Abstract
Carbohydrate-protein conjugates are utilized extensively in basic research and as immunogens in a variety of bacterial vaccines and cancer vaccines. As a result, there have been significant efforts to develop simple and reliable methods for the construction of these conjugates. While direct coupling via reductive amination is an appealing approach, the reaction is typically very inefficient. In this paper, we report improved reaction conditions providing an approximately 500% increase in yield. In addition to optimizing a series of standard reaction parameters, we found that addition of 500 mM sodium sulfate improves the coupling efficiency. To illustrate the utility of these conditions, a series of high mannose BSA conjugates were produced and incorporated into a carbohydrate microarray. Ligand binding to ConA could be observed and apparent affinity constants (Kds) measured using the array were in good agreement with values reported by surface plasmon resonance. The results show that the conditions are suitable for microgram scale reactions, are compatible with complex carbohydrates, and produce biologically active conjugates.
INTRODUCTION
Carbohydrate-protein conjugates play a key role in basic and applied research [for some recent reviews, see (1-5)]. Several polysaccharide-protein conjugate vaccines such as Prevnar, Menactra, and HIBTiter are FDA approved and used routinely for the prevention of invasive bacterial infections. Many others are in clinical trials for the prevention of infections and for treatment of cancer (1, 6). In basic research, carbohydrate-BSA (bovine serum albumin) conjugates have been used for years to study molecular recognition of carbohydrates (7, 8). As a result, there has been significant effort to develop simple and efficient methods to covalently couple carbohydrate molecules to proteins (7, 9); however, existing methods are not ideal and pursuit of improved methods remains an active area of research.
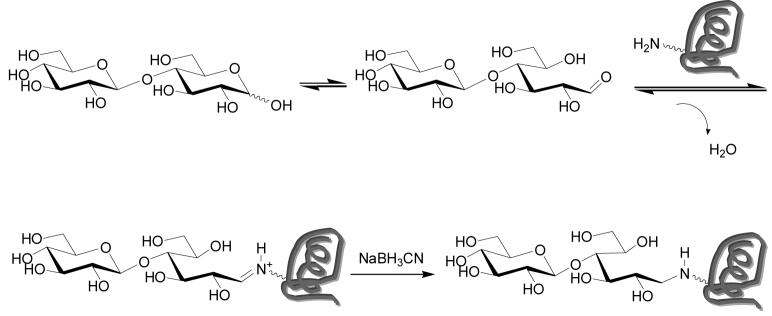
Carbohydrate-protein conjugates are frequently made via multi-step procedures involving modification of the carbohydrate to introduce reactive functionality (e.g. addition of a linker, oxidation of hydroxyls) followed by coupling to the protein. One of the most appealing strategies is direct coupling of oligosaccharides to proteins in a single step. Glycans containing a free reducing end can be covalently attached to protein amino- groups via reductive amination (10). The reducing end of a sugar exists as an equilibrium mixture composed of the cyclic hemiacetal form (lactol) and the open chain aldehyde form (see Scheme 1). Under suitable conditions, amine groups will condense with the aldehyde to form an iminium ion which can be reduced to an amine. The process results in ring opening of the reducing end monosaccharide. When coupling to proteins, this reaction is carried out in aqueous buffer.
Scheme 1.
Reductive Amination of Proteins
Several factors make reductive amination of sugars to proteins challenging. First, the reducing agent must be stable in water, unreactive with aldehydes, and yet reactive with iminium ions. Sodium cyanoborohydride meets these requirements and is by far the most common reducing reagent used for reductive amination of aldehydes to proteins. Second, the equilibrium between aldehyde and lactol form of sugars highly favors the lactol; therefore, the concentration of free aldehyde is low. Third, the high concentration of water in aqueous buffer sets up an equilibrium that disfavors release of a water molecule, a process that is required to form the iminium ion. Due to the second and third features, reductive amination of carbohydrates directly to proteins is slow and inefficient. As a result, one typically uses large excesses of sugar (∼300 equivalents), high concentrations (∼60 mM), and long reaction times. Therefore, direct reductive amination has been limited to sugars that are readily available in large quantities.
