Summary
In Arabidopsis, two floral homeotic genes APETALA2 (AP2) and AGAMOUS (AG) specify the identities of perianth and reproductive organs, respectively, in flower development. The two genes act antagonistically to restrict each other to their proper domains of action within the floral meristem. In addition to AG, which antagonizes AP2, miR172, a microRNA, serves as a negative regulator of AP2. In this study, we showed that AG and miR172 have distinct functions in flower development and that they largely act independently in the negative regulation of AP2. We uncovered functions of miR172-mediated repression of AP2 in the regulation of floral stem cells and in the delineation of the expression domain of another class of floral homeotic genes. Given the antiquity of miR172 in land plants, our findings have implications for the recruitment of a microRNA in the building of a flower in evolution.
Keywords: miR172, flower development, stem cells, B function, APETALA2, AGAMOUS
Introduction
The shoot apical meristem (SAM) formed during embryo-genesis is the ultimate source of all above-ground structures of the plant. The dynamic balance between differentiation and stem cell renewal in the SAM is achieved through a negative feedback loop involving the CLAVATA (CLV)-1, -2 and -3 genes, and a homeodomain transcription factor, WUSCHEL (WUS) (reviewed in Brand et al., 2001; Clark, 2001; Sharma et al., 2003). CLV3 encodes a small, secreted protein, presumably a ligand for the potential transmembrane receptors CLV1 and CLV2. WUS RNA is found in a small number of cells underneath the stem cells. While WUS specifies the overlying cells as stem cells, CLV-mediated signaling from the stem cells in turn restricts the WUS expression domain. APETALA2 (AP2), a transcription factor known to act in floral patterning and seed development (Jofuku et al., 1994), regulates stem cell maintenance in the SAM through the CLV–WUS pathway (Würschum et al., 2006).
The CLV–WUS pathway also functions in the floral meristem to regulate floral stem cells (reviewed in Sharma et al., 2003). Unlike the SAM, however, the floral meristem terminates upon production of the final whorl of organs, carpels. The timely termination of the floral meristem requires AGAMOUS (AG), a MADS-domain transcription factor (Lenhard et al., 2001; Lohmann et al., 2001; Yanofsky et al., 1990). In severe ag mutants, the floral meristem continues to put out floral organs to produce a flowers-within-flower phenotype (Bowman et al., 1989).
In Arabidopsis flowers, two types of perianth organs, sepals and petals, are found in the outer two whorls and the reproductive organs, stamens and carpels, are found in the inner two whorls. The identities of the four types of floral organs are specified by three classes of regulatory genes, the A, B and C genes (reviewed in Jack, 2004; Krizek and Fletcher, 2005). The class A genes, APETALA1 (AP1) and AP2, specify sepal identity in whorl 1 and petal identity in whorl 2 together with the class B genes APETALA3 (AP3) and PISTILLATA (PI). The B genes specify stamen identity in whorl 3 together with the C gene AG. AG determines carpel identity in whorl 4. Consistent with the spatially restricted activities of the ABC genes in the floral meristem, transcripts of the ABC genes, with the exception of AP2, are found in two adjacent whorls. Studies on flower development in other plant species suggest that the role of A function in specifying perianth identity is restricted to Brassicaceae (reviewed in Litt, 2007). Even in Brassicaceae, it is possible that the sole role of the A function is to specify floral meristem identity, and perianth organs (such as sepals) represent the ground state of floral organs (reviewed in Litt, 2007). Whether AP2 plays a direct or indirect role in specification of perianth identity does not influence the interpretations of our studies in this paper.
Antagonism between AP2 and AG is crucial in floral patterning. Loss-of-function ap2 alleles have reproductive organs in place of perianth organs, indicating the expansion of AG activity into the outer two whorls (Bowman et al., 1991b). Loss-of-function ag alleles have petals in place of stamens and a new flower in place of carpels (Bowman et al., 1991b). The replacement of all reproductive organs by perianth organs suggests that AP2 is active throughout the flower in ag mutants. At the molecular level, AP2 restricts AG activity to the inner two whorls by preventing AG transcription in the outer two whorls (Drews et al., 1991; Sieburth and Meyerowitz, 1997). AG, however, does not repress AP2 expression and AP2 RNA is found throughout the flower (Jofuku et al., 1994). It is unknown how AG counteracts AP2 activity in the inner two whorls. Ectopic AG expression in the outer two whorls results in phenotypes resembling those of ap2 mutants, indicating that ectopic AG is able to counteract AP2 activity in the outer two whorls (Mizukami and Ma, 1992).
In recent years, a microRNA, miR172, has been demonstrated to be another negative regulator of AP2 (Aukerman and Sakai, 2003; Chen, 2004). miR172 is initially present throughout the floral meristem but is concentrated in the inner two whorls after floral stage 7 (Chen, 2004). Misexpression of MIR172 genes with the 35S promoter results in flowers resembling ap2 mutants (Aukerman and Sakai, 2003; Chen, 2004). Expression of a miR172-resistant version of AP2 cDNA, AP2m3, but not wild-type AP2 cDNA, leads to dramatic floral phenotypes, indicating that miR172 acts in the flower to repress AP2 (Chen, 2004). Aspects of the 35S::AP2m3 phenotypes are similar to those of ag mutants (Bowman et al., 1991b), such as the loss of floral determinacy and the transformation of reproductive organs into perianth organs. This raises the possibility that miR172 mediates the negative regulation of AP2 by AG. However, these data do not preclude miR172 and AG acting as independent negative regulators of AP2 in flower development.
