Abstract
The WAVE/SCAR complex promotes actin nucleation through the Arp2/3 complex, in response to Rac signaling. We show that loss of WVE-1/GEX-1, the only C. elegans WAVE/SCAR homolog, by genetic mutation or by RNAi, has the same phenotype as loss of GEX-2/Sra1/p140/PIR121, GEX-3/NAP1/HEM2/KETTE, or ABI-1/ABI, the three other components of the C. elegans WAVE/SCAR complex. We find that the entire WAVE/SCAR complex promotes actin-dependent events at different times and in different tissues during development. During C. elegans embryogenesis loss of CED-10/Rac1, WAVE/SCAR complex components, or Arp2/3 blocks epidermal cell migrations despite correct epidermal cell differentiation. 4D movies show that this failure occurs due to decreased membrane dynamics in specific epidermal cells. Unlike myoblasts in Drosophila, epidermal cell fusions in C. elegans can occur in the absence of WAVE/SCAR or Arp2/3. Instead we find that subcellular enrichment of F-actin in epithelial tissues requires the Rac-WAVE/SCAR- Arp2/3 pathway. Intriguingly, we find that at the same stage of development both F-actin and WAVE/SCAR proteins are enriched apically in one epithelial tissue and basolaterally in another. We propose that temporally and spatially regulated actin nucleation by the Rac-WAVE/SCAR- Arp2/3 pathway is required for epithelial cell organization and movements during morphogenesis.
Keywords: morphogenesis, cell migration, actin nucleation, epithelial polarity
INTRODUCTION
Actin nucleation through the Arp2/3 complex provides the mechanical force to drive diverse cellular processes including cell motility (Pollard, 2007). During the dynamic movements of morphogenesis, actin nucleation by Arp2/3 is required for cell movements, but the exact role of actin nucleation in these movements is not well understood. In C. elegans, morphogenetic movements of the epidermis include a convergent-extension-like movement called dorsal intercalation that requires polarized microtubules and actin (Priess and Hirsh, 1986; Williams-Masson et al., 1998). Actin regulation is also required for the movements of the epidermis to enclose the embryo, or epiboly (Priess and Hirsh, 1986; Costa et al., 1997; Williams-Masson et al., 1997, 1998; Reviewed in Chin-Sang and Chisholm, 2000; Simske and Hardin 2001). During these movements actin nucleation may be contributing to cellular protrusions, to cell-cell adhesion, and to the overall apical/basal polarity of the moving cells.
The Arp2/3 complex must first be activated before it becomes an efficient nucleator of dendritic, branched actin. Motile cells are proposed to receive extracellular signals that pass through cell surface receptors to activate small GTPases, which in turn activate the WASP and WAVE/SCAR nucleation promoting factors (Pollard, 2007). The WASP and WAVE/SCAR protein families act as powerful switches that lead to maximal actin nucleation through the Arp2/3 complex (Takenawa and Miki, 2001). Once actin is polymerized and reorganized the cell can initiate movements. Screens for C. elegans mutants that fail to initiate morphogenesis despite correctly specified cell fates have identified actin nucleation regulators as key components in this process (Soto et al., 2002; Fig. 1A). C. elegans embryos can still initiate morphogenetic movements when they are depleted of adhesion molecules including E-cadherin/HMP-1, alpha and beta integrins (ina-1, pat-2 and pat-3) and the actin regulators Ena/Vasp/UNC-34 and WSP-1 (Costa et al., 1998; Baum and Garriga, 1997; Williams and Waterston, 1994; Withee et al., 2004). In contrast, C. elegans mutants with the unique Gex (gut on the exterior) phenotype fail to initiate any of the epidermal cell movements of morphogenesis (Soto et al., 2002). We previously described the essential role of two C. elegans WAVE/SCAR components, GEX-2/Sra1/p140/PIR121 and GEX-3/NAP1/HEM2/KETTE, in embryonic morphogenesis. Loss of gex-2 or gex-3 leads to a 100% penetrant maternal effect embryonic lethality due to a complete failure in morphogenesis (Soto et al., 2002). By comparison, the single C. elegans Wasp homolog, wsp-1, is mostly dispensable for embryonic development due to redundancy with other actin nucleation promoting proteins (Withee et al., 2004). Eden and colleagues proposed that the mammalian homologs of GEX-2 (Sra1/p140/PIR121) and GEX-3 (NAP1/HEM2/KETTE) are negative regulators of WAVE1. In their model Rac signaling causes the GEX-2 and GEX-3 homologs to release WAVE1, allowing it to interact with Arp2/3 to promote actin nucleation (Eden et al., 2002). However, subsequent studies have suggested alternate models for WAVE/SCAR regulation including positive roles for GEX-2 and GEX-3 homologs (Reviewed in Blagg and Insall, 2004; Stradal et al., 2004).
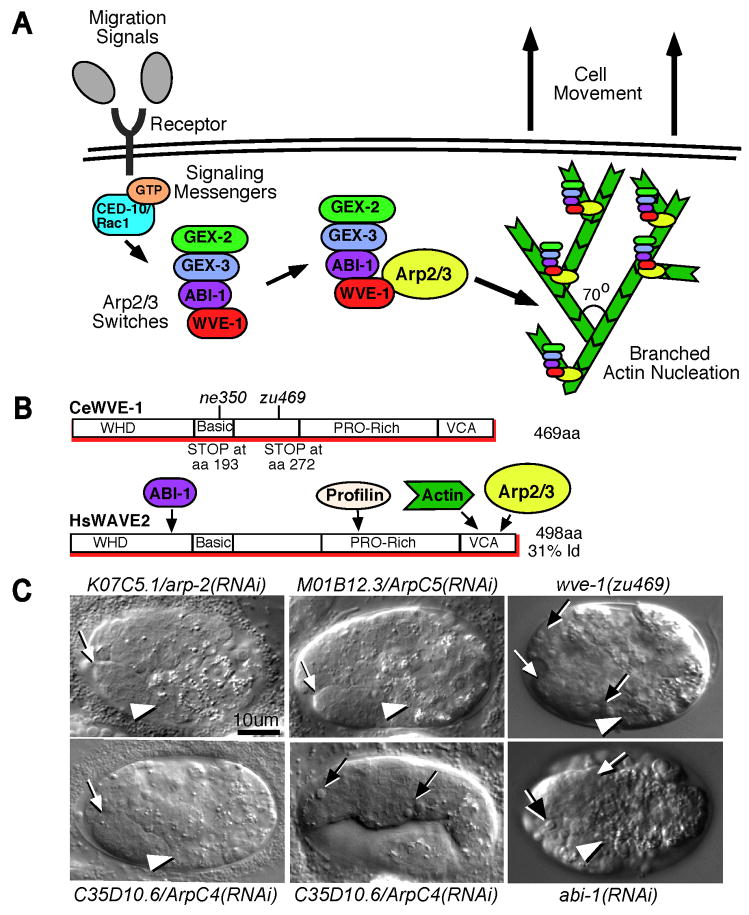
Fig. 1. Mutations in wve-1 disrupt the C. elegans WAVE/SCAR homolog.
A. The Rac-WAVE/SCAR-Arp2/3 pathway.
A signaling pathway that connects migration signals or cues, through the small GTPase CED-10/Rac1, through the WAVE/SCAR nucleation promoting complex, to the Arp2/3 complex is thought to promote branched polymerization of actin. The known C. elegans homologs are shown. Plants and humans contain a fifth component of the WAVE/SCAR complex, HSPC300/BRICK (Eden et al., 2002; Frank et al., 2003; Le et al., 2006; Cascon et al., 2007) but homology searches have not identified a homolog in C. elegans.
B. C. elegans WVE-1/WAVE has similar domains to human WAVE.
WVE-1 shows a high degree of homology to all WAVEs and a conserved domain structure. C. elegans WVE-1/WAVE is 31% identical to human WAVE2 over its entire length, and shows similar homology to WAVE1 and WAVE3. Like other WAVEs it contains the Wave Homology Domain (WHD), including the basic region, a Proline-rich region thought to mediate profilin binding, and the verprolin homology, cofilin homology and acidic (VCA) region through which WAVEs are thought to bind actin and the Arp2/3 complex. The two wve-1 mutations, ne350 and zu469, are predicted to lead to early truncations of the protein.
