Abstract
Background
Previous studies have shown important effects of stromal elements in carcinogenesis. To explore the tumor-stromal relationship in esophageal neoplasia, we examined methylation of COX-2 (PTGS2), a gene etiologically associated with the development of gastrointestinal cancers, in adjacent foci of epithelium, subepithelial lymphocytes and non-lymphocytic stromal cells found in sections of normal squamous epithelium, squamous dysplasia and invasive esophageal squamous cell carcinoma.
Methods
Adjacent foci of epithelium, subepithelial lymphocytic aggregates and non-lymphocytic stromal tissues were laser microdissected from six fully embedded, ethanol fixed, esophagectomy samples from Shanxi, China, a high-risk region for esophageal cancer. Promoter CpG site-specific hypermethylation status of COX-2 was determined using real-time methylation specific PCR (qMS-PCR) based on Taqman Chemistry. The methylation status of a subset of samples was confirmed by pyrosequencing.
Results
Forty-nine microdissected foci were analyzed. COX-2 gene methylation was significantly more common in subepithelial lymphocytes (12/16 (75% of all foci)) than in epithelial foci (3/16 (19%)) or foci of non-lymphocytic stromal tissues (3/17 (18%)) (Fisher’s Exact p=0.05). Two of three epithelial samples and all three stromal samples that showed COX-2 methylation were adjacent to foci of methylated subepithelial lymphocytes. Pyrosequencing confirmed the methylation status in a subset of samples.
Conclusions
In these esopohageal cancer patients, COX-2 gene methylation was more common in subepithelial lymphocytes than in adjacent epithelial or stromal cells in both grades of dysplasia and in foci of invasive cancer. These findings raise the possibility that methylation of subepithelial lymphocytes may be important for tumorigenesis. Future studies of gene methylation should consider separate evaluation of epithelial and non-epithelial cell populations.
Condensed abstract
COX2 (PTGS2) gene methylation increases with disease severity and is more common in subepithelial lymphocytes than in adjacent epithelial or stromal cells in dysplastic and early invasive esophageal squamous cell cancer foci.
Keywords: esophagus, neoplasms, cancer, cyclooxgenase-2, precancerous conditions, methylation, lymphocytes, squamous cell cancer
1. Introduction
Understanding the influence of stromal elements on carcinogenesis continues to unfold. In vitro studies have shown significantly faster tumor growth when tumor cells are mixed with carcinoma-associated fibroblasts, and it has been hypothesized that observed epithelial cellular responses to cytokines may be secondary to the bioactivity of matrix-associated macromolecules whose expression is itself under primary cytokine control [1–3]. In addition, mucosal associated lymphoid tissue, which is particularly abundant along the aerodigestive tract and is thought to be important for defending against pathogenic organisms, may also play a role in neoplastic progression by modulating pro-carcinogenic and anti-carcinogenic immunologic responses to oncogenically transformed cells [4]. Consequently, improved understanding of the interactions between supporting and epithelial tissue constituents may enhance our insight into the mechanisms involved in neoplastic progression.
Esophageal Squamous Cell Carcinoma (ESCC) continues to be a significant cause of morbidity and mortality worldwide, and it is the fourth most common cause of cancer death in China [5]. Within China, the Taihang mountain region in Hebei, Henan and Shanxi Provinces has the highest incidence and mortality rates, reaching more than 100/100,000 persons/year [6]. Efforts are ongoing to understand the etiology of this cancer in such high-risk areas, and to develop more effective cancer prevention, screening, and treatment strategies.
Many recent epigenetic investigations have focused on gene methylation, the addition of methyl groups to cytosines in CpG islands, which can silence gene transcription. CpG islands are 0.5–2 kilobase regions rich in cytosine-guanine dinucleotides that are present in the 5’ region of about half of all human genes [7]. Methylation of these CpG islands have been shown to cause silencing of several genes in GI cancers, including those involved in transcription, apoptosis, and calcium signaling [8]. Despite these advances, the role of gene methylation in the neoplastic progression of ESCC remains poorly understood.
One important gene that is susceptible to gene methylation and is etiologically implicated in the development of gastrointestinal cancers is COX-2 (PTGS2) [9]. Inhibition of COX-2 by non-specific NSAIDS or specific inhibitors of COX-2 causes cell death in gastrointestinal cancer cells [10,11].