Our group has been using carbohydrate-protein conjugates and glycoproteins for the construction of carbohydrate microarrays (11-14). Carbohydrate microarrays contain diverse sets of glycans immobilized on a solid support in an spatially defined arrangement (15-21). Using a carbohydrate microarray, one can evaluate binding of a protein, cell, or virus to all of the carbohydrate ligands simultaneously. While reductive amination is an attractive approach for obtaining glycoconjugates, many biologically interesting carbohydrates such as N-linked sugars are very difficult to obtain. Microgram amounts of these sugars (20-200 μg) can be very expensive or require considerable time and effort to isolate and purify from natural sources; therefore, one cannot afford to use 300 equivalents and waste >95% of the material in the coupling process. Moreover, the volume of buffer required to achieve high concentrations can be completely impractical when working with micrograms of material. For example, to obtain a 60 mM solution starting with 100 μg of Man9, one would require a final reaction volume of only 1 μL. Clearly, methods to improve the efficiency and facilitate coupling on small scale were needed. Improved conditions would also be beneficial for the development and production of carbohydrate-protein conjugate vaccines.
MATERIALS AND METHODS
General
Unless indicated otherwise, all chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MI) and used without further purification. LNT, LSTa, and Man8D1D3 were purchased from Glycotech (Gaithersburg, MD). Tetra-N-acetylchitotetraose, Man3, Man7D1, and Man7D3 were purchased from Dextra labs (Reading, UK). Man6 was purchased from IsoSep (Tullinge, Sweden). Man5 and Man9 were generous gifts from Dr. Lai-Xi Wang.
General procedure for reductive amination
Bovine serum albumin (BSA; 2 μL of a 150 mg/mL solution; fraction V), sodium borate (5.5 μL of a 400 mM solution, pH 8.5), sodium sulfate (3.7 μL of a 3 M solution, 50°C), oligosaccharide (3.3 μL of a 20 mM solution for 15eq), H2O (5.3 μL), and sodium cyanoborohydride (2.2 μL of a 3 M solution) were combined in a 200 μL PCR tube. The tube was capped and then incubated in a PCR thermal cycler at 56°C for 96 h with a heated lid. The reaction was diluted with H2O (100 μL), transferred to a 500 μL dialysis tube (MWCO 10,000, Slide-A-Lyzer MINI Dialysis Unit, Pierce, Rockford, IL) and then dialyzed against H2O (2.5 L) three times. Conjugates were diluted with H2O to a final concentration of 1 mg/mL and then analyzed by matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) to determine the extent of conjugation.
Mass Spectrometric analysis of glycoconjugates
MALDI-TOF mass spectrometric analysis of the glycoconjugates was performed on an Applied Biosystems Voyager-DE Pro time-of-flight mass spectrometer. The accelerating voltage was 25kV, guide wire 0.15% and grid voltage 91.5%. The instrument was operated in linear mode under positive ion conditions. A nitrogen laser was used at 337 nm with 250 laser shots averaged per spectrum. Sinapinic acid (3,5-dimethoxy-4-hydroxycinnamic acid, Aldrich Chemicals, St. Louis, MO) was used as the matrix for all experiments. Matrix was prepared as a saturated solution in 50% acetonitrile / 0.1% trifluoroacetic acid (v/v). Sample preparation was a modified “dried droplet” procedure (22), whereby 0.3 μL of sample was spotted onto the MALDI target followed by 0.3 μL of matrix. The mixture was then allowed to air dry prior to analysis. External calibration was performed using bovine serum albumin as a standard. Data analysis was carried out using “Data Explorer” software resident on the Voyager mass spectrometer. Representative spectra can be found in the supplementary material.