C. Loss of wve-1, abi-1 or Arp2/3 complex components leads to the Gex phenotype.
Embryos from mothers injected with RNA corresponding to Arp2/3 components ARP-2/K07C5.1, ArpC5/M01B12.3 and ArpC4/C35D10.6 show the Gex-like epidermal enclosure defect. Animals produced early when RNAi is not fully penetrant still have maternal product so they display zygotic phenotypes including a Ced (cell death defective) corpse engulfment defect. The Ced phenotype is also seen in wve-1 genetic mutants and abi-1 RNAi embryos. The Ced phenotype is well studied for ced-10 and has been seen in gex-2 and gex-3 mutant embryos (Reddien and Horvitz, 2000; Kinchen et al., 2005; Soto et al., 2002). White arrows: anterior of the pharynx; white arrow heads: anterior of the intestine; black arrows: unengulfed apoptotic cells. Embryos are shown at a late stage (at least 700 minutes after first cleavage), when wild-type larvae have few unengulfed corpses (Soto et al., 2002). Embryos in all figures are oriented with anterior at left and dorsal up unless otherwise indicated. Scale bars in all figures represent 10um.
In this report we show that all components of the WAVE/SCAR actin nucleation complex contribute to specific actin-nucleation dependent movements during C. elegans embryogenesis. We remove the components of the proposed Ced-10/Rac-WAVE-Arp2/3 pathway to determine how regulators of actin nucleation contribute to the earliest movements of epithelial cells in the embryo (Fig. 1A). The WAVE/SCAR complex in C. elegans has only one copy of each component including one highly conserved homolog of WAVE/SCAR called WVE-1/GEX-1/R06C1.3 (Fig. 1B) (Sawa et al., 2003; Sheffield et al., 2007; Shakir et al., 2008). Using genetic mutations and RNAi we show conclusively that WVE-1 is as important for embryonic morphogenesis as GEX-2 and GEX-3. Loss of wve-1 causes the same complete failure in morphogenesis and 100% embryonic lethality as loss of gex-2 or gex-3. We remove the only copy of the fourth WAVE/SCAR component, the ABI homolog abi-1/B0336.6, and find the same phenotype. We find that loss of the CED-10/Rac1, any WAVE/SCAR component, or any Arp2/3 component leads to the same phenotype: arrested migration of the embryonic epidermis. 4D movies show that loss of wve-1, gex-2, gex-3 or arp-2 blocks cell migrations. Four epidermal cells that are essential to the migration show smaller and fewer cellular protrusions. Surprisingly, cell fusions occur in the absence of WAVE/SCAR or Arp2/3 in C. elegans epidermal cells, in contrast to results from Drosophila myoblast fusion studies. Localization studies show that the WAVE/SCAR complex is enriched subcellularly in the same regions where filamentous actin (F-actin) is enriched, and that loss of WAVE/SCAR disrupts F-actin subcellular enrichment. Our results suggest that localized actin nucleation regulates epithelial cell organization during the dynamic events of morphogenesis.
MATERIALS and METHODS
Strains
C. elegans strains were cultured as described in Brenner (1974). The following strains were used in this analysis: wve-1(ne350), wve-1(zu469), gex-2(ok1603)/dpy-9 (out-cross 3), gex-3(zu196); ced-10(tm597), rrf-3(pk1426), nhr-25::yfp, xnIs16 (DLG-1::GFP), jcIs1 (AJM-1::GFP), VJ402 fgEx11[ERM-1::GFP], OX84 nhr-25::yfp rrf-3(pk1426), OX1 wve-1(zu469) unc-101(m1); Ex[GFP::WVE-1], WM58 unc-24 gex-3(zu196); Ex[GFP::GEX-3], OX169 gex-3(zu196)/DnT1; nhr-25::yfp. wve-1(zu469) was discovered by Jim Priess. wve-1(ne350) was discovered by M. Soto.
Cloning of wve-1
We cloned wve-1 by positional cloning and SNP mapping. It maps to approximately +11 m.u. on linkage group I. Sequencing the wve-1/R06C1.3 gene revealed that zu469 has a C to T transition that creates a STOP codon at aa272 while ne350 has a C to T transition that creates a STOP codon at aa193. Both mutations are predicted to lead to early truncations of the 469 aa protein (Fig. 1B).
RNAi of WAVE/SCAR genes
The term “Gex embryos” is used in this report to describe animals that die due to morphogenesis defects from the loss of ced-10/Rac1, WAVE/SCAR or Arp2/3 components by genetic mutation or RNAi. Genetic mutations in wve-1, gex-2 and gex-3 have the same embryonic phenotype as loss of function through RNAi, but RNAi results in lower penetrance. While genetic mutations lead to 100% embryonic lethality with complete epidermal enclosure failure, feeding dsRNA leads to 40%, 90% and 100% penetrant embryonic lethality, respectively, with RNAi escaper animals able to undergo varying degrees of enclosure. We use RNAi to remove abi-1 instead of the two existing putative deletion strains of abi-1, ok640 and tm494, because these are not true deletions of abi-1. They contain both the deletion and also a wild-type copy and are homozygous viable (unpublished results). Injection or feeding of gex-2 or gex-3 RNA results in 90–100% embryonic lethality, similar to the 100% embryonic lethality seen in genetic mutations (our unpublished observations). However, wve-1 and abi-1 are partially resistant to RNAi. Feeding wve-1 or abi-1 dsRNA to the N2 wild-type strain results in 40% and 60% embryonic lethality, respectively. This may explain why previous investigators failed to detect a phenotype for wve-1 by RNAi (Fraser et al., 2000; Sawa et al., 2003). All of the RNAi embryos shown in this paper were generated in the N2 wild type genetic background. For biochemical studies we used the RNAi hypersensitive strain rrf-3(pk1426) (Simmer et al., 2003) and obtained 49% and 92% embryonic lethality for wve-1 and abi-1, respectively (Fig. 6C,D). Since rrf-3(pk1426) animals show increasing sterility at higher temperatures, animals were cultured at 22°C. The 7–15% of embryonic lethality we noted at 22°C (Fig. 6C) is distinct from the Gex embryonic phenotype. For the experiments in Fig. 7C on intestinal lumen expansion, we performed feeding RNAi in the N2 background and used animals in which RNAi was not yet fully penetrant to find animals that enclosed and then died. This allowed us to measure the width of the lumen in animals with intestines of WT or near WT length. In contrast, animals that fail to enclose, like 100% of wve-1, gex-2, and gex-3 genetic mutants, do not elongate and have a shorter intestine.
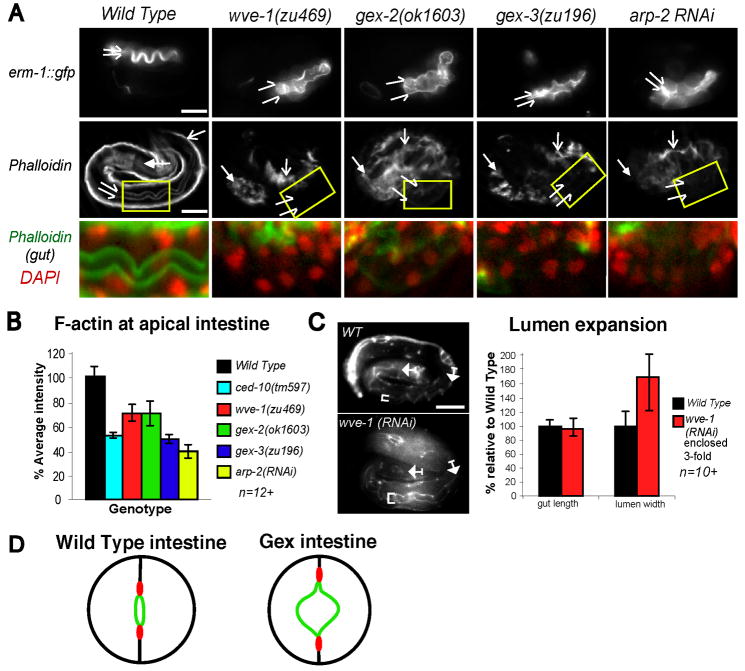
Fig. 6. Analysis of GEX protein localization and dependence on other GEX proteins.
A. F- actin and WAVE/SCAR proteins are enriched in the basolateral epidermis and apical intestine.
GFP::WVE-1 and GFP::GEX-3 rescuing transgenes are expressed in the cytoplasm and enriched at membrane regions. This localization pattern resembles the enrichment of F-actin visualized with phalloidin. In epidermal cells, where apical is outside and basolateral is inside, GFP::WVE-1, GFP::GEX-3 and F-actin (green) are enriched basolaterally at the bean stage (380 minutes, top panels) as shown by their localization basal to the adherens junction associated proteins DLG-1 and AJM-1. In intestinal and pharyngial cells, where apical is inside and basolateral is outside, GFP::WVE-1, GFP::GEX-3 and F-actin are enriched at apical regions by the 1.5-fold stage (440 minutes, bottom panels). Arrows point to the developing nerve ring.
B. & C. WVE-1 and GEX-3 require other WAVE/Scar components for abundance.
The rescuing GFP::WVE-1 and GFP::GEX-3 transgenic strains were grown on control bacteria (no RNAi, WT) or bacteria expressing double stranded wve-1, gex-2, gex-3 or abi-1 RNA.