In this study, we evaluated the role of miR172-mediated regulation of AP2 in flower development by analyzing the effects of expressing miR172-resistant AP2 under its own promoter. We found that this regulation is crucial for floral patterning in the inner two whorls. We uncovered functions of miR172, and thereby probably of AP2, in the control of floral stem cell fate and in the delineation of the inner boundary of the B gene expression domain. Furthermore, we showed that miR172 and AG have distinct roles in flower development and act independently as negative regulators of AP2.
Results
Effects of impaired AP2 regulation by miR172 on flower development
Previous studies demonstrated that miR172 is a negative regulator of AP2 in the flower but failed to delineate the role of this regulation in flower development (Aukerman and Sakai, 2003; Chen, 2004). Expression of AP2m3 with the strong 35S promoter led to two major types of floral defect (Figure 1a–c; Chen, 2004). Type I flowers had numerous petals surrounding an indeterminate floral meristem (Figure 1b) while type II flowers had numerous stamens flanking an indeterminate floral meristem (Figure 1c). It was unclear which phenotype, if any, reflected the outcome of loss of miR172 function because the strong 35S promoter may have contributed to aspects of the phenotypes. miR172 is encoded by at least five genes in the Arabidopsis genome. A mir172a-1 mir172a-2 double mutant does not exhibit any floral defects (LZ and XC, unpubl. result). A quintuple mutant in the MIR172 genes may be necessary to evaluate the developmental role of miR172. In the absence of such a mutant, we sought to evaluate the function of miR172-mediated regulation of AP2 in flower development by analyzing lines that express AP2m3 under the control of the AP2 regulatory sequences including the promoter, 5′ untranslated region (UTR) and 3′ UTR.
Figure 1.
Floral phenotypes of 35S::AP2m3 and pAP2::AP2m3 transgenic lines.
(a) Wild type.
(b) 35S::AP2m3, type I.
(c) 35S::AP2m3, type II.
(d) pAP2::AP2m3.
(e, f) Scanning electron micrographs of pAP2::AP2m3 flowers. (e) A close-up of the center of the flower showing the indeterminate floral meristem and the spiral arrangement of the emerging stamen primordia. (f) Occasional carpelloid organs with stigmatic papillae (arrow) are found in pAP2::AP2m3 flowers. Scale bars: 300 μm in (e) and 1 mm in (f).
We cloned a 5.4-kb fragment upstream of the AP2 cDNA as the AP2 promoter. This fragment encompassed the upstream sequence previously used in an AP2 clone to rescue ap2 mutant phenotypes (Jofuku et al., 1994). When fused to the GUS reporter and introduced into wild-type plants, this sequence led to ubiquitous GUS expression in the flower (LZ and XC, unpubl. result) in agreement with the ubiquitous presence of AP2 RNA detected by in situ hybridization (Jofuku et al., 1994). We fused this fragment to the full-length wild-type (AP2WT) or AP2m3 cDNAs and introduced the pap2::AP2WT or pAP2::AP2m3 constructs into wild-type Arabidopsis plants. None of the pAP2::AP2m3 lines had flowers with the type I 35S::AP2m3 phenotypes, suggesting that the type I phenotypes were partly because of the 35S promoter. Over 30% of the independent T1 pAP2::AP2m3 transgenic lines, but none of the pAP2::AP2WT lines, showed floral defects similar to the type II 35S::AP2m3 lines. In brief, the outer two floral whorls were normal in organ identity, but numerous stamens were found internal to the petals (Figure 1d). Scanning electron microscopy showed that the stamen primordia arose in a spiral, rather than whorled, phyllotaxy (Figure 1e). In some lines, large numbers of unfused carpels were found internal to the numerous stamens, or occasional unfused carpels were found among the numerous stamens (Figure 1f). Similar phenotypes were also present in some 35S::AP2m3 type II lines.
As pAP2::AP2m3 transgenic lines mainly displayed type II 35S::AP2m3 phenotypes, we conclude that the type II phenotype is a better measure of the consequence of impaired miR172-mediated repression of AP2 in flower development. To our knowledge, phenotypes similar to those of pAP2::AP2m3 flowers have not been found in known floral mutants, suggesting that miR172-mediated regulation of AP2 performs a previously unknown function in flower development. In fact, the presence of stamens and sometimes carpels in pAP2::AP2m3 flowers suggests that miR172, in contrast to AG, does not play a major role in the specification of reproductive organ identities.
