Abstract
Kynurenine 3-monooxygenase (KMO) catalyses the hydroxylation of kynurenine to 3-hydroxykynurenine. KMO has a key role in tryptophan catabolism and synthesis of ommochrome pigments in mosquitoes. The gene encoding this enzyme in the yellow fever mosquito, Aedes aegypti, is called kynurenine hydroxylase (kh) and a mutant allele that produces white eyes has been designated khw. A number of cDNA clones representative of wild-type and mutant genes were isolated. Sequence analyses of the wild-type and mutant cDNAs revealed a deletion of 162 nucleotides in the mutant gene near the 3′-end of the deduced coding region. RT-PCR analyses confirm the transcription of a truncated mRNA in the mutant strain. The in-frame deletion results in a loss of 54 amino acids, which disrupts a major α-helix and which probably accounts for the loss of activity of the enzyme. Recombinant Ae. aegypti KMO showed high substrate specificity for kynurenine with optimum activity at 40 °C and pH = 7.5. Kinetic parameters and inhibition of KMO activity by Cl− and pyridoxal-5-phosphate were determined.
Keywords: Aedes aegypti, kynurenine 3-monooxygenase, 3-hydroxykynurenine, kynurenine, kynurenine hydroxylase, white eye
Introduction
Kynurenine 3-monooxygenase (KMO, EC 1.14.13.9), a flavin-containing enzyme, catalyses the hydroxylation of kynurenine to 3-hydroxykynurenine (3-HK) in the tryptophan oxidation pathway. In our previous studies of tryptophan catabolism in Aedes aegypti, we showed that oxidation of tryptophan to xanthurenic acid (XA), via kynurenine and 3-HK intermediates, is the major pathway of tryptophan catabolism during larval and egg development (Li & Li, 1997). In this species, XA is derived from 3-HK by 3-HK transaminase (Han et al., 2002); consequently, KMO is obligatory in the pathway that converts tryptophan to XA. 3-HK also is a direct precursor for the production of ommochromes, which are major eye pigments in Ae. aegypti (Li et al., 1999) (Fig. 1). The gene encoding this enzyme in the yellow fever mosquito, Ae. aegypti, has been named kynurenine hydroxylase (Cornel et al., 1997), and a mutation in the gene that results in a white-eye phenotype has been renamed khw (Cornel et al., 1997; Bhalla, 1968).
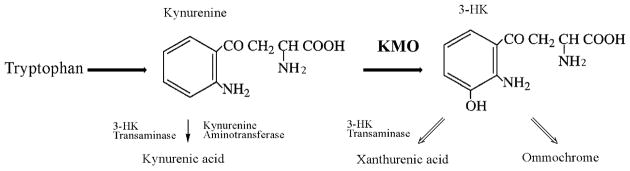
Figure 1.
Tryptophan metabolism in Aedes aegypti. The metabolic pathway in which the KMO enzyme participates starts with the conversion of tryptophan to kynurenine (left). KMO catalyses the conversion of kynurenine to 3-HK (right) that can be either converted to xanthurenic acid or oxidized to ommochrome pigments. Abbreviations: 3-HK, 3-hydroxykynurenine; KMO, kynurenine monooxygenase.
Early studies of KMOs were concerned primarily with their role in the eye pigmentation of some insects, including Drosophila melanogaster (Howells et al., 1977; Paton & Sullivan, 1978; Howells, 1979), Lucilia cuprina (Summers & Howells, 1978) and Musca domestica (Akaboshi, 1979). In D. melanogaster, the cinnabar (cn) gene encodes the KMO enzyme (Warren et al., 1996–1997), and this name is used by many contemporary authors to refer to the gene. Recent efforts in the genetic manipulation of mosquitoes with the goal of controlling mosquito-transmitted diseases have raised considerable interest in the gene because it can serve as an excellent marker for the successful generation of transgenic Ae. aegypti (Coates et al., 1998; Jasinskiene et al., 1998). The Ae. aegypti white-eye mutant strain (Bhalla, 1968) was shown to be complemented by a wild-type copy of the D. melanogaster cinnabar gene (Cornel et al., 1997), providing genetic evidence that the product encoded by the white-eye gene was KMO. We continue to use the designation KMO for the protein, kh for the wild-type gene and khw for the mutant allele. Although the D. melanogaster gene complements various white-eye and other mutations in Diptera, it does not function well outside this order (Atkinson et al., 2001). However, the general application of the eye-colour genes as broad-spectrum transformation markers in the Coleoptera species also has been discussed in a recent article (Lorenzen et al., 2002).
The KMO-encoding gene in mosquitoes is also of interest to vector biologists because one of the end products, XA, plays a role in the exflagellation of gametes of malaria parasites (Billker et al., 1998; Garcia et al., 1998). The source of XA for this process is likely to come from both the mosquito and the vertebrate host (Arai et al., 2001).
Genes encoding KMO have been cloned from several insect species and for a few the biochemical and kinetic parameters of the enzyme have been analysed (Quan et al., 2002; Hirai et al., 2002). The genetic lesion leading to the phenotype in a mutant strain of Bombyx mori has been identified (Quan et al., 2002). In this study, we isolated a cDNA corresponding to the Ae. aegypti kh gene, expressed its open reading frame (ORF) in an insect cell/baculovirus expression system and characterized its recombinant protein. Using gene amplification procedures, we compared expression products from mutant and wild-type animals. We provide data that describe the basic biochemical characteristics of the Ae. aegypti KMO (AeKMO) and the nature of the genetic lesion that produces the white-eye phenotype. We also discuss the importance of this enzyme during mosquito development.
Results
Primary structure of cDNAs derived from wild-type and mutant mosquito strains
A 1790 bp cDNA clone derived from the wild-type Liverpool strain contained a 1428 bp ORF encoding a polypeptide of 476 amino acid residues with a predicted molecular mass of 54 000 (Fig. 2). Comparison of its predicted product with other animal KMOs revealed the presence of the conserved sequence motif xhxhGxGxxGxxxhxxh(x)8hxhE(D), where x is any residue and h is a hydrophobic residue, a common motif among flavin adenine dinucleotide (FAD)- and NAD(P)H-dependent oxidoreductases (Dym & Eisenberg, 2001), indicating that the AeKMO is a member of the glutathione reductase structural family. This consensus is known as the dinucleotide-binding motif (Wierenga et al., 1983), and therefore KMO is probably an FAD-containing protein. AeKMO is a hydrophobic protein and it has two predicted transmembrane segments, residues 18–46 and residues 431–459. AeKMO showed 60%, 56%, 44%, 46% and 50% amino acid identity with KMOs from Bombyx mori, D. melanogaster, Sus scrofa, Ratus norvegicus and Homo sapiens, respectively.