Evaluation of neoglycoconjugate activity by enzyme-linked lectin assay (ELLA)
BSA conjugates (50 μL of a 10 μg/mL solution in PBS) were added to wells of a 96 well plate (white Maxisorp, Nunc, Rochester, NY) and incubated for 2 h at room temperature. Liquid was removed and the wells were blocked with BSA (200 μL/well of a 3% solution in PBS). After removal of the liquid, the wells were incubated with various concentrations of biotinylated wheat germ agglutinin (WGA; Vector labs, Burlingame, CA) diluted in 3% BSA/PBS solution. After 2 h, wells were emptied and washed 3 times with PBS. Next, wells were incubated with streptavidin-alkaline phosphatase (2 μg/mL in 3% BSA/PBS) for 1h and then washed 7 times with PBS. Wells were then incubated with methyl umbelliferryl phosphate (50 μL of a 100 μM solution in Tris buffer at pH 9.0). After 15 minutes of incubation, the fluorescence intensities for each well were measured using an FLX800 plate reader.
Carbohydrate microarray fabrication
Carbohydrate arrays were printed with Stealth Micro Spotting Pins (SMP3; TeleChem, Sunnyvale, CA) using a robotic array spotter (Genomic Solutions, BioRobotics MicroGrid II 600/610). Conjugates and glycoproteins were distributed into 384-well plates at 4 wells per sample and 20 μL per well. Each component was prepared at 125 μg/mL in print buffer (1X PBS: 140 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), 2.5% glycerol, 0.006% Triton-X 100) onto ArrayIt® SuperEpoxy 2 Protein glass slides. The pins were dipped into the sample wells and blotted 10 times before printing. The humidity level in the arraying chamber was maintained at about 65% during printing. Each of the components was printed in duplicate in a grid of 110 μm diameter spots with 220 μm pitch. Sixteen complete arrays were printed on each slide. Printed slides were allowed to dry, protected from light, for about 14 hours at room temperature then stored at -20°C until use.
Carbohydrate microarray binding assay
The binding experiment was carried out in triplicate. Slides were assembled on 16-well slide holders (ProPlate, Grace Bio-Labs) and blocked with 3% BSA/PBS overnight at 4°C, then washed 6 × 200 μL PBST0.05 (PBS with 0.05% Tween 20). A dilution series of biotinylated Concanavalin A (ConA) was prepared in 25 mM Tris, 124 mM NaCl, pH 7.6 at concentrations ranging from 0.03 nM to 48 μM. Lectin solutions were added to arrays, covered tightly with a seal strip, and allowed to incubate for 1 h at room temperature. After washing unbound lectin with 3 × 200 μL PBST0.05, detection of bound lectin was carried out by incubating with Cy3-streptavidin in 3% BSA/PBS (5 μg/mL) for 30 mins at room temperature. Slides were then washed 7 × 200 μL PBST0.05, removed from holders, immersed in wash buffer for 5 min then centrifuged at 453g for 5 min.
Image processing and calculation of apparent dissociation constant
Slides were scanned with a Genepix 4000B microarray scanner (Molecular Devices Corporation, Union City, CA) at PMT voltage settings where no saturated pixels were obtained. Image analysis was carried out with Genepix Pro 6.0 analysis software (Molecular Devices Corporation, Union City, CA). Spots were defined as circular features with maximum diameter of 100 μm. Local background subtraction was performed and features were allowed to be resized to 70 μm as needed. The background-corrected median feature intensity, F532median-B532, was used for initial data processing, which was performed with Microsoft Excel. The mean for replicate spots for each array component was calculated, and then the mean and standard deviations for the 3 replicate experiments were obtained and used for subsequent Kd determination.
Apparent dissociation constants were determined using the scientific graphing and analysis software Origin (OriginLab). For each glycoconjugate, the mean intensity of 3 replicate experiments was plotted as a function of ConA concentration on a logarithmic scale. Nonlinear line-fitting to the equation
was performed, where Fobs is the mean intensity of the replicate spots for ConA binding at a particular concentration, Fmax is the maximum fluorescence intensity, Kd is the apparent dissociation constant for interaction between lectin and immobilized glycoconjugate, and [L] is the concentration of ConA. For each glycoconjugate, the reported values are the apparent Kd of the mean ± the standard deviation of 3 replicate experiments
RESULTS AND DISCUSSION
The protocol most frequently used for direct reductive amination of carbohydrates was developed by Roy et al (23). Typical reaction conditions involve coupling protein (200 μM) with sugar (60 mM) in the presence of NaBH3CN (300 mM) and aqueous sodium borate buffer (200 mM, pH 9.0) for 10-24 h at 37-50 °C. Borate buffer appears to significantly enhance the yield of coupling when compared to other buffers, although the exact role of borate is not well understood.