The current study explores tumor-stromal relationships in esophageal neoplasia by examining methylation of COX-2 (PTGS2) in adjacent microdissected foci of epithelium, subepithelial lymphocytic aggregates and non-lymphocytic stromal cells in sections of normal squamous epithelium, squamous dysplasia, and invasive esophageal squamous cell carcinoma.
2. Materials and methods
Patients presenting in 1998 to the Shanxi Cancer Hospital in Taiyuan, Shanxi province, People’s Republic of China who were diagnosed with ESCC and were considered candidates for curative surgical resection were identified and recruited to participate in the study. The study was approved by the Institutional Review Boards of the Shanxi Cancer Hospital and the United States National Cancer Institute. For this study, a total of six patients (5 males and a female) who underwent esophagectomy for ESCC were selected. None of the patients received pre-operative chemo or radiotherapy.
Each esophageal resection specimen was fixed in ethanol and then cut entirely into 0.4-cm wide columns and into 2.0-cm long rows suitable for histologic processing. Each section was histologically reviewed and given a diagnosis based on the worst epithelial change present, according to criteria described previously (low-grade dysplasia (LGD) = mild dysplasia; high-grade dysplasia (HGD) = moderate or severe dysplasia) [12].
Laser Capture Microdissection
Sections from normal, LGD, HGD, and invasive cancer were selected from each case based on the ease with which the lesional tissue could be microdissected and analyzed. Serial 10-micron sections were then cut from each tissue block and adjacent foci of epithelium, subepithelial lymphocytic aggregates and non-lymphocytic stromal tissues were laser microdissected (Arcturus Microdissection System, Molecular Devices, Sunnyvale, CA) under direct light microscopic visualization, using methods described previously [13].
Bisulfite modification and real-time methylation-specific PCR
Gene-specific hypermethylation status was determined using methylation-specific PCR (qMS-PCR) based on Taqman Chemistry (Applied Biosystem, Foster City, CA) as previously described [14, 15]. Bisulfite modification was performed as previously described, using a primer and hybridization probe specifically designed to bind bisulfite-converted sequences of CpG islands in the promoter region of the gene [15]. The primers and probe sequences for the internal reference gene, ß-actin, have been previously published [14]. The sequences for the PTGS2 assay are as follows: sense: 5’GGA AGC GTT CGG GTA AAG ATT-3’; anti-sense: 5’-CGC CCC AAA CGC ACA A-3’; and probe: 6FAM-AAG AAG AAA AGA TAT TTG GCG GAA-3’-MGB (molecular grove binding).
The caps from the dissection were incubated at 37C overnight in 25 µL of lysis buffer (10mM Tris, PH8.0, 1mM EDTA, 1% Tween-20, and 1 mg/mL proteinase K) and boiled at 95°C for 10 min to deactivate the proteinase K. The prepared lysate was used directly for subsequent DNA methylation assays. The qMS-PCR reactions were carried out in a volume of 10 µl using Taqman Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems, Foster City, CA). Each PCR reaction mixture contained 300nM of primer, 100nM of probe, and 1X Taqman Buffer. Amplification and detection were carried out using the following profile: one step at 50°C for 2 min, one step at 95°C for 10 min, and 50 cycles at 95°C for 15 sec and 60°C for 1 min. The degree of gene methylation of each sample was assessed by extrapolation of standard curves generated with serial dilutions with known amounts of methylated DNA ranging from 10 ng to 0.01 ng and unmethylated DNA. Percent methylation was derived using the following formula: (ng gene/ng β-actin) X 100. The samples were categorized as unmethylated, low methylation (1%–50%), or high methylation (51%–100%). All but five samples had sufficient DNA and were well within the assay’s range of sensitivity and reproducibility based on the internal reference standard (β-actin) (Ct scores below 38 for input DNA). All samples were run in duplicate and the average values used.