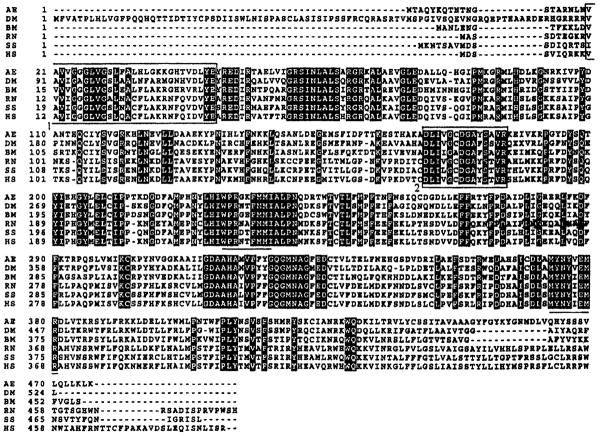
Figure 2.
Multiple alignment of the amino acid sequence of AeKMO with KMOs from diverse animals. AE, AeKMO (GenBank accession no. AF325508); DM, Drosophila melanogaster KMO (GenBank accession no. U56245); BM, Bombyx mori KMO (GenBank accession no. AB063490); RN, Rattus norvegicus KMO (GenBank accession no. AF056031); SS, Sus scrofa KMO (GenBank accession no. AF163971); HS, Homo sapiens KMO (GenBank accession no. Y13153). The white letters with black background indicate identical amino acids. The underlined sequences are those used to design degenerate primers. The first boxed fragment is the dinucleotide-binding motif of ‘xhxhGxGxxGxxxhxxh(x)8hxhE(D)’, where x is any residue and h is a hydrophobic residue. The second boxed fragment is a putative consensus domain for both FAD and NAD(P)H binding.
In order to compare the predicted Liverpool strain KMO protein with the KMO from two other Ae. aegypti strains, the sequences of the full-length KMO cDNAs from the wild-type Rockefeller and khw mutant strains were determined by RT-PCR and 3′ and 5′ RACE reactions (data not shown). The two cDNAs are 1636 and 1481 bp in length, respectively, excluding the poly-A tail. Both cDNAs showed a 5′-UTR of twenty-six nucleotides and a putative polyadenylation signal, AATAAA occurring thirty-five nucleotides to the 5′-end of the insertion of the poly-A domains. A short sequence, CCTTCTTT, is present at the end of the 3′-end nonpolydenylated sequence of the khw gene product and is not found in the cDNA derived from the wild-type strain. A deletion of 162 nucleotides, located at positions 1120–1282, and corresponding to fifty-four amino acids, is located near the predicted carboxyl terminal portion of the khw translated protein. The alignment of the predicted proteins encoded by these sequences with the Liverpool KMO indicates that whereas Rockefeller and Liverpool strains encode for almost identical proteins (476 amino acids), differing in only eight amino acids, the mutant khw cDNA encodes for a shorter protein (422 amino acids) (Fig. 3A). The deletion in the khw product results in a disruption of a predicted α-helical domain near the carboxyl terminal region of the protein (Fig. 3B).
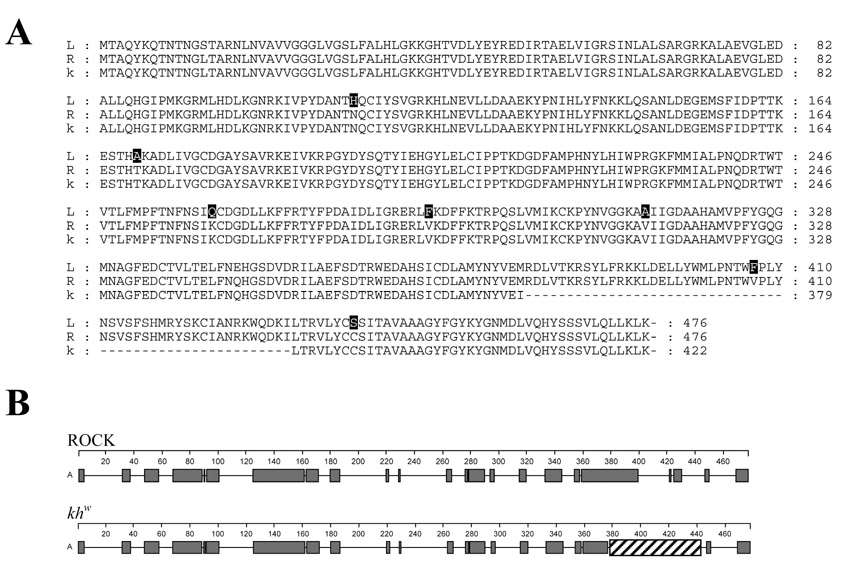
Figure 3.
Comparisons of the predicted KMO proteins of the Rockefeller, khw and Liverpool strains. (A) Amino acid alignment of the predicted KMO proteins. The Rockefeller (R) and khw (k) proteins are identical except for a fifty-four amino acid deletion denoted by the dashes (−). The Liverpool (L) and Rockefeller have eight different amino acids indicated in bold (GenBank accession no. Rockefeller AY194225; khw AY194224). (B) The large deletion in the khw KMO disrupts the α-helixal structure in the carboxyl terminal region of the protein. The predicted α-helicies are shown in red and the deletion as a cross-hatched box. The Garnier–Robson model (Persson, 2000) was used to predict the structure.
Expression profile of the wild-type and mutant genes during development
The expression profiles of the Rockefeller kh and khw genes were evaluated by RT-PCR and Northern blotting analyses. The gene amplification analysis using the KMOgap-for and KMOgap-rev primers detected RNA in all stages examined for both wild-type and khw mosquitoes (Fig. 4). The presence of a deletion in the khw genome was confirmed by this analysis as evidenced by the smaller product amplified by the primers.
