Abstract
T cell large granular lymphocyte leukemia (T-LGL) is a chronic clonal lymphoproliferation of CTL. In many ways, T-LGL clones resemble terminal effector CTL, including down-modulation of CD28 and overexpression of perforin, granzymes, and CD57. We studied the transcriptome of T-LGL clones and compared it with healthy CD8+CD57+ effector cells as well as CD8+CD57− populations. T-LGL clones were sorted based on their TCR variable β-chain restriction, and controls were obtained by pooling cell populations from 14 donors. Here, we focus our analysis on immunological networks, as immune mechanisms play a prominent role in the etiology of bone marrow failure in T-LGL. Informative genes identified by expression arrays were studied further in an independent cohort of patients using Taqman PCR, ELISA assays, and FACS analysis. Despite a strikingly similar gene expression profile between T-LGL clones and their healthy counterparts, important phenotypic differences were identified, including up-modulation of TNFRS9, myeloid cell leukemia sequence 1, IFN-γ, and IFN-γ-related genes, and several integrins/adhesion molecules. In addition, T-LGL clones were characterized by an overexpression of chemokines and chemokine receptors that are typically associated with viral infections (CXCL2, Hepatitis A virus cellular receptor 1, IL-18, CCR2). Our studies suggest that immunodominant LGL clones, although phenotypically similar to effector CTL, show significantly altered expression of a number of genes, including those associated with an ongoing viral infection or chronic, antigen-driven immune response.
Keywords: autoimmune, antigen-driven, CTL expansion, viral infection
INTRODUCTION
Immunodominant expansion of CTL with effector phenotype is a common pathologic feature of various conditions, including autoimmune diseases [1–4], viral infections [5–7], or solid and hematologic malignancies [8–13]. The transition from naive to antigen-primed effector CTL phenotype is characterized by loss of CD27 and CD28 markers, down-regulation of homing receptors CCR7 and CD62 ligand, and up-regulation of effector proteins such as perforin, granzymes, and NKG2D receptor [14–18]. In addition, terminally differentiated, antigen-specific CTL are characterized by expression of CD57 (HNK1 antigen), an adhesion molecule specific for terminally differentiated effector CTL [19, 20]. Under physiologic conditions, after antigen encounter, CTL clonally expand, and when the pathogen is cleared, the effector population down-regulates bcl-2 and initiates apoptosis [21]. It has been hypothesized that disruption of this homeostatic mechanism may result in persistence of clonal expansion and give rise to malignant or autoimmune T cell proliferation. Clonal CTL expansion in T cell large granular lymphocyte leukemia (T-LGL) represents a pathologic counterpart of the expanded CTL effectors [22]; the pathologic CTL population typically expresses CD3, CD8, and CD57 antigens and is TCR variable β-chain (VB)-restricted [23–28]. In T-LGL, one or two CTL clones with a unique, productively rearranged TCR expand and persist in the clonal state.
Clinically, T-LGL is indolent and in most cases, does not behave as a true leukemia, but rather reflects an autoimmune, semiautonomous process [23–25, 29, 30]. Frequent association with several autoimmune diseases further supports this notion. Patients present with various degrees of isolated or combined cytopenias, and neutropenia occurs most frequently. In vitro experiments have indicated that bone marrow cytotoxicity in LGL is mediated in two ways: The LGL clone recognizes hematopoietic progenitors and directly inhibits hematopoiesis, or it exerts suppressive effects via secretion of inhibitory cytokines/chemokines [9, 24, 31–33]. In analogy to the features of the expanded pathologic CTLs in T-LGL, CD8+CD57+ cells from healthy, elderly adults exhibit inhibitory activity on hematopoiesis [34]. Recent findings suggest that although T-LGL clones evolve in a nonrandom manner, they show dysregulation of signaling pathways similar to virally transformed T cells [35, 36]. Despite expressing high levels of Fas T-LGL clones are resistant to Fas-dependent apoptosis, which may explain persistent clonal expansion; such resistance can be overcome by in vitro activation, suggesting disruption of Fas signaling in T-LGL clones [37–41]. Previous investigations aimed at the characterization of surface phenotype, signal transduction, or gene expression in T-LGL used various techniques, including RNA microarrays performed on total PBMC [31–33, 39, 42, 43].
In this study, we chose to investigate the expression profile of monoclonal LGL populations purified based on their VB restriction; we compared them to their physiologic counterparts: terminally differentiated effector CTL from healthy control subjects. For this purpose, we applied Affymetrix U133 Plus 2.0 whole genome arrays covering over 47,000 transcripts. The goal of this investigation was to identify a unique genetic profile specific for LGL clones that might have resulted from, e.g., virus-induced transformation or an exuberant, unopposed antigenic drive. For that purpose, we focused our investigations on gene networks functionally involved with inflammatory response (particularly viral infection), such as, e.g., adhesion markers and soluble factors.
MATERIALS AND METHODS
Patients and controls
Informed consent for the sample collection was signed by the individuals in accordance with protocols approved by the Institutional Review Board of the Cleveland Clinic Foundation (Cleveland, OH, USA). Peripheral blood specimens were obtained from a total of 48 patients with LGL and 14 healthy controls (Table 1). The diagnosis of LGL leukemia was based on clinical and laboratory parameters as described previously [44].
TABLE 1.
Clinical and Laboratory Data of LGL Patients
| Pat | Method | Sex | Age | PRES | TCR | VB | VB Exp. | JB | CDR3 Clone/VB | CDR3 Clone/CD8+ |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | taq/el | F | 76 | RCA | y | 17 | 83% | 2.3 | 100% | 83% |
| 2 | array/taq/el | F | 42 | PAN | y | 20 | 25% | 2.7 | 33% | 8% |
| 3 | taq/el/ics | M | 63 | RCA | y | 17 | 45% | 2.3 | 100% | 45% |
| 4 | el | F | 30 | NEU | y | 18 | 42% | 2.7 | 40% | 17% |
| 5 | el | F | 61 | NEU | y | 6 | 51% | 2.1 | 95% | 49% |
| 6 | el | M | 56 | NEU | y | 13 | 65% | 2.7 | 31% | 20% |
| 7 | taq/el | F | 77 | PAN | y | 9 | 25% | 2.5 | 100% | 25% |
| 8 | taq/el | M | 72 | NEU | y | 5 | 27% | 2.7 | 33% | 6% |
| 9 | el | M | 70 | NEU | y | 22/3 | 60%/10% | 1.2/2.7 | 100%/100% | 60%/10% |
| 10 | taq/el/ics | M | 56 | NEU | y | 13 | 89% | 2.5 | 100% | 89% |
| 11 | taq/el | F | 75 | RCA | y | 2 | 73% | — | — | — |
| 12 | el | M | 50 | RCA | y | 2 | 64% | — | — | — |
| 13 | el/ics | F | 66 | NEU | y | 13/18 | 7%/78% | 2.3/2.5 | 26%/19% | 1.82%/14.9% |
| 14 | taq/el | F | 55 | NEU | y | 13 | 55% | — | — | — |
| 15 | taq/el | M | 52 | PAN | y | 14 | 28% | 2.1 | 100% | 27% |
| 16 | taq/el/ics | M | 74 | RCA/NEU | y | 1 | 98% | — | — | — |
| 17 | taq/el | M | 84 | RCA | y | 13 | 88% | — | — | — |
| 18 | el | M | 31 | RCA | y | NIP | 43%a | — | — | — |
| 19 | el | M | 71 | NEU | y | NIP | 73.4%a | — | — | — |
| 20 | el/ss/ics | M | 46 | NEU | y | 4/13 | 11%/7% | — | — | — |
| 21 | taq/el/ics | F | 59 | NEU | y | 3 | 63% | 1.1 | 100% | 63% |
| 22 | array/taq/el/ics | M | 53 | NEU | y | 1 | 95% | 1.6 | 100% | 95% |
| 23 | taq/el | M | 76 | NEU | y | 7 | 67% | — | — | — |
| 24 | el | M | 47 | RCA | n | NIP | 77%a | — | — | — |
| 25 | taq | M | 67 | PAN | y | 14 | 65% | — | — | — |
| 26 | array/el | M | 75 | RCA | y | 13 | 95% | — | — | — |
| 27 | taq | M | 72 | PAN | y | 3 | 88% | 2.4 | 61% | 54% |
| 28 | el | F | 70 | NEU | y | 2 | 16% | — | — | — |
| 29 | el | F | 72 | PAN | y | 12 | 8% | 2.1 | 51% | — |
| 30 | el | F | 65 | PAN | y | 7 | 76% | 2.7 | 10% | — |
| 31 | el/ics | M | 75 | RCA | y | 21 | 16% | — | — | — |
| 32 | el | M | 37 | NEU | y | 23/14 | 15%/16% | 1.1/2.7 | 8% | — |
| 33 | el | F | 72 | RCA | y | 18/3 | 27%/10% | 2.7/2.7 | 33% | — |
| 34 | el | M | 56 | RCA/NEU | y | NIP | 97.7%a | — | — | — |
| 35 | el | F | 63 | RCA | y | NIP | 65%a | 2.1/2.3 | 25% | — |
| 36 | el | F | 60 | NEU | y | NIP | 82.4%a | — | — | — |
| 37 | el | M | 43 | NEU | y | 13 | 14% | 2.7/2.5 | 54% | — |
| 38 | el/ics | F | 58 | RCA | y | 8/13 | 11%/8% | — | — | — |
| 39 | el | M | 62 | NEU | y | 13/18/22 | 43%, 25%/NA | 1.1/2.7/2.7 | 27% | — |
| 40 | el | F | 51 | NEU | n | NA | NA | 1.1 | 52% | — |
| 41 | el | M | 32 | PAN | y | 2 | 80% | 2.7 | 24% | — |
| 42 | el | F | 70 | NEU | NA | 1 | 70% | 1.5 | 49% | — |
| 43 | ss | M | 81 | RCA | y | 13/18 | 19%/19% | — | — | — |
| 44 | ss/ics | M | 55 | PAN | y | 20 | 10% | — | — | — |
| 45 | ss/ics | M | 72 | RCA/NEU | y | 20 | 91% | — | — | — |
| 46 | ss | F | 63 | PAN | y | 8 | 88% | — | — | — |
| 47 | ics | M | 74 | RCA/NEU | y | 13 | 98% | — | — | — |
| 48 | ics | F | 76 | NA | y | 14 | 35% | — | — | — |
PAT, Patient number; PRES, clinical presentation; TCR, T cell receptor gamma chain clonality; VB EXP., expansion of the VB chain within the CD8+ population as measured by VB flow cytometry; JB, joining β-chain of the expanded clonotype; CDR3 clone/VB, expansion of a unique clonotypic complementarity-determining region 3 (CDR3) within the expanded VB chain; CDR3 clone/CD8+, expansion of CDR3 within total CD8+ population as calculated by multiplication of CDR3 expansion within a VB family by expansion of the respective VB family within the CD8+ population; taq, Taqman PCR; el, ELISA assay; array, microarray; ics, intracellular staining for IFN-γ; ss, surface-staining for CD31, CD38, CD40, and CD86; RCA, red cell aplasia; PAN, pancytopenia; NEU, neutropenia; NA, not available; NIP, not in panel: CD8+ cells not covered by available antibodies.