Pyrosequencing
To confirm methylation status of PTGS2, we used a second technique, pyrosequencing, on a subset of the samples (3 epithelial, 4 lymphocytes, and 3 stromal samples). Pyrosequencing (Biotage, Charlottesville, VA, USA) was performed following described protocols [16.17]. A PCR product from each sample was generated in a 25 µl reaction containing 1× AmpliTaq Gold Master Mix, 2 U of AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA), 250 µM dNTPs, 0.25 µM forward primer PTGS2-F-5- GGA GAT TAG TTT AGA ATT GGT TTT-3, 0.25 µM reverse primer PTGS2-R-5’-biotin- AAT CCC CAC TCT CCT ATC TAA TCC -3’, and 4 µl of bisulfite-modified sample DNA. The amplifications were carried out at 95°C for 10 min, followed by a six-cycle touchdown PCR protocol of 95°C for 1 min, 63°C for 1 min with −1 °C for each cycle to 58°C, and 72°C for 1 min. This was followed by 44 cycles at 95°C for 1 min, 60°C for 1 min, 72°C for 1 min, and a 10 min extension at 72°C. Single-stranded DNA from 10µl of each PCR sample was generated using the PSQ 96 sample preparation vacuum filtration device following the manufacturer’s instructions (Biotage, Charlottesville, VA). The single-stranded product was annealed to 0.3 µM of the sequencing primer 5-ATT AGT TTA GAA TTG GTT TT-3 and placed at 85°C for 2 min and cooled to room temperature for 5 min. Pyrosequencing was performed on a PSQ96 HS system (Biotage) with the Biotage reagent kit (Biotage) according to the manufacturer’s instructions. Methylated DNA at different percentages (100%, 50%, 10% and 0%) was employed as controls. The raw data were analyzed with the allele quantification algorithm of the software provided (Biotage). The assay spans four CpG sites and percent methylation was averaged across the four sites and the data categorized as unmethylated, low methylation (1%–50%), or high methylation (51–100%).
Statistical Analysis
All statistical analyses were performed on STATA 8.0 (College Station, TX). The Fisher’s exact test was used to test for differences in the frequency of methylation among foci of epithelium, subepithelial lymphocytes and non-lymphocytic stromal tissues.
3. Results
Fifty-four microdissected ethanol fixed foci from six esophageal resections were evaluated, and 49 foci had sufficient DNA for analysis. The full histologic spectrum was not present in every case. The final analytical group contained 2 foci of normal epithelium, 5 of low-grade dysplasia, 6 of high-grade dysplasia and 5 invasive cancers (Table I).
Table I.
COX-2 (PTGS2) Methylation in adjacent Epithelium (E), Lymphocytes (L), and Stromal cells (S), by epithelial histologic severity, in six ESCC patients
| Case No | Histology | E | L | S |
|---|---|---|---|---|
| 3904 | Low-grade dysplasia | |||
| Low-grade dysplasia | ||||
| High-grade dysplasia | ND | |||
| Invasive Cancer | ||||
| 3564 | Normal | ND | ||
| Low-grade dysplasia | ||||
| Invasive Cancer | ||||
| 3696 | High-grade dysplasia | |||
| Invasive Cancer | ||||
| 3776 | Low-grade dysplasia | |||
| High-grade dysplasia | ND | |||
| Invasive Cancer | ND | |||
| 3348 | Normal | |||
| Low-grade dysplasia | ||||
| High-grade dysplasia | ||||
| Invasive Cancer | ND | |||
| 4066 | High-grade dysplasia | |||
| High-grade dysplasia |
ND=insufficient DNA
White=0 %, Gray=1–50%, Black=51+% Methylation.
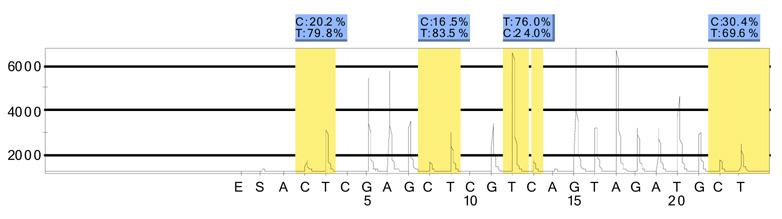
Real-time methylation-specific PCR showed COX-2 (PTGS2) gene methylation to be significantly more common in subepithelial lymphocytes (12/16 (75% of all foci)) than in epithelial foci (3/16 (19%)) or foci of non-lymphocytic stromal tissues (3/17 (18%)) (Fishers Exact p=0.05) (Table II). Two of three epithelial samples and all three stromal samples that showed COX2 methylation were adjacent to foci of methylated subepithelial lymphocytes. Pyrosequencing of a subset of samples was concordant with qMS-PCR for the presence of methylation (10/10 samples (100%)) (Figure 1).
Table II.