In order to further characterize the novel phenotypes of pAP2::AP2m3 flowers to better understand the role of miR172 in flower development, it was necessary to compare the effects of the transgene in various known floral mutant backgrounds. This required crossing the transgene from an established pAP2::AP2m3 line into various mutants so that transgene location and number would remain the same across different genotypes to warrant phenotypic comparison. However, it was not possible to do so for pAP2::AP2m3 lines as they were male and female sterile. Although numerous stamens were present, they seldom bore pollen. In contrast, type II 35S::AP2m3 lines, which were almost identical to pAP2::AP2m3 in floral patterning defects, were fully male fertile (but female sterile). The difference was probably because of the 35S promoter not being as active as the AP2 promoter in the pollen. As we were only focusing on early patterning roles of miR172, type II 35S::AP2m3 lines were an excellent genetic resource to represent impaired AP2 regulation by miR172. We first established several type II 35S::AP2m3 lines in which the transgene was in a single locus. The lines were maintained by crossing 35S::AP2m3 to wild type such that the transgene was always in a hemizygous configuration. The transgene was introduced into other genetic backgrounds by two consecutive crosses into various recessive mutants such that the transgene was hemizygous in homozygous mutant backgrounds.
To confirm that the phenotypes observed for 35S::AP2m3 in various floral mutant backgrounds truly reflected those of pAP2::AP2m3 in the corresponding genetic backgrounds, we also transformed various floral mutants with the pAP2::AP2m3 construct and examined multiple T1 transgenic lines. pAP2::AP2m3 was introduced into wus-1, lfy-6, ufo-2, ap3-3, pi-1, and clv3-1. The phenotypes of pAP2::AP2m3 in these mutant backgrounds (Supplementary Figure S1) were nearly identical to those of 35S::AP2m3 in the corresponding mutant backgrounds. Our analyses as described below were conducted largely with the 35S::AP2m3 lines. From this point on, we simply refer to the established type II 35S::AP2m3 lines as 35S::AP2m3, which we use to represent impaired miR172-mediated regulation of AP2.
Role of miR172 in the regulation of floral stem cells
At least two parallel genetic pathways regulate floral stem cells through WUS. CLV signaling restricts the number of WUS-expressing cells. Loss-of-function mutations in CLV genes result in an enlarged WUS expression domain in the floral meristem and an enlarged floral meristem that produces a larger number of floral organs (Schoof et al., 2000). AG, however, does not affect the domain of WUS expression but shuts down WUS expression at around stage 7 when carpel primordia are formed (Lenhard et al., 2001; Lohmann et al., 2001). The CLV and AG pathways probably act in parallel as the clv1 ag double mutant has greatly enlarged and often fasciated floral meristems, a phenotype that is more severe than that of either single mutant (Clark et al., 1993).
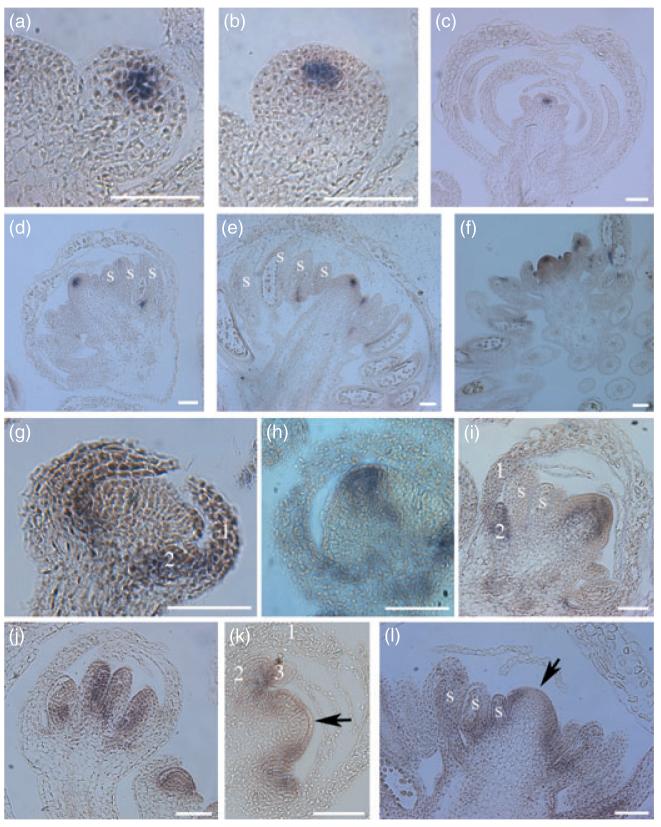
The fact that 35S::AP2m3 flowers had numerous stamens flanking an indeterminate floral meristem suggested that miR172-mediated repression of AP2 played a role in the control of floral stem cells. Introducing the wus-1 mutation into the 35S::AP2m3 background completely abolished the indeterminate phenotype (Figure 2a,b). In fact, 35S::AP2m3 wus-1 flowers were identical to wus-1 flowers in that the floral meristem terminated in a single stamen. The complete epistasis of wus-1 to 35S::AP2m3 indicated that the effect of the transgene is mediated by WUS. A probable underlying mechanism is that de-repressed AP2 expression in 35S::AP2m3 results in reduced AG expression and secondarily to prolonged WUS expression. Indeed, the onset and the initial domain of WUS expression in 35S::AP2m3 flowers were similar to wild type (Figure 3a,b), but WUS remained expressed in 35S::AP2m3 throughout flower development as in ag mutants (Figure 3c–e). As in wild type, AG RNA was found in the center of stages 3–4 35S::AP2m3 floral meristems (Figure 3j and data not shown; Drews et al., 1991). AG RNA continues to be present in the inner two whorls throughout flower development in wild type (Figure 3j; Bowman et al., 1991a). In 35S::AP3m3 flowers, however, AG RNA was much reduced, but apparently not totally abolished, at the very apical end of the meristem starting from stage 6 (Figure 3k,l). These observations were consistent with AP2m3 promoting stem cell fate, at least in part, through the repression of AG.