Percent of CD8+ cells not covered by the standard VB antibody panel and thus, most likely containing the expanded populations. (In healthy controls, maximal expansions of CD8+ cells not covered by available antibodies generally do not exceed 30%.)
VB cytometry (VB skewing)
VB flow cytometry analysis was performed on fresh peripheral blood according to the manufacturer’s instructions (IOTest Beta Mark kit, Beckman Coulter, Fullerton, CA, USA) with the following modifications: phycoerythrin-cyanine 5 (PC5)-conjugated CD4 (5 μl), 5 μl phycoerythrin-Texas red (ECD) CD8 mAb, and 20 μl anti-VB mAb (VB1–5, VB7–9, VB11–14, VB16–18, VB20–23) were added. An additional anti-VB6.7 mAb (not included in the kit) was used. A four-color protocol was applied, and the lymphocyte gate was set according to the size and forward-scatter properties. The individual contribution of each of the 19 VB subfamilies detectable by specific mAb was determined as described previously, and results were expressed in percent of α/β CD4+ or CD8+ cells [45, 46]. VB over-representation was established when contribution of a particular VB family was greater than the mean + 2 SD of values found in healthy volunteers. Immunodominant clonotypes were characterized by sequencing of a number of bacterial colonies harboring subcloned TCR amplification products as described previously [35, 47].
Sample preparation for microarray hybridization
Mononuclear cells were separated from peripheral blood by density gradient sedimentation (Mediatech, Herndon, VA, USA). LGL cells were separated by flow cytometric sorting using anti-VB and CD8 mAb as described previously [46]. Healthy, donor-derived CD8+CD57+ cells were isolated by flow cytometric sorting using CD3, CD8, and CD57 mAb. As a result of the relatively low size of the CD57+ population in healthy individuals, sorted CD8+CD57+ CTL were pooled from 14 healthy donors (pool was composed of equal amounts of cells from each individual). Total RNA was extracted from cells using Trizol (Invitrogen, Carlsbad, CA, USA) and Phase-Lock gel tubes (Eppendorf, Hamburg, Germany), cleaned up using RNAeasy columns (Qiagen, Valencia CA, USA), and dissolved in diethylpyrocarbonate water. The purity of RNA was confirmed with spectrophotometry. Total cRNA was prepared using the in vitro-transcribed protocol (Affymetrix, Santa Clara, CA, USA) and hybridized to U133 Plus 2.0 arrays, according to the manufacturer’s instructions (Affymetrix). All of the microarrays were examined for surface defects, grid placement, background intensity, housekeeping gene expression, and a 3′:5′ ratio of probe sets from genes of varying length (signal 3′:5′ ratio<3).
Data analysis and reduction
Expression analysis was conducted using standard Affymetrix analysis software algorithms (Microarray Suite 5.0). Comparative analysis between expression profiles of CD8+CD57−, CD8+CD57+ cells from controls and LGL clones was carried out on GeneSpring™ software, Version 7.1 (Agilent, Santa Clara, CA, USA). Scanned images of Affymetrix chips were converted to spreadsheet numbers using Affymetrix proprietary GeneChip Operating software (GCOS). Signal log ratios were converted to fold changes in MS Excel. Spreadsheet data were imported into the MS Access database manager. Data were mined for credible changes. Fold changes of absolute value ≥2 were considered (e.g., alteration between CD8+CD57+ and CD8+CD57−, more than twofold; P ≤ 0.02). Gene expression data were normalized in two ways: “per-gene normalization” and “per-sample normalization”. This approach has been previously described in detail [48, 49]. In the per-sample normalization, specific samples were normalized to one another: Each measurement for each gene in those specific samples was divided by the mean of that gene’s measurements in the corresponding control samples. Gene expression in LGL patients was normalized to the expression values obtained from the CD8+CD57+ population and graphically represented in a hierarchical clustering. (Fig. 1C, II.). Analogous GeneSpring clustering analysis was performed on healthy effector versus noneffector cell populations (Fig. 1B). In the per-gene normalization, imported data were normalized to the median value of the six combined Affymetrix probe sets representing CD8 antigen and then normalized to the median value per gene across the sample set by dividing by the 50.0th percentile of all measurements in that sample (Fig. 1C, I.). The NETAFFX gene ontology mining tool was used to study alterations in gene expression with regard to their significance in biological processes, molecular function, and cellular component [50].
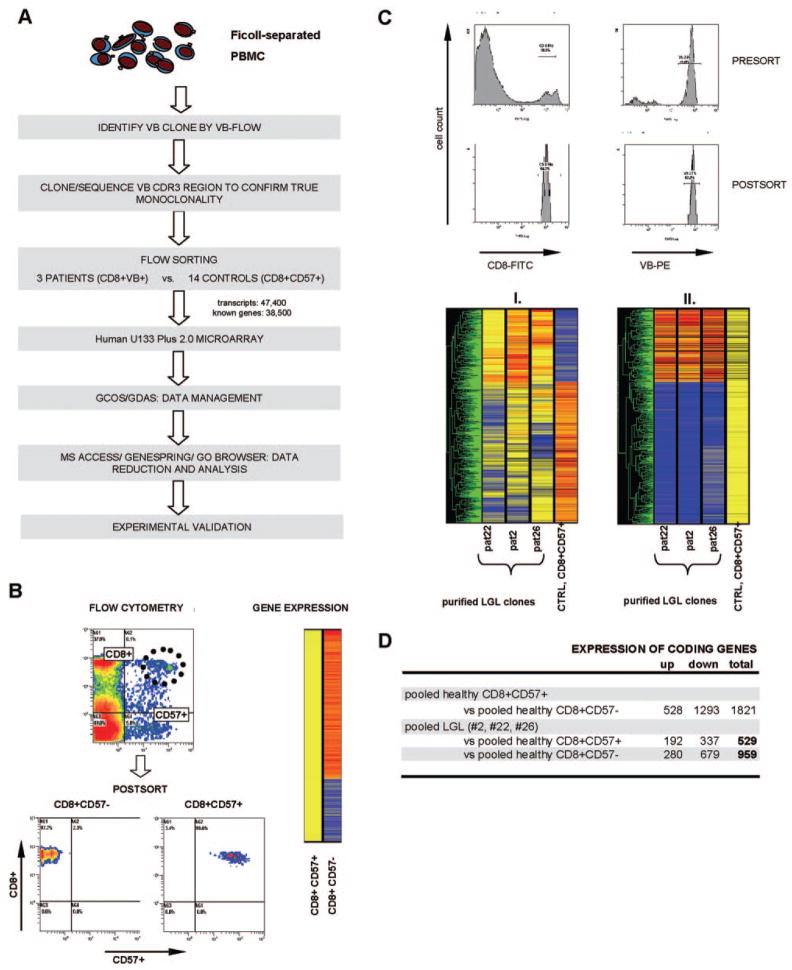
Fig. 1.
Detection of pathologically altered gene expression in LGL patients. (A) Rational experimental approach for the analysis of differential gene expression in purified LGL clones and healthy control effector cells. Initially, VB family expansions were identified by VB flow cytometry, and monoclonal TCR CDR3 regions were confirmed by cloning and sequencing. Control CD8+CD57+ cells and patients VB+ LGL clones and/or CD8+ cells were sorted using standard flow cytometry. Purified LGL clones and control CD8+CD57+ populations were run on a microarray, and the data were reduced and analyzed using GCOS/GeneChip DNA Analysis software (GDAS), MS Access, Genespring, and Go Browser. Finally, experimental validation was performed on the validation cohort consisting of additional LGL patients and healthy controls. (B) Flow cytometric sorting and Genespring hierarchical clustering of the gene expression profile of healthy effector cells expressing CD57+. Purity of cells was analyzed after sorting. (C) The upper panel shows exemplary flow cytometric sorting based on the expression of a monoclonal VB chain and postsort purity analysis on one selected patient. The lower panel illustrates Genespring analysis between LGL patients (pat) and controls (CTRL), which was graphically represented in hierarchical clustering in two ways: (I.) Data were normalized to the expression values of the control CD8+CD57+ population (per-sample normalization); (II.) to the median value of the six combined probe sets for CD8 antigen (per-gene normalization). (D) Differential expression of healthy effector CD8+CD57+ versus CD8+CD57− and of purified VB clones versus healthy CD8+CD57+/CD8+CD57− is shown.
Taqman PCR
To decrease the risk of nonspecific DNA amplification, total RNA was treated with TurboDNase (Ambion, Austin, TX, USA) and inactivation mix was added to remove the enzyme. cDNA was generated from 5 μl total DNase-treated RNA by first-strand cDNA synthesis using the SuperScript III RT kit (Invitrogen). Gene sequences derived from the University of California, San Francisco (San Francisco, CA, USA) genome browser were used to design a Taqman assay using the gene-specific Taqman probe and a combination of forward and reverse primers (Table 2). Products of 150 bp or less were amplified as follows: 15 μl of Taqman Universal Master Mix (2×, Applied Biosystems, Foster City, CA, USA) was used with 0.6 μl (50 μM) forward and reverse primers, 0.66 μl (10 μM) Taqman probe, 6 μl cDNA (1:3 dilution with water), and 1 μl water. Samples were run on a 7500 real-time PCR system (Applied Biosystems) at 95°C for 10 min followed by 55 cycles of 15 s at 95°C and 60 s at 60°C. All reactions were performed in duplicates (endogenous control GAPDH was measured in triplicates) in the Model 7500 Taqman thermocycler (PE Applied Biosystems). The values obtained for the target gene expression were normalized to GAPDH and quantified relative to the expression in control samples. For the calculation of relative quantification, the 2−ΔΔCT formula was used, where −ΔΔCT = (CT,target−CT,GAPDH) experimental sample − (CT,target−CT,GAPDH) control sample.