B. Embryos fed the control or WAVE/SCAR bacteria are shown. In all cases, loss of other WAVE/SCAR components leads to lower expression of GFP::WVE-1 and GFP::GEX-3. Remaining spots are produced by autofluorescent gut granules. DAPI staining indicates the stage (at least 500 minutes) and orientation (widely spaced intestinal nuclei are ventral).
C. The graph indicates the percentage of embryos expressing GFP when the GFP::WVE-1 and GFP::GEX-3 transgenic strains are grown on wild type bacteria or bacteria expressing WAVE/SCAR components.
D. GEX-2, GEX-3 and ABI-1 are required to maintain WVE-1 protein levels.
Embryonic lysates were depleted of wve-1, gex-2, gex-3 or abi-1 by feeding control bacteria or bacteria expressing wve-1, gex-2, gex-3, or abi-1 dsRNA to the RNAi sensitive strain rrf-3(pk1426). Western blots were probed with antibodies to WVE-1, GEX-2 and GEX-3. The effectiveness of the RNAi food was measured as percent embryonic lethality. The bottom of each gel was cut and probed with antibodies to tubulin as a loading control. 20ug of embryonic lysates were loaded per lane.
Fig. 7. GEX proteins regulate lumen morphology and apical F-actin enrichment in the intestine.
A. The apical intestine is altered in Gex embryos.
Top row: An erm-1::gfp transgene (Gobel et al, 2004) is enriched apically in wild-type and mutant intestines, although the apical domain is wider in the mutants (split arrows). Second row: Phalloidin staining shows that wild-type embryos are enriched for F-actin at the apical region of the pharynx (anterior arrows), and intestine (split arrows). Dorsal arrows indicate the body wall muscles. Third row: Enlarged image of intestine (yellow box). In Gex embryos phalloidin levels (green) drop in the apical intestine. DAPI (red) identifies the intestinal nuclei.
B. Apical F-actin shows a pronounced drop in the embryonic intestine of Gex embryos.
The phalloidin signal, in the intestine of wild-type and Gex embryos, was measured using ImageJ (n=at least 12). Using the DAPI signal to confirm intestinal cell location, an equally sized area of each embryo was tested for average fluorescence intensity.
C. Enclosed wve-1(RNAi) animals show increased lumen area.
Wild-type animals and wve-1(RNAi) escaper animals that enclose and reach the 3-fold stage were compared. Intestine length (between arrows) and lumen width (brackets) were marked by the MH27 antibody to the junctional protein AJM-1 (n= at least 10).
D. Summary of the gex intestinal epithelial defects.
Cartoons of cross-sections through the intestine summarize apical/basal changes in Gex embryos. The lumen is shown in green, adherens junctions are shown in red and basolateral regions are shown in black. Loss of Arp2/3-dependent actin nucleation leads to an expansion of the apical domain, a basal shift of the junctional domains, and cell shape changes in the basolateral regions.
Feeding RNAi
cDNAs for wve-1, gex-2, gex-3, abi-1 and arp-2 were inserted into L4440 vector and transformed into HT115 cells. Saturated overnight cultures were diluted 1:250 and grown for 6–7 hours until the OD600was close to 1. Bacteria were resuspended in LB Amp 100ug/ml. 1mM IPTG was added to the bacteria and Amp plates before use. C. elegans animals were synchronized by hypochlorite treatment followed by hatching in M9 Buffer. For embryonic lysates synchronous L1 larvae were fed either control HT115 E. Coli, or HT115 containing the L4440 plasmid carrying the gene of interest. Worms were grown at 22°C for 3 days, and lysates were made from the embryos obtained by hypochlorite treatment.
Lysates for biochemistry
Embryos were ground in Lysis Buffer (150mM NaCl, 1mM DTT, 0.1% Triton and protease inhibitor cocktail - Roche #11836153001) in a stainless steel homogenizer (Wheaton #357572). Lysates were spun at 5K and the supernatant fraction was flash frozen in liquid nitrogen for western blots.
Antibodies
Antibodies to WVE-1 were generated by immunizing rabbits with the C-terminus of WVE-1 (aa 387–469) (Capralogics, MA). The antibodies recognize the WVE-1 70KD band in wild-type lysates but not lysates depleted of wve-1 by feeding RNAi.
Rescue of wve-1 with GFP::WVE-1 fusion strain
The strain wve-1(zu469) unc-101/hIn1 was injected with a plasmids carrying genomic wve-1 including 1.4 kb of upstream and 400 bp of downstream DNA and the injection marker pRF4 (Mello et al., 1991). WVE-1 with N-terminal tags could rescue, but not WVE-1 with C-terminal GFP. Lines that supported the viability of the progeny of homozygous wve-1 animals were studied for their GFP localization.
In vivo analysis of membrane protrusions in epidermal cells
Live embryos were attached to poly-L-lysine-coated coverslips and mounted in embryo buffer atop vacuum-grease “feet” on a standard microscope slide, with the coverslip edges sealed by silicone oil. Embryos were imaged using a 40×1.3 NA oil immersion lens on a Nikon TE2000 inverted microscope fitted with a Yokogawa CSU21 spinning-disk confocal scanhead (Perkin Elmer), a Melles-Griot argon laser (514 nm excitation) controlled by a Neos programmable AOTF. Multidimensional datasets were acquired using Molecular Dynamics MetaMorph software on a Hamamatsu Orca-AG cooled CCD camera, and stereo QuickTimeVR movies were constructed from the raw data using a custom-written plugin for the Java program ImageJ (http://rsb.info.nih.gov/ij/).
Quantitation of protrusive extension in the 4D movies
Embryos expressing nhr-25::YFP were filmed at an interval of 2.5 minutes. Stacks were projected into 4D QuickTime movies that allowed viewing of the embryos from multiple angles (http://fsbill.vcell.uchc.edu/gloworm/Patel_et_al_2008_Data/). Stills were extracted from the movies, converted to TIFF files, and analyzed using ImageJ software. The protrusive extension of both leading cells and pocket cells was measured by taking the protrusion perimeter (traced with the Freehand Tool to measure path length) and dividing it by the cell width (measured using the Segmented Line tool) as in Sheffield et al., 2007 (Fig. 4B). To compare the protrusive extension of the leading cells relative to the pocket cells we divided the protrusive extension of the leading cells by the protrusive extension of the pocket cells to generate a ratio (Fig. 4C). This morphometry analysis was performed on 5 time points 10 minutes apart, spanning a 40-minute time interval when ventral enclosure occurs in wild type.
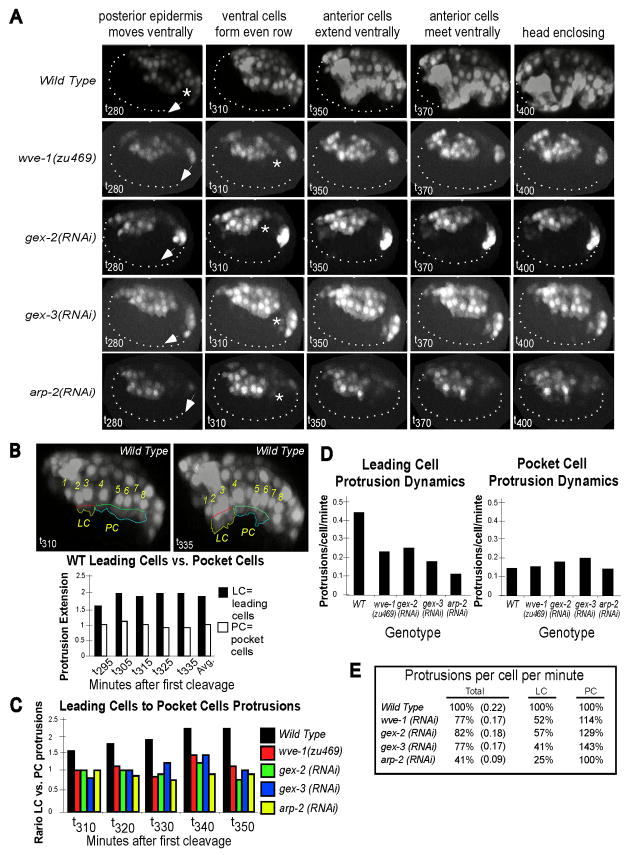
Fig. 4. 4D movies reveal loss of gex gene function disrupts epidermal movements in specific epidermal cells.