Figure 2.
35S::AP2m3 or pAP2::AP2m3 in various genetic backgrounds. In all cases, the trans-gene is hemizygous and is in a homozygous mutant background.
(a) The 35S::AP2m3 line that was used to cross with various mutants. Its phenotype serves as a reference for comparison with that of 35S::AP2m3 in various genetic backgrounds.
(b) 35S::AP2m3 wus-1.
(c) A flower from a pAP2::AP2m3 clv3-1 T1 transgenic plant. Note the mass of apparently undifferentiated cells in the center.
(d, e) An ag-1 flower (d) and a 35S::AP2m3 ag-1 flower (e) at the same magnification.
(f) 35S::AP2m3 pi-1.
Figure 3.
In situ hybridization for the detection of WUS, AP1, and AG RNAs in flowers of various genotypes.
(a–f) Detection of WUS RNA. (a) A wild-type stage 2 flower. (b) A 35S::AP2m3 stage 2 flower. (c) An old ag-1 flower. (d–f) 35S::AP2m3 flowers of increasingly advanced stages in development. (g–i) Detection of AP1 RNA. (g) A wild-type stage 6 flower. (h,i) 35S::AP2m3 stage 6 (h) and older (i) flowers.
(j–l) Detection of AG RNA. (j) wild-type stage 8 (left) and stage 4 (right) flowers. (k,l) 35S::AP2m3 stage 6 (k) and older (l) flowers. The arrows mark the center of the meristems where AG RNA levels are low compared with the surrounding areas. The numbers indicate the floral whorls. ‘s’ represents stamens or stamen primordia. Scale bars: 50 μm.
Our genetic evidence indicated that AP2m3 also acted independently of AG in the regulation of floral stem cells. In the ag-1 background, 35S::AP2m3 still led to a dramatic increase in floral organ number (Figure 2d,e) and floral meristem size (compare Figure 3c with 4e), indicating that AP2m3 exerted a large effect on floral stem cells through an AG-independent, perhaps the CLV, pathway. In fact, 35S::AP2m3 and clv mutants exhibited a number of similarities. Firstly, 35S::AP2m3 floral meristems were taller than wild type starting at stage 3 (Figure 3g,h), a phenotype similar to clv mutants (Clark et al., 1993). Although the initial domain of WUS expression was normal in young 35S::AP2m3 flowers (Figure 3b), an obvious expansion of the WUS expression domain was often observed in later flowers, and very late stage flowers showed an expansion of WUS expression to the entire meristem and to young organ primordia (Figure 3f). Secondly, 35S::AP2m3 and clv1-4 flowers showed near identical patterns of AG and AP1 expression. AG RNA was reduced in the center of the meristem in 35S::AP2m3 flowers (Figure 3k,l) as well as in clv1-4 flowers (Clark et al., 1993). While AP1 RNA is normally restricted to the outer two floral whorls after stage 3 in wild-type flowers (Figure 3g; Mandel et al., 1992), it was found in the center of the floral meristems in 35S::AP2m3 (Figure 3h,i) and in clv1-4 (Clark et al., 1993) plants. Finally, introduction of ag-1 into 35S::AP2m3 resulted in a synergistic interaction such that the floral meristem was greatly expanded (compare Figure 4d,e), which was similar to the effects of combining ag and clv mutations (Clark et al., 1993). These observations support but do not prove that miR172-mediated repression of AP2 regulates floral stem cells through the CLV pathway. As AP2 was shown to regulate stem cell maintenance in the SAM through the CLV pathway (Würschum et al., 2006), it is likely that AP2 has a similar function in floral meristems.
Figure 4.
In situ hybridization for the detection of AP3 and PI RNAs in flowers of various geno-types.
(a–e) AP3 in situ hybridization.
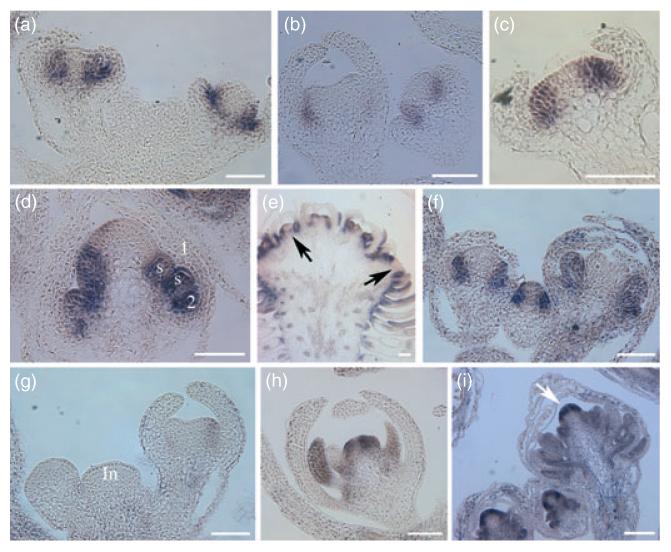
(a) Stage 6 (left) and stage 4 (right) wild-type flowers.
(b) Stage 6 (left) and stage 4 (right) ap2-2 flowers.