Summary data for COX-2 (PTGS2) methylation
| Histology | Epithelium | Lymphocytes | Stroma |
|---|---|---|---|
| Foci+/Total Foci (%) | Foci+/Total Foci (%) | Foci+/Total Foci (%) | |
| Normal | 1/2 (50%) | 0/1 ( 0%) | 0/2 ( 0%) |
| Low-grade dysplasia | 0/5 ( 0%) | 3/5 ( 60%) | 0/5 ( 0%) |
| High-grade dysplasia | 1/4 (25%) | 6/6 (100%) | 3/6 (50%) |
| Cancer | 1/5 (12%) | 3/4 ( 75%) | 0/4 ( 0%) |
| Total | 3/16 (19%) | 12/16 (75%) | 3/17 (18%) |
Figure 1.
Example of a pyrosequencing analysis for COX-2 (PTGS2) showing percent methylation (C) at four individual CpG sites.
4. Discussion
In the current study, COX-2 (PTGS2) gene methylation was more common in subepithelial lymphocytes than in adjacent epithelial or stromal cells in both grades of squamous dysplasia and in foci of invasive esophageal squamous cell carcinoma. Moreover, two of the three epithelial foci and all three of the non-lymphocytic stromal foci that showed COX-2 methylation were adjacent to foci of subepithelial lymphocytes that were also methylated. These findings raise the possibility that methylation of subepithelial lymphocytes may affect methylation of cells in the other two tissue compartments, and may be important in tumorigenesis.
This is the first evaluation of gene methylation in separate tissue compartments in esophageal neoplasia. In fact, relatively little work has been done to evaluate methylation in supporting tissues adjacent to any cancers, even though the importance of such tissues in cancer initiation and progression is increasingly well documented. Previous studies have evaluated methylation in stromal tissues adjacent to prostate cancer and have found gene methylation in tumor-associated stromal cells [18, 19]. The current study provides additional evidence for the potential importance of such stromal-epithelial interactions, and identifies subepithelial lymphocytic aggregates as a stromal component that may be particularly important in this interaction.
It is unclear why there is apparent preferential methylation of COX-2 in the subepithelial lymphocytes. COX-2 is an important inflammatory mediator, and its suppression results in lower levels of prostaglandin E2 and tumor-promoting cytokines, enhanced cell-mediated immunity, and prevention of tumor-induced suppression of dendritic cell activity [4, 20–22]. Thus, COX-2 methylation of subepithelial lymphocytes may represent an adaptive host defense against tumorigenesis [23].
The current study evaluated methylation in a single gene in a small number of tissue sections. The interesting findings must be confirmed in additional studies evaluating more genes and larger numbers of samples. More generally, our results imply the need to further evaluate methylation separately in different tissue compartments, to better understand the mechanisms and overall impact of gene methylation on carcinogenesis.
In conclusion, COX-2 (PTGS2) gene methylation was more common in subepithelial lymphocytes than in adjacent epithelial or stromal cells in a histologic spectrum of esophageal squamous disease from low-grade squamous dysplasia to invasive esophageal squamous cell carcinoma. These findings raise the possibility that methylation of stromal lymphocytes or other stromal tissue compartments may directly or indirectly affect tumorigenesis. Future studies of gene methylation should evaluate non-epithelial as well as epithelial tissue components.
Abbreviations
- COX-2
cyclooxygenase-2
- EDTA
ethylenediaminetetraacetic acid
- ESCC
esophageal squamous cell carcinoma
- HGD
high-grade squamous dysplasia
- LGD
low-grade squamous dysplasia
- NSAIDS
non-steroidal antiinflammatory drugs
- PTGS2
prostaglandin-endoperoxide synthase 2
- qMS-PCR
quantitative methylation specific polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None.
References
- 1.Kim JB, Stein R, O'Hare MJ. Tumour-stromal interactions in breast cancer: the role of stroma in tumourigenesis. Tumour Biol. 2005;26:173–185. doi: 10.1159/000086950. [DOI] [PubMed] [Google Scholar]
- 2.Shekhar MP, Werdell J, Santner SJ, Pauley RJ, Tait L. Breast stroma plays a dominant regulatory role in breast epithelial growth and differentiation: implications for tumor development and progression. Cancer Res. 2001;61:1320–1326. [PubMed] [Google Scholar]
- 3.Schor SL. Cytokine control of cell motility: modulation and mediation by the extracellular matrix. Prog Growth Factor Res. 1994;5:223–248. doi: 10.1016/0955-2235(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 4.Sharma RA, Browning MJ. Mechanisms of the self/non-self-survey in the defense against cancer: potential for chemoprevention? Crit Rev Oncol Hematol. 2005;56:5–22. doi: 10.1016/j.critrevonc.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Parkin DM, Li LD, Chen YD, Bray F. Estimation and projection of the national profile of cancer mortality in China: 1991–2005. Br J Cancer. 2004;90:2157–2166. doi: 10.1038/sj.bjc.6601813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ke L. Mortality and incidence trends from esophagus cancer in selected geographic areas of China circa 1970–90. Int J Cancer. 2002;102:271–274. doi: 10.1002/ijc.10706. [DOI] [PubMed] [Google Scholar]
- 7.Rashid A, Issa JP. CpG island methylation in gastroenterologic neoplasia: a maturing field. Gastroenterology. 2004;127:1578–1588. doi: 10.1053/j.gastro.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 9.Kikuchi T, Itoh F, Toyota M, Suzuki H, Yamamoto H, Fujita M, et al. Aberrant methylation and histone deacetylation of cyclooxygenase 2 in gastric cancer. Int J Cancer. 2002;97:272–277. doi: 10.1002/ijc.1612. [DOI] [PubMed] [Google Scholar]
- 10.Elder DJ, Halton DE, Crew TE, Paraskeva C. Apoptosis induction and cyclooxygenase-2 regulation in human colorectal adenoma and carcinoma cell lines by the cyclooxygenase-2-selective non-steroidal anti-inflammatory drug NS-398. Int J Cancer. 2000;86:553–560. doi: 10.1002/(sici)1097-0215(20000515)86:4<553::aid-ijc18>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Sheng H, Shao J, Morrow JD, Beauchamp RD, DuBois RN. Modulation of apoptosis and Bcl-2 expression by prostaglandin E2 in human colon cancer cells. Cancer Res. 1998;58:362–366. [PubMed] [Google Scholar]
- 12.Dawsey SM, Lewin KJ, Liu FS, Wang GQ, Shen Q. Esophageal morphology from Linxian, China. Squamous histologic findings in 754 patients. Cancer. 1994;73:2027–2037. doi: 10.1002/1097-0142(19940415)73:8<2027::aid-cncr2820730803>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 14.Woodson K, Hayes R, Wideroff L, Villaruz L, Tangrea J. Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55:199–205. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 15.Woodson K, Gillespie J, Hanson J, Emmert-Buck M, Phillips JM, Linehan WM, et al. Heterogeneous gene methylation patterns among pre-invasive and cancerous lesions of the prostate: a histopathologic study of whole mount prostate specimens. Prostate. 2004;60:25–31. doi: 10.1002/pros.20013. [DOI] [PubMed] [Google Scholar]
- 16.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 17.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 18.Kekeeva TV, Popova OP, Shegai PV, Alekseev BI, Adnreeva I, Zaletaev DV, et al. Abberant methylation of p16, HIC1, N33 and GSTP1 genes in tumor epitelium and tumor-associated stromal cells of prostate cancer. Mol Biol (Mosk) 2001;41:79–85. [PubMed] [Google Scholar]
- 19.Rodriguez-Canales J, Hanson JC, Tangrea MA, Erickson HS, Albert PS, Wallis BS, et al. Identification of a unique epigenetic sub-microenvironment in prostate cancer. J Pathol. 2007;211:410–419. doi: 10.1002/path.2133. [DOI] [PubMed] [Google Scholar]
- 20.Weinberg JB. Nitric oxide synthase 2 and cyclooxygenase 2 interactions in inflammation. Immunol Res. 2000;22:319–341. doi: 10.1385/IR:22:2-3:319. [DOI] [PubMed] [Google Scholar]
- 21.DeLong P, Tanaka T, Kruklitis R, Henry AC, Kapoor V, Kaiser LR, et al. Use of cyclooxygenase-2 inhibition to enhance the efficacy of immunotherapy. Cancer Res. 2003;63:7845–7852. [PubMed] [Google Scholar]
- 22.Sharma S, Stolina M, Yang SC, Baratelli F, Lin JF, Atianzar K, et al. Tumor cyclooxygenase 2-dependent suppression of dendritic cell function. Clin Cancer Res. 2003;9:961–968. [PubMed] [Google Scholar]
- 23.Ohno S, Ohno Y, Suzuki N, Inagawa H, Kohchi C, Soma G, et al. Multiple roles of cyclooxygenase-2 in endometrial cancer. Anticancer Res. 2005;25:3679–3687. [PubMed] [Google Scholar]