Abstract
Background
While genetic variation at chromosome 9p21.3 is associated with incident cardiovascular disease (CVD), it is unclear whether screening for this polymorphism improves risk prediction.
Objective
To determine whether knowledge of variation at chromosome 9p21.3 provides predictive information beyond that from other readily available risk factors.
Design
Prospective cohort study
Setting
United States
Patients
22,129 female Caucasian health professionals participating in the Women's Genome Health Study, initially free of any major chronic disease, who were prospectively followed over a median of 10.2 years for incident CVD.
Measurements
Polymorphism at rs10757274 in chromosome 9p21.3 and additional CVD risk factors (blood pressure, smoking status, diabetes, blood levels of cholesterol, high-sensitivity C-reactive protein, and family history of premature myocardial infarction).
Results
Polymorphism at rs10757274 was associated with an adjusted hazard ratio for incident CVD of 1.25 (95% CI 1.04 − 1.51) for the AG genotype and 1.32 (95% CI 1.07 − 1.63) for the GG genotype. However, the addition of the genotype to a prediction model based on traditional risk factors, high-sensitivity C-reactive protein and family history of premature myocardial infarction had no effect on model discrimination as measured by the c-index (0.807 to 0.809) nor showed improvement of the Net Reclassification Improvement (−0.2%, p=0.59) or the Integrated Discrimination Improvement (0.0, p=0.18).
Limitations
Results are limited to Caucasian women.
Conclusions
In this large prospective cohort of Caucasian women, genetic variation in chromosome 9p21.3 was associated with incident CVD but did not improve on the discrimination or classification of predicted risk achieved with traditional risk factors, high-sensitivity C-reactive protein and family history of premature myocardial infarction.
Introduction
The success of risk prediction models for cardiovascular disease (CVD) reflects an increasing understanding of the molecular basis of atherothrombosis. Aside from age, which integrates many biological activities and environmental exposures at once, other important components of risk prediction include plasma biomarkers for lipid metabolism, inflammation, thrombosis, and metabolic status.(1) Current prediction models, however, remain unable to anticipate many cases of incident CVD, thus motivating imperatives for identifying new risk factors and optimizing analytic methods for their use. Traditionally aggregated by the notion of family history, individual genetic variants may represent a class of risk factors with new prognostic information; they may be more distal to underlying disease processes than plasma risk factors or other clinical variables, but they may also be more general. For example, a recent exploration of the influence of lipid associated variation on prediction of incident CVD found residual predictive value for the genetic variation after adjustment for plasma lipid levels.(2).
Genetic variation at the chromosome 9p21.3 region is a good candidate for adding additional information to risk prediction. Variation at this locus has consistently been found to have an association with coronary artery disease(3-5) as well as diabetes.(6-9) Additionally the risk allele is carried by almost 75% of the Caucasian population, and the lack of correlation between any of the disease-associated 9p21.3 genetic variants and major cardiovascular risk factors suggests novel influences on disease progression. To assess whether knowledge of variation at 9p21.3 improves global risk prediction, we examined the effect of adding genetic information from a single nucleotide polymorphism (SNP) in the 9p21.3 region to previously published prediction models in the Women's Health Study, a large, prospective cohort of initially healthy American women followed over a 10 year period for incident cardiovascular events.
Methods
Study Population
Study participants were members of the Women's Genome Health Study (WGHS),(10) an ongoing prospective genetic evaluation study being conducted among initially healthy American women who enrolled in the Women's Health Study (WHS), which was a trial of aspirin and vitamin E for the primary prevention of cardiovascular disease and cancer in women aged 45 years or older.(11) The WHS recruited U.S. female health professionals, beginning in 1992, who were free of any major chronic disease including cancer and CVD at baseline and then were followed prospectively for incident myocardial infarction, stroke, coronary revascularization, and cardiovascular death. The study was approved by the institutional review board of the Brigham and Women's Hospital (Boston, Massachusetts).
Among the WHS participants, 28,345 provided blood samples that were stored in liquid nitrogen until the time of analysis, as well as consent for ongoing analyses linking blood-derived observations with baseline risk factor profiles and incident disease events. Of these women, 23,226 had standard risk factor information available and were genotyped for the rs10757274 polymorphism. To reduce the potential for population stratification to impact upon our results, this analysis included only the 22,129 (95.3%) women of Caucasian ancestry
Risk Factor Ascertainment
Baseline information on age, diabetes, smoking status, parental history of myocardial infarction (MI) before 60 years, blood pressure, and hypertension treatment were collected at study initiation. Plasma biomarkers were analyzed in a core laboratory facility, certified by the National Heart, Lung and Blood Institute/Centers for Disease Control and Prevention Lipid Standardization Program, for total cholesterol, high density lipoprotein cholesterol, apolipoprotein B 100, apolipoprotein A-I, high-sensitivity C-reactive protein (hsCRP), lipoprotein(a), and hemoglobin A1C.
Genotypes for rs10757274 in the WGHS participants were determined by an oligonucleotide ligation procedure that combined polymerase chain reaction amplification of target sequences from 3ng of genomic DNA with subsequent allele-specific oligonucleotide ligation as previously described.(12) The ligation products of the two alleles were separated by hybridization to product-specific oligonucleotides, each coupled to spectrally distinct Luminex100 ×MAP microspheres (Luminex, Austin, TX). The captured products were fluorescently labeled with streptavidin R-phycoerythrin (Prozyme, San Leandro, CA), sorted on the basis of microsphere spectrum, and detected by a Luminex100 instrument.(12)
Outcome Ascertainment
Study participants were followed through March 2004 for total CVD, which was comprised of incident MI, ischemic stroke, coronary revascularization, and cardiovascular deaths. Events were adjudicated by an end-points committee using medical record review. Morbidity data were available on nearly all the women through 8 years of follow-up
Risk Prediction Models
In order to assess the effect of variation at rs10757274 on global CVD risk prediction, covariates from two non-genetic risk prediction models were considered. The first included the covariates from the Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (ATP III) risk score as well as history of diabetes (noted as a high-risk equivalent).(13) The second used the covariates from the Reynolds Risk Score, a previously published model which includes additional biomarker information as well as data on family history.(1) To provide direct comparability and the highest level of internal validity, we elected on an a priori basis to model all covariates, including rs10757274, in the same base population, rather than using beta coefficients for the non-genetic covariates derived from previously published models. By so doing, we avoid the potential for bias that might occur by modeling the effect of the genetic data within the test cohort but using estimates of effect for the other covariates from a second unrelated cohort.
Statistical Analysis
Cox proportional hazards models were used to generate crude and adjusted hazard ratios across genotypes and to test for trend. Models with separate effects for each genotype were used for all analyses. Adjusted survival curves were generated by stratifying Cox proportional hazards models by genotype. Cox models were also used to generate estimates of predicted risk with and without genotype information, which were then assessed for accuracy. No evidence for departures from proportionality was seen for any of the models used. The primary measure of discrimination used was Harrell's c-index,(14) a generalization of the area under the receiver operator characteristic curve that allows for censored data. The c-index assesses the ability of the risk score to rank women who develop incident CVD higher than women who do not. General calibration was assessed across deciles of predicted risk using the Hosmer-Lemeshow goodness-of-fit test (15) to compare the average predicted risk to the Kaplan-Meier risk estimate within each decile, with a chi-square value of 20 or higher (p < 0.01) considered poor calibration.(16)
Risk reclassification (1, 17) was assessed by categorizing the predicted 10-year risk for each model into categories of less than 5%, 5% to less than 10%, 10% to less than 20%, and 20% or higher. We then compared the assigned categories for a pair of models. For each pair, we calculated the proportion of participants who were reclassified by the comparison model as compared to the reference model. Reclassification was considered correct if the Kaplan-Meier risk estimate for the reclassified group was closer to the comparison category than the reference. We computed the Hosmer-Lemeshow statistic for the reclassification tables, (18) which assesses agreement between the Kaplan-Meier risk estimate and predicted risk within the reclassified categories. We also computed the Net Reclassification Improvement, (19) which compares the shifts in reclassified categories by observed outcome, and the Integrated Discrimination Improvement, (19) which directly compares the average difference in predicted risk for women who go on to develop CVD with women who do not for the two models, on the women who were not censored prior to 8 years.
Role of the Funding Sources
This study was supported by grants from the National Institutes of Health, the Donald W Reynolds Foundation and the Leducq Foundation. Genotyping of the 9p21.3 variant was performed by Celera, Alameda, CA. The funding agencies had no role in the design, conduct or reporting of this study or in the decision to submit the manuscript for publication.
Results
Of the 22,129 Caucasian women genotyped for this analysis, 5793 (26.2%) had no risk (G) alleles at rs10757374, 10952 (49.5%) had one risk allele, and 5384 (24.3%) had two risk alleles. The number of risk alleles had a significant association with family history of premature MI (p for trend < 0.001) and a modest association with a history of diabetes, driven by the higher proportion of diabetics in those with two risk alleles (p for trend = 0.04) (Table 1). No association was seen for the other standard CVD risk factors, nor for the newer biomarkers. Additionally, there was no association between genotype and other lipid measures, including lipoprotein (a), apolipoprotein A-I and apolipoprotein B100 (data not shown).
Table 1.
Baseline Characteristics by rs10757274 Genotype*
| rs10757274 Genotype | ||||
|---|---|---|---|---|
|
AA (n=5793) |
AG (n=10952) |
GG (n=5384) |
P for trend | |
| Age (years) | 53 (48 − 59) | 52 (48 − 58) | 52 (48 − 59) | 0.095 |
| Systolic Blood Pressure (mm Hg) | 125 (115 − 135) | 125 (115 − 135) | 125 (115 − 135) | 0.77 |
| Total Cholesterol | ||||
| (mmol\L) | 5.38 (4.76 − 6.10) | 5.38 (4.73 − 6.08) | 5.38 (4.76 − 6.08) | 0.81 |
| (mg\dL) | 208 (184 − 236) | 208 (183 − 235) | 208 (184 − 235) | |
| High Density Lipoprotein | ||||
| Cholesterol | ||||
| (mmol\L) | 1.35 (1.12 − 1.62) | 1.34 (1.12 − 1.61) | 1.33 (1.10 − 1.61) | 0.166 |
| (mg\dL) | 52.2 (43.4 − 62.8) | 51.9 (43.3 − 62.3) | 51.6 (42.7 − 62.4) | |
| hsCRP (mg\dL) | 1.99 (0.79 − 4.30) | 2.02 (0.81 − 4.36) | 2.00 (0.77 − 4.31) | 0.98† |
| Current Smoker | 11.4 (663) | 11.1 (1221) | 12.2 (656) | 0.24 |
| Antihypertensive Use | 12.5 (722) | 12.1 (1324) | 11.9 (643) | 0.40 |
| Family History of MI | 11.7 (677) | 13.1 (1431) | 13.9 (751) | <0.001 |
| History of Diabetes | 2.5 (142) | 2.4 (258) | 3.1 (166) | 0.039 |
| HbA1c if Diabetic | 7.0 (6.1 − 8.9) | 6.9 (5.8 − 8.3) | 7.0 (5.9 − 8.2) | 0.26 |
hsCRP=high sensitivity C-reactive protein, MI=myocardial infarction, HbA1c= hemoglobin A1c
Median (25th percentile – 75th percentile) for continuous characteristics, percent answering yes (number) for categorical characteristics
natural log used for linear regression trend test
The crude association between rs10757274 genotype and total CVD was not affected by adjustment for age or additional risk factors (Table 2) and the cardiovascular hazard associated with increasing number of risk alleles was consistent across all components of the primary study endpoint. Specifically, polymorphism at rs10757274 was associated with an adjusted HR with incident CVD of 1.25 (95% CI 1.04 − 1.51) for the AG genotype and 1.32 (95% CI 1.07 − 1.63) for the GG genotype. Similar associations, though with less power, were seen for coronary heart disease (AG: HR 1.28, 95%CI 1.02 − 1.62; GG: HR 1.30, 95% CI 1.00 − 1.69), for myocardial infarction (MI) (AG: HR 1.33, 95%CI 0.93 − 1.91; GG: HR 1.28, 95% CI 0.85 − 1.92), and for stroke (AG: HR 1.33, 95%CI 0.96 − 1.85; GG: HR 1.54, 95% CI 1.07 − 2.21). The stratified mean survival curves (Figure) show attenuation in the absolute risk from the use of the mean covariate values in the adjusted models, but no change in the relative risk between groups.
Table 2.
Adjusted Relative Hazards and 95 % Confidence Intervals by rs10757274 Genotype
| rs10757274 Genotype | ||||
|---|---|---|---|---|
|
AA (n=5793) |
AG (n=10952) |
GG (n=5384) |
Additive Model | |
| Total CVD (n) | 158 | 362 | 195 | |
| Crude | 1.0 | 1.22 (1.01 − 1.46) | 1.33 (1.08 − 1.65) | 1.15 (1.04 − 1.28) |
| Age | 1.0 | 1.25 (1.03 − 1.50) | 1.38 (1.12 − 1.70) | 1.17 (1.06 − 1.30) |
| ATPIII Covariates† | 1.0 | 1.25 (1.04 − 1.51) | 1.32 (1.07 − 1.63) | 1.15 (1.03 − 1.27) |
| CHD* (n) | 102 | 241 | 126 | |
| Crude | 1.0 | 1.25 (0.99 − 1.58) | 1.33 (1.03 − 1.73) | 1.15 (1.01 − 1.31) |
| Age | 1.0 | 1.28 (1.02 − 1.61) | 1.36 (1.05 − 1.77) | 1.16 (1.02 − 1.32) |
| ATPIII Covariates† | 1.0 | 1.28 (1.02 − 1.62) | 1.30 (1.00 − 1.69) | 1.13 (1.00 − 1.29) |
| MI (n) | 42 | 103 | 51 | |
| Crude | 1.0 | 1.30 (0.91 − 1.86) | 1.31 (0.87 − 1.97) | 1.14 (0.93 − 1.38) |
| Age | 1.0 | 1.33 (0.93 − 1.91) | 1.34 (0.89 − 2.02) | 1.15 (0.95 − 1.40) |
| ATPIII Covariates† | 1.0 | 1.33 (0.93 − 1.91) | 1.28 (0.85 − 1.92) | 1.12 (0.92 − 1.36) |
| Stroke (n) | 50 | 123 | 72 | |
| Crude | 1.0 | 1.30 (0.94 − 1.81) | 1.55 (1.08 − 2.23) | 1.24 (1.04 − 1.48) |
| Age | 1.0 | 1.34 (0.97 − 1.86) | 1.61 (1.12 − 2.31) | 1.26 (1.06 − 1.50) |
| ATPIII Covariates† | 1.0 | 1.33 (0.96 − 1.85) | 1.54 (1.07 − 2.21) | 1.23 (1.04 − 1.47) |
CVD=cardiovascular disease, CHD=coronary heart disease, MI=myocardial infarction
CHD is comprised of MI, coronary revascularization, and CHD deaths.
ATP III covariates include: natural log of age, systolic blood pressure, total and high density lipoprotein cholesterol, and smoking status, antihypertensive use and history of diabetes.
Figure.
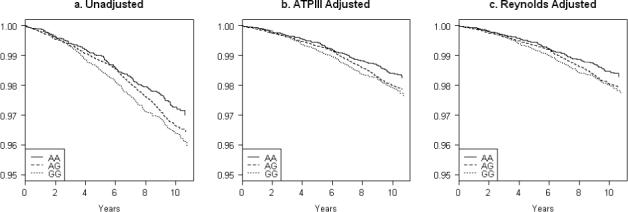
Stratified Survival Curves by rs10757274 Genotype
Knowledge of genotype had little effect on the coefficients for the other risk factors for either the ATP III or Reynolds Risk Score covariates (Table 3). As also shown in Table 3, the addition of genotype information to either the ATP III covariates or the Reynolds Risk Score covariates had essentially no effect on discrimination (c-index 0.803 to 0.805 and 0.807 to 0.809, respectively). All models were calibrated as measured by the Hosmer-Lemeshow chi-square.
Table 3.
Beta-Coefficients, Calibration and Discrimination for Cardiovascular Risk Prediction Cox Models with and without rs10757274 Genotype (N= 21644)
| Predictors | ATP III Covariates | ATP III Covariates Plus Genotype | Reynolds Risk Score Covariates | Reynolds Risk Score Covariates Plus Genotype | ||||
|---|---|---|---|---|---|---|---|---|
| Beta (SE) | P value | Beta (SE) | P value | Beta (SE) | P value | Beta (SE) | P value | |
| Age* (years) | 4.092 (0.287) | <0.001 | 4.108 (0.287) | <0.001 | 0.074 (0.005) | <0.001 | 0.074 (0.005) | <0.001 |
| Systolic Blood Pressure† (mm Hg) | 3.578 (0.381) | <0.001 | 3.569 (0.381) | <0.001 | 3.653 (0.353) | <0.001 | 3.648 (0.353) | <0.001 |
| Total Cholesterol† (mmol/L or mg\dL) | 1.174 (0.198) | <0.001 | 1.173 (0.198) | <0.001 | 0.997 (0.200) | <0.001 | 0.996 (0.200) | <0.001 |
| High Density Lipoprotein Cholesterol† (mmol/L or mg\dL) | −1.114 (0.143) | <0.001 | −1.117 (0.143) | <0.001 | −0.978 (0.145) | <0.001 | −0.979 (0.145) | <0.001 |
| Current Smoker | 0.888 (0.091) | <0.001 | 0.887 (0.091) | <0.001 | 0.880 (0.092) | <0.001 | 0.876 (0.092) | <0.001 |
| Antihypertensive Use | 0.243 (0.093) | 0.009 | 0.243 (0.093) | 0.009 | - | - | - | - |
| History of Diabetes | 1.340 (0.110) | <0.001 | 1.335 (0.110) | <0.001 | - | - | - | - |
| HbAlc if Diabetic | - | - | - | - | 0.163 (0.014) | <0.001 | 0.163 (0.014) | <0.001 |
| hsCRP‡ (mg\dL) | - | - | - | - | 0.181 (0.036) | <0.001 | 0.180 (0.036) | <0.001 |
| Family History of MI | - | - | - | - | 0.423 (0.100) | <0.001 | 0.415 (0.101) | <0.001 |
| rs10757274 genotype AG | 0.223 (0.095) | 0.019 | 0.222 (0.095) | 0.020 | ||||
| rs10757274 genotype GG | - | - | 0.280 (0.107) | 0.009 | - | - | 0.274 (0.107) | 0.011 |
| Harrell's C-Index§ (SE) | 0.803 (0.019) | 0.805 (0.019) | 0.807 (0.019) | 0.809 (0.019) | ||||
| Hosmer-Lemeshow Chi-square∥ (p-value) | 6.24 (0.62) | 5.96 (0.65) | 7.75 (0.46) | 7.43 (0.49) | ||||
hsCRP=high-sensitivity C-reactive protein, MI=myocardial infarction, HbA1c= Hemoglobin A1c
The natural logarithm was used for the ATP II models only
The natural logarithm was used for all models
The natural logarithm was used for the Reynolds models only
Harrell's c-index measures model discrimination
Hosmer-Lemeshow chi-square measures the calibration of the model; chi-square greater than 20 suggests lack of calibration.
When reclassification was examined, a modest improvement was seen with the addition of genotype to the ATP III covariates (Table 4). The addition of genotype information to the ATP III covariates reclassified 2.7% (606) of the women. Of those, 86.9% (526) were reclassified correctly, corresponding to a Net Reclassification Improvement of 2.7% (p = 0.02) and an Integrated Discrimination Improvement of 0.001 (p = 0.11). Both the original ATP III covariates and the ATP III covariates with the genotype information remained calibrated within the reclassification table (reclassification Hosmer-Lemeshow chi-square 17.9 and 14.9 respectively).
Table 4.
Reclassification of Predicted 10-Year Cardiovascular Disease Risk Categories with and without rs10757274 Genotype*
| a) | ||||||
|---|---|---|---|---|---|---|
| ATP III Covariates†Plus Genotype - Categories of Predicted 10 Year Risk | % Reclassified Correctly / % Reclassified | |||||
| ATP III Covariates† - Categories of Predicted 10 Year Risk | <5% | 5% to <10% | 10% to <20% | >20% | ||
| N | 18609 | 205 | ||||
| <5% | % Reclassified | - | 1.1% | - | - | 1.1 / 1.1 |
| K-M Estimate | 1.5% | 8.0% | ||||
| N | 181 | 1933 | 83 | |||
| 5% to <10% | % Reclassified | 8.2% | - | 3.8% | - | 12.0 / 12.0 |
| K-M Estimate | 4.9% | 8.0% | 19.3% | |||
| N | 80 | 697 | 31 | |||
| 10% to <20% | % Reclassified | - | 9.9% | - | 3.8% | 3.8 / 13.7 |
| K-M Estimate | 10.9% | 12.9% | 23.6% | |||
| N | 26 | 284 | ||||
| >20% | % Reclassified | - | - | 8.4% | - | 8.4 / 8.4 |
| K-M Estimate | 15.0% | 31.0% | ||||
| b) | ||||||
|---|---|---|---|---|---|---|
| Reynolds Covariates‡Plus Genotype - Categories of Predicted 10-Year Risk | % Reclassified Correctly / % Reclassified | |||||
| Reynolds Covariates‡- Categories of Predicted 10-Year Risk | <5% | 5% to <10% | 10% to <20% | >20% | ||
| N | 18527 | 188 | ||||
| <5% | % Reclassified | - | 1% | - | - | 0.0 / 1.0 |
| K-M Estimate | 1.5% | 2.7% | ||||
| N | 183 | 1960 | 75 | |||
| 5% to <10% | % Reclassified | 8.3% | - | 3.4% | - | 8.3 / 11.6 |
| K-M Estimate | 1.4% | 7.7% | 8.3% | |||
| N | 85 | 761 | 31 | |||
| 10% to <20% | % Reclassified | - | 9.7% | - | 3.5% | 3.5 / 13.2 |
| K-M Estimate | 10.6% | 15.2% | 21.4% | |||
| N | 23 | 296 | ||||
| >20% | % Reclassified | - | - | 7.2% | - | 0 / 7.2 |
| K-M Estimate | 31.5% | 30.4% | ||||
Each cell lists total number of people, percent of row reclassified, and Kaplan-Meier estimated event rate for the cell
ATP III covariates include: natural log of age, systolic blood pressure, total and high density lipoprotein cholesterol, and smoking status, antihypertensive use and history of diabetes.
Reynolds covariates include: age, smoking status, family history of myocardial infarction, hemoglobin A1c levels if diabetic and the natural log of systolic blood pressure, total and high density lipoprotein
By contrast, when genotype information was added to the Reynolds Risk Score covariates, 2.6% (585) of the women were reclassified. However, of those, only 36.6 % (214) were reclassified correctly, corresponding to a Net Reclassification Improvement of −0.2% (p = 0.59) and an Integrated Discrimination Improvement of 0.0 (p = 0.18). Both the original Reynolds Risk Score covariates and the Reynolds Risk Score covariates with the genotype information remained calibrated within the reclassification table (reclassification Hosmer-Lemeshow chi-square 12.6 and 12.7 respectively). Virtually identical results were observed in analyses adding genotype information to the published Reynolds Risk Score, rather than the Reynolds Risk Score covariates.
Discussion
Using recently developed prediction models and performance metrics, we have examined the incremental contribution of 9p21.3 variation to cardiovascular risk among more than 22,000 initially healthy Caucasian participants in the Women's Genome Health Study. Like a previous report examining this same variation among Caucasian men, (20) we find a strong and significant association of 9p21.3 variation with CVD in Caucasian women. However, in our data, knowledge of this genetic variation only marginally improved the classification of risk prediction in a model based on ATP-III covariates and did not improve classification in a model that included family history and CRP (Reynolds Risk Score covariates). In both settings, the addition of genotype information had no appreciable effect on the c-index
The rs10757274 SNP is one of a cluster of tightly linked SNPs associated with coronary artery disease, (3-5) MI, (3, 6) and stroke, (21) abdominal aortic aneurysm,(6) and intracranial aneurysm (6) at 9p21.3. The linkage disequilibrium block spanning these SNPs extends 28kb from rs10757274 toward the telomere about 55.7kb from the 3’ end of the CDKN2B transcript, although weaker linkage disequilibrium continues through the CDKN2A/CDKN2B gene region. The major transcript of a third gene, MTAP, corresponds with an entirely separate linkage disequilibrium block still further toward the telomere, but an alternatively spliced variant may be transcribed into the linkage disequilibrium block containing CDKN2A/CDKN2B. Toward the centromere, the linkage disequilibrium block including the associations with the arterial diseases extends 28.3kb bases from rs10757274 and is juxtaposed with a smaller linkage disequilibrium block of about 6.9kb including a SNP associated with diabetes (rs10822661).(6-9)
While the mechanism of the association between the rs10757274 genotype and CVD remains unclear, the magnitude of association observed in our study of women is consistent with previously reported associations for 9p21.3 SNPs in studies largely comprised of men.(5) Our data also extend prior work demonstrating significant association with incident stroke.(21) As we were unable to find correlations between polymorphisms in rs10757274 and standard lipids, apolipoprotein B 100, apolipoprotein A-I, lipoprotein(a), or hsCRP, our data provide further evidence that the effect of this genetic variation is unlikely to reflect genetic differences in lipid levels or biomarkers of hemostasis and inflammation.
Despite high prevalence of the risk allele and consistent association of rs10757274 with vascular events, inclusion of genotype information did not lead to any substantive improvement in global risk prediction in our study. Instead, the addition of genotype information to a model which included CRP and family history of CVD actually worsened classification. Thus, while knowledge of variation in chromosome 9p21.3 may yield important pathophysiologic insights into atherothrombosis, it appears unlikely that screening for SNPs in this region will have clinical utility for daily practice.
Improved cardiovascular risk prediction has clear potential for public health, but has been difficult to achieve, and the potential role of genetic information is just beginning to be explored. Whether or not a precisely measured gene panel will prove superior to knowledge of family history of CVD is uncertain, though combination scores have had promising effects on prediction.(2) As shown in this paper, and as expected from statistical work by Pepe (22) and Cook,(17) single polymorphisms alone, even common ones with consistent modest associations, are unlikely to improve prediction.
Our study benefits from a large sample size and extensive prospective follow-up. However, because of the small numbers of non-Caucasian participants, our results are limited to Caucasian women. Further studies are needed in other populations, both of the association between the rs10757274 genotype and CVD and the resultant effect on risk prediction. Our study also evaluated only variation at chromosome 9p21.3 and thus does not exclude the possibility that multigene panels might result in larger proportions of individuals reclassified correctly. Our study did not examine the additional effect of genotype relative to the previously published risk scores, but rather examined the additional effect after refitting all covariates in the same population. While this comparison potentially limits the generalizability of our results, it preserves their validity.
In conclusion, we confirmed the association of 9p21.3 with total CVD as well as coronary heart disease, MI and stroke in a large prospective cohort of Caucasian women. However, addition of the 9p21.3 genetic variation did not improve discrimination or classification of predicted risk achieved with traditional risk factors, CRP and family history of CVD.
Acknowledgments
Grant Support: The Women's Health Study and the Women's Genome Health Study are supported by funds from the National Heart Lung and Blood Institute and National Cancer Institute (Bethesda, MD) grants HL 043851, HL 080467, and CA 047988, the Donald W Reynolds Foundation (Las Vegas, NV), and the Leducq Foundation, Paris FR. Genotyping of the 9p21.3 variant was performed by Celera (Alameda, CA). Additional support for DNA extraction, reagents, and data analysis was provided by Roche Diagnostics (Indianapolis, IN) and Amgen, Inc (Thousand Oaks, CA).
Footnotes
Potential Financial Conflicts of Interest: Employment: D. Shiffman (Celera); Stock Ownership: D. Shiffman (Celera); Grants Received: P.M Ridker and J.E. Buring (Investigator Initiated from NHLBI, NCI, the Donald W Reynolds Foundation, the Leducq Foundation, Roche Diagnostics, and Amgen, Inc.)
Reproducible Research Statement: Study Protocol: Available to interested readers by contacting Dr. Paynter at npaynter@partners.org; Statistical Code: Available to interested readers by contacting Dr. Paynter at npaynter@partners.org; Data: Not Available
References
- 1.Ridker PM, Buring JE, Rifai N, Cook NR. Development and Validation of Improved Algorithms for the Assessment of Global Cardiovascular Risk in Women: The Reynolds Risk Score. JAMA: The Journal of the American Medical Association. 2007;297(6):611–619. doi: 10.1001/jama.297.6.611. [DOI] [PubMed] [Google Scholar]
- 2.Kathiresan S, Melander O, Anevski D, et al. Polymorphisms Associated with Cholesterol and Risk of Cardiovascular Events. N Engl J Med. 2008;358(12):1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 3.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 4.McPherson R, Pertsemlidis A, Kavaslar N, et al. A Common Allele on Chromosome 9 Associated with Coronary Heart Disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schunkert H, Gotz A, Braund P, et al. Repeated Replication and a Prospective Meta-Analysis of the Association Between Chromosome 9p21.3 and Coronary Artery Disease. Circulation. 2008;117(13):1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helgadottir A, Thorleifsson G, Magnusson KP, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40(2):217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 7.Diabetes Genetics Initiative of Broad Institute of Harvard and Mit LUaNIoBR. Saxena R, Voight BF, et al. Genome-Wide Association Analysis Identifies Loci for Type 2 Diabetes and Triglyceride Levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 8.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A Genome-Wide Association Study of Type 2 Diabetes in Finns Detects Multiple Susceptibility Variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of Genome-Wide Association Signals in UK Samples Reveals Risk Loci for Type 2 Diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridker PM, Chasman DI, Zee RYL, et al. Rationale, Design, and Methodology of the Women's Genome Health Study: A Genome-Wide Association Study of More Than 25 000 Initially Healthy American Women. Clin Chem. 2008;54(2):249–255. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 11.Ridker PM, Cook NR, Lee IM, et al. A randomized trial of low-dose aspirin in the primary prevention of cardiovascular disease in women. The New England Journal of Medicine. 2005;352(13):1293–1304. doi: 10.1056/NEJMoa050613. [DOI] [PubMed] [Google Scholar]
- 12.Shiffman D, O'Meara ES, Bare LA, et al. Association of gene variants with incident myocardial infarction in the Cardiovascular Health Study. Arterioscler Thromb Vasc Biol. 2008;28(1):173–9. doi: 10.1161/ATVBAHA.107.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) Final Report. Circulation. 2002;106(25):3143. [PubMed] [Google Scholar]
- 14.Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in Medicine. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Lemeshow S, Hosmer DW., Jr. A review of goodness of fit statistics for use in the development of logistic regression models. American Journal of Epidemiology. 1982;115(1):92–106. doi: 10.1093/oxfordjournals.aje.a113284. [DOI] [PubMed] [Google Scholar]
- 16.D'Agostino RB, Sr., Grundy S, Sullivan LM, Wilson P. Validation of the Framingham coronary heart disease prediction scores: results of a multiple ethnic groups investigation. JAMA: The Journal of the American Medical Association. 2001;286(2):180–187. doi: 10.1001/jama.286.2.180. [DOI] [PubMed] [Google Scholar]
- 17.Cook NR. Use and Misuse of the Receiver Operating Characteristic Curve in Risk Prediction. Circulation. 2007;115(7):928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 18.Cook NR. Statistical evaluation of prognostic versus diagnostic models: beyond the ROC curve. Clin Chem. 2008;54(1):17–23. doi: 10.1373/clinchem.2007.096529. [DOI] [PubMed] [Google Scholar]
- 19.Pencina MJ, D'Agostino RBS, D'Agostino RBJ, Vasan RS. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat.Med. 2007 doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 20.Talmud PJ, Cooper JA, Palmen J, et al. Chromosome 9p21.3 Coronary Heart Disease Locus Genotype and Prospective Risk of CHD in Healthy Middle-Aged Men. Clin Chem. 2008;54(3):467–74. doi: 10.1373/clinchem.2007.095489. [DOI] [PubMed] [Google Scholar]
- 21.Matarin M, Brown WM, Singleton A, Hardy JA, Meschia JF. Whole Genome Analyses Suggest Ischemic Stroke and Heart Disease Share an Association With Polymorphisms on Chromosome 9p21. Stroke. 2008 doi: 10.1161/STROKEAHA.107.502963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. American Journal of Epidemiology. 2004;159(9):882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]