TABLE 2.
Sequences of Primer and Probes used in Taqman PCR
| Gene | Forward primer | Reverse primer | Taqman probe |
|---|---|---|---|
| ccr2 | gagtaactgtgaaagcaccagtcaa | gcagtgagtcatcccaagagtct | tggaccaagccacgcaggtgac |
| cd302 | ttctgattctcgtttcaccacagt | tccaactaccaaaacacagtcttca | ttcaaccgcaccccaatcacc |
| cd31 | catatgcagacctcagaatctacca | cagttcgggcttggaaaatagt | tgaactggtcaccgtgacggcag |
| cd36 | cactgcgacatgattaatggtaca | tgcaatacctggcttttctcaac | tgcagcctcatttccaccttt |
| cd40 | cccatcatcttcgggatcct | ttggccacctttttgataaagac | tttgccatcctcttggtgct |
| cd86 | gggccgcacaagttttgat | gcccttgtccttgatctgaaga | cggacagtggaccctgagacttc |
| cxcl2 | cccctggccactgaactg | tggatgttcttgaggtgaattcc | ccagtgcttgcagaccctgca |
| havcr2 | ggtcatcaaaccagcaaggt | tggaaaggctgcagtgaagtc | acccctgcaccgactcggca |
| mcl1 | gaaatcgttgtctcgagtgatgat | cacaatcctgccccagtttg | catgttttcagcgacggcgt |
| tnfrs9 | tgttttcaggaccaggaagga | aaaccctggagtgcagtcaca | tgttcctccaccagcaatgca |
| gapdh | catccatgacaactttggtatcgt | cagtcttctgggtggcagtga | aggactcatgaccacagtc |
| ifngamma | tattcggtaactgacttgaatgtcc | aggcaggacaaccattactgg | agggaagcgaaaaaggagtcagatgctg |
Primers and probes are shown in the 5′ to 3′ direction. All probes are 5′-6-carboxyfluorescein (FAM) and 3′-black hole quencher 1 (BHQ1)-labeled.
ELISA assays
Blood plasma was obtained by centrifugation at 1800 rpm for 10 min, and 50 – 200 ul was used in ELISA tests for IFN-γ, IL-18 [IFN-γ-inducing factor (IGIF), IL-18, BD Biosciences, San Jose, CA, USA], IL-8 (CXCL8), IFN-γ-inducible, 10 kD protein (IP10; Raybiotech, Norcross, GA, USA), and MCP-1/CCL2 (BD Biosciences). In each assay, plasma samples and freshly solubilized standards were run in duplicates, according to the manufacturer’s instructions. Absorbance was measured at 450 nm with wavelength correction (OD readings at 570 nm were subtracted from readings at 450 nm).
Flow cytometry for surface and intracellular marker expression
Immunophenotyping was performed on the whole blood by indirect immunofluorescence with a panel of mAb including anti-CD57, anti-CD3, anti-CD8, anti-CD31, anti-CD40 (Beckman Coulter), and anti-CD38 and anti-CD86 (PharMingen, San Diego, CA, USA), according to the manufacturer’s instructions. Following 20 min incubation in the dark, erythrocytes were lysed and fixed, and samples were washed twice with FACS buffer (PBS supplemented with 1% FCS and NaN3). Multiparametric four-color flow cytometry was performed using a Coulter Epics XL-MCL sequence flow cytometer (Beckman Coulter). At least 20,000 events were acquired for each sample; events were analyzed using the EXPO32 Advanced Digital Compensation software (Beckman Coulter). Lymphocytes were initially gated by forward/side-scatter; secondary gates were set on a basis of staining with isotypic control mAb, and further analysis included additional gates set on CD3+ and CD8+ cells.
Intracellular staining for IFN-γ was performed using the Cytofix/Cytoperm Plus (containing GolgiStop) intracellular staining kit (PharMingen). Briefly, PBMC were isolated by a standard procedure using Ficoll density gradient centrifugation (density, 1.077 g/ml). After isolation, PBMC were resuspended in RPMI 1640 containing 10% FBS, 1% glutamine, and 1% penicillin/streptomycin at a concentration of 1 × 106 cells/ml and cultured overnight at 37°C and 5% CO2. On the next day, cells remained unstimulated (control) or were stimulated with PMA (20 ng/ml) and ionomycin (1.4 μM) for 5 h at 37°C and 5% CO2. To promote the accumulation of de novo-synthesized cytokines in the Golgi apparatus, monensin was added according to the manufacturer’s instruction. Following stimulation, cells were washed with FACS buffer, and surface staining was performed with 5 ul each anti-CD3, anti-CD8, and anti-CD4 mAb (all Beckman Coulter). Following two washing steps, cells were permeabilized by means of a saponin-based method (PharMingen). Finally, cells were incubated with 10 ul anti-IFN-γ (Beckman Coulter) at 4°C in the dark for 30 min. Intracellular IFN-γ production was determined within the CD3+ gate on CD8+ and CD4+ cells using a Coulter Epics XL MCL flow cytometer (Beckman Coulter). Threshold for cytokine positivity was set using irrelevant isotypic control mAb. Results for cytokine production were expressed as a percentage of the respective subpopulation.
RESULTS
General approach for gene expression analysis in LGL leukemia using high-density expression microarrays
For our analysis, we have selected a cohort of patients with a typical T-LGL leukemia (Table 1). Whole genome expression microarray was performed on Patients #2, #22, and #26 (microarray cohort), and the remaining patients (validation cohort) were analyzed to selected results obtained from the expression array (Table 1 and Fig. 1). To determine gene expression pattern specific to T-LGL cells, LGL cells were sorted according to the clonal expression of pathognomonic VB chain. As expected, sorted clonal cells showed a high expression of the CD57 marker. As controls, we have used flow-sorted CD8+CD57+ CTL effector cells derived from healthy controls. This effector population represents the physiologic counterpart of clonal CTL derived from T-LGL. To obtain a sufficient amount of control RNA and abolish the noise of individual measurements, RNA extracted from sorted cells of 14 healthy donors was pooled (Fig. 1).
Microarray fidelity and differential gene expression of healthy CD8+CD57+ and CD8+CD57− cells
The fidelity of the microarray platform was tested using two approaches. First, we selected a set of genes that is known to be characteristically up-/down-regulated in clonal LGL CTL. After sorting for CD8 and CD57 antigens, RNA extraction, and array analysis, results from a comparison between LGL clones and controls were compared with findings reported in literature (Table 3). Second, we compared the differences of healthy effector CTL versus noneffector populations: Several genes that were described previously to be up-regulated in healthy effector cells were also overexpressed in our healthy, pooled CD8+CD57+ population. These examples include β-1,3-glucuronyltransferase 1 (glucuronosyltransferase P/CD57 transferase), granzyme B, CD38, HLA-DQ, HLA-DR, and serine protease 23. Similarly, genes known to be down-regulated in CD8+ lymphocytes undergoing transition to an effector CTL, were also underexpressed in the CD8+CD57+ control population: CD27, CD28, IL-7R, BCL2, and CCR7 (Table 4).
TABLE 3.
Previous Reports of Genes/Proteins Differentially Expressed between LGL Patients and Healthy Controls Displayed in Comparison with Data from Microarray Cohort
| Method/ref. number | Differentially expressed genes in previous reports | Differentially expressed genes in microarray cohort | |
|---|---|---|---|
| Oligonucleotide/cDNA microarray [33] | Up-regulated: serine proteinases (granzymes A, B, H, and K), proteinases (cathepsin C/W), calpain subunit, caspase-8, perforin, NKG2 gene family (KLRC family) | Granzyme A/H: | n.c. |
| Granzyme B: | ↑11/3 | ||
| Granzyme K: | ↓23/3 | ||
| Cathepsin A: | ↑11/3 | ||
| Cathepsin B: | ↑12/3 | ||
| Cathepsin D: | ↑12/3 | ||
| Cathepsin L: | ↑12/3 | ||
| Cathepsin K: | ↑11/3 | ||
| Cathepsin H: | ↑12/3 | ||
| Cathepsin S: | ↑13/3 | ||
| Calpain subunit: | ↑11/3 | ||
| Caspase 8: | ↑13/3 | ||
| Perforin: | n.c. | ||
| KLRC4: | ↑11/3 | ||
| Down-regulated: proteolytic inhibitors (cystatin C, A, α-1 antitrypsin) | Cystatin A: | ↑11/3 | |
| Cystatin C: | ↑11/3 | ||
| Serpin A1 (antitrypsin): | ↑12/3 | ||
| RT-PCR [31] | Constitutive expression of FasL | FasL: | ↑11/3 |
| ELISA/RNAse protection assay [32] | Constitutive expression of proinflammatory cytokines RANTES (CCL5), MIP-1β (CCL4), IL-18, IL-8, and IL-1RA | RANTES: | n.c. |
| Mip-1β: | n.c. | ||
| IL-18: | ↑11/3 | ||
| IL-8 | ↑12/3 | ||
| IL-1RA: | ↑12/3 | ||
| ELISA [42] | Increased cytokine expression in a portion of LGL patients: sIL-2R, TNF-α (TNFSF2), IL-6, IL-10, IL-8 | IL-2R: | n.c. |
| TNF-α: | n.c. | ||
| IL-6ST (sign. transducer): | ↑12/3 | ||
| IL-10: | ↑12/3 | ||
| IL-8: | ↑12/3 | ||
| Western blot [39] | Constitutive expression of STAT3 | STAT3 | ↑13/3 |
n.c., No statistically significant change in expression values between patients and controls; ↑, increase in expression; ↓, decrease in expression; 1/3, 2/3, 3/3, significant differential expression in one, two, and three out of three patients, respectively. FasL, Fas ligand; sIL-2R, soluble IL-2R; TNFSF2, TNF superfamily 2.
TABLE 4.