We began by evaluating reactions of several sugars with bovine serum albumin (BSA) under the Roy conditions but with only 15-30 equivalents of sugar (3-6 mM). In addition, the reaction volume was limited to 20-50 μL to assess couplings on small scale. The extent of conjugation was evaluated by MALDI-TOF MS. Initially, reactions were carried out in eppendorf tubes heated in either a water bath or on a heat block. Due to the small volumes, evaporation with condensation on the lid became a serious problem resulting in highly variable results. To avoid this problem, all subsequent reactions were carried out in PCR tubes and heated in a PCR thermal cycler with a heated lid. Under these conditions, results were much more reproducible and evaporation was not observed.
After surveying a variety of oligosaccharides, the yield was found to be highly dependent on the identity of the reducing end sugar. Cummings group has also observed substrate dependent coupling efficiency (24). Saccharides with a reducing end glucose residue such as maltose, LNT, and LSTa (see Figure 1) coupled efficiently. For example, reactions utilizing 30 equivalents of sugar produced conjugates with an average of 10 oligosaccharide chains per molecule of BSA (after 24 h at 50 °C). In contrast, sugars with mannose (Man), N-acetylglucosamine (GlcNAc), or galactose (Gal) at the reducing end coupled much less efficiently yielding conjugates with an average of 0-1 oligosaccharide chain per molecule of BSA. These results were disheartening since many interesting N-linked oligosaccharides have a GlcNAc at the reducing end (see Man3-9, Figure 1); therefore, efforts to improve efficiency were investigated.
Figure 1.
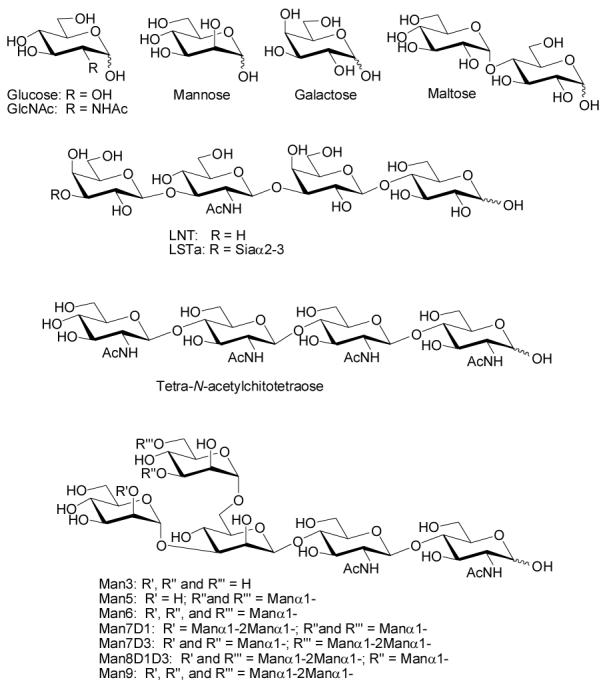
Representative Oligosaccharide Structures
To increase the yield, a number of reaction parameters such as the temperature, time, and pH were systematically varied. While conceptually straightforward, it was not known if changes such as higher temperatures and longer reaction times would compromise the biological activity of the resulting conjugates. Tetra-N-acetylchitotetraose, a sugar containing GlcNAc at the reducing end (see Figure 1), was used as a model oligosaccharide in couplings to BSA. Differences in pH ranging from 8.5 to 10.0 had no effect on the ratio (data not shown), but changes to the time and temperature led to improvements. Increasing the reaction time from 24 h to 96 h improved the ratio about 2 fold (see Table 1). Raising the temperature from 50 °C to 56 °C led to a 1.5 fold increase. Further increases in temperature resulted in precipitate formation, presumably due to denaturation of the protein. Changes to the concentration of NaBH3CN from 50-500 mM had no effect.
Table 1.
Efficiency of Reductive Amination Using Various Reaction Conditions.a
| # | Eq. | Temp (°C) | Salt | Time (h) | Ratiob |
|---|---|---|---|---|---|
| 1 | 15 | 50 | none | 24 | 1.0 |
| 2 | 15 | 50 | none | 96 | 2.2 |
| 3 | 15 | 56 | none | 96 | 3.0 |
| 4 | 15 | 56 | 0.5 M NaCl | 96 | 3.3 |
| 5 | 15 | 56 | 1 M NaCl | 96 | 4.1 |
| 6 | 15 | 56 | 2 M NaCl | 96 | 3.8 |
| 7 | 15 | 56 | 1M KCl | 96 | 3.3 |
| 8 | 15 | 56 | 1M NaBr | 96 | 2.6 |
| 9 | 15 | 56 | 1M MgCl2 | 96 | ppt |
| 10 | 15 | 56 | 1M LiCl | 96 | ppt |
| 11 | 15 | 56 | 1 M NaOAc | 96 | 3.8 |
| 12 | 15 | 56 | 0.5 M Na3Citrate | 96 | 3.8 |
| 13 | 15 | 56 | 0.5 M MgSO4 | 96 | 4.5 |
| 14 | 15 | 56 | 1M MgSO4 | 96 | 4.5 |
| 15 | 15 | 56 | 0.5 M Na2SO4 | 96 | 5.1 |
| 16 | 15 | 56 | 1M Na2SO4 | 96 | ppt |
| 17 | 15 | 60 | 0.5 M Na2SO4 | 96 | ppt |
| 18 | 30 | 56 | 0.5 M Na2SO4 | 96 | 8.2 |
| 19 | 30 | 37 | No salt | 96 | 1.0 |
| 20 | 30 | 37 | 0.5 M Na2SO4 | 96 | 2.7 |
| 21 | 300 | 50 | No salt; Roy conditionsc | 24 | 9.2 |
Tetra-N-acetylchitotetraose (3-6 mM) was coupled with BSA (200 μM) in 100 mM sodium borate buffer (pH 8.5).
The number of sugars attached to BSA after reductive amination as determined by MALDI-MS. ppt = protein precipitated and the ratio could not be determined.
See reference 23.
Formation of the key imine intermediate requires expulsion of a molecule of water. In an aqueous solution, this equilibrium is highly disfavored. It was anticipated that high salt concentrations might make the expulsion of water somewhat less unfavorable, facilitating formation of the imine. In addition, high salt concentrations can have a substantial effect on protein structure and solubility. For example, high salt concentrations are reported to reduce the rate of aggregation of BSA (25); therefore, a series of salt additives was examined next.
As a first test case, NaCl was added at concentrations of 0.5 M, 1.0 M, and 2.0 M. A measurable increase in conjugation ratio was observed, with 1.0 M giving the best result. A number of other salt additives were investigated (see Table 1) and many improved the yield of the reaction. Overall, addition of 0.5 M Na2SO4 produced the best results. A 1.0 M Na2SO4 solution occasionally produced even higher ratios but frequently resulted in precipitate formation and unrecoverable protein. The added salt also improved the conjugation efficiency at lower temperatures; at 37 °C, added salt produced a nearly 3 fold increase in conjugation efficiency relative to the absence of salt (compare entries 19 and 20, Table 1). This result indicates that addition of salt may be especially useful when coupling sugars to proteins that are sensitive to high temperatures.
Overall, the combination of higher temperatures, longer reaction times, and added salt produced about a 5 fold increase in the amount of sugar conjugated to BSA and an overall yield of about 33% with 15 equivalents of sugar. Higher ratios could be obtained with 30 equivalents of sugar, albeit with slightly lower yield (based on sugar). This level of incorporation and efficiency was judged to be suitable for making a variety of BSA conjugates for the array.
To determine whether the altered reaction conditions affect the biological activity or binding capacity of the final conjugate, binding of biotinylated wheat germ agglutinin (WGA) was measured over a range of concentrations using an ELLA assay. Binding could be detected down to concentrations of 10 ng/mL (277 pM). For comparison, a conjugate with a similar ratio of tetra-N-acetylchitotetraoses to BSA (9.2/BSA) was prepared using the Roy conditions (entry 21, Table 1). Both conjugates displayed similar binding by ELLA (see Figure 2) indicating that the modified reaction conditions do not reduce the biological activity of the conjugate.
Figure 2.
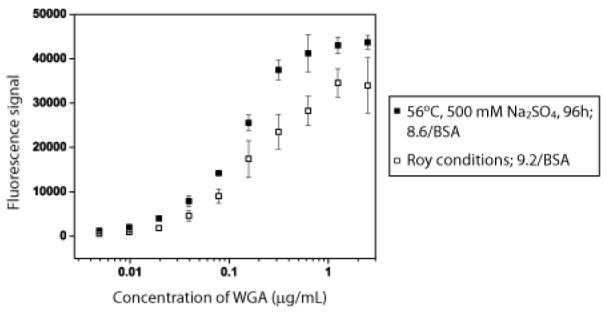
Comparison of Wheat Germ Agglutinin Binding to N-Acetylchitotriose-BSA prepared using the Roy conditions (open squares) vs 56 °C for 96 h with 500 mM Na2SO4 (closed squares).
Next, the modified reaction conditions were applied to a series of biologically interesting high mannose oligosaccharides. 30 equivalents (200-300 μg, 100-200 nmol) of Man3, Man5, Man6, Man7D1, Man7D3, Man8D1D3, and Man9 were coupled to BSA (250-400 μg, 3.5-6 nmol) in 20-25 μL reactions containing 100 mM sodium borate at pH 8.5, 300 mM sodium cyanoborohydride, and 500 mM sodium sulfate. Reactions were incubated in the PCR thermal cycler at 56 °C for 96 h and then dialyzed extensively with water. MALDI-TOF MS analysis (see Supplementary Material) showed that the resulting glycoconjugates contained an average of 7 oligosaccharide chains per molecule of BSA. The results verify that the improved conditions for reductive amination can be successfully applied to a variety of expensive, complex oligosaccharides on a microgram scale. Using our current printing conditions for the carbohydrate microarrays, 250 μg of BSA conjugate is sufficient for over 100,000 array experiments.
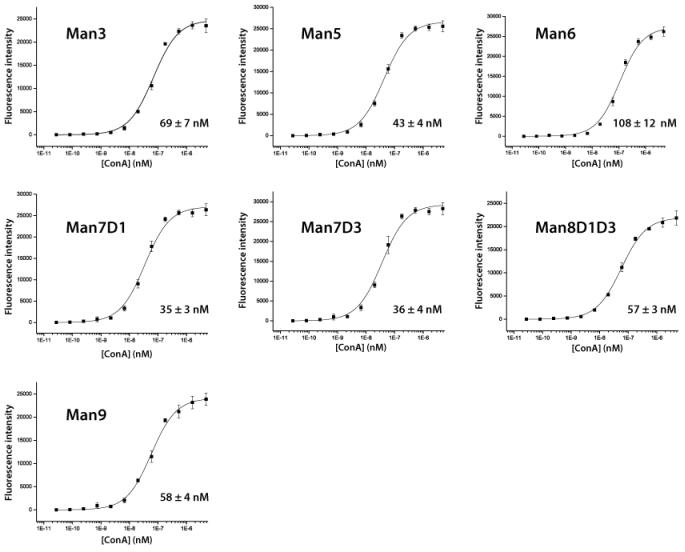
To verify that the high mannose sugars were structurally intact and suitable for use in a carbohydrate microarray, binding to concanavalin A was examined. Con A is a mannose binding lectin that has been used extensively as a research tool for detection of mannose containing glycans. It has been studied at length and is frequently used as a test lectin to validate assays. Mannose conjugates were printed on epoxide coated glass microscope slides as reported previously (11-13). Binding was assessed by incubating the array with a range of concentrations of biotinylated ConA, washing away excess lectin, and incubating the array with Cy3-labeled streptavidin. After washing, fluorescent spots were detected and intensities quantitated using a DNA microarray scanner with GenePix software. Apparent Kds were determined on the array using the method of MacBeath (26). Kd values are shown in Figure 3 and are in good agreement with values previously obtained for ConA with related sugars measured by surface plasmon resonance (27), frontal affinity chromatography (28), and carbohydrate microarray (29). Thus, the neoglycoconjugates appear to retain appropriate bioactivity.
Figure 3. Binding of ConA to High Mannose Oligosaccharides on the Carbohydrate Microarray.
Biotinylated ConA was incubated on the carbohydrate microarray at various concentrations. After washing, the array was incubated with Cy3-labeled streptavidin and fluorescence intensities were measured using a Genepix microarray scanner. The apparent Kds for each high mannose sugar are listed in the bottom right corner of each panel.
CONCLUSIONS
In summary, improved conditions for the direct conjugation of oligosaccharides to proteins using reductive amination are described. The conditions are compatible with microgram quantities of sugar and provide biologically active conjugates suitable for use in ELISA/ELLA assays and incorporation into microarrays. The conditions were successfully applied to a series of mannose containing oligosaccharides and these neoglycoconjugates were shown to bind to ConA. Improved conditions for reductive amination should prove useful for the preparation of glycoconjugates for both basic and applied research.
Supplementary Material
ACKNOWLEDGMENT
We thank Lai-Xi Wang for the generous gift of high mannose sugars. This research was supported by the Intramural Research Program of the NIH, NCI. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- (1).Vliegenthart JFG. Carbohydrate based vaccines. FEBS Letters. 2006;580:2945. doi: 10.1016/j.febslet.2006.03.053. [DOI] [PubMed] [Google Scholar]
- (2).Seeberger PH, Werz DB. Synthesis and medical applications of oligosaccharides. Nature. 2007;446:1046. doi: 10.1038/nature05819. [DOI] [PubMed] [Google Scholar]
- (3).Oppenheimer SB, Alvarez M, Nnoli J. Carbohydrate-based experimental therapeutics for cancer, HIV/AIDS and other diseases. Acta Histochemica. 2008;110:6. doi: 10.1016/j.acthis.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cipolla L, Peri F, Airoldi C. Glycoconjugates in cancer therapy. Anti-cancer agents in medicinal chemistry. 2008;8:92. doi: 10.2174/187152008783330815. [DOI] [PubMed] [Google Scholar]
- (5).Pozsgay V, Coxon B, Glaudemans CPJ, Schneerson R, Robbins JB. Towards an oligosaccharide-based glycoconjugate vaccine against Shigella dysenteriae type 1. Synlett. 2003:743. [Google Scholar]
- (6).Freire T, Bay S, Vichier-Guerre S, Lo-Man R, Leclerc C. Carbohydrate antigens: Synthesis aspects and immunological applications in cancer. Mini Rev. Med. Chem. 2006;6:1357. doi: 10.2174/138955706778992996. [DOI] [PubMed] [Google Scholar]
- (7).Bovin NV. Neoglycoconjugates: Trade and art. Biochemical Society Symposium. 2002;69:143. doi: 10.1042/bss0690143. [DOI] [PubMed] [Google Scholar]
- (8).Wong SYC. Neoglycoconjugates and their applications in glycobiology. Curr. Opin. Struct. Biol. 1995;5:599. doi: 10.1016/0959-440x(95)80050-6. [DOI] [PubMed] [Google Scholar]
- (9).Brinkley M. A brief survey of methods for preparing protein conjugates with dyes, haptens, and cross-linking reagents. Bioconjugate Chem. 1992;3:2. doi: 10.1021/bc00013a001. [DOI] [PubMed] [Google Scholar]
- (10).Gray GR. The direct coupling of oligosaccharides to proteins and derivatized gels. Arch. Biochem. Biophys. 1974;163:426. doi: 10.1016/0003-9861(74)90495-0. [DOI] [PubMed] [Google Scholar]
- (11).Manimala J, Li Z, Jain A, VedBrat S, Gildersleeve JC. Carbohydrate Array Analysis of Anti-Tn Antibodies and Lectins Reveals Unexpected Specificities: Implications for Diagnostic and Vaccine Development. ChemBioChem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- (12).Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-Throughput Carbohydrate Microarray Analysis of 24 Lectins. Angew. Chem. Int. Ed. Engl. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- (13).Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- (14).Hsu K-L, Gildersleeve JC, L.K. M.A simple strategy for the creation of a recombinant lectin microarray Mol. BioSyst 2008, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).de Paz JL, Seeberger PH. Recent advances in carbohydrate microarrays. QSAR Comb. Sci. 2006;25:1027–1032. [Google Scholar]
- (16).Dyukova VI, Shilova NV, Galanina OE, Rubina AY, Bovin N. Design of carbohydrate multiarrays. Biochem. Biophys. Acta. 2006;1760:603–609. doi: 10.1016/j.bbagen.2005.12.005. [DOI] [PubMed] [Google Scholar]
- (17).Feizi T, Fazio F, Chai W, Wong C-H. Carbohydrate Microarrays- A New Set of Technologies at the Frontiers of Glycomics. Curr. Opin. Struct. Biol. 2003;13:637–45. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- (18).Oyelaran O, Gildersleeve JC. Application of carbohydrate array technology to antigen discovery and vaccine development. Expert Rev. Vaccines. 2007;6:957. doi: 10.1586/14760584.6.6.957. [DOI] [PubMed] [Google Scholar]
- (19).Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat. Chem. Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- (20).Shin I, Park S, Lee MR. Carbohydrate microarrays: An advanced technology for functional studies of glycans. Chem. Eur. J. 2005;11:2894–2901. doi: 10.1002/chem.200401030. [DOI] [PubMed] [Google Scholar]
- (21).Liang P-H, Wu C-Y, Greenberg WA, Wong C-H. Glycan arrays: biological and medical applications. Curr. Opin. Chem. Biol. 2008;12:86. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- (23).Roy R, Katzenellenbogen E, Jennings HJ. Improved Procedures for the Conjugation of Oligosaccharides to Protein by Reductive Amination. Can. J. Chem. 1984;62:270–275. doi: 10.1139/o84-037. [DOI] [PubMed] [Google Scholar]
- (24).Stowell SR, Dias-Baruffi M, Penttila L, Renkonen O, Nyame AK, Cummings RD. Human galectin-1 recognition of poly-N-acetyllactosamine and chimeric polysaccharides. Glycobiology. 2004;14:157. doi: 10.1093/glycob/cwh018. [DOI] [PubMed] [Google Scholar]
- (25).Bagger HL, Ogendal LH, Westh P. Solute effects on the irreversible aggregation of serum albumin. Biophys. Chem. 2007;130:17. doi: 10.1016/j.bpc.2007.06.014. [DOI] [PubMed] [Google Scholar]
- (26).Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- (27).Smith EA, Thomas WD, Kiessling LL, Corn RM. Surface Plasmon Resonance Imaging Studies of Protein-Carbohydrate Interactions. J. Am. Chem. Soc. 2003;125:6140–6148. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- (28).Ohyama Y, Kasai K, Nomoto H, Inoue Y. Frontal affinity chromatography of ovalbumin glycoasparagines on a concanavalin A-sepharose column. A quantitative study of the binding specificity of the lectin. J. Biol. Chem. 1985;260:6882–6887. [PubMed] [Google Scholar]
- (29).Liang PH, Wang SK, Wong CH. Quantitative Analysis of Carbohydrate-Protein Interactions Using Glycan Microarrays: Determination of Surface and Solution Dissociation Constants. J. Am. Chem. Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.