A. wve-1, gex-2, gex-3 and arp-2 mutants display similar defects of morphogenesis.
Time points were extracted from 180 minute 4D movies (270 to 450 minutes after first cleavage) of wild-type and Gex embryos (Supplemental Movie 1). The nhr-25::yfp transgene is expressed in both the nucleus and cytoplasm of epidermal cells, but not in the underlying cells of the embryo (dotted line). Wild-type embryos undergo dramatic morphogenetic changes during the two hours of development shown (from 280 to 400 minutes). The first epidermal movement detected with nhr-25::yfp is a ventral extension of the posterior epidermal cells, probably due to cell division along the anterior/posterior axis. This movement occurs in Gex embryos (left panel, arrows). The next movement detected is the ventral cell migration of epidermal cells to form an even ventral row. In Gex embryos ventral movements fail, leading to an uneven lateral row of cells (asterisks). In wild-type embryos the epidermal cells go on to enclose the embryo. In Gex embryos ventral epidermal cells display arrested migration, despite protrusions that lasts at least 5 hours. In wild-type embryos the epidermis undergoes circumferential constriction at 700 minutes while in Gex embryos epidermal cells collapse inwardly at the equivalent time point (Fig. 2B, 2C; unpublished movies). At least two movies were analyzed for each genotype. For the analysis below we chose movies where the Gex phenotype is confirmed by following the embryos for at least 6 hours of morphogenesis and made 4D QuickTime projections which permit viewing of the embryos from multiple angles (http://fsbill.vcell.uchc.edu/gloworm/Patel_et_al_2008_Data/). Measurements were done on one to two movies for each genotype. The movies were staged by DIC analysis and by noting the first appearance of the nhr-25::yfp transgene, at approximately 280 minutes after first cleavage.
B. The leading cells are more protrusive than the pocket cells in wild-type embryos. The ventral epidermal cells in a wild-type animal carrying the nhr-25::yfp transgene show how protrusions in leading cells (#2,3) vs. pocket cells (#4–8) were measured. The perimeter of the protrusive edge of the leading cells was traced using the Freehand Tool and the width of the leading cells was measured using the Segmented Line tool (ImageJ). Protrusion extension at the leading edge is represented as the ratio of the perimeter of the leading edge cells relative to their width. The same analysis was performed on the pocket cells. The analysis was performed on 5 time points, 10 minutes apart, spanning a 40-minute time interval when ventral enclosure occurs in wild type.
C. The ratio of leading cell to pocket cell protrusions changes in Gex embryos. Protrusions in leading cells and pocket cells were measured as above at five time points when ventral enclosure occurs in wild type. To compare the protrusion extenstion of the leading cells to that of the pocket cells we divided the protrusive extension of the leading cells by the protrusive extension of the pocket cells and plotted this ratio. The ratio of leading cell to pocket cell protrusions is shown for wild-type and Gex embryos.
D. The number of protrusions per cell per minute drops in Gex embryos. The number of protrusions made by leading cells and pocket cells during 17 time points 2.5 minutes apart (from 300 to 340 minutes) was counted in wild-type and Gex embryos.
E. Table of protrusion number comparisons. The number of protrusions made by the ventral epidermal cells (cells 1–8, Fig. 4B) was counted over a period of 40 minutes as in 4D. The average protrusions/cell/minute, relative to wild type, is shown for all cells (Total, #1–8), or for subsets of the cells (LC: leading cells, #2,3; PC: pocket cells, #4–8).
RESULTS and DISCUSSION
To determine how cytoskeletal polarity is regulated during morphogenesis we characterize here the complete C. elegans WAVE/SCAR complex, a major regulator of cell migration initiation in C. elegans embryos (Soto et al., 2002).
Two mutations resulting in the Gex phenotype are nonsense mutations in WVE-1
Our genetic screens have identified new gex mutants. Genetic mutations in gex-1 lead to 100% embryonic lethality with complete failure in morphogenesis (Fig. 2A). Sequencing the wve-1 gene, the only C. elegans homolog of the WAVE/SCAR gene family, revealed that two alleles of gex-1, ne350 (Shakir et al., 2008) and zu469, have point mutations that create STOP codons less than half way through the protein (Fig. 1B). Hereafter gex-1 is referred to as wve-1. A PCR product encoding the wve-1 genomic coding region plus 1.4 kb of upstream and 400 bp of downstream DNA rescues the embryonic lethality of the wve-1(zu469) allele. We had previously identified two other components of the C. elegans WAVE/SCAR complex with the same phenotype, gex-2 and gex-3 (Soto et al., 2002). To further analyze the C. elegans WAVE/SCAR complex, we removed the only C. elegans ABI homolog, abi-1/B0336.6, by RNAi and found the same defects in morphogenesis seen in gex mutants in 60% of the abi-1(RNAi) embryos (Fig. 2A). Thus, the four members of the C. elegans WAVE/SCAR complex are required for normal epidermal enclosure.
Fig. 2. ced-10/Rac1, WAVE/SCAR and Arp2/3 mutants show similar morphogenesis phenotypes.
A. Gex terminal embryonic phenotypes. Left panel: DIC images. In wild-type embryos the pharynx (arrows) and intestine (arrow heads) are maintained internally by epidermal enclosure. In Gex embryos these tissues lie outside at the end of embryogenesis. Center panel: mAb MH46 to LET-805/Myotactin (Francis and Waterston, 1991; Hresko et al., 1999) shows that the pharynx (left arrows) is short and disorganized in Gex embryos while body-wall muscles (right arrows) fail to elongate in Gex embryos. Right panel: mAb MH33 to IFB-2 (Francis and Waterston, 1985; Bossinger et al., 2004) shows the width of the intestinal lumen (split arrows), which expands in Gex embryos. Embryos are pictured at late embryogenesis.
B. Morphogenesis defects in epithelial tissues of Gex embryos using DLG-1::GFP.
Comparison of wild-type embryos carrying the DLG-1::GFP transgene (Firestein and Rongo, 2001; Totong et al., 2007) and the same strain fed RNAi food for wve-1, gex-2, gex-3 or arp-2 shows disruption of morphogenetic movements. Top row: Dorsal intercalation fails in Gex embryos. Second row: In wild type (lateral view), cells of the intercalated dorsal row expand laterally, then fuse (asterisk) to form syncytial hyp7 (Podbilewicz and White, 1994). In Gex embryos (dorsal view) dorsal cells fail to expand laterally, yet they fuse as in wild type (asterisks). Third row: In wild-type embryos the ventral-most rows (open arrow) migrate and meet up at the ventral surface (Chin-Sang and Chisholm, 2000; Simske and Hardin, 2001). The ventral-most cells (open arrows) in Gex embryos arrest migration at or close to where they are born. Dotted lines indicate the unenclosed regions of each embryo. Fourth row: A wild-type embryo (ventral view) shows the epidermal cells meeting at the ventral surface and enclosed internal organs are not visible. Gex embryos (3/4 side view) show arrested epidermal cells and internal organs exposed on the ventral surface. Split arrows show width of intestinal DLG-GFP domain.
C. A summary of the morphogenesis defects caused by loss of Rac, WAVE/SCAR or Arp2/3. Loss of ced-10/Rac1, wve-1, gex-2, gex-3, abi-1 or arp-2 leads to the distinctive Gex (gut on the exterior) phenotype due to failures in cell movement and cell shape changes. The epidermis is shown in blue, the pharynx in green and the intestine in red. Lateral view: Gex embryos fail to initiate epidermal ventral movements. By 400 minutes after first cleavage, wild-type embryos initiate circumferential constrictions to squeeze the embryo into a worm. Gex embryos undergo constriction that leads the epidermis to collapse inwardly. The internal organs (pharynx and intestine) end morphogenesis exposed on the ventral surface by 700 minutes. Dorsal view: in Gex embryos dorsal intercalation fails (330 minutes), but this does not prevent fusion of the dorsal epidermis (by 450 minutes).
Loss of CED-10/Rac1, or any component of the WAVE/SCAR or Arp2/3 complexes leads to similar phenotypes
The most likely activator of the embryonic C. elegans WAVE/SCAR complex is the small GTPase CED-10/Rac1which had previously been implicated in the regulation of embryonic morphogenesis (Lundquist et al., 2001; Soto et al., 2002) (Fig. 1A). The expected target of the WAVE/SCAR complex is the Arp2/3 actin nucleation complex. The C. elegans Arp2/3 complex has been described and has been suggested to function in embryonic morphogenesis (Severson et al., 2002; Sawa et al., 2003). If the C. elegans WAVE/SCAR complex is the main regulator of embryonic actin nucleation through Arp2/3, then the loss of Arp2/3 complex should be similar to the loss of WAVE/SCAR components (Fig. 1A). We therefore compared the morphogenesis role of ced-10/Rac1, the four components of the C. elegans WAVE/Scar complex, and the components of the C. elegans Arp2/3 complex.
Embryonic defects
We find that just as in gex-2 and gex-3 mutants, the epidermis fails to enclose the embryo in ced-10/Rac1(tm597), wve-1(zu469), abi-1(RNAi), and arp-2(RNAi) animals while the pharynx and intestine show similar tissue disorganization (Fig. 2A). The penetrance of the enclosure defect is 70%, 100%, 60% and 100% in these embryos, respectively (n=500+ for all genotypes; see Methods regarding RNAi of abi-1). All of the 8 members of the C. elegans Arp2/3 complex, including the two homologs for ArpC5 (M01B12.3 and C45H11.3) show a Gex phenotype with at least 40% penetrance when removed by RNAi (n=100+ for all genotypes). For example, loss of ARP-2/K07C5.1, ArpC4/C35D10.6 or ArpC5/M01B12.3 results in failed epidermal enclosure so that the pharynx and intestine are found at the outside at the end of embryogenesis (Fig. 1C, 2A).
We used antibodies to specific tissues to check the morphology of internal organs in Gex embryos. MH46 antibody recognizes LET-805/myotactin that is deposited between epidermal cells and both pharynx and body wall muscles (Francis and Waterston, 1991; Hresko et al., 1999). In wild-type animals the pharynx develops into a long tube and the body wall muscles span the length of the worm. Gex animals have a short, disorganized pharynx and the muscles end development compressed at the dorsal side, beneath the collapsed epidermis (Fig. 2A). MH33 antibody recognizes the intermediate filaments that form the terminal web at the apical region of the intestine (Francis and Waterston, 1985; Bossinger et al., 2004). In wild-type animals the terminal web surrounds the thin lumen at the apical intestine. Gex animals end development with a disorganized intestine with an expanded lumen and expanded MH33 domain (Fig. 2A, double arrows indicate intestinal lumen boundaries).
We used live imaging to better understand the cellular defects underlying the Gex phenotype. DLG-1::GFP marks adherens junctions in the migrating epidermis, pharynx and intestine (Firestein and Rongo, 2001; Koppen et al., 2001; McMahon et al., 2001; Totong et al., 2007). In wild-type embryos DLG-1::GFP is first visible as the epidermal cells align into rows prior to cell movements. In Gex animals the earliest DLG-1::GFP pattern appears normal but once morphogenesis begins defects are seen in dorsal intercalation (Fig. 2B, top row) and in ventral cell migrations of the epidermis (Fig. 2B, third row, open arrow). Dorsal cells fail to intercalate and then fail to lengthen laterally as in wild type. While wve-1, gex-2, gex-3 and arp-2 embryos are fully penetrant for these phenotypes, ced-10(tm597) deletion null animals from homozygous mothers die, but 25% undergo at least partial enclosure (n=300). Using DIC analysis we had previously reported that ced-10 null animals appear to undergo dorsal intercalation (Soto et al., 2002). We used DLG-1::GFP to monitor dorsal intercalation in ced-10(tm597) deletion null animals from homozygous mothers and find that indeed most of these embryos undergo at least some dorsal intercalation. However, DLG-1::GFP reveals that 48% show abnormal dorsal cell organization (n=40). We interpret the 75% penetrant enclosure defect of ced-10(tm597) null embryos, and the 48% penetrant dorsal misorganization phenotype as evidence for redundancy of ced-10 with the two other C. elegans Racs, mig-2 and rac-2. However, among the Racs only ced-10/Rac1 is embryonic lethal (Lundquist et al., 2001).
At the end of development, the epidermis, which has failed to migrate, collapses and constricts thereby forcing the internal organs to extrude to the ventral side of the embryo (Fig. 2B, bottom row; unpublished movies). Loss of ced-10/Rac1, WAVE/SCAR or Arp2/3 complex genes all produce a similar phenotype. In contrast, loss of the other C. elegans Rac homologs, mig-2 and rac-2, does not produce these phenotypes (Soto et al., 2002; our unpublished observations). Other C. elegans morphogenesis mutants, such as mutations in the cadherin/catenin complex, have a much milder phenotype in which the epidermal cells do begin cell migrations (Costa et al., 1998). We propose that the earliest events of epidermal enclosure require a signaling pathway that activates the components of the C. elegans WAVE/SCAR complex through the CED-10/Rac1 GTPase to activate Arp2/3.
Embryonic and post-embryonic cell fusions occur in the absence of WAVE/SCAR and Arp2/3
Dorsal intercalation fails in embryos missing WAVE/SCAR and Arp2/3 components, yet fusion of the dorsal hyp7 epidermal cells proceeds (asterisks in Fig. 2B, second row). We observed fusions of hyp7 cells in all embryos examined (n=20+ for each genotype). It was known that dorsal intercalation is not required for cell fusion, since other mutations that disrupt dorsal intercalation, like die-1, also show dorsal cell fusion (Heid et al., 2001). However, this result is in striking contrast to studies showing that the Drosophila WAVE/SCAR and Arp2/3 complexes are required for cell fusion during embryonic myogenesis (Schroter et al., 2004; Kim et al., 2007; Massarwa et al., 2007; Schafer et al., 2007; Richardson et al., 2007; Berger et al., 2008).
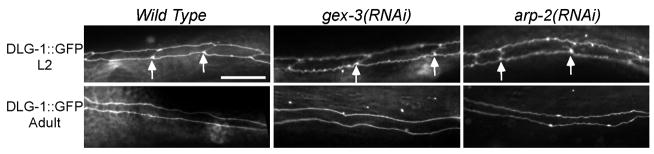
Fusion of the embryonic hyp7 cells requires the EFF-1 fusogen (Mohler et al., 2002). To test if other cells at other times in development can fuse, we used DLG-1::GFP larvae raised on gex-3 and arp-2 dsRNA to observe post-embryonic epidermal seam cell fusions. Fusion of the seam cells requires a different fusogen, AFF-1 (Sapir et al., 2007). To monitor post-embryonic RNAi in these animals, we followed another phenotype where we have documented that the RNAi defect matches what we see in genetic mutants. Gex animals have zygotic phenotypes when we remove WAVE/SCAR components by mutation or by RNAi. For example, homozygous gex mutants are alive due to maternal rescue, and they have a fully penetrant zygotic egg laying defective phenotype (Egl) despite the presence of an apparently wild-type L3 and L4 stage vulva (Soto et al., 2002). Our two alleles of wve-1, zu469 and ne350, display this completely penetrant egg-laying defect in which animals lay at most 10 eggs before becoming Egl (data not shown). In addition, loss of any WAVE/SCAR component, or arp-2 by RNAi leads to Egl adults (n=300+ for all genotypes). Using the DLG-1::GFP transgene we detected epidermal seam cell fusions after the L4 to adult molt, as expected, in wild-type adults (Ambros, 1989). DLG-1::GFP animals depleted of gex-3 and arp-2 by RNAi, as confirmed by their Egl phenotype, also showed adult seam cell fusions (n=30+ for each genotype) (Fig. 3). While these animals may have residual gex-3 and arp-2, they went on to make 100% and 96% dead embryos, respectively, with the Gex phenotype (n=200+). We did not detect differences in the timing of the fusions. Multiple models have been proposed to explain epidermal cell fusions in C. elegans and other organisms (Reviewed by Chen and Olson, 2005; Podbilewicz, 2006). Our results suggest that in contrast to Drosophila myoblasts, fusion of epidermal cells requires a different actin nucleator besides Arp2/3. Alternatively, epidermal cells may be able to undergo fusion using a mechanism that does not require actin nucleation factors. Since both EFF-1 and AFF-1 dependent cell fusions can occur, neither of these molecularly distinct cell fusion mechanisms in the worm appears to require WAVE/SCAR or Arp2/3.
Fig. 3. Epidermal cells can fuse in the absence of WAVE/SCAR components or ARP-2.
Larvae carrying the DLG-1::GFP transgene (Totong et al., 2007) were cultured from the first larval stage (L1) on either control food (no RNAi) or food containing gex-3 or arp-2 dsRNA. During larval stages (L2 shown here) epidermal seam cells in wild-type, gex-3(RNAi) and arp-2(RNAi) animals are unfused (white arrows indicate cell boundaries) (Podbilewizc and White, 1994). After the L4 to adult molt, wild-type (Ambros, 1989), gex-3(RNAi) and arp-2(RNAi) animals undergo seam cell fusions. The efficiency of post-embryonic RNAi was monitored by checking adults for the Gex Egl phenotype and their progeny for the Gex Mel phenotype. The slight twist in the seam cells is due to the dominant rol-6(su1006) mutation used to mark the DLG-1::GFP transgene (Totong, et al., 2007).
The C. elegans WAVE/SCAR complex regulates membrane dynamics in migrating epidermal cells
Gex embryos have properly differentiated epidermal cells that express epidermal markers (Soto et al., 2002), yet they do not initiate migrations. To determine if the loss of cell migrations is due to aberrant epidermal membrane protrusions we visualized cell shapes using the nhr-25::yfp transgene, which is only expressed in the epidermis (Baugh et al., 2003). nhr-25::yfp is visible in the nucleus and cytoplasm of the epidermal cells at the point when the dorsal sheet of epidermal cells first begins migrating (Fig. 4). 4D movies of nhr-25::yfp embryos with images taken every 2.5 minutes allow us to follow the ventral migration of cells in the two ventral epidermal rows to enclose the embryo. Movies of nhr-25::yfp expression (uncompressed time-animated rotating projection QuickTime VR format) in wild-type and Gex embryos are available at: http://fsbill.vcell.uchc.edu/gloworm/Patel_et_al_2008_Data/
Gex embryos fail in ventral enclosure despite some protrusions
We fed nhr-25::yfp animals bacteria expressing double stranded RNA for wve-1, gex-2, and gex-3 or crossed the wve-1(zu469) and gex-3(zu196) mutations into the nhr-25::yfp strain. We analyzed the ventral enclosure defect and found that in Gex embryos the cells in the ventral-most epidermal rows maintain dynamic protrusions along the leading edge. These protrusions extend and retract continually for hours, with no associated migration of the cells (Fig. 4A, Supplemental Movie 1). The overall number of protrusions revealed by nhr-25::yfp is roughly similar in wild-type and Gex embryos with, for example, a 23% drop in wve-1(zu469) mutant embryos (Fig. 4E). The remaining protrusions may be due to redundancy between the WAVE/SCAR complex and WSP-1, another activator of Arp2/3 actin nucleation. However, removal of arp-2 by RNAi also blocked migration but not protrusion (Fig. 4A, Supplemental Movie 1). These results suggest that the WAVE/SCAR and Arp2/3 complexes are required for epidermal cell migrations, but are not required for the general ability of the ventral epidermal cells to extend membrane protrusions. Other actin assembly factors, like one or more of the six C. elegans formin homologs, may drive the remaining protrusions (Swan et al. 1998; Chhabra and Higgs, 2007). However, remaining factors are not sufficient to initiate cell migrations when Arp2/3 is missing.
Gex embryos have fewer and smaller protrusions in four cells essential for the migration
We measured the protrusiveness of the two ventral leading cells on each side, comparing them to the more posterior pocket cells, during a 40 minute period in the early part of the migration. We measured the leading edge perimeter of the ventral protrusions relative to the width of the cells as in Sheffield et al. 2007, except we focused on lateral views (Fig. 4B). We find that the extent of pocket cell protrusions does not change significantly during these time points. However, by 330 min. after first cleavage, the two leading cells on each side become twice as protrusive as the pocket cells. In contrast, the leading cells in Gex embryos do not show this increased protrusiveness (Fig. 4C). In addition, the rate of protrusions measured as protrusions/cell/minute relative to wild type drops specifically in the leading cells but not the pocket cells of Gex embryos (Fig. 4D). The leading cells are crucial for the ventral migration. They are enriched in actin and their ablation blocks ventral enclosure (Williams-Masson et al., 1997). Further analysis of the relative protrusiveness of the leading cells and the pocket cells using a transgene that expresses the filamentous actin-binding domain of VAB-10 in epidermal cells (plin-26::vab-10 ABD::gfp; Bosher et al. 2003; Diogon et al. 2007) supports this conclusion (Fig. 5A). Live imaging with this F-actin-binding transgene shows significantly increased protrusiveness in the leading cells compared to the pocket cells, but not when arp-2 is depleted (Fig. 5B). In addition, the amount of F-actin-binding protein accumulation at the leading edge of the leading cells drops by 50% in arp-2 mutants (Figure 5B). These results suggest that failure of ventral enclosure in Gex embryos is due to altered protrusive activity, associated with decreased filamentous actin accumulation, specifically in the four cells that lead the migration. Previous reports suggest that Arp2/3 is cell autonomously required in the epidermis (Sawa et al., 2003). Our results thus support that GEX proteins are required for these four cells to initiate cell migration.
Fig. 5. ARP-2 regulates actin dynamics at the leading edge of leading cells.
A. Live imaging with a filamentous actin-binding transgene.
A plin-26::vab-10 ABD (actin binding domain)::gfp transgene was used to investigate protrusion extension at the leading edge. This line is not integrated, so care was taken to analyze only those embryos that expressed the transgene evenly in the epidermis. Protrusion extension was analyzed over 25 minutes as described for Fig. 4B. Wild-type leading cells are up to 3.4 times as protrusive as the pocket cells when analyzed with this transgene. In contrast, the leading cells of arp-2(RNAi) embryos are only slightly more protrusive than the pocket cells.
B. Actin binding protein enrichment at the leading edge of leading cells requires ARP-2.
A plin-26::vab-10 ABD::gfp transgene was used to compare filamentous actin enrichment at the leading edge of wild-type and arp-2(RNAi) epidermal cells over a 20-minute period (while ventral enclosure is proceeding). Average fluorescence intensity within an equally sized region at the leading edge of the leading cells was compared. This transgene also illustrates changes in cell shape that occur in arp-2(RNAi) embryos.
WAVE/SCAR proteins and F-actin are enriched basolaterally in the epidermis and apically in the intestine during morphogenesis
If WAVE/SCAR complex proteins are regulating actin polymerization in migrating cells, their localization patterns may reflect their function. We therefore generated a WVE-1 GFP transgenic line that rescues the wve-1(zu469) mutation and monitored WVE-1 expression. GFP::WVE-1 is diffusely cytoplasmic and is enriched at cell boundaries in all cells, with particularly high expression in the larval nerve ring, which includes glial cells and axonal projections. This pattern is highly reminiscent of the localization pattern of GFP::GEX-3 (Fig. 6A, B). We focused on the subcellular localization of WAVE/SCAR proteins at the stage when we first detect severe morphogenesis defects in Gex embryos. At the bean stage we found that the rescuing GFP::WVE-1 and GFP::GEX-3 proteins are basolateral in epidermal cells, basal to the junctional component AJM-1 (Fig. 6A). We previously showed similar basolateral localization of GEX-2 in epidermal cells at the bean stage (Soto et al., 2002). Epidermal dorsal cells have been shown to extend basolateral membrane protrusions during dorsal intercalation (Williams-Masson et al., 1998; Heid et al., 2001), but the enrichment of F-actin in these cells has not been reported. Using phalloidin we find that epidermal cells at this stage contain strong accumulation of F-actin in basolateral regions, and much fainter accumulation in the apical regions (Fig. 6A). This shared enrichment of F-actin and WAVE/SCAR proteins in the basolateral domains of epidermal cells during dorsal intercalation coupled with the failure of WAVE/SCAR mutants to undergo this process suggests that WAVE/SCAR proteins are at the correct place and time to be contributing to F-actin recruitment, and that F-actin contributes to the basolateral protrusions that have been described.
We then examined the subcellular distribution of WAVE/SCAR proteins and F-actin in another tissue that shows apical/basal polarity, the intestine. At the bean stage, and later throughout development, internal organs show highest GFP-WVE-1 and GFP::GEX-3 enrichment in another epithelial domain, the region apical to the junctional component AJM-1 (Fig. 6A). This pattern of enrichment in internal organs also matches the pattern of F-actin. Like GEX proteins, F-actin is enriched at cell-cell junctions in early embryos and by the time tissue morphogenesis is underway it becomes enriched in the apical regions of the pharynx and intestine (Waddle et al., 1994; Leung et al., 1999). In the intestine Gex embryos display apical defects including an enlarged intestinal lumen (Fig. 2A). Three components of the WAVE/SCAR complex show polarized apical/basal distribution in epithelial cells and this enrichment temporally matches F-actin accumulation. Therefore, polarized membrane enrichment of WAVE/SCAR proteins may reflect where actin nucleation is most needed within a developing tissue.
GEX-2, GEX-3 and ABI-1 are required to maintain protein levels of WVE-1
When we removed gex-2, gex-3 and abi-1 by RNAi, we found that GFP::WVE-1 fluorescence levels were strongly reduced. Likewise, GFP::GEX-3 fluorescence was reduced when wve-1, gex-2 and abi-1 were removed (Fig. 6B). Since these studies suggested that other components of the complex are required to maintain the wild-type levels of WVE-1 and GEX-3, we tested this directly on our wild-type and gex-depleted lysates. Overall, the data suggest that all members of the complex are required for other members of the complex to be maintained at wild-type levels (Fig. 6D). We found that there is a drop of at least 50% in the levels of WVE-1 when embryos are depleted of gex-2, gex-3 or abi-1 and that loss of wve-1, gex-2, gex-3 or abi-1 leads to lower levels of the rest of the complex (Fig. 6D). Loss of GEX-3 leads to significantly lower levels of WVE-1, and GEX-2. Loss of GEX-2 leads to lower levels of WVE-1 and GEX-3. Depleting abi-1 lowers the levels of WVE-1, GEX-2 and GEX-3. Therefore the C. elegans WAVE/SCAR complex, like the WAVE/SCAR complex in Drosophila and Dictyostelium (Blagg et al., 2003; Rogers et al., 2003; Kunda et al., 2003; Ibarra et al., 2006) appears to be regulated coordinately at the level of protein abundance. Our in vivo results complement those Drosophila studies (Schenck et al. 2004; Qurashi et al. 2007) showing that in embryos all components of the WAVE/SCAR complex can regulate each other’s levels.
CED-10/Rac1, WAVE/SCAR and Arp2/3 proteins regulate apical F-actin enrichment in the intestine
Apical lumen expansion
The lumen of the intestine becomes increasingly expanded in Gex embryos as can be seen with DIC optics and by using the terminal web marker MH33 (Fig. 2A) (Francis and Waterston, 1985; Bossinger et al., 2004). In all Gex embryos other apical markers, including erm-1::gfp (Gobel et al. 2004), show an expanded domain around the expanded lumen (Fig. 7A). An expanded lumen may be an indirect effect of failure of epidermal enclosure. However, we noticed that embryos that do enclose, such as 25% of ced-10(tm597) null embryos that enclose and then die, still show an expanded lumen (Fig. 2A). To obtain enclosed Gex embryos we took advantage of the weak penetrance of RNAi when wve-1 dsRNA is fed to N2 worms and focused on embryos that enclosed before dying. We used an antibody to the apical junction protein AJM-1 to visualize two dimensions of the intestine. We measured the length of the entire intestine (between arrows, Fig. 7C) and also measured the lumen width (brackets, Fig. 7C). We found that wild-type embryos and enclosed wve-1(RNAi) embryos that reach the three-fold stage show no significant difference in intestinal length, yet consistently observed that the lumen width is enlarged at least 1.6-fold (Fig. 7C, n=10+). Therefore, a shortened intestine or failure to enclose does not explain the enlarged lumen. An alternative explanation is that defects in actin nucleation are altering lumen morphogenesis.
Loss of apical F-actin
Phalloidin recognizes all five species of filamentous actin found in C. elegans and in particular shows the dramatic accumulation of filamentous actin that is normally found at the apical regions of epithelial cells in the embryonic pharynx and intestine (Leung et al., 1999; Willis et al., 2006; Labouesse 2006) (Fig. 7A). The apical intestine contains both filamentous microvillar actin and basal to this, filamentous belt actin that is thought to associate with adherens junctions (Labouesse, 2006; Bossinger et al., 2004). We found that ced-10, wve-1, gex-2, gex-3 and arp-2 mutant and RNAi embryos are compromised in their enrichment of apical actin in the pharynx and intestine. In the intestine we saw at least a 30% drop in the enrichment of apical filamentous actin around the lumen as compared to wild type (Fig. 7A, B). The actin in the pharynx is highly disorganized in WAVE/SCAR and arp-2 mutant embryos, in contrast to the apically enriched filamentous actin seen in wild-type embryos (anterior arrow in Fig. 7A, second row). We conclude that apical enrichment of actin in these epithelial cells requires ced-10/Rac1, the WAVE/SCAR complex and the Arp2/3 complex.
Conclusion
In Dictyostelium, null mutations in GEX-2/PIR121 and in GEX-3/Nap1 have the same phenotype as loss of WAVE/Scar (Ibarra et al. 2006; Pollitt et al. 2006). In Drosophila, tissue culture experiments are at odds with genetic experiments regarding the nature of the active WAVE/Scar complex (Blagg & Insall, 2004). Our in vivo imaging studies reveal that the loss of any WAVE/SCAR component or ARP-2 leads to the same morphogenesis defects including fewer overall protrusions and smaller protrusions in the four cells that lead the epidermal migration. It has been suggested that differences among WAVE/SCAR complex components between organisms are due to difference in the rates of protein degradation or of cell movements or in the overall levels of WAVE/Scar (Blagg & Insall, 2004). We observe similar loss of functions phenotypes at different times in development and in different tissues and conclude that the WAVE/SCAR complex is spatially and temporally regulated in a similar fashion throughout C. elegans embryogenesis. In contrast to Drosophila myoblast fusion studies, we show that fusion of epidermal cells, by either EFF-1 or AFF-1 fusogens, does not require actin nucleation by Arp2/3. This may indicate that muscle cells have special requirements for cytoskeletal regulators, or that different cytoskeletal regulators regulate fusion in different cell types. Instead we find that polarized sub-cellular enrichment of F-actin requires the Rac1-WAVE/SCAR-Arp2/3 proteins. WAVE/SCAR proteins show apical enrichment in C. elegans intestinal cells that is required for apical F-actin enrichment (Fig. 6A, 7A,B). One explanation for the spectrum of functions described here is that Rac-WAVE/SCAR-Arp2/3 signaling is required to regulate the strength of adhesion at cell-cell contacts: between the tips of protrusive extensions of epidermal cells and the neuroblasts they migrate on, and between neighboring cells within a polarized epithelium.
Supplementary Material
Loss of ARP-2 by RNAi results in similar defects as loss of WAVE/SCAR components wve-1, gex-2 or gex-3. Staged embryos are shown from the earliest appearance of nhr-25::yfp expression until 180 minutes later in development. This corresponds to approximately 270 to 450 minutes after first cleavage. The nhr-25::yfp expression marks only the epidermal cells, not the underlying cells including the neuroblasts that the epidermal cells crawl over to enclose the embryo. Wild-type embryos accomplish most of the events of epidermal enclosure by 400 min. after first cleavage, but the epidermal cells in Gex mutant embryos arrest their migration and completely fail to enclose the embryo. However, the arrest is accompanied by on-going protrusions and retractions that last for at least five hours after the onset of nhr-25::yfp expression. We obtained similar results when we deplete gex gene products by RNAi or using genetic mutations.
Acknowledgments
We thank the NCRR-funded Caenorhabditis Genetics center, Erik Lundquist, Barth Grant, Chris Rongo, Jeremy Nance, Craig Hunter, Ryan Baugh, Verena Gobel for strains, the NICHD-funded University of Iowa Hybridoma Bank for the MH27, MH33 and MH46 antibodies, Kozo Kaibuchi and Hiroshi Qadota for antibodies to GEX-2 and GEX-3 and Jim Priess for the wve-1(zu469) allele. We thank Craig Mello and Daniel Brownell for supporting early stages of this project and Soto Lab undergraduates for mapping new gex genes: Veronica Zappi, Agnieska Zolnierowska, Holly Patel, Dipal Soni. We thank our colleagues for helpful suggestions on the manuscript: William Wadsworth, Peter Yurchenco, Don Winkelman, Barth Grant, Andy Singson, Elizabeth Ryder, Maureen Barr and Adam Steinberg. This research was funded by NIH Grant HD43156 to W.A.M and by grants from the American Heart Association (0435424T), the National Science Foundation (0641123) and the Robert Wood Johnson Medical School Foundation to M.C.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult development switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130(5):889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Baum PD, Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19(1):51–62. doi: 10.1016/s0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- Berger S, Schäfer G, Kesper DA, Holz A, Eriksson T, Palmer RH, Beck L, Klämbt C, Renkawitz-Pohl R, Onel SF. WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J Cell Sci. 2008;121(Pt 8):1303–1313. doi: 10.1242/jcs.022269. [DOI] [PubMed] [Google Scholar]
- Blagg SL, Insall RH. Solving the WAVE function. Nat Cell Biol. 2004;6(4):279–81. doi: 10.1038/ncb0404-279. [DOI] [PubMed] [Google Scholar]
- Blagg SL, Stewart M, Sambles C, Insall RH. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Current Biology. 2003;13:1480–1487. doi: 10.1016/s0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- Bosher JM, Hahn BS, Legouis R, Sookhareea S, Weimer RM, Gansmuller A, Chisholm AD, Rose AM, Bessereau JL, Labouesse M. The Caenorhabditis elegans vab-10 spectraplakin isoforms protect the epidermis against internal and external forces. J Cell Biol. 2003;161(4):757–68. doi: 10.1083/jcb.200302151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossinger O, Fukushige T, Claeys M, Borgonie G, McGhee JD. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev Biol. 2004;268(2):448–456. doi: 10.1016/j.ydbio.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascon A, Escobar B, Montero-Conde C, Rodriguez-Antona C, Ruiz-Llorente S, Osorio A, Mercadillo F, Leton R, Campos JM, Garcia-Sagredo JM, Benitez J, Malumbres M, Robledo M. Loss of the actin regulator HSPC300 results in clear cell renal cell carcinoma protection in Von Hippel-Lindau patients. Hum Mutat. 2007;28(6):613–621. doi: 10.1002/humu.20496. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Revealing the Mechanisms of Cell-Cell Fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9(10):1110–21. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, Chisholm AD. Form of the worm: genetics of epidermal morphogenesis in C. elegans. Trends Genet. 2000;16(12):544–51. doi: 10.1016/s0168-9525(00)02143-0. [DOI] [PubMed] [Google Scholar]
- Costa M, Draper BW, Priess JR. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev Biol. 1997;184:373–384. doi: 10.1006/dbio.1997.8530. [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogon M, Wissler F, Quintin S, Nagamatsu Y, Sookhareea S, Landmann F, Hutter H, Vitale N, Labouesse M. The RhoGAP RGA-2 and LET-502/ROCK achieve a balance of actomyosin-dependent forces in C. elegans epidermis to control morphogenesis. Development. 2007;134(13):2469–79. doi: 10.1242/dev.005074. [DOI] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418(6899):790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Firestein BL, Rongo C. DLG-1 is a MAGUK similar to SAP97 and is required for adherens junction formation. Mol Biol Cell. 2001;12(11):3465–3475. doi: 10.1091/mbc.12.11.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101(4):1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114(3):465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Cartwright HN, Smith LG. Three Brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development. 2003;130(4):753–762. doi: 10.1242/dev.00290. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408(6810):325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gobel V, Barrett PL, Hall DH, Fleming JT. Lumen morphogenesis in C. elegans requires the membrane-cytoskeleton linker erm-1. Dev Cell. 2004;6(6):865–873. doi: 10.1016/j.devcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Heid PJ, Raich WB, Smith R, Mohler WA, Simokat K, Gendreau SB, Rothman JH, Hardin J. The zinc finger protein DIE-1 is required for late events during epithelial cell rearrangement in C. elegans. Dev Biol. 2001;236(1):165–180. doi: 10.1006/dbio.2001.0315. [DOI] [PubMed] [Google Scholar]
- Hresko MC, Schriefer LA, Shrimankar P, Waterston RH. Myotactin, a novel hypodermal protein involved in muscle-cell adhesion in Caenorhabditis elegans. J Cell Biol. 1999;146(3):659–672. doi: 10.1083/jcb.146.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra N, Blagg SL, Vazquez F, Insall RH. Nap1 regulates Dictyostelium cell motility and adhesion through SCAR-dependent and -independent pathways. Curr Biol. 2006;16(7):717–22. doi: 10.1016/j.cub.2006.02.068. [DOI] [PubMed] [Google Scholar]
- Kim S, Shilagardi K, Zhang S, Hong SN, Sens KL, Bo J, Gonzalez GA, Chen EH. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev Cell. 2007;12(4):571–586. doi: 10.1016/j.devcel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3(11):983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Current Biology. 2003;13(21):1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Labouesse M. Epithelial junctions and attachments. WormBook, The C. elegans Research Community, WormBook. 2006 doi: 10.1895/wormbook.1.56.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Legouis R, Gansmuller A, Sookhareea S, Bosher JM, Baillie DL, Labouesse M. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat Cell Biol. 2000;2:415–422. doi: 10.1038/35017046. [DOI] [PubMed] [Google Scholar]
- Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216(1):114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- Massarwa R, Carmon S, Shilo BZ, Schejter ED. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev Cell. 2007;12(4):557–569. doi: 10.1016/j.devcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- McMahon L, Legouis R, Vonesch JL, Labouesse M. Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J Cell Sci. 2001;114(12):2265–2277. doi: 10.1242/jcs.114.12.2265. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, Scranton V, Assaf N, White JG, Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2(3):355–62. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B. Cell fusion WormBook. The C. elegans Research Community, WormBook. 2006 doi: 10.1895/wormbook.1.52.1. http://www.wormbook.org. [DOI]
- Podbilewicz B, White JG. Cell fusions in the developing epithelial of C. elegans. Dev Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollitt AY, Blagg SL, Ibarra N, Insall RH. Cell motility and SCAR localisation in axenically growing Dictyostelium cells. Eur J Cell Biol. 2006;85(9–10):1091–8. doi: 10.1016/j.ejcb.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Priess JR, Hirsh DI. Caenorhabditis elegans morphogenesis: The role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- Qurashi A, Sahin HB, Carrera P, Gautreau A, Schenck A, Giangrande A. HSPC300 and its role in neuronal connectivity. Neural Develop. 2007;2:18. doi: 10.1186/1749-8104-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Nowak SC, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134(24):4357–67. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162(6):1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir A, Choi J, Leikina E, Avinoam O, Valansi C, Chernomordik LV, Newman AP, Podbilewicz B. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev Cell. 2007;12(5):683–98. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Suetsugu S, Sugimoto A, Miki H, Yamamoto M, Takenawa T. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J Cell Sci. 2003;116(8):1505–1518. doi: 10.1242/jcs.00362. [DOI] [PubMed] [Google Scholar]
- Schenck A, Qurashi A, Carrera P, Bardoni B, Diebold C, Schejter E, Mandel JL, Giangrande A. WAVE/SCAR, a multifunctional complex coordinating different aspects of neuronal connectivity. Dev Biol. 2004;274(2):260–70. doi: 10.1016/j.ydbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Schroter RH, Lier S, Holz A, Bogdan S, Klambt C, Beck L, Renkawitz-Pohl R. kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development. 2004;131(18):4501–4509. doi: 10.1242/dev.01309. [DOI] [PubMed] [Google Scholar]
- Schafer G, Weber S, Holz A, Bogdan S, Schumacher S, Muller A, Renkawitz-Pohl R, Onel SF. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev Biol. 2007;304(2):664–674. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and Profilin are required for Cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Current Biology. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- Shakir MA, Jiang K, Struckhoff EC, Demarco RS, Patel F, Soto MC, Lundquist EA. The Arp2/3 activators WAVE and WASP have distinct genetic interactions with Rac GTPases in C. elegans axon guidance. Genetics. 2008;179(4):1957–71. doi: 10.1534/genetics.108.088963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield M, Loveless T, Hardin J, Pettitt J. C. elegans Enabled exhibits novel interactions with N-WASP, Abl, and cell-cell junctions. Curr Biol. 2007;17(20):1791–6. doi: 10.1016/j.cub.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1(1):E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simske JS, Hardin J. Getting into shape: epidermal morphogenesis in Caenorhabditis elegans embryos. Bioessays. 2001;23(1):12–23. doi: 10.1002/1521-1878(200101)23:1<12::AID-BIES1003>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Simske JS, Koppen M, Sims P, Hodgkin J, Yonkof A, Hardin J. The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nat Cell Biol. 2003;5(7):619–25. doi: 10.1038/ncb1002. [DOI] [PubMed] [Google Scholar]
- Soto MC, Qadota H, Kasuya K, Inoue M, Tsuboi D, Mello CC, Kaibuchi K. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 2002;16(5):620–32. doi: 10.1101/gad.955702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal TE, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14(6):303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Swan KA, Severson AF, Carter JC, Martin PR, Schnabel H, Schnabel R, Bowerman B. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J Cell Sci. 1998;111(Pt 14):2017–27. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci. 2001;114(10):1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- Totong R, Achilleos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134(7):1259–1268. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- Waddle JA, Cooper JA, Waterston RH. Transient localized accumulation of actin in Caenorhabditis elegans blastomeres with oriented asymmetric divisions. Development. 1994;120(8):2317–2328. doi: 10.1242/dev.120.8.2317. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124(4):475–90. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Masson EM, Heid PJ, Lavin CA, Hardin J. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol. 1998;204:263–276. doi: 10.1006/dbio.1998.9048. [DOI] [PubMed] [Google Scholar]
- Williams-Masson EM, Malik AN, Hardin J. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development. 1997;124(15):2889–2901. doi: 10.1242/dev.124.15.2889. [DOI] [PubMed] [Google Scholar]
- Willis JH, Munro E, Lyczak R, Bowerman B. Conditional dominant mutations in the Caenorhabditis elegans gene act-2 identify cytoplasmic and muscle roles for a redundant actin isoform. Mol Biol Cell. 2006;17(3):1051–64. doi: 10.1091/mbc.E05-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withee J, Galligan B, Hawkins N, Garriga G. Caenorhabditis elegans WASP and Ena/VASP proteins play compensatory roles in morphogenesis and neuronal cell migration. Genetics. 2004;167(3):1165–1176. doi: 10.1534/genetics.103.025676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Loss of ARP-2 by RNAi results in similar defects as loss of WAVE/SCAR components wve-1, gex-2 or gex-3. Staged embryos are shown from the earliest appearance of nhr-25::yfp expression until 180 minutes later in development. This corresponds to approximately 270 to 450 minutes after first cleavage. The nhr-25::yfp expression marks only the epidermal cells, not the underlying cells including the neuroblasts that the epidermal cells crawl over to enclose the embryo. Wild-type embryos accomplish most of the events of epidermal enclosure by 400 min. after first cleavage, but the epidermal cells in Gex mutant embryos arrest their migration and completely fail to enclose the embryo. However, the arrest is accompanied by on-going protrusions and retractions that last for at least five hours after the onset of nhr-25::yfp expression. We obtained similar results when we deplete gex gene products by RNAi or using genetic mutations.