Confirmation of Array Results Based on Expected Gene Expression in Effector CTL (CD8+CD57+)
| Gene symbol | Gene name | Function | Fold change CD57+ versus CD57− |
|---|---|---|---|
| B3GAT1 | β-1,3-Glucuronyltransferase (CD57 transferase) | Cell adhesion | 56 |
| GZMB | Granzyme B | Proteolysis | 4.6 |
| CD38 | CD38 antigen (p45) | Cell death regulation | 5 |
| HLA-DQ | MHC class II | Cell adhesion/antigen processing | 3.5 |
| HLA-DR | MHC class II | Cell adhesion/antigen processing | 5.2 |
| PRSS23 | Serine protease 23 | Effector cells, antiviral | 8.4 |
| TNFRS7 (CD27) | TNFRSF | Lymphocyte homeostasis | −2.2 |
| CD28 | CD28 antigen (Tp44) | Cell death regulation | −11.3 |
| IL7R(CD127) | IL-7R | Signal transduction | −2.2 |
| BCL2 | B cell lymphoma 2 | Cell death regulation | −2.8 |
| CCR7 | Chemokine receptor 7 | Homing marker | −2.8 |
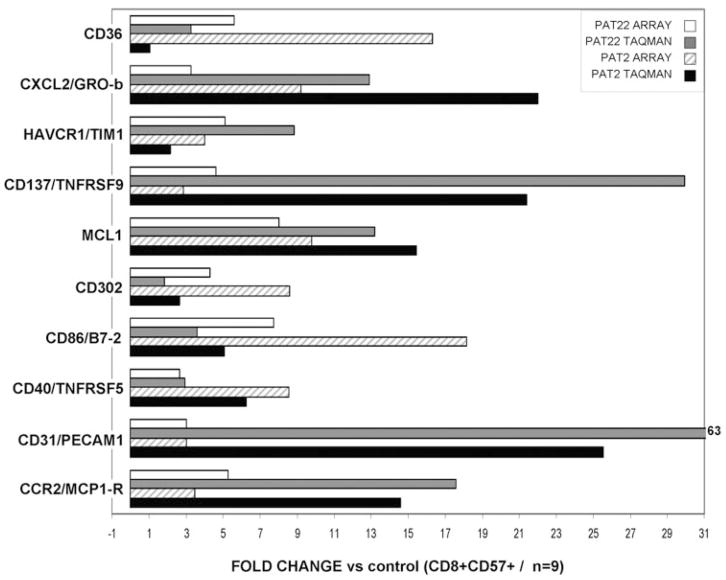
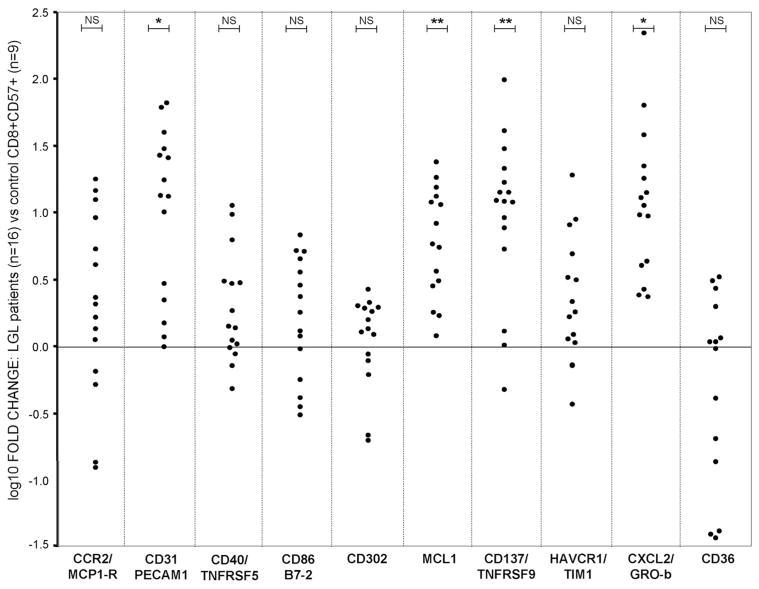
Next, we studied the expression of 10 indicator genes by Taqman PCR in two of the three LGL patients originally analyzed by microarrays and compared the results for concordance (Fig. 2). For most genes, Taqman PCR experiments verified respective microarray results, and in general, a high concordance between the two methods was observed. Relative expression of CD36 in Patient #22 was, using Taqman PCR, in contrast to microarray analysis, not notably elevated [1.07-fold (log 10 0.03) increase], and other patients showed a great heterogeneity with regard to this marker (Fig. 3). Analysis by quantitative PCR confirmed overexpression of most genes in the extended cohort of patients.
Fig. 2.
Validation of microarray data. Relative expression of 10 representative genes was determined in two patients (previously studied by microarray) using Taqman PCR. Mean difference of comparative threshold (dCT; d of the threshold cycle) values of nine control CD8+CD57+ samples was used as a calibrator to calculate relative expression of LGL patients. GRO-b, Growth-related oncogene-β; HAVCR1/TIM1, Hepatitis A virus cellular receptor 1/T cell Ig and mucin domain-containing protein.
Fig. 3.
Expression of genes in LGL patients as compared with sorted healthy control CD8+CD57+ by Taqman PCR. Ten genes initially detected as concordantly overexpressed in three LGL patients by microarray analysis were selected for further studies. Taqman PCR was performed on an independent validation group of 16 LGL patients. Dots indicate expression of the respective gene in patients in relation to the averaged CD8+CD57+ control population (dCt values of nine controls were averaged and used as calibrator). NS, Not significant (P>0.05); *, P < 0.01; **, P ≤ 0.001, by Student’s t-test.
Comparison of terminally differentiated healthy effector cells and clonal LGL CTL
Initially, we compared mature effector cells derived from controls with their less-differentiated counterparts characterized by lack of CD57 expression. Based on our microarray analysis, healthy effector CD8+CD57+ and CD8+CD57− cell populations discordantly expressed 1821 genes: 528 up- and 1293 down-regulated (change greater than twofold; P ≤ 0.02; multiplicate probes for a single gene and noninformative probes were excluded from this calculation; Fig. 1, B and D). Various genes that were described previously in literature to be expressed specifically in antigen-experienced effector cells were concordantly up- or down-regulated in our CD8+CD57+ pool (Table 4). Subsequently, we compared expression patterns of purified LGL clones with healthy CD8+CD57+ and CD8+CD57− lymphocytes (Fig. 1D, Table 5, and Supplementary data).
TABLE 5.
Common Genes Differentially Expressed between LGL Clones and Healthy Pooled CD57+ and CD57+ Populations
| Fold change versus control CD8+CD57+ |
Fold change versus control CD8+CD57− |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene symbol | Gene title | #22 | #2 | #26 | #22 | #2 | #26 | I/D |
| ARRB1 | arrestin, β-1 | 4.59 | 6.06 | 2.83 | 13.45 | 15.23 | 7.36 | I |
| ATF3 | activating transcription factor 3 | 13.93 | 25.99 | 10.56 | 11.31 | 32.00 | 8.00 | I |
| BCL3 | B cell CLL/lymphoma 3 | 2.64 | 4.59 | 4.00 | 2.46 | 3.73 | 4.00 | I |
| CASP1 | caspase 1, apoptosis-related cysteine peptidase | 2.30 | 2.30 | 2.30 | 2.28 | 2.27 | 2.37 | I |
| CD38 | CD38 antigen (p45) | 2.38 | 4.45 | 7.43 | 6.06 | 5.28 | 9.85 | I |
| CTNNA1 | catenin (cadherin-associated protein), α 1, 102 kDa | 5.16 | 4.72 | 3.05 | 6.41 | 5.97 | 6.94 | I |
| FCGR3A/B (CD16) | Fc fragment of IgG, low-affinity IIIa/IIIb, receptor (CD16a/CD16b) | 8.57 | 10.56 | 3.25 | 16.97 | 18.86 | 6.76 | I |
| GLUL | glutamate-ammonia ligase (glutamine synthetase) | 6.96 | 9.52 | 2.30 | 7.57 | 9.15 | 2.79 | I |
| HAVCR2 | hepatitis A virus cellular receptor 2 | 4.80 | 3.71 | 5.78 | 6.27 | 5.63 | 6.91 | I |
| HDGFRP3 | hepatoma-derived growth factor, related protein 3 | 12.21 | 9.75 | 13.91 | 3.25 | 3.48 | 5.13 | I |
| HIST2H2AA | histone 2, H2aa | 4.00 | 5.47 | 3.14 | 4.15 | 5.66 | 3.16 | I |
| HLA-DQA1 | MHC, class II, DQ α 1 | 24.11 | 47.10 | 7.73 | 51.98 | 84.45 | 17.15 | I |
| HLA-DQB1 | MHC, class II, DQ α 1 | 13.50 | 17.15 | 2.81 | 17.78 | 31.08 | 4.92 | I |
| IFIT2 | IFN-induced protein with tetratricopeptide repeats 2 | 3.73 | 9.19 | 2.64 | 3.04 | 7.06 | 2.87 | I |
| IFIT3 | IFN-induced protein with tetratricopeptide repeats 3 | 3.56 | 2.73 | 3.41 | 4.29 | 4.59 | 5.66 | I |
| IFNG | IFN-γ | 12.13 | 13.00 | 25.99 | 45.25 | 45.25 | 78.79 | I |
| ITGAX | integrin, α × [antigen CD11C (p150), α polypeptide] | 3.25 | 6.50 | 4.29 | 6.06 | 9.85 | 6.96 | I |
| JUN | v-jun sarcoma virus 17 oncogene homolog (avian) | 3.92 | 13.48 | 3.62 | 3.48 | 11.75 | 2.65 | I |
| LAT2 | linker for activation of T cells family, member 2 | 3.48 | 3.25 | 3.48 | 6.96 | 6.50 | 6.50 | I |
| LGALS3 | lectin, galactoside-binding, soluble, 3 (galectin 3) | 5.66 | 10.56 | 3.73 | 4.00 | 8.00 | 2.64 | I |
| LYN | v-yes-1 Yamaguchi sarcoma viral-related oncogene homolog | 2.00 | 3.25 | 2.30 | 5.17 | 7.82 | 8.60 | I |
| MIRN21 | microRNA 21 | 5.28 | 8.57 | 3.73 | 4.00 | 5.28 | 2.46 | I |
| MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | 8.04 | 9.81 | 2.74 | 2.25 | 2.71 | NC | I |
| NCR1 | natural cytotoxicity triggering receptor 1 | 5.28 | 3.03 | 4.29 | 12.31 | 14.96 | 13.20 | I |
| NOTCH2 | Notch homolog 2 (Drosophila)///Notch homolog 2 (Drosophila) | 2.64 | 3.73 | 2.46 | 2.83 | 4.00 | 2.64 | I |
| NUAK1 | NUAK family, SNF1-like kinase, 1 | 2.64 | 2.00 | 2.14 | 45.25 | 36.76 | 36.76 | I |
| PARD6G | par-6 partitioning defective 6 homolog γ (Caenorhabditis elegans) | 3.25 | 2.64 | 3.48 | 17.15 | 13.93 | 19.70 | I |
| PECAM1 (CD31) | platelet/endothelial cell adhesion molecule (CD31 antigen) | 3.02 | 3.02 | 2.53 | 5.91 | 5.88 | 9.28 | I |
| PLSCR1 | phospholipid scramblase 1 | 2.15 | 3.25 | 2.15 | 2.00 | 3.03 | 9.28 | I |
| PRSS23 | protease, serine, 23 | 3.04 | 2.22 | 3.74 | 21.98 | 17.32 | 28.13 | I |
| RHOB | ras homolog gene family, member B | 9.19 | 18.38 | 3.25 | 21.11 | 48.50 | 8.57 | I |
| SLC1A4 | Solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 3.62 | 2.52 | 5.78 | 9.73 | 5.83 | 25.17 | I |
| TNS3 | Tensin 3 | 7.46 | 16.00 | 19.70 | 4.59 | 10.56 | 16.00 | I |