(c) A stage 4-5 ap2-2 ag-1 flower.
(d) A stage 6 35S::AP2m3 flower.
(e) An old 35S::AP2m3 ag-1 flower in which the meristem has fasciated and has been subdivided into many second-order floral meristems. The arrows indicate that the center of the second-order meristems has little AP3 RNA.
(f–i) PI in situ hybridization. (f) Three wild-type flowers of various stages with strong PI expression in the second and third whorls. (g) A longitudinal section through the inflorescence (In) of ap2-2. PI expression is barely detectable in the stage 2 (left) and stage 6 (right) flowers. (h) An ap2-2 ag-1 flower showing PI expression. (i) Three 35S::AP2m3 flowers of various stages showing strong and ectopic (arrows) PI expression. The numbers indicate the floral whorls. ‘s’ represents stamen primordia. Scale bars: 50 μm.
Assuming that 35S::AP2m3 acts in the CLV pathway, we tested whether it regulated floral stem cells exclusively through the CLV pathway. If this were the case, we would expect the phenotypes of 35S::AP2m3 flowers to be identical to those of 35S::AP2m3 clv3-1 flowers. We generated transformants of pAP2::AP2m3 in the clv3-1 background. Flowers of T1 pAP2::AP2m3 clv3-1 plants differed from pAP2::AP2m3 flowers in that they accumulated a mass of apparently undifferentiated tissue in the center (Figure 2c). The fact that pAP2::AP2m3 clv3-1 flowers have more severe phenotypes than either pAP2::AP2m3 or clv3-1 flowers indicates that de-repressed AP2 does not act exclusively through the CLV pathway. In summary, our analysis shows that miR172 is a crucial factor in the regulation of floral stem cells and it acts through an AG-dependent pathway and an AG-independent, perhaps CLV, pathway and that these two pathways converge on WUS.
35S::APm3 leads to an expansion of B gene expression domain
We examined the unexpected role of miR172-mediated repression of AP2 on stamen number in flower development. Specification of stamen identity requires AP3 and PI. The presence of numerous stamens in 35S::AP2m3 suggests that AP3 and PI are expressed in a larger domain covering all the stamen primordia. Introduction of either ap3-3 or pi-1 loss-of-function mutations into 35S::AP2m3 transformed the stamens to sepal-like organs or filaments (Figure 2f and Supplementary Figure S1e), indicating that AP3 and PI were active in a domain containing all the stamens in 35S::AP2m3 flowers. We examined the accumulation of AP3 and PIRNAs in developing flowers by in situ hybridization. In stages 3–5 35S::AP2m3 flowers, AP3 RNA appeared to occupy whorls 2 and 3 as in wild type (data not shown and Figure 4a; Jack et al., 1992). In later 35S::AP2m3 flowers, AP3 RNA continued to be present in all internal stamen primordia (Figure 4d). In wild-type flowers, PIRNA is initially present in whorls 2–4 but the whorl 4 expression soon abates and a whorl 2–3 pattern is established and maintained throughout flower development (Figure 4f; Goto and Meyerowitz, 1994). In 35S::AP2m3 flowers, the initiation of PI expression appeared normal, but PI RNA never disappeared from the center of the meristem (Figure 4i). In addition, PI RNA was present in all stamen primordia (Figure 4i). These data suggested that de-repressed AP2 expression led to an expansion of the AP3/PI expression domain towards the center of the meristem.
Floral meristem identity genes LEAFY (LFY) and UNUSUAL FLORAL ORGANS (UFO) are required for the initiation of B gene expression (Levin and Meyerowitz, 1995; Parcy et al., 1998). We asked whether the expansion of the B gene expression domain caused by 35S::AP2m3 requires the activity of the floral meristem identity genes. Severe mutant alleles in LFY and UFO were introduced into 35S::AP2m3. 35S::AP2m3 lfy-6 and 35S::AP2m3 ufo-2 flowers had sepal-like organs or filaments rather than stamens (Supplementary Figure S1f,g), suggesting that LFY and UFO are necessary for B gene activation by 35S::AP2m3. Indeed, in situ hybridization showed that AP3 and PI signals were patchy and reduced in intensity in 35S::AP2m3 lfy-6 and 35S::AP2m3 ufo-2 flowers (Supplementary Figure S1h,i and data not shown). Therefore, de-repressed AP2 expression was not sufficient to induce AP3 and PI expression. Our data are more consistent with a function of 35S::AP2m3 in affecting the inner boundary of the B gene expression domain rather than activating B gene expression.
Mutations in the SUPERMAN (SUP) gene lead to an increased number of stamens (Bowman et al., 1992). It is possible that the increase in stamen number in 35S::AP2m3 was caused by reduced SUP expression. However, this was unlikely because the 35S::AP2m3 phenotype is much more severe than that of sup loss-of-function alleles, which show a modest number of extra stamens (Bowman et al., 1992). Indeed, SUP expression was not affected in 35S::AP2m3 (Figure 5b).
Figure 5.
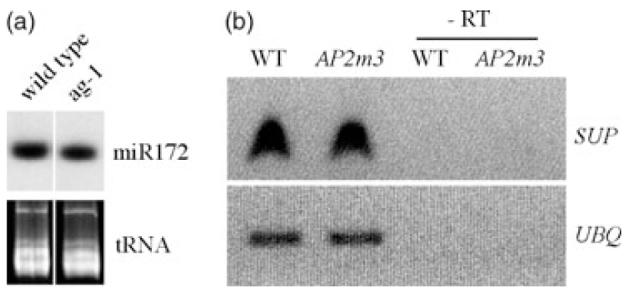
miR172 and SUP mRNA levels in various genotypes.
(a) miR172 levels in inflorescences of wild-type and ag-1 plants as determined by RNA filter hybridization. The region of the gel where tRNAs migrate is shown below the hybridization image to indicate the amount of RNAs loaded.
(b) SUP RNA levels in inflorescences of wild-type and 35S::AP2m3 plants as determined by RT-PCR. UBQ RNA serves as an internal control. Thirty-five and 25 cycles of PCR are shown for SUP and UBQ, respectively, although shorter and longer cycles were performed for each gene to ensure that these numbers of cycles did not result in saturation of the reactions. -RT, no reverse transcription.
AP2 and AG control B gene expression domain in the floral meristem
We asked whether a potential role for AP2 in the control of the B gene expression domain was supported by loss-of-function ap2 mutant defects. Although a role for AP2 in the control of the B gene expression domain had not been proposed, earlier observations (Jack et al., 1992) were consistent with such a role. Jack et al. found that AP3 RNA was at a much lower level and occupied a smaller area in ap2-2 flowers compared with wild type. Furthermore, they reported that AP3 expression was restored by introducing an ag mutation into AP2-2. We confirmed these observations by comparing the intensity and the domain of AP3 RNA among wild-type, ap2-2, and ap2-2 ag-1 flowers on the same glass slide (Figure 4a–c). In addition, we found that PI expression was drastically reduced in ap2-2 (Figure 4f,g) and restored in ap2-2 ag-1 (Figure 4h). Therefore, AP2 promotes the levels of AP3 and PI expression and establishes the size of the expression domain by counteracting AG. The antagonistic interaction between AP2 and AG defines the domain of B function. By repressing AP2 in the inner two whorls, miR172 plays a key role in defining the inner boundary of the B gene expression domain.
The AP2-AG antagonistic interaction appeared to affect AP3 and PI expression somewhat differently. While PI RNA was present in the center of the meristem of 35S::AP2m3 flowers (Figure 4i), AP3 RNA was not (Figure 4d). In fact, AP3 RNA was also absent from the center of the meristem in ag-1 (Figure 4c) or 35S::AP2m3 ag-1 flowers (Figure 4e). This indicates that a gene(s) other than AP2, AG or MIR172 causes the absence of AP3 expression from the center of the meristem.
miR172 and AG act independently on AP2 in flower development
As negative regulators of ap2 in flower development, miR172 and AG may act in the same genetic pathway or in parallel genetic pathways. We evaluated the relationship between AG and miR172 in the negative regulation of AP2. We asked whether one is functional in the absence of the other. Introduction of ag-1 into 35S::AP2m3, whose pheno-type represented impaired AP2 regulation by miR172, resulted in the transformation of stamens into petals (Figure 2a,e), indicating that AG was still active in antagonizing AP2 in the absence of miR172. Conversely, introducing 35S::AP2m3 into ag-1 resulted in a great increase in organ number (Figure 2d,e) and floral meristem size (Figure 3c and 4e), indicating that miR172 was still repressing AP2 in an ag mutant. Consistent with these genetic observations, AG RNA was still present in 35S::AP2m3 flowers (Figure 3k,l) and miR172 was still present in ag-1 inflorescences (Figure 5a). Therefore, miR172 and AG act independently on AP2 in flower development. The drastic floral defects of either 35S::AP2m3 or ag-1 flowers underscore the crucial function of both miR172 and AG as negative regulators of AP2. Previous experiments to mis-express AG with the 35S promoter led to the conclusion that AG is sufficient to counteract AP2 when ectopically present in the outer two whorls. One might extrapolate to assume that AG is normally sufficient to counteract AP2 in the inner two whorls. However, our data show that AG is not sufficient to counteract AP2 in the inner two whorls. Without miR172, elevated AP2 levels led to severe consequences for flower development.
Discussion
miR172-mediated repression of AP2 is crucial for the patterning of the inner two whorls
Our detailed characterization of the pAP2::AP2m3 and type II 35S::AP2m3 phenotypes led to the following conclusions concerning the role of miR172-mediated repression of AP2 in floral patterning (Figure 6). Firstly, miR172-mediated repression of AP2 is crucial for maintaining floral meristem size and for the timely termination of floral stem cells. The role of miR172 on floral meristems is achieved through both an AG-dependent and an AG-independent pathway. Secondly, miR172-mediated repression of AP2 defines the inner boundary of the domain of B gene expression. This role of miR172 is probably achieved through an AG-dependent pathway. Thirdly, miR172-mediated repression of AP2 plays little role in the specification of reproductive organ identities as does AG,as 35S::AP2m3 flowers still have stamens and sometimes carpels. It appears that de-repressed AP2 expression has the most adverse impact on floral patterning in the center of the floral meristem (which would become the fourth whorl in wild-type flowers) such that the meristem continues to proliferate and that the B gene expression domain expands into the center. De-repressed AP2 expression does not affect the identity of the third whorl organs (stamens). Many of the effects on floral patterning by de-repressed AP2 expression, such as prolonged WUS expression and expansion of B gene expression towards the center of the meristem, can be explained by compromised AG expression in the fourth whorl of the flower. However, de-repressed AP2 expression affects floral meristem size independently of AG.
Figure 6.
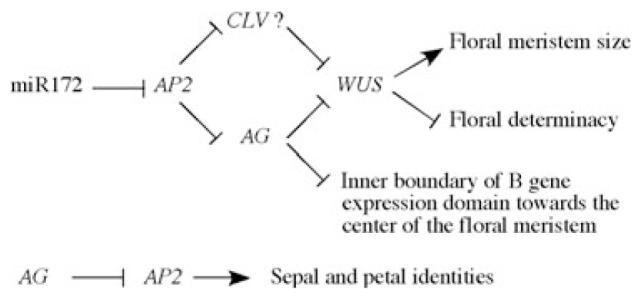
A diagram summarizing the functions of miR172 and AG in flower development as negative regulators of AP2.
miR172-mediated repression of AP2 acts through an AG-dependent pathway to promote floral determinacy and to define the inner boundary of the B gene expression domain. miR172-mediated repression of AP2 also acts in an AG-independent, perhaps the CLV, pathway to restrict floral meristem size. AG antagonizes AP2 to prevent sepal and petal identities in the inner two whorls. Note that miR172-mediated repression of AP2 does not play a role in specification of organ identity.
Role of AP2 in flower development
It is necessary to consider to what extent the consequences of de-repressed AP2 in floral patterning reflect the normal role of AP2. While de-repressed AP2 expression leads to an expansion of the B gene domain, prolonged WUS expression, and increased meristem size, does AP2 normally regulate the B gene expression domain, timing of WUS expression, and floral meristem size? It is possible that AP2 does not normally act in these processes, but elevated levels of AP2 allow it to feed into other pathways that do. The key to addressing the question lies in whether ap2 loss-of-function mutations affect these developmental processes. Evidence in the literature (Jack et al., 1992) and from this study supports the conclusion that AP2 promotes the B gene expression domain by antagonizing AG. Our preliminary studies on WUS expression in ap2-2 mutant flowers did not reveal any difference from wild type (LZ and XC, unpubl. results). Therefore, at present, we do not have evidence from ap2 loss-of-function alleles that AP2 regulates the timing of WUS expression. We have not compared the sizes of ap2 mutant and wild-type floral meristems and thus cannot conclude on whether AP2 plays a role in the regulation of floral meristem size. However, AP2 has been shown to play a role in stem cell maintenance in the SAM through the CLV pathway (Würschum et al., 2006). It is possible that AP2 has a similar function in the control of floral stem cells.
miR172 and AG have distinct roles in floral patterning
What is the relationship between AG and miR172 in floral patterning? AG has long been known to counteract AP2 activity in the inner two whorls, but the underlying mechanism is unknown. Accumulation of AP2 RNA is ubiquitous in the four floral whorls and is similar in wild type and ag mutants (Jofuku et al., 1994), suggesting that AG does not repress AP2 at the transcriptional level. miR172 was found to serve as a translational repressor of AP2 mRNA (Aukerman and Sakai, 2003; Chen, 2004). One attractive hypothesis is that AG regulates the transcription of MIR172 genes and miR172 leads to reduced AP2 protein levels in the inner two whorls. Our studies show that this model is incorrect. Firstly, miR172 levels in wild type and ag-1 flowers are similar, indicating that AG is not required to turn on MIR172 genes. Secondly, the fact that 35S::AP2m3 ag-1 flowers are phenotypically different from either 35S::AP2m3 or ag-1 flowers demonstrates that miR172 is active in ag-1 and that AG is active in 35S::AP2m3. Therefore, miR172 and AG act independently in floral patterning.
miR172 has an ancient origin in land plants (Axtell and Bartel, 2005) and must therefore have been recruited by angiosperms to build the flower. Angiosperm flowers display the basic types of floral organs but vary dramatically in the number of the organs among species. While some species have a small number of stamens in the flower, others, such as representatives of the Magnoliaceae, have numerous stamens arranged in a spiral phyllotaxy as seen in 35S::AP2m3 flowers. One cannot help but wonder whether miR172-mediated repression of AP2 plays a role in the evolution of floral morphology.
Experimental procedures
Constructs and transgenic lines
For the generation of pAP2::AP2m3 and pAP2::AP2WT lines, an AP2 promoter was amplified by PCR from Col genomic DNA with the primers AP2p41 (5′-AAG CTG CAG TTA GGC CCG ACC TAT CGT CCA TC-3′) and AP2p42 (5′-AAA CTG CAG CTA AAG AGA GAG AGA GAA GAA AAT AAA ATA-3′). The PCR product was digested by PstI and ligated into the pPZP211 vector to generate pPZP211-pAP2. The full-length AP2WT and AP2m3cDNAs were obtained by digestion of the plasmids pBSSK-AP2WT and pBSSK-AP2m3 (Chen, 2004) with SacI and were ligated downstream of the AP2 promoter in pPZP211-pAP2. The resulting constructs pAP2::AP2WT and pAP2::AP2m3 were delivered into wild-type Arabidopsis plants of the Landsberg erecta (Ler) ecotype by agroinfiltration.
pAP2::AP2m3 was also transformed into clv3-1, ufo-2, lfy-6, wus-1, ap3-3, and pi-1 mutant backgrounds. For clv3-1 and ufo-2, homozygous plants were used for transformation. For lfy-6, wus-1, ap3-3, and pi-1, populations that segregated each of these mutations were used for transformation.
Introduction of 35S::AP2m3 into various mutant backgrounds
35S::AP2m3 lines in which the transgene was inserted into a single locus were first obtained by crossing 18 independent T1 lines with type II phenotypes as the pollen donor with Ler and following the segregation of the phenotypes in the F1 populations. Ones with a 1:1 segregation of 35S::AP2m3 versus wild-type phenotypes were retained. Because of the female sterility of the lines, they were maintained in the form of F1 seeds of crosses to Ler.
The 35S::AP2m3 transgene from the single-locus lines was introduced into ap3-3, pi-1, ag-1, wus-1, ufo-2, and lfy-6 backgrounds by two consecutive crosses with the homozygous mutants (in the case of ap3-3, pi-1, ufo-2, and lfy-6) or heterozygous mutants when the homozygous mutants were female sterile (in the case of ag-1 and wus-1). With the homozygous mutants, a cross was first performed with an 35S::AP2m3 line and F1 plants exhibiting 35S::AP2m3 phenotypes (they would be hemizygous for the 35S::AP2m3 transgene and heterozygous for the mutation) were crossed to the corresponding homozygous mutants again. F1 plants from the second crosses were screened to identify the homozygous mutants containing the transgene. In the crosses to ag-1/+ and wus-1/+, F1 plants with 35S::AP2m3 phenotypes were first genotyped to identify ones that were also ag-1/+ or wus-1/+. These plants were then crossed again with ag-1/+ or wus-1/+ plants. 35S::AP2m3 in ap3-3, pi-1, ufo-2, ag-1 and lfy-6 backgrounds was recognized by novel phenotypes distinguishable from both parents. The 35S::AP2m3 crosses to wus-1/+ did not give any phenotypes other than those of 35S::AP2m3 or wus-1. 35S::AP2m3 wus-1 plants were identified by PCR genotyping plants with wus-1 phenotypes for the presence of 35S::AP2m3.
In situ hybridization, RNA filter hybridization, and RT-PCR
In situ hybridization was carried out as described (Chen et al., 2002) and RNA filter hybridization to detect miR172 was performed as described (Park et al., 2002). For RT-PCR, total RNA was first isolated from wild-type and 35S::AP2m3 inflorescences as described (Chen et al., 2002). 2.5 μg of total RNA from each genotype was treated with DNaseI and subjected to reverse transcription. A portion of the reverse transcription reaction was amplified by PCR with SUP (SUP-F: 5′-CTT GGA GCT TGA GAT TGG-3′ and SUP-R: 5′-CGG TAA CAA GCG CAT ACA-3′) and UBQ (NUBQ: 5′-GGT GCT AAG AAG AGG AAG AAT-3′ and CUBQ: 5′-CTC CTT CTT TCT GGT AAA CGT-3′) primers. Twenty, 25, and 30 cycles of PCR were performed for UBQ and 30, 35, and 40 cycles of PCR were performed for SUP to ensure that the reactions did not reach saturation at the number of cycles presented in Figure 5.
Scanning electron microscopy
Scanning electron microscopy was performed with fresh flowers using a Hitachi TM1000 scanning electron microscope (http://www.hitachi.com/).
Supplementary Material
Acknowledgements
We thank Dr Randall Kerstetter for helpful discussions, Dr David Carter for help with scanning electron microscopy, Drs Zhiyong Yang and Bin Yu for critical reading of the manuscript, and NIH (GM61146 to XC) for support. TTD is supported by a National Science Foundation IGERT grant (DGE0504249).
References
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM. Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell. 1991a;3:749–758. doi: 10.1105/tpc.3.8.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991b;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM. SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development. 1992;114:599–615. doi: 10.1242/dev.114.3.599. [DOI] [PubMed] [Google Scholar]
- Brand U, Hobe M, Simon R. Functional domains in plant shoot meristems. Bioessays. 2001;23:134–141. doi: 10.1002/1521-1878(200102)23:2<134::AID-BIES1020>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE. Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2001;2:276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Jack T. Molecular and genetic mechanisms of floral control. Plant Cell. 2004;16(Suppl):S1–S17. doi: 10.1105/tpc.017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, Montagu MV, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet. 2005;6:688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Levin JZ, Meyerowitz EM. UFO:an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell. 1995;7:529–548. doi: 10.1105/tpc.7.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A. An evaluation of A-function: evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 2007;168:73–91. [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mizukami T, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. A genetic framework for floral patterning. Nature. 1998;395:561–566. doi: 10.1038/26903. [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Sharma VK, Carles C, Fletcher JC. Maintenance of stem cell populations in plants. Proc. Natl Acad. Sci. USA. 2003;100(Suppl 1):11823–11829. doi: 10.1073/pnas.1834206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Gross-Hardt R, Laux T. APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell. 2006;18:295–307. doi: 10.1105/tpc.105.038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.