechnol Appl Biochem. 1999;30(Pt 3):267–275. [PubMed] [Google Scholar]
- 28.Van Loy CP, Sokurenko EV, Samudrala R, Moseley SL. Identification of amino acids in the Dr adhesin required for binding to decay-accelerating factor. Mol Microbiol. 2002;45:439–452. doi: 10.1046/j.1365-2958.2002.03022.x. [DOI] [PubMed] [Google Scholar]
- 29.Powell RM, Ward T, Evans DJ, Almond JW. Interaction between echovirus 7 and its receptor, decay-accelerating factor (CD55): evidence for a secondary cellular factor in A-particle formation. J Virol. 1997;71:9306–9312. doi: 10.1128/jvi.71.12.9306-9312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Anderson KL, Billington J, Pettigrew D, Cota E, Simpson P, et al. An atomic resolution model for assembly, architecture, and function of the Dr adhesins. Mol Cell. 2004;15:647–657. doi: 10.1016/j.molcel.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Carnoy C, Moseley SL. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol Microbiol. 1997;23:365–379. doi: 10.1046/j.1365-2958.1997.2231590.x. [DOI] [PubMed] [Google Scholar]
- 33.Pettigrew D, Anderson KL, Billington J, Cota E, Simpson P, et al. High resolution studies of the Afa/Dr adhesin DraE and its interaction with chloramphenicol. J Biol Chem. 2004;279:46851–46857. doi: 10.1074/jbc.M409284200. [DOI] [PubMed] [Google Scholar]
- 34.Tan K, Zelus BD, Meijers R, Liu JH, Bergelson JM, et al. Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. Embo J. 2002;21:2076–2086. doi: 10.1093/emboj/21.9.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boehm MK, Mayans MO, Thornton JD, Begent RH, Keep PA, et al. Extended glycoprotein structure of the seven domains in human carcinoembryonic antigen by X-ray and neutron solution scattering and an automated curve fitting procedure: implications for cellular adhesion. J Mol Biol. 1996;259:718–736. doi: 10.1006/jmbi.1996.0353. [DOI] [PubMed] [Google Scholar]
- 36.Bos MP, Hogan D, Belland RJ. Homologue scanning mutagenesis reveals CD66 receptor residues required for neisserial Opa protein binding. J Exp Med. 1999;190:331–340. doi: 10.1084/jem.190.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hefta LJ, Chen FS, Ronk M, Sauter SL, Sarin V, et al. Expression of carcinoembryonic antigen and its predicted immunoglobulin-like domains in HeLa cells for epitope analysis. Cancer Res. 1992;52:5647–5655. [PubMed] [Google Scholar]
- 38.Hunter I, Sawa H, Edlund M, Obrink B. Evidence for regulated dimerization of cell-cell adhesion molecule (C-CAM) in epithelial cells. Biochem J. 1996;320(Pt 3):847–853. doi: 10.1042/bj3200847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krop-Watorek A, Oikawa S, Oyama Y, Nakazato H. Oligomerization of N-terminal domain of carcinoembryonic antigen (CEA) expressed in Escherichia coli. Biochem Biophys Res Commun. 1998;242:79–83. doi: 10.1006/bbrc.1997.7920. [DOI] [PubMed] [Google Scholar]
- 40.Taheri M, Saragovi U, Fuks A, Makkerh J, Mort J, et al. Self recognition in the Ig superfamily. Identification of precise subdomains in carcinoembryonic antigen required for intercellular adhesion. J Biol Chem. 2000;275:26935–26943. doi: 10.1074/jbc.M909242199. [DOI] [PubMed] [Google Scholar]
- 41.Markel G, Gruda R, Achdout H, Katz G, Nechama M, et al. The critical role of residues 43R and 44Q of carcinoembryonic antigen cell adhesion molecules-1 in the protection from killing by human NK cells. J Immunol. 2004;173:3732–3739. doi: 10.4049/jimmunol.173.6.3732. [DOI] [PubMed] [Google Scholar]
- 42.Watt SM, Teixeira AM, Zhou GQ, Doyonnas R, Zhang Y, et al. Homophilic adhesion of human CEACAM1 involves N-terminal domain interactions: structural analysis of the binding site. Blood. 2001;98:1469–1479. doi: 10.1182/blood.v98.5.1469. [DOI] [PubMed] [Google Scholar]
- 43.Kansau I, Berger C, Hospital M, Amsellem R, Nicolas V, et al. Zipper-like internalization of Dr-positive Escherichia coli by epithelial cells is preceded by an adhesin-induced mobilization of raft-associated molecules in the initial step of adhesion. Infect Immun. 2004;72:3733–3742. doi: 10.1128/IAI.72.7.3733-3742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das M, Hart-Van Tassell A, Urvil PT, Lea S, Pettigrew D, et al. Hydrophilic domain II of Escherichia coli Dr fimbriae facilitates cell invasion. Infect Immun. 2005;73:6119–6126. doi: 10.1128/IAI.73.9.6119-6126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jouve M, Garcia MI, Courcoux P, Labigne A, Gounon P, et al. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3 gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect Immun. 1997;65:4082–4089. doi: 10.1128/iai.65.10.4082-4089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plancon L, Du Merle L, Le Friec S, Gounon P, Jouve M, et al. Recognition of the cellular beta1-chain integrin by the bacterial AfaD invasin is implicated in the internalization of afa-expressing pathogenic Escherichia coli strains. Cell Microbiol. 2003;5:681–693. doi: 10.1046/j.1462-5822.2003.00308.x. [DOI] [PubMed] [Google Scholar]
- 47.Selvarangan R, Goluszko P, Popov V, Singhal J, Pham T, et al. Role of decay-accelerating factor domains and anchorage in internalization of Dr-fimbriated Escherichia coli. Infect Immun. 2000;68:1391–1399. doi: 10.1128/iai.68.3.1391-1399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 49.McCaw SE, Liao EH, Gray-Owen SD. Engulfment of Neisseria gonorrhoeae: revealing distinct processes of bacterial entry by individual carcinoembryonic antigen-related cellular adhesion molecule family receptors. Infect Immun. 2004;72:2742–2752. doi: 10.1128/IAI.72.5.2742-2752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]