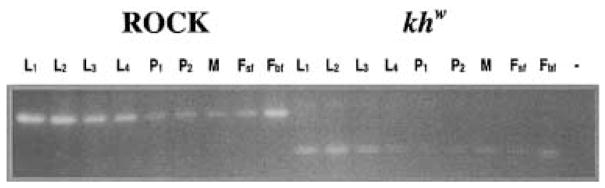
Figure 4.
Developmental expression profiles of wild-type and mutant genes. RT-PCR analyses of total RNA from first (L1), second (L2), third (L3) and fourth (L4) larval instars, early (P1) and late-stage (P2) pupae, adult males (M), and sugar-(Fsf) and blood-fed (Fbf), adult females from the wild-type strain, Rockefeller (Rock), and the mutant khw strain.
Recombinant AeKMO (rAeKMO) expression and purification
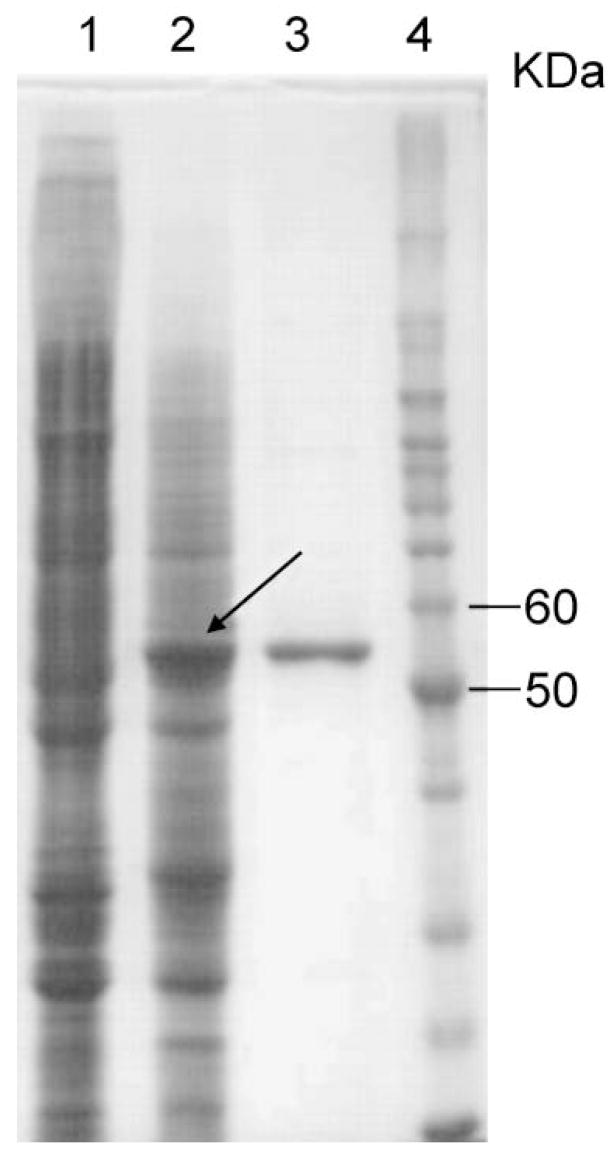
Cotransfection of the linearized AcMNPV DNA and baculovirus transfer vectors containing an AeKMO coding sequence resulted in the successful isolation of recombinant viruses. Analysis of the soluble protein from Sf9 cells infected with the AeKMO recombinant virus by SDS-PAGE detected a major protein species with a relative molecular mass (Mr) of 54 000 (Fig. 5). This same moiety was not observed in the soluble cell protein of uninfected control cells. KMO activity was detected in the soluble cell protein of infected cells. The recombinant enzyme was purified from viral infected cells by an Ni2+-charged His-Bind affinity column. The Mr of the rAeKMO (54 000) is similar to the calculated molecular mass of the rAeKMO (54 000 plus 1000 for the His-tag). These data indicate the successful expression of the recombinant KMO.
Figure 5.
Purification of rAeKMO. Lane 1 shows the SDS-PAGE protein profiles of the supernatant of cell lysate obtained from uninfected Sf9 cells; Lane 2 shows the protein profiles of the supernatant of cell lysate obtained from Sf9 cells infected with an AeKMO recombinant virus; Lane 3 shows the purified rAeKMO; and Lane 4 shows the protein molecular mass standards on the same polyacrylamide gel.
Biochemical characterization of rAeKMO
Substrate specificity and kinetic parameters
Kinetic analysis determined that the purified rAeKMO had a Vmax of 3550 ± 87, 3821 ± 188 and 1616 ± 110 nmol/min/mg, for kynurenine, NADPH and NADH, respectively (Table 1). The Kcat/Km for NADPH and NADH are 149 ± 26/min/mM and 10 ± 2.6/min/mM, respectively, indicating that NADPH is the preferred electron donor for the enzyme. When kynurenic acid, anthranilic acid, tryptophan, p-hydroxybenzoate or D-kynurenine was mixed with rAeKMO in the presence of either NADH or NADPH, no hydroxylation of these compounds was detected (data not shown), indicating that AeKMO is highly specific for L-kynurenine.
Table 1.
Kinetic parameters of rAeKMO*
| Substrates/cofactors | Km (mM) | Vmax (nmol/min/mg) | Kcat (/min) | Kcat/Km (min/mM) |
|---|---|---|---|---|
| Kynurenine | 0.89 ± 0.07 | 3550 ± 87 | 113 ± 2.8 | 127 ± 12 |
| NADPH | 0.82 ± 0.10 | 3821 ± 188 | 122 ± 6.0 | 149 ± 26 |
| NADH | 5.17 ± 0.97 | 1616 ± 110 | 51 ± 3.5 | 10 ± 2.6 |
The biochemical activities were measured as described in the Experimental procedures. The kinetic parameters of rAeKMO were calculated by fitting the experimental data to the Michaelis–Menten equation using the Enzyme Kinetics Module (SPSS Science).