| TMEM49 | transmembrane protein 49 | 6.06 | 9.19 | 4.59 | 4.59 | 8.00 | 2.83 | I |
| TNFRSF9 | TNFRSF, member 9 | 4.61 | 2.87 | 6.00 | 9.85 | 4.00 | 17.15 | I |
| ZNRF1 | zinc and ring finger 1 | 8.57 | 5.28 | 12.13 | 16.00 | 8.57 | 21.11 | I |
| BACH2 | basic leucine zipper transcription factor 2 | −3.86 | −2.80 | −4.31 | −31.24 | −17.70 | −34.14 | D |
| BCL2 | B cell CLL/lymphoma 2 | −9.57 | −14.16 | −5.87 | −16.98 | −22.25 | −12.28 | D |
| CD2 | CD2 antigen (p50) | −2.14 | −3.48 | −3.03 | −2.00 | −3.48 | −2.83 | D |
| CD28 | CD28 antigen (Tp44) | −2.46 | −8.57 | −256 | −5.88 | −16.21 | −414 | D |
| CD44 | CD44 antigen (homing function and Indian blood group system) | −5.03 | −3.87 | −4.29 | −8.93 | −6.08 | −6.73 | D |
| CD5 | CD5 antigen (p56-62) | −3.25 | −2.64 | −5.28 | −3.48 | −3.73 | −5.66 | D |
| CXCR3 | chemokine (C-X-C motif) receptor 3 | −13.00 | −7.46 | −16.00 | −9.74 | −6.64 | −28.47 | D |
| FOXP1 | Forkhead box P1 | −7.87 | −6.51 | −6.96 | −21.13 | −11.21 | −25.94 | D |
| IL18R1 | IL-18R1 | −6.50 | −9.19 | −3.25 | −3.48 | −4.92 | −2.14 | D |
| IL7R | IL-7R | −3.04 | −8.66 | −9.39 | −8.00 | −16.00 | −18.38 | D |
| PTEN | phosphatase and tensin homolog | −2.64 | −4.00 | −2.46 | −3.25 | −4.59 | −2.83 | D |
| TNFRSF25 | TNFRSF, member 25 | −9.85 | −6.06 | −13.00 | −12.31 | −22.50 | −42.85 | D |
| TNFRSF7 | TNFRSF, member 7 | −7.46 | −13.93 | −45.25 | −14.93 | −34.30 | −90.51 | D |
| TNFSF8 | TNFSF (ligand), member 8 | −2.46 | −2.30 | −6.96 | −5.25 | −4.66 | −17.15 | D |
| XCL1/XCL2 | chemokine (C motif) ligand 1/chemokine (C motif) ligand 2 | −22.68 | −5.86 | −19.59 | −9.52 | −2.65 | −8.08 | D |
I/D, Increase/decrease in gene expression; CLL, chronic lymphocytic leukemia; SNF1, sucrose-nonfermenting-1. Fold-change values for each gene were averaged from multiplicate Affymetrix probes specific for the respective gene (for individual fold-change values for each probe, see Supplemental data).
Distinct phenotype of LGL clones as compared with their healthy counterparts
The finding that clonal LGL cells are phenotypically more homologous to normal terminal effector cells than to healthy, less-differentiated CTL (lacking CD57 expression) was expected and not surprising. For that reason, we assumed that the comparison focusing on differences between LGL clones and their normal counterparts derived from healthy individuals would be most insightful. One could expect a great deal of similarity between these cells, but discrete differences may be indicative of mechanisms governing clonal transformation or clinical features. Second, these changes could explain LGL-specific effector pathways leading to common sequelae of T-LGL. To narrow the search for candidate genes specifically associated with LGL clones, we analyzed genes concordantly altered across all LGL patients studied by microarray as compared with the healthy effector population. LGL clones differed from a healthy CD8+CD57+ population in the expression of 529 coding genes (including genes coding for hypothetical proteins; multiplicates and noninformative probes were omitted; for complete list of probes, see Supplementary data), with 192 up- and 337 down-regulated (Fig. 1, C and D). By comparison, this difference in expression pattern was relatively modest as compared with the drastically altered expression between the LGL population and healthy CD8+CD57− lymphocyte fraction (959 genes: 280 up- and 679 down-regulated).
Clonal CTL show marked similarity to normal T cell populations but also distinct differences reminiscent of changes observed in responses to viral pathogens
To identify the “signature” expression pattern that distinguishes LGL clones from all healthy populations, we then selected sets of up- and down-regulated genes in LGL as compared with healthy CD8+CD57+ or CD8+CD57− populations (Table 5). Examples of differentially expressed genes that are altered in a similar direction when LGL clones are compared with control effector (CD57+) or noneffector (CD57−) cell populations include: PLSCR1, ATF3, HAVCR2, CD2, CXCR3, TNFRSF9, CD38, JUN, and PTEN. Conversely, several genes are much more discordantly expressed when LGL patients’ profiles are juxtaposed with one of the two control populations: FOXP1, HDGFRP3, ARRB1, PRSS23, ITGAX, and NUAK1 (Table 5).
Expression of selected genes in the validation cohort
Gene expression patterns measured in a few individual patients allow for selection of genes, which can be separately studied in a larger cohort of patients. Of particular interest to us were genes involved in immune/inflammatory response during acute or chronic viral infections, for example, adhesion molecules and cyto/chemokines. We selected a number of genes that were differentially expressed in the microarray cohort and determined their expression in a larger cohort of patients (validation cohort) using various methods. Using Taqman PCR, we studied the expression of 10 various genes in 16 LGL patients versus nine flow-sorted, healthy CD8+CD57+ clones (Fig. 3). Expression values obtained from control CD57+ duplicate samples were averaged and used as a calibrator to determine relative gene expression. In concordance with the array results, a significant increase in the expression of CD31/PECAM1, MCL1, CD137/TNFRS9, and CXCL2/GRO-b was observed in almost all LGL. Moreover, in many LGL patients, elevated expression of CCR2, CD40, CD86, CD302, and HAVCR2 was detected.
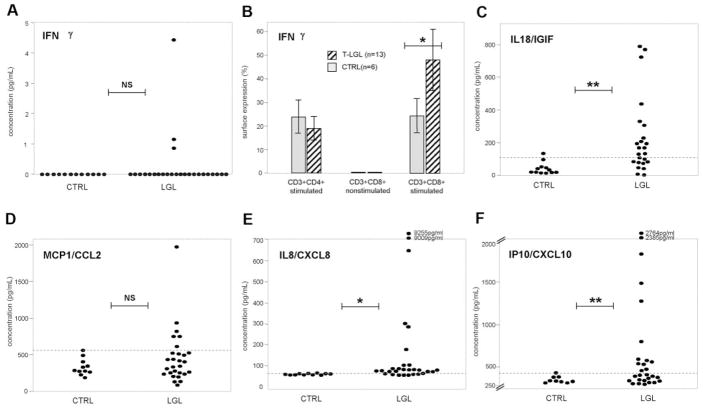
Next, to validate our findings in vivo, we measured the levels of selected cytokines in plasma of LGL patients and compared them with healthy controls (Fig. 4). Expression values greater than 2 SD from the mean levels of cytokine expression in controls were defined as pathologic. Although increased production of IFN-γ by LGL cells (>2 SD) resulted in elevated plasma levels of this cytokine, only in three of 25 patients, overexpression of IL-18, MCP-1, and IL-8 resulted in increased plasma levels in 14 of 25, six of 29, and 21 of 29 patients, respectively; IP10 plasma levels were elevated (>2 SD; after exclusion of outliers, >5000 pg/ml) in 13 of 28 patients.
Fig. 4.
Quantitation of soluble factors in LGL and healthy controls by ELISA and FACS. (A and C–F), Determination of expression levels of IFN-γ, IL-18/IGIF, MCP1/CCL2, IL-8/CXCL8, and IP10/CXCL10 in plasma samples from 25 to 29 LGL patients and 11 healthy controls. The mean absorbance was calculated for each set of duplicate standards, controls, and patient samples. The concentration was calculated based on the values on the standard curve. Dashed lines symbolize the average + 2 SD of values obtained from healthy controls and serve as a threshold for defining pathologic expansions in LGL patients. For IP10/CXCL10, outliers (>5000 pg/ml) were removed from analysis. NS, P > 0.05. *, P ≤ 0.05; **, P < 0.01, by Student’s unpaired t-test. (B) Quantitation of intracellular IFN-γ levels using FACS. Average values obtained from 13 LGL (hatched bars) patients and six controls (shaded bars) are shown for stimulated CD3+CD4+ and CD3+CD8+ populations as well as for nonstimulated CD3+CD8+ cells serving as negative controls. Error bars represent SD; *, P ≤ 0.05, by Student’s unpaired t-test.
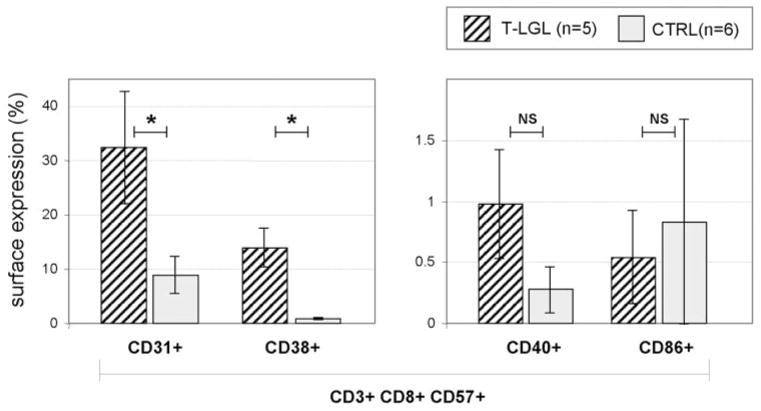
Finally, for additional confirmation of the microarray results, we analyzed surface expression of selected markers in patients (n=5) and healthy controls (n=6; Fig. 5). Average surface expression in patients versus controls was for CD31, 32.38 ± 20.76% versus 8.92 ± 6.71%; for CD38, 14.02 ± 12.35% versus 0.82 ± 0.49%; and for CD40, 0.98 ± 0.90% versus 0.28 ± 0.38%. There was no increase in CD86 expression in patients versus controls (0.54±0.76% vs. 0.83±1.69%).
Fig. 5.
Surface expression of selected antigens in LGL patients and healthy controls. Hatched bars illustrate average expression values of selected four surface markers (CD31, CD38, CD40, and CD86) on the CD3+CD8+CD57+ cell population in five LGL patients, and shaded bars represent the values obtained from six healthy controls. Error bars represent SD. NS, P > 0.05. *, P < 0.05, by Student’s unpaired t-test.
DISCUSSION
Analysis of global gene expression patterns using high-density microarrays is a hypothesis-forming tool. Genes described previously in association with T-LGL suggest that immune processes may be involved in the pathogenesis of this disease. This study was designed to elucidate mechanisms that are triggered during the transition process from the reactive lymphoproliferations to the extreme monoclonal process that resembles true malignancy and second, to identify uniquely expressed genes that distinguish T-LGL clones from their healthy counterparts. We attempted to overcome some of the inherent limitations of microarray expression studies, and therefore, we used highly purified, flow-sorted cell populations in our experiments. Although comparisons with various control populations such as total PBMC or normal CD8+ lymphocytes are of promising potential, we chose to base our data on evaluation of selected LGL clones in relation to the phenotypically equivalent, antigen-primed and terminally differentiated effector CTL from healthy controls. This CD8+CD57+ lymphocyte population is difficult to obtain as a result of its low frequency in the healthy population. Previously, expression analysis was limited to the comparison between nonpurified PBMC of patients and controls, run on cDNA/oligonucleotide microarrays [33]. Our results extend the finding of these initial studies.
Historically, one weakness of microarray analysis was poor reproducibility. To overcome this limitation, we have used a large cohort of controls that was pooled for the microarray experiment to decrease noise. In addition, we introduced various levels of validation in the original microarray/analyzed patients as well as in an independent cohort of patients and controls. Nevertheless, clinical heterogeneity may result in partially discordant results. In this respect, microarray analysis performed on a limited number of patients provides firm clues to be studied further in a targeted manner in an independent cohort of patients. Clearly, a weak point of our study is the low number of patients analyzed using microarrays, which inadvertently, could result in a high rate of false-positive results. Therefore, we carefully scrutinized our microarray data before performing validation.
In the current study, differences in gene expression between T-LGL and healthy effector CTL provide insights into the mechanisms of clonal expansion underlying the development of LGL leukemia. We intentionally focused on genes involved in the immune response, and we noted a striking pattern of dysregulation of genes associated with the CTL response to viral pathogens and generalized immune overactivation. For rational analysis and for verification of the microarray data, we selected a number of genes that are known to be linked to viral infections of tissues or to CTL themselves. For example, various cytokines that were significantly altered in expression in LGL patients versus healthy controls are produced typically by CD8+ cells during viral infections, and some of them are almost invariably associated with viral processes. Examples of such cytokines include IFN-γ and chemokines CXCL10 (IP10) and CXCL8 (IL-8) [51–55]. Interestingly, up-regulation of phosphorylated AKT and ERK, previously found in LGL [36, 56], can be caused by various chemokines such as CXCL8 (IL-8), which may prevent homeostatic apoptosis. Furthermore, it is known that viruses can modulate the PI-3K-Akt signaling pathway during acute and chronic, persistent viral infections as well as during viral transformation, leading to inhibition of apoptosis [57].
The role of causative viral culprits in T-LGL has been postulated and studied intensively. Although to date, no particular virus has been singled out, human T lymphotropic virus type 1 (HTLV-1) or a related lymphotropic retrovirus and the γ-herpesvirus family [especially human herpesvirus 8 (HHV-8)] are interesting candidates [58–62]. These viruses are clinically associated with hematologic malignancies, show lymphotropic activity, and most importantly, modulate a number of genes that were differentially expressed in our cohort of LGL patients [52, 53, 57, 63]. For example, HHV-8 up-regulates CXCL8 (IL-8), and this chemokine was also elevated in LGL clones, a finding clearly distinct from normal CD57+ CTL effector cells. Similarly, overactivated CD8+ T cells overexpress CCR2 (e.g., as a response to a virus), which during antiviral response, facilitates T cell migration to sites of infection [64]. CCR2 was strongly overexpressed in most LGL clones. Interestingly, Patient #26 showed an exception to this pattern: Unlike most of the other LGL cases associated with neutropenia, this patient presented with reticulocytopenic anemia. In the extended cohort of patients tested specifically for CCR2, 11 out of 16 patients showed up-regulation of this receptor. It is important to point out that patients with no CCR2 overexpression may belong to a subgroup combining other etiologies than viral. In such cases, it is possible that the initial step involving antigen-driven expansion is followed by a crucial “second hit” affecting genes that regulate homeostatic apoptosis.
LGL patients studied by microarray showed a considerable increase (average, 4.75-fold increase) of CD38 transcript when compared with the healthy effector CTL population; an even higher discordance in the expression of this gene was found between LGL patients and healthy noneffector cells (average, 7.06-fold increase). These microarray results were confirmed in an additional flow cytometry experiment (Fig. 5). In a recent report, persistent parvovirus B19 infection was associated with a CTL response characterized by overexpression of CD38, perforin, and CD57+ and in analogy to typical LGL, down-regulation of CD28 and CD27 markers [6]. These virus-infected cells retained strong effector function and intact proliferative capacity over a prolonged period of time. As CD38 up-regulation on CTL appears to be a general mechanism also present in other viral infections [65, 66], it has been proposed as a marker of viral replication in acute or untreated chronic infection [67]. Another surface protein overexpressed in LGL patients was CD32a (also known as FcγRII). High levels of activating CD32a were consistently found in LGL patients. Ligation of this receptor results in T cell stimulation and enhanced cytokine secretion [68, 69].
IFN and IFN-stimulated genes (ISG), including IFN-γ, IFN-induced protein with tetratricopeptide repeats 2 (IFIT2), IFIT3, IFN regulatory factor 4, IFN-inducible 27 (IFI27), IFI30, CXCL10 (small inducible cytokine B/IP10), and CXCL9 (monokine induced by IFN-γ), were generally up-modulated in LGL clones when compared with healthy effector CD57+ cells (Supplementary data). Although high expression of ISG is characteristic of antigen-induced T cell activation, in LGL clones, this feature appears to be much more pronounced. This finding signifies that the LGL clone is phenotypically related to healthy effector CTL but shows an even higher degree of cytotoxic activation, presumably as a result of persistent antigenic drive enhanced by an intrinsic dysregulation of homeostasis.
CD137 and CD31 are examples of genes that point toward exaggerated activation of memory cells, resulting in clonal expansion. CD137 (TNFRS9, 4-1BB) was originally isolated from a library constructed from activated HTLV-1-transformed lymphocytes; it was reported that this molecule contributes to clonal expansion and development of the Tc1 phenotype after antigen encounter in an inflammatory environment [70, 71]. In addition, its role as a main player in the etiology of autoimmune disease has been discussed [70–73]. Although antigen-specific effector cells transiently up-regulate CD137, its constitutive over-expression is mainly limited to clonally transformed cells. CD31, also known as PECAM, is the ligand for CD38 and stimulates integrin-dependent adhesion and transmigration of leukocytes through vascular cells. Its expression is generally elevated in TCR-stimulated lymphocytes [74–77]. Elevated levels of CD137 and CD31 in clonal CTL of LGL patients as compared with their healthy counterpart further support the notion that LGL cells are clonally transformed autoimmune CTL that are antigen-primed and pushed toward terminal differentiation.
In addition to genes associated with antiviral effector function, we found altered expression of various proteins involved in signal transduction pathways that could potentially explain clonal expansion/transformation and persistence of immunodominance in T-LGL. For example, decreased expression of PTEN in LGL clones is consistent with resistance to apoptosis. PTEN, an inhibitor of PI-3K, counterbalances activation via the AKT pathway, previously shown to be up-modulated in LGL [36]. The PI-3K-AKT pathway, if overactivated, antagonizes the ability of Fas to initiate apoptosis. Interestingly, this mechanism of apoptotic inhibition plays an important role in viral oncogenesis [57]. In a previous report elucidating the mechanism of apoptotic resistance in LGL leukemia, elevated levels of STAT3 were found, and activated STAT3 was shown to bind to the MCL-1 promoter, and MCL-1 is an antiapoptotic Bcl-2 family protein that promotes lymphomagenesis in human and murine B cell lymphoma and is required for the survival of clonal B and T cells [78, 79]. In our study, we found an increase of MCL-1 expression in LGL patients when compared with healthy effector cells (average fold change, 6.86 by array and 10.88 by Taqman). Further studies are required to clarify the prosurvival function of MCL-1 in clonally expanded LGL cells.
The relatively low detection rate of IFN-γ and IL-18 in plasma of LGL patients can be explained in different ways. For example, in many settings, assays that use antibodies for detection of proteins such as ELISA may have lower sensitivity than mRNA-based assays, including Taqman RT-PCR or expression array. Additionally, in our study, we have tested plasma derived from nonstimulated peripheral blood samples, and in previous reports, which focused on cytokine detection in LGL, supernatants from stimulated cell cultures were used.
In summary, leukemic LGL clones are more similar in their expression profile to healthy effector CD57+ CTL than to noneffector CTL. However, when focused on immune networks, significant dissimilarities exist between the phenotypes of LGL clones and healthy effector lymphocytes. These differences suggest that various features of the LGL transcriptome show a strong relationship with a CTL response, suggestive of the response to a chronic stimulus, such as viral infection or persistent inflammatory state. We must, however, acknowledge that viral infection may not be the sole cause of differential expression of some of the aforementioned genes; LGL leukemia is a heterogeneous disease and can hypothetically combine various etiologies. Our findings reveal important new aspects of the phenotype of expanded CTL clones and should provide an exploratory basis for a better understanding of the CTL-mediated autoimmune processes.
Supplementary Material
Acknowledgments
We thank Alexander Rodriguez for excellent technical assistance.
This work was supported in part by a grant from the National Institutes of Health, RO1 HL073429-01A1, awarded to J. P. M. M. W. W. designed the research, performed experiments, analyzed data, and wrote the manuscript; Z. N. performed experiments and analyzed data; A. J. performed experiments and analyzed data; J. P. performed experiments; N. B. and H-D.V. provided crucial reagents and helped in data analysis; P. L. performed preliminary analysis of data; and J. P. M. enrolled patients, designed the research, and wrote the manuscript.
References
- 1.Martin A, Barbesino G, Davies TF. T-cell receptors and autoimmune thyroid disease—signposts for T-cell-antigen driven diseases. Int Rev Immunol. 1999;18:111–140. doi: 10.3109/08830189909043021. [DOI] [PubMed] [Google Scholar]
- 2.Neumann H, Medana IM, Bauer J, Lassmann H. Cytotoxic T lymphocytes in autoimmune and degenerative CNS diseases. Trends Neurosci. 2002;25:313–319. doi: 10.1016/s0166-2236(02)02154-9. [DOI] [PubMed] [Google Scholar]
- 3.Posnett DN, Gottlieb A, Bussel JB, Friedman SM, Chiorazzi N, Li Y, Szabo P, Farid NR, Robinson MA. T cell antigen receptors in autoimmunity. J Immunol. 1988;141:1963–1969. [PubMed] [Google Scholar]
- 4.Ehl S, Hombach J, Aichele P, Rulicke T, Odermatt B, Hengartner H, Zinkernagel R, Pircher H. Viral and bacterial infections interfere with peripheral tolerance induction and activate CD8+ T cells to cause immunopathology. J Exp Med. 1998;187:763–774. doi: 10.1084/jem.187.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maini MK, Gudgeon N, Wedderburn LR, Rickinson AB, Beverley PC. Clonal expansions in acute EBV infection are detectable in the CD8 and not the CD4 subset and persist with a variable CD45 phenotype. J Immunol. 2000;165:5729–5737. doi: 10.4049/jimmunol.165.10.5729. [DOI] [PubMed] [Google Scholar]
- 6.Isa A, Kasprowicz V, Norbeck O, Loughry A, Jeffery K, Broliden K, Klenerman P, Tolfvenstam T, Bowness P. Prolonged activation of virus-specific CD8+T cells after acute B19 infection. PLoS Med. 2005;2:e343. doi: 10.1371/journal.pmed.0020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronsson B, Troye-Blomberg M, Smedman L. Increase of circulating CD8+CD57+ lymphocytes after measles infection but not after measles vaccination. J Clin Lab Immunol. 2004;53:1–12. [PubMed] [Google Scholar]
- 8.Dummer W, Niethammer AG, Baccala R, Lawson BR, Wagner N, Reisfeld RA, Theofilopoulos AN. T cell homeostatic proliferation elicits effective antitumor autoimmunity. J Clin Invest. 2002;110:185–192. doi: 10.1172/JCI15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handgretinger R, Geiselhart A, Moris A, Grau R, Teuffel O, Bethge W, Kanz L, Fisch P. Pure red-cell aplasia associated with clonal expansion of granular lymphocytes expressing killer-cell inhibitory receptors. N Engl J Med. 1999;340:278–284. doi: 10.1056/NEJM199901283400405. [DOI] [PubMed] [Google Scholar]
- 10.Risitano AM, Kook H, Zeng W, Chen G, Young NS, Maciejewski JP. Oligoclonal and polyclonal CD4 and CD8 lymphocytes in aplastic anemia and paroxysmal nocturnal hemoglobinuria measured by V β CDR3 spectratyping and flow cytometry. Blood. 2002;100:178–183. doi: 10.1182/blood-2002-01-0236. [DOI] [PubMed] [Google Scholar]
- 11.Carlson JA, Grabowski R, Chichester P, Paunovich E, Malfetano J. Comparative immunophenotypic study of lichen sclerosus: epidermotropic CD57+ lymphocytes are numerous—implications for pathogenesis. Am J Dermatopathol. 2000;22:7–16. doi: 10.1097/00000372-200002000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Kook H, Zeng W, Guibin C, Kirby M, Young NS, Maciejewski JP. Increased cytotoxic T cells with effector phenotype in aplastic anemia and myelodysplasia. Exp Hematol. 2001;29:1270–1277. doi: 10.1016/s0301-472x(01)00736-6. [DOI] [PubMed] [Google Scholar]
- 13.Sze DM, Giesajtis G, Brown RD, Raitakari M, Gibson J, Ho J, Baxter AG, Fazekas de St Groth B, Basten A, Joshua DE. Clonal cytotoxic T cells are expanded in myeloma and reside in the CD8(+)CD57(+)CD28(−) compartment. Blood. 2001;98:2817–2827. doi: 10.1182/blood.v98.9.2817. [DOI] [PubMed] [Google Scholar]
- 14.Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol. 2003;4:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 15.Hamann D, Baars PA, Rep MH, Hooibrink B, Kerkhof-Garde SR, Klein MR, van Lier RA. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med. 1997;186:1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigouret V, Hoffmann T, Arlettaz L, Villard J, Colonna M, Ticheli A, Gratwohl A, Samii K, Chapuis B, Rufer N, Roosnek E. Monoclonal T-cell expansions in asymptomatic individuals and in patients with large granular leukemia consist of cytotoxic effector T cells expressing the activating CD94:NKG2C/E and NKD2D killer cell receptors. Blood. 2003;101:3198–3204. doi: 10.1182/blood-2002-08-2408. [DOI] [PubMed] [Google Scholar]
- 17.Trimble LA, Kam LW, Friedman RS, Xu Z, Lieberman J. CD3ζ and CD28 down-modulation on CD8 T cells during viral infection. Blood. 2000;96:1021–1029. [PubMed] [Google Scholar]
- 18.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 19.Abo T, Balch CM. A differentiation antigen of human NK and K cells identified by a monoclonal antibody (HNK-1) J Immunol. 1981;127:1024–1029. [PubMed] [Google Scholar]
- 20.Kern F, Ode-Hakim S, Vogt K, Hoflich C, Reinke P, Volk HD. The enigma of CD57+ Clin Exp Immunol. 1996;104:180–184. doi: 10.1046/j.1365-2249.1996.d01-635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opferman JT, Korsmeyer SJ. Apoptosis in the development and maintenance of the immune system. Nat Immunol. 2003;4:410–415. doi: 10.1038/ni0503-410. [DOI] [PubMed] [Google Scholar]
- 22.Mollet L, Fautrel B, Leblond V, Bergeron F, Merle-Beral H, Baumelou E, Hubert P, Debre P, Autran B. Leukemic CD3+ LGL share functional properties with their CD8+ CD57+ cell counterpart expanded after BMT. Leukemia. 1999;13:230–240. doi: 10.1038/sj.leu.2401266. [DOI] [PubMed] [Google Scholar]
- 23.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993;82:1–14. [PubMed] [Google Scholar]
- 24.Lamy T, Loughran TP., Jr Current concepts: large granular lymphocyte leukemia. Blood Rev. 1999;13:230–240. doi: 10.1054/blre.1999.0118. [DOI] [PubMed] [Google Scholar]
- 25.Loughran TP, Jr, Starkebaum G. Large granular lymphocyte leukemia. Report of 38 cases and review of the literature. Medicine (Baltimore) 1987;66:397–405. [PubMed] [Google Scholar]
- 26.Lima M, Almeida J, Santos AH, dos Anjos Teixeira M, Alguero MC, Queiros ML, Balanzategui A, Justica B, Gonzalez M, San Miguel JF, Orfao A. Immunophenotypic analysis of the TCR-Vβ repertoire in 98 persistent expansions of CD3(+)/TCR-αβ(+) large granular lymphocytes: utility in assessing clonality and insights into the pathogenesis of the disease. Am J Pathol. 2001;159:1861–1868. doi: 10.1016/s0002-9440(10)63032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loughran TP, Jr, Starkebaum G, Aprile JA. Rearrangement and expression of T-cell receptor genes in large granular lymphocyte leukemia. Blood. 1988;71:822–824. [PubMed] [Google Scholar]
- 28.Melenhorst JJ, Eniafe R, Follmann D, Molldrem J, Kirby M, El Ouriaghli F, Barrett AJ. T-cell large granular lymphocyte leukemia is characterized by massive TCRBV-restricted clonal CD8 expansion and a generalized overexpression of the effector cell marker CD57. Hematol J. 2003;4:18–25. doi: 10.1038/sj.thj.6200212. [DOI] [PubMed] [Google Scholar]
- 29.Sokol L, Loughran TP., Jr Large granular lymphocyte leukemia. Oncologist. 2006;11:263–273. doi: 10.1634/theoncologist.11-3-263. [DOI] [PubMed] [Google Scholar]
- 30.Lamy T, Loughran TP., Jr Clinical features of large granular lymphocyte leukemia. Semin Hematol. 2003;40:185–195. doi: 10.1016/s0037-1963(03)00133-1. [DOI] [PubMed] [Google Scholar]
- 31.Perzova R, Loughran TP., Jr Constitutive expression of Fas ligand in large granular lymphocyte leukemia. Br J Haematol. 1997;97:123–126. doi: 10.1046/j.1365-2141.1997.d01-2113.x. [DOI] [PubMed] [Google Scholar]
- 32.Kothapalli R, Nyland SB, Kusmartseva I, Bailey RD, McKeown TM, Loughran TP., Jr Constitutive production of proinflammatory cytokines RANTES, MIP-1β and IL-18 characterizes LGL leukemia. Int J Oncol. 2005;26:529–535. [PubMed] [Google Scholar]
- 33.Kothapalli R, Bailey RD, Kusmartseva I, Mane S, Epling-Burnette PK, Loughran TP., Jr Constitutive expression of cytotoxic proteases and down-regulation of protease inhibitors in LGL leukemia. Int J Oncol. 2003;22:33–39. [PubMed] [Google Scholar]