Optimal pH and temperature
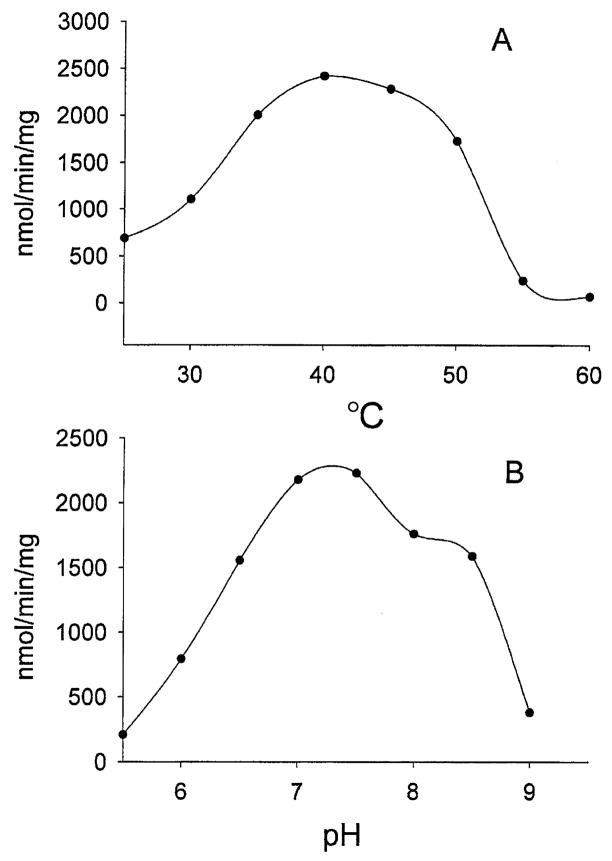
Both temperature and pH had major effects on the activity of rAeKMO. The protein showed the highest activity at a temperature near 40 °C (Fig. 6), and was rapidly inactivated at temperatures above 55 °C. The rAeKMO was more active at neutral conditions with an optimum pH between 7.0 and 7.5. Analysis of the pH profile using Enzyme Kinetics (SPSS Science) derived a pKa = 6.4 and a pKb = 8.5 for the enzyme, respectively.
Figure 6.
Effects of temperature and pH on rAeKMO activity. The detailed assay methods are described in the Experimental procedures. The biochemical activity was assayed with kynurenine (2 mM) as a hydroxyl group acceptor and NADPH (0.8 mM) as an electron donor. (A) rAeKMO activity at temperatures from 35 to 70 °C; (b) the pH profile of rAeKMO, analysed using Enzyme Kinetics Module (SPSS Science).
Inhibition by pyridoxal-5-phosphate (PLP) and Cl−1
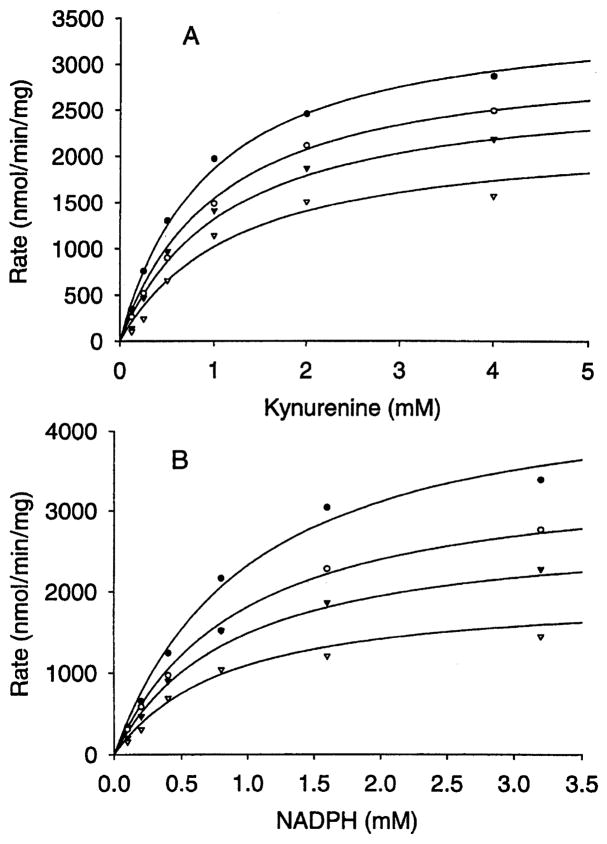
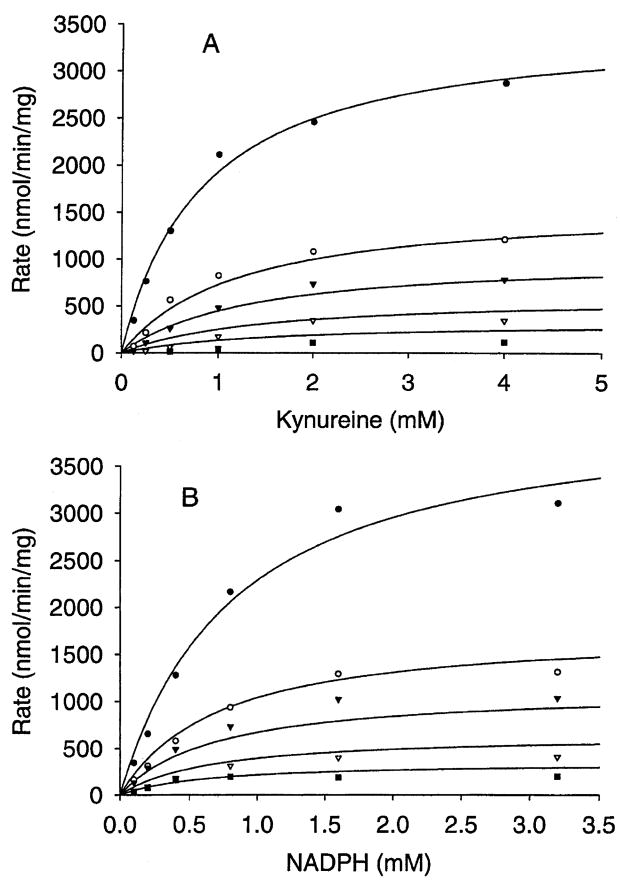
Inhibition of hydroxylase activity by PLP and Cl−1 was investigated by the overall reaction rate in the presence of varying concentrations of either PLP or Cl−1 (PLP: 0, 0.05, 0.1 and 0.2 mM; KCl: 0, 25, 50, 100 and 200 mM) at saturating concentration of one substrate while varying the concentration of the other. The Michaelis–Menten curves of the PLP inhibition study using the Enzyme Kinetics program showed that PLP inhibition of rAeKMO fitted a noncompetitive inhibition equation with respect to both substrates (kynurenine: R2 = 0.987, AICc = 227, Sy.x = 98; NADPH: R2 = 0.989, AICc = 230, Sy.x = 104) (Fig. 7). The Ki of PLP inhibition to rAeKMO with respect to kynurenine and NADPH was 266 μM and 170 μM, respectively. The Michaelis–Menten curves of Cl−1 inhibition revealed that it was a mixed type with respect to both NADPH (R2 = 0.982, AICc = 294, Sy.x = 116) and kynurenine (R2 = 0.988, AICc = 250, Sy.x = 88), which means it behaves in both competitive and noncompetitive manners (Fig. 8). The Ki of Cl−1 inhibition of rAeKMO with respect to kynurenine and NADPH was 11.2 mM and 24.5 mM, respectively.
Figure 7.
Inhibition of KMO by PLP. The Michaelis–Menten curves of different concentrations of PLP: zero (closed circles), 0.05 mM (open circles), 0.1 mM (closed triangles) and 0.2 mM (open triangles). (A) Activity was assayed at a fixed concentration of NADPH (3.2 mM) and different concentrations of kynurenine (0.125, 0.25, 0.5, 1, 2, 4 mM); (B) activity was assayed at a fixed concentration of kynurenine (4 mM) and different concentrations of NADPH (0.1, 0.2, 0.4, 0.8, 1.6, 3.2 mM).
Figure 8.
Inhibition of KMO by Cl−1. The Michaelis–Menten curves of different concentrations of Cl−1: zero (closed circles), 25 mM (open circles), 50 mM (closed triangles), 100 mM (open triangles) and 200 mM (closed squares). (A) Activity was assayed at a fixed concentration of NADPH (3.2 mM) and different concentrations of kynurenine (0.125, 0.25, 0.5, 1, 2, 4 mM); (B) activity was assayed at a fixed concentration of kynurenine (4 mM) and different concentrations of NADPH (0.1, 0.2, 0.4, 0.8, 1.6, 3.2 mM).
Discussion
In this study, we isolated cDNA clones encoding KMO from wild-type and mutant Ae. aegypti, expressed recombinant protein using an insect/baculovirus expression system, and determined the basic biochemical characteristics of the expressed enzyme. Our data show that the AeKMO shares considerable sequence identity with KMOs described in other animals and that the enzyme is highly substrate specific to kynurenine and prefers NADPH as reducing agent, which are properties similar to those described for human KMO (Breton et al., 2000).
3-HK is oxidized easily under physiological conditions and this stimulates the production of reactive oxygen species (Li & Li, 1997). Recent studies of the kynurenine to 3-HK pathway focused primarily on its toxic impact on living organisms. For example, accumulation of 3-HK in the central nervous system leads to neurodegeneration (Ceresoli et al., 1997; Guidetti & Schwarcz, 1999; Wu et al., 2000). Levels of 3-HK are often elevated in disease conditions (Chiarugi et al., 2001). By contrast, inhibition of human KMO reduced somewhat neuronal death in brain ischaemia or excitotoxicity (Cozzi et al., 1999; Moroni, 1999). Inhibition of KMO also led to a contemporary increase in kynurenic acid, an antagonist of neuroexcitotoxin that plays an important role in protecting neurons from being overstimulated by excitotoxins. Accordingly, KMO, the enzyme responsible for the production of 3-HK, has been considered as an attractive pharmacological target.
Although studies concerning the kynurenine to 3-HK pathway indicate a considerable side-effect on the organism, our work on tryptophan catabolism in mosquitoes suggests that the kynurenine to 3-HK pathway is important in mosquitoes. In mammals, both kynurenine and 3-HK can be hydrolysed by kynureninase to anthranilic acid and 3-hydroxyanthranilic acid, respectively, which is the necessary step for kynurenine and 3-HK to be completely oxidized to CO2 and H2O or used towards the synthesis of NAD+ or NADP+ via a quinolinic acid intermediate (Saito et al., 1993). By contrast, mosquitoes do not have a kynureninase, which blocks the above pathway for kynurenine and 3-HK. Our previous data show that hydroxylation of kynurenine to 3-HK is the major pathway of tryptophan catabolism (Li & Li, 1997), but the fate of 3-HK is quite different in mosquitoes during different stages of development. During larval development, 3-HK is transaminated to XA, whereas during the pupal and adult stages, 3-HK is transported to the compound eyes for eye-pigmentation (Li et al., 1999). The lack of eye pigment in Ae. aegypti white-eye mutant has been confirmed to be due to a mutation in the KMO gene (Cornel et al., 1997). Apparently, the kynurenine to 3-HK pathway is an important step in the tryptophan catabolic pathway in mosquitoes.
The catalytic mechanism of KMOs is suggested to be similar to bacterial p-hydroxybenzoate hydroxylase (PHBH) (Breton et al., 2000). Sequence comparison between AeKMO and Pseudomonas putida PHBH, an extensively characterized flavin monooxygenase, revealed some positive relation between the two enzymes. Although AeKMO and P. putida PHBH share only limited overall sequence identity (14–16%), AeKMO contains the sequence motif xhxh-GxxGxxxhxxh(x)8hxhE(D), a well-known fingerprint for dinucleotide binding domain, where x is any residue and h is a hydrophobic residue (Eppink et al., 1997). Moreover, the amino acid sequence, 153DYIAGCDGFHGISR166, in PHBH, which has been reported to play a dual function in both FAD and NAD(P)H binding, also is highly conserved in AeKMO (amino acids 172DLIVGCDGAYSAVR185). Based on these observations, the catalytic mechanism of KMO may be similar to that of PHBH, a prototype for FAD-dependent monooxygenase. However, some sequence fragments that are highly conserved in KMOs are not present in PHBH. For example, the residues 60–68, 225–230, 233–240 and 326–332 in AeKMO are likely to play relevant roles in either fold stability or catalysis. In order fully to understand the structure/function relationship of KMOs, determination of its three-dimensional (3-D) structure is needed. The lack of detailed 3-D structural data for these enzymes has been limited by the availability of adequate amounts of active protein. Moreover, it must also be considered that the enzyme is poorly soluble in aqueous solutions owing to the presence of the fairly hydrophobic regions near its carboxyl end and N-terminus, which probably functions as a transmembrane domain through which the protein is associated to the outer mitochondrial membrane. By using a baculovirus/insect cell expression system, we have been able to produce soluble and active recombinant AeKMO (10% of the total soluble protein), which makes it possible to use this recombinant material to produce the conditions for crystallization and subsequent 3-D structural analysis.
Experimental procedures
Materials
The Ae. aegypti black-eyed Liverpool strain used in this study was reared at UIUC according to a previously described method (Christensen et al., 1984). The wild-type Rockefeller and the mutant khw strains were reared at the UCI insectary facility using similar procedures. Chemicals were purchased from Sigma (St Louis, MO, USA) unless otherwise specified.
cDNA cloning
Degenerate oligonucleotide primers (5′-TGGCCKMGDAARCCT-TYATGATGAT-3′) and (5′-CGCATCTCHAYRTCATTGTACAT-3′) were designed based on the conserved amino acid sequences, WPRN(T/A)FMMI and MYNYVEMR (Fig. 2), in genes derived from Bombyx mori (GenBank accession no. AB063490), D. melanogaster (GenBank accession no. U56245), Sus scrofa (GenBank accession no. AF163971), Rattus norvegicus (GenBank accession no. AF056031) and Homo sapiens (GenBank accession no. Y13153). These primers were used for gene amplification of the first strand cDNA synthesized from the total RNA of 3-day-old Liverpool strain Ae. aegypti larvae (Life Technologies). A 460 bp fragment was amplified, sequenced and used as a probe to screen a larval cDNA library (constructed using a library construction kit from Stratagene). A total of 5 × 105 plaques were screened at high stringency. Two cDNA clones were isolated and sequenced. Sequence data were analysed using a Biology WorkBench 3.2 program (http://workbench.sdsc.edu) and the software package from Genetics Computer Group, Inc (GCG, University of Wisconsin, Madison).
Total RNA from late-stage pupae from a wild-type strain, Rockefeller, and the mutant khw strain was isolated using TRIZOL reagent (Gibco-BRL) according to the manufacturer’s instructions. Approximately 1 μg of total RNA from both strains was used as templates for RT-PCR reactions and primed with the gene-specific primer pairs: KMO-for (5′-ATTGGAGATGACTGCCCAG-3′) and KMO-rev (5′-GTAGAGTTTGAATCATCACCGATTG-3′) using the Superscript One Step RT-PCR Platinum System (Invitrogen). The amplified products were cloned into the plasmid TOPO-Zero blunt (Invitrogen) and sequenced. The 3′- and 5′-ends of cDNAs were obtained using the primers (KMO-5RACE 5′-GCATCGGGGAAG-TATGTCCGGAAGAAC-3′ and KMO-3RACE 5′-CGGCCGGGT-TATGACTATAGTCAG-3′) following a RACE protocol (Smart RACE cDNA kit, Clontech). The gene amplification cycling included an initial denaturation for 2 min at 94 °C, thirty cycles of 1 min at 94 °C, 1 min at 65 °C and 2 min at 72 °C. The final step was extended for 7 min at 72 °C. The amplified DNA fragments were cloned into the TOPO-TA vector (Invitrogen) and submitted for DNA sequencing. Independent sequence determinations from nine or more reactions confirmed the accuracy of the final sequence.
Expression analysis of wild-type and mutant genes
Total RNA from first, second, third and fourth larval instars, early-and late-stage pupae, adult males and sugar- and blood-fed adult females from the wild-type strain, Rockefeller, and the mutant khw strain was isolated using TRIZOL reagent (Gibco-BRL) according to the manufacturer’s instructions. Approximately 1 μg of total RNA from all stages of both strains was primed with the gene-specific primer pairs: KMOgap-for (5′-AGTGAATACGCGTT-GGGAGGATG-3′) and KMOgap-rev (5′-GAACAGAGGAAGAG-TAGGTTGCACGAG-3′) using the Superscript One Step RT-PCR Platinum System (Invitrogen). These primers were designed to flank the deletion in the mutant strain and result in an ~150 bp size difference between the products amplified from the wild-type and mutant cDNA templates. The gene amplification cycling included an initial denaturation for 2 min at 94 °C, thirty cycles of 1 min at 94 °C, 1 min at 65 °C and 2 min at 72 °C. The final step was extended for 7 min at 72 °C. Select amplified DNA fragments were cloned into the TOPO-TA vector (Invitrogen) and sequenced to verify their primary structure.
rAeKMO expression and purification
Recombinant transfer vector construction
The open reading frame (ORF) of the Liverpool cDNA was amplified using a primer (5′-GGCTCTAGATGACTGCCCAGTACAA-3′) containing an XbaI site (underlined), and a primer (5′-GAAGCTTAATGATGATGAT-GATGATGTTTTAGCTTAAGGAATTA-3′) containing a HindIII site (underlined), and an His-tag sequence (italic letters). The amplified DNA fragments were propagated in a TA cloning vector and then subcloned into a baculovirus transfer vector pBlueBac4.5 (Invitrogen) between the XbaI and HindIII restriction sites.
Recombinant baculoviruses isolation
Recombinant transfer vectors were sequenced to verify that the cDNA insert encoding KMO was in-frame and adjacent to the 3′-end of the polyhedrin promoter. The recombinant AeKMO pBlueBac4.5 transfer vector was co-transfected with linearized Bac-N-Blue™ (AcMNPV, Autographa californica multiple nuclear polyhedrosis virus) viral DNA in the presence of InsectinPlus™ insect cell-specific liposomes into Spodoptera frugiperda (Sf9) insect cells (Invitrogen). The recombinant baculoviruses were purified by a plaque assay procedure. Blue putative recombinant plaques were transferred to twelve-well microtitre plates and amplified in Sf9 cells. Viral DNA was isolated for gene amplification analysis to determine the purity of the recombinant viruses. A high-titre viral stock (HTS) of a pure recombinant virus was generated by amplification in suspension cultured Sf9 cells. The titre of HTS and the time-course at a multiplicity of infection (MOI) of 6 were established according to the manufacturer’s instructions.
rAeKMO expression and purification
Sf9 cells were cultured in spinner flasks in TNM-FH medium containing 10% fetal bovine serum (Gibco BRL). HTS of pure recombinant virus was inoculated into the culture at a cell density of 2.5 × 106 cells/ml. Sf9 cells were harvested at day 4 postinoculation by centrifugation (800 g for 15 min at 4 °C), and the cell pellets were dissolved in 50 mM phosphate buffer, pH 7.5, containing 0.1% Triton X-100 and 1 mM phenylmethylsulphonyl fluoride (PMSF). After sonication in cold water for 30 min, cell lysate was centrifuged at 20 000 g for 20 min at 4 °C and the supernatant obtained was used as the starting material for rAeKMO purification. The supernatant was mixed with an equal volume of 2 × Ni2+ binding buffer and purified using an Ni2+-charged His-Bind resin column (1.5 × 2.5 cm, Novagen) according to the supplied protocol. The purified protein was dialysed against 100 mM phosphate buffer (pH 7.5) containing 40 μM FAD, 0.1% Triton X-100 for 10 h at 4 °C. The purity of the protein was assessed by SDS-PAGE analysis. Protein concentration was determined by a Bio-Rad protein assay kit using bovine serum albumin (BSA) as a standard.
Biochemical characterization of rAeKMO
The rAeKMO enzymatic assays were based on a previously described method (Breton et al., 2000). Briefly, a 50-μl reaction mixture, containing 2 mM kynurenine, 0.8 mM NADPH and a varying amount of purified rAeKMO, was prepared in 100 mM potassium phosphate (pH 7.5) consisting of 2 mM MgCl2, and the mixture was incubated at 37 °C. An equal volume of 8% trichloro-acetic acid was added to the reaction mixture at 10 min after incubation to stop the reaction. The supernatant was obtained by centrifugation of the reaction mixture at 15 000 g at 4 °C for 10 min and analysed by high-performance liquid chromatogrpahy (HPLC) with UV detection at 340 nm to separate and quantify 3-HK. To determine the effect of temperature on rAeKMO activity, the reaction mixture was incubated at different temperatures for 5 min prior to the addition of 0.4 μg rAeKMO in 4 μl of enzyme preparation and continuously incubated for 10 min after enzyme addition. The effect of pH on rAeKMO activity was assayed by preparation of the typical reaction mixtures in either 100 mM citrate buffer (pH 5.5), phosphate buffer (pH 6.0–7.5) or borate buffer (pH 8–9). The effects of pH and temperature on rAeKMO were determined by the specific activities. The possibility of NADH as an electron donor and the inhibition of Cl−1 (KCl) and PLP on rAeKMO activity were also studied. To determine the substrate specificity for rAeKMO, kynurenine was replaced with either kynurenic acid, anthranilic acid, tryptophan, p-hydroxybenzoate or the D-isomer of kynure-nine in the above reaction mixture and the absorbance was continuously monitored spectrophotometrically at 340 nm for NADPH concentration decrease.
Acknowledgments
This work was supported by grants from the National Institutes of Health, AI44399 (J.L.) and AI29746 (A.A.J.). We thank Lynn Olson for help in preparing the manuscript.
References
- Akaboshi E. Kynurenine hydroxylase in Musca domestica L. Comp Biochem Physiol B. 1979;62:549–555. doi: 10.1016/0305-0491(79)90132-9. [DOI] [PubMed] [Google Scholar]
- Arai M, Billker O, Morris HR, Panico M, Delcroix M, Dixon D, Ley SV, Sinden RE. Both mosquito-derived xanthurenic acid and a host blood-derived factor regulate gametogenesis of Plasmodium in the midgut of the mosquito. Mol Biochem Parasitol. 2001;116:17–24. doi: 10.1016/s0166-6851(01)00299-7. [DOI] [PubMed] [Google Scholar]
- Atkinson PW, Pinkerton AC, O’Brochta DA. Genetic transformation systems in insects. Annu Rev Entomol. 2001;46:317–346. doi: 10.1146/annurev.ento.46.1.317. [DOI] [PubMed] [Google Scholar]
- Bhalla SC. White eye, a new sex-linked mutant of Aedes aegypti. Mosquito News. 1968;28:380–385. [Google Scholar]
- Billker O, Lindo V, Panico M, Etienne AE, Paxton T, Dell A, Rogers M, Sinden RE, Morris HR. Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito. Nature. 1998;392:289–292. doi: 10.1038/32667. [DOI] [PubMed] [Google Scholar]
- Breton J, Avanzi N, Magagnin S, Covini N, Magistrelli G, Cozzi L, Isacchi A. Functional characterization and mechanism of action of recombinant human kynurenine 3-hydroxylase. Eur J Biochem. 2000;267:1092–1099. doi: 10.1046/j.1432-1327.2000.01104.x. [DOI] [PubMed] [Google Scholar]
- Ceresoli G, Guidetti P, Schwarcz R. Metabolism of [5-(3) H]kynurenine in the developing rat brain in vivo: effect of intrastriatal ibotenate injections. Dev Brain Res. 1997;100:73–81. doi: 10.1016/s0165-3806(97)00029-1. [DOI] [PubMed] [Google Scholar]
- Chiarugi A, Cozzi A, Ballerini C, Massacesi L, Moroni F. Kynurenine 3-mono-oxygenase activity and neurotoxic kynurenine metabolites increase in the spinal cord of rats with experimental allergic encephalomyelitis. Neuroscience. 2001;102:687–695. doi: 10.1016/s0306-4522(00)00504-2. [DOI] [PubMed] [Google Scholar]
- Christensen BM, Sutherland DR, Gleason LN. Defense reactions of mosquitoes to filarial worms: comparative studies on the response of three different mosquitoes to inoculated Brugia pahangi and Dirofilaria immitis microfilariae. J Invertebr Pathol. 1984;44:267–274. doi: 10.1016/0022-2011(84)90024-7. [DOI] [PubMed] [Google Scholar]
- Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci USA. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornel AJ, Benedict MQ, Rafferty CS, Howells AJ, Collins FH. Transient expression of the Drosophila melanogaster cinnabar gene rescues eye color in the white eye (WE) strain of Aedes aegypti. Insect Biochem Mol Biol. 1997;27:993–997. doi: 10.1016/s0965-1748(97)00084-2. [DOI] [PubMed] [Google Scholar]
- Cozzi A, Carpenedo R, Moroni F. Kynurenine hydroxylase inhibitors reduce ischemic brain damage: studies with (m-nitrobenzoyl)-alanine (mNBA) and 3,4-dimethoxy-[-N-4-(nitrophenyl) thiazol-2yl]benzenesulfonamide (Ro 61–8048) in models of focal or global brain ischemia. J Cereb Blood Flow Metab. 1999;19:771–777. doi: 10.1097/00004647-199907000-00007. [DOI] [PubMed] [Google Scholar]
- Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001;10:1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppink MH, Schreuder HA, van Berkel WJ. Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD (P) H binding. Protein Sci. 1997;6:2454–2458. doi: 10.1002/pro.5560061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia GE, Wirtz RA, Barr JR, Woolfitt A, Rosenberg R. Xanthurenic acid induces gametogenesis in Plasmodium, the malaria parasite. J Biol Chem. 1998;273:12003–12005. doi: 10.1074/jbc.273.20.12003. [DOI] [PubMed] [Google Scholar]
- Guidetti P, Schwarcz R. 3-Hydroxykynurenine potentiates quinolinate but not NMDA toxicity in the rat striatum. Eur J Neurosci. 1999;11:3857–3863. doi: 10.1046/j.1460-9568.1999.00806.x. [DOI] [PubMed] [Google Scholar]
- Han Q, Fang J, Li J. 3-hydroxykynurenine transaminase, identity with alanine glyoxylate transaminase, a probable detoxification protein in Aedes aegypti. J Biol Chem. 2002;277:15781–15787. doi: 10.1074/jbc.M201202200. [DOI] [PubMed] [Google Scholar]
- Hirai M, Kiuchi M, Wang J, Ishii A, Matsuoka H. cDNA cloning, functional expression and characterization of kynurenine 3-hydroxylase of Anopheles stephensi (Diptera: Culicidae) Insect Mol Biol. 2002;11:497–504. doi: 10.1046/j.1365-2583.2002.00358.x. [DOI] [PubMed] [Google Scholar]
- Howells AJ. Isolation and biochemical analysis of a temperature-sensitive scarlet eye color mutant of Drosophila melanogaster. Biochem Genet. 1979;17:149–158. doi: 10.1007/BF00484480. [DOI] [PubMed] [Google Scholar]
- Howells AJ, Summers KM, Ryall RL. Developmental patterns of 3-hydroxykynurenine accumulation in white and various other eye color mutants of Drosophila melanogaster. Biochem Genet. 1977;15:1049–1059. doi: 10.1007/BF00484496. [DOI] [PubMed] [Google Scholar]
- Jasinskiene N, Coates CJ, Benedict MQ, Cornel AJ, Rafferty CS, James AA, Collins FH. Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. Proc Natl Acad Sci USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Beerntsen BT, James AA. Oxidation of 3-hydroxkynurenine to produce xanthommatin for eye pigmentation: a major branch pathway of tryptophan catabolism during pupal development in the Yellow Fever Mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1999;29:329–338. doi: 10.1016/s0965-1748(99)00007-7. [DOI] [PubMed] [Google Scholar]
- Li J, Li G. Transamination of 3-HK to produce xanthurenic acid: a major branchpathway of tryptophan metabolism in the mosquito, Aedes aegypti, during larval development. Insect Biochem Mol Biol. 1997;27:859–867. doi: 10.1016/s0965-1748(97)00068-4. [DOI] [PubMed] [Google Scholar]
- Lorenzen MD, Brown SJ, Denell RE, Beeman RW. Cloning and characterization of the Tribolium castaneum eye-color genes encoding tryptophan oxygenase and kynurenine 3-monooxygenase. Genetics. 2002;160:225–234. doi: 10.1093/genetics/160.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni F. Tryptophan metabolism and brain function: focus on kynurenine and other metabolites. Eur J Pharmacol. 1999;375:87–100. doi: 10.1016/s0014-2999(99)00196-x. [DOI] [PubMed] [Google Scholar]
- Paton DR, Sullivan DT. Mutagenesis at the cinnabar locus in Drosophila melanogaster. Biochem Genet. 1978;16:855–865. doi: 10.1007/BF00483738. [DOI] [PubMed] [Google Scholar]
- Persson B. Bioinformatics in protein analysis. EXS. 2000;88:215–231. doi: 10.1007/978-3-0348-8458-7_14. [DOI] [PubMed] [Google Scholar]
- Quan GX, Kim I, Komoto N, Sezutsu H, Ote M, Shimada T, Kanda T, Mita K, Kobayashi M, Tamura T. Characterization of the kynurenine 3-monooxygenase gene corresponding to the white egg 1 mutant in the silkworm Bombyx mori. Mol Genet Genomics. 2002;267:1–9. doi: 10.1007/s00438-001-0629-2. [DOI] [PubMed] [Google Scholar]
- Saito K, Quearry BJ, Saito M, Nowak TS, Jr, Markey SP, Heyes MP. Kynurenine 3-hydroxylase in brain: species activity differences and effect of gerbil cerebral ischemia. Arch Biochem Biophys. 1993;307:104–109. doi: 10.1006/abbi.1993.1567. [DOI] [PubMed] [Google Scholar]
- Summers KM, Howells AJ. Xanthommatin biosynthesis in wild-type and mutant strains of the Australian sheep blowfly Lucilia cuprina. Biochem Genet. 1978;16:1153–1163. doi: 10.1007/BF00484536. [DOI] [PubMed] [Google Scholar]
- Warren WD, Palmer S, Howells AJ. –1997) Molecular characterization of the cinnabar region of Drosophila melanogaster: identification of the cinnabar transcription unit. Genetica. 1996;98:249–262. doi: 10.1007/BF00057589. [DOI] [PubMed] [Google Scholar]
- Wierenga RK, Drenth J, Schulz GE. Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J Mol Biol. 1983;167:725–739. doi: 10.1016/s0022-2836(83)80106-5. [DOI] [PubMed] [Google Scholar]
- Wu HQ, Guidetti P, Goodman JH, Varasi M, Ceresoli-Borroni G, Speciale C, Scharfman HE, Schwarcz R. Kynurenergic manipulations influence excitatory synaptic function and excitotoxic vulnerability in the rat hippocampus in vivo. Neuroscience. 2000;97:243–251. doi: 10.1016/s0306-4522(00)00030-0. [DOI] [PubMed] [Google Scholar]