- 34.Coakley G, Iqbal M, Brooks D, Panayi GS, Lanchbury JS. CD8+, CD57+ T cells from healthy elderly subjects suppress neutrophil development in vitro: implications for the neutropenia of Felty’s and large granular lymphocyte syndromes. Arthritis Rheum. 2000;43:834–843. doi: 10.1002/1529-0131(200004)43:4<834::AID-ANR14>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 35.Wlodarski MW, O’Keefe C, Howe EC, Risitano AM, Rodriguez A, Warshawsky I, Loughran TP, Jr, Maciejewski JP. Pathologic clonal cytotoxic T-cell responses: nonrandom nature of the T-cell-receptor restriction in large granular lymphocyte leukemia. Blood. 2005;106:2769–2780. doi: 10.1182/blood-2004-10-4045. [DOI] [PubMed] [Google Scholar]
- 36.Schade AE, Powers JJ, Wlodarski MW, Maciejewski JP. Phosphatidylinositol-3-phosphate kinase pathway activation protects leukemic large granular lymphocytes from undergoing homeostatic apoptosis. Blood. 2006;107:4834–4840. doi: 10.1182/blood-2005-08-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamy T, Liu JH, Landowski TH, Dalton WS, Loughran TP., Jr Dysregulation of CD95/CD95 ligand-apoptotic pathway in CD3(+) large granular lymphocyte leukemia. Blood. 1998;92:4771–4777. [PubMed] [Google Scholar]
- 38.Liu JH, Wei S, Lamy T, Li Y, Epling-Burnette PK, Djeu JY, Loughran TP., Jr Blockade of Fas-dependent apoptosis by soluble Fas in LGL leukemia. Blood. 2002;100:1449–1453. [PubMed] [Google Scholar]
- 39.Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, Li Y, Wang JM, Yang-Yen HF, Karras J, Jove R, Loughran TP., Jr Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107:351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Epling-Burnette PK, Loughran TP., Jr Survival signals in leukemic large granular lymphocytes. Semin Hematol. 2003;40:213–220. doi: 10.1016/s0037-1963(03)00135-5. [DOI] [PubMed] [Google Scholar]
- 41.Melenhorst JJ, Brummendorf TH, Kirby M, Lansdorp PM, Barrett AJ. CD8+ T cells in large granular lymphocyte leukemia are not defective in activation- and replication-related apoptosis. Leuk Res. 2001;25:699–708. doi: 10.1016/s0145-2126(01)00010-8. [DOI] [PubMed] [Google Scholar]
- 42.Shvidel L, Duksin C, Tzimanis A, Shtalrid M, Klepfish A, Sigler E, Haran M, Eilat E, Berrebi A. Cytokine release by activated T-cells in large granular lymphocytic leukemia associated with autoimmune disorders. Hematol J. 2002;3:32–37. doi: 10.1038/sj.thj.6200149. [DOI] [PubMed] [Google Scholar]
- 43.Kothapalli R, Yoder SJ, Mane S, Loughran TP., Jr Microarray results: how accurate are they? BMC Bioinformatics. 2002;3:22. doi: 10.1186/1471-2105-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP., Jr The lymphoproliferative disease of granular lymphocytes: updated criteria for diagnosis. Blood. 1997;89:256–260. [PubMed] [Google Scholar]
- 45.Langerak AW, van den Beemd R, Wolvers-Tettero IL, Boor PP, van Lochem EG, Hooijkaas H, van Dongen JJ. Molecular and flow cytometric analysis of the Vβ repertoire for clonality assessment in mature TCRαβ T-cell proliferations. Blood. 2001;98:165–173. doi: 10.1182/blood.v98.1.165. [DOI] [PubMed] [Google Scholar]
- 46.O’Keefe CL, Plasilova M, Wlodarski M, Risitano AM, Rodriguez AR, Howe E, Young NS, Hsi E, Maciejewski JP. Molecular analysis of TCR clonotypes in LGL: a clonal model for polyclonal responses. J Immunol. 2004;172:1960–1969. doi: 10.4049/jimmunol.172.3.1960. [DOI] [PubMed] [Google Scholar]
- 47.Wlodarski MW, Gondek LP, Nearman ZP, Plasilova M, Kalaycio M, Maciejewski JP. Molecular strategies for detection and quantitation of clonal cytotoxic T cell responses in aplastic anemia and myelodysplastic syndrome. Blood. 2006;108:2632–2641. doi: 10.1182/blood-2005-09-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng W, Miyazato A, Chen G, Kajigaya S, Young NS, Maciejewski JP. Interferon-γ-induced gene expression in CD34 cells: identification of pathologic cytokine-specific signature profiles. Blood. 2006;107:167–175. doi: 10.1182/blood-2005-05-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng W, Kajigaya S, Chen G, Risitano AM, Nunez O, Young NS. Transcript profile of CD4+ and CD8+ T cells from the bone marrow of acquired aplastic anemia patients. Exp Hematol. 2004;32:806–814. doi: 10.1016/j.exphem.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Cheng J, Sun S, Tracy A, Hubbell E, Morris J, Valmeekam V, Kimbrough A, Cline MS, Liu G, Shigeta R, Kulp D, Siani-Rose MA. NetAffx gene ontology mining tool: a visual approach for microarray data analysis. Bioinformatics. 2004;20:1462–1463. doi: 10.1093/bioinformatics/bth087. [DOI] [PubMed] [Google Scholar]
- 51.Lewis MJ, Gautier VW, Wang XP, Kaplan MH, Hall WW. Spontaneous production of C-C chemokines by individuals infected with human T lymphotropic virus type II (HTLV-II) alone and HTLV-II/HIV-1 coinfected individuals. J Immunol. 2000;165:4127–4132. doi: 10.4049/jimmunol.165.7.4127. [DOI] [PubMed] [Google Scholar]
- 52.Cook WJ, Kramer MF, Walker RM, Burwell TJ, Holman HA, Coen DM, Knipe DM. Persistent expression of chemokine and chemokine receptor RNAs at primary and latent sites of herpes simplex virus 1 infection. Virol J. 2004;1:5. doi: 10.1186/1743-422X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melchjorsen J, Sorensen LN, Paludan SR. Expression and function of chemokines during viral infections: from molecular mechanisms to in vivo function. J Leukoc Biol. 2003;74:331–343. doi: 10.1189/jlb.1102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 55.Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu Rev Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. [DOI] [PubMed] [Google Scholar]
- 56.Epling-Burnette PK, Bai F, Wei S, Chaurasia P, Painter JS, Olashaw N, Hamilton A, Sebti S, Djeu JY, Loughran TP. ERK couples chronic survival of NK cells to constitutively activated Ras in lymphoproliferative disease of granular lymphocytes (LDGL) Oncogene. 2004;23:9220–9229. doi: 10.1038/sj.onc.1208122. [DOI] [PubMed] [Google Scholar]
- 57.Cooray S. The pivotal role of phosphatidylinositol 3-kinase-Akt signal transduction in virus survival. J Gen Virol. 2004;85:1065–1076. doi: 10.1099/vir.0.19771-0. [DOI] [PubMed] [Google Scholar]
- 58.Loughran TP, Jr, Hadlock KG, Perzova R, Gentile TC, Yang Q, Foung SK, Poiesz BJ. Epitope mapping of HTLV envelope seroreactivity in LGL leukemia. Br J Haematol. 1998;101:318–324. doi: 10.1046/j.1365-2141.1998.00691.x. [DOI] [PubMed] [Google Scholar]
- 59.Loughran TP, Jr, Coyle T, Sherman MP, Starkebaum G, Ehrlich GD, Ruscetti FW, Poiesz BJ. Detection of human T-cell leukemia/lymphoma virus, type II, in a patient with large granular lymphocyte leukemia. Blood. 1992;80:1116–1119. [PubMed] [Google Scholar]
- 60.Tagawa S, Mizuki M, Onoi U, Nakamura Y, Nozima J, Yoshida H, Kondo K, Mukai T, Yamanishi K, Kitani T. Transformation of large granular lymphocytic leukemia during the course of a reactivated human herpesvirus-6 infection. Leukemia. 1992;6:465–469. [PubMed] [Google Scholar]
- 61.Swa S, Wright H, Thomson J, Reid H, Haig D. Constitutive activation of Lck and Fyn tyrosine kinases in large granular lymphocytes infected with the γ-herpesvirus agents of malignant catarrhal fever. Immunology. 2001;102:44–52. doi: 10.1046/j.1365-2567.2001.01154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sokol L, Agrawal D, Loughran TP., Jr Characterization of HTLV envelope seroreactivity in large granular lymphocyte leukemia. Leuk Res. 2005;29:381–387. doi: 10.1016/j.leukres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 63.Bangham CR, Osame M. Cellular immune response to HTLV-1. Oncogene. 2005;24:6035–6046. doi: 10.1038/sj.onc.1208970. [DOI] [PubMed] [Google Scholar]
- 64.Nansen A, Marker O, Bartholdy C, Thomsen AR. CCR2+ and CCR5+ CD8+ T cells increase during viral infection and migrate to sites of infection. Eur J Immunol. 2000;30:1797–1806. doi: 10.1002/1521-4141(200007)30:7<1797::AID-IMMU1797>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 65.Jiang JQ, Balasubramanian S, Hawley-Foss NC, Badley AD, Rosenthal KL, Copeland KF. Production of CD8+ T cell nonlytic suppressive factors by CD28, CD38, and HLA-DR subpopulations. AIDS Res Hum Retroviruses. 2003;19:497–502. doi: 10.1089/088922203766774540. [DOI] [PubMed] [Google Scholar]
- 66.Benito JM, Lopez M, Lozano S, Martinez P, Gonzalez-Lahoz J, Soriano V. CD38 expression on CD8 T lymphocytes as a marker of residual virus replication in chronically HIV-infected patients receiving antiretroviral therapy. AIDS Res Hum Retroviruses. 2004;20:227–233. doi: 10.1089/088922204773004950. [DOI] [PubMed] [Google Scholar]
- 67.Mocroft A, Bofill M, Lipman M, Medina E, Borthwick N, Timms A, Batista L, Winter M, Sabin CA, Johnson M, Lee CA, Phillips A, Janossy G. CD8+, CD38+ lymphocyte percent: a useful immunological marker for monitoring HIV-1-infected patients. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:158–162. doi: 10.1097/00042560-199702010-00009. [DOI] [PubMed] [Google Scholar]
- 68.Sandilands GP, MacPherson SA, Burnett ER, Russell AJ, Downie I, MacSween RN. Differential expression of CD32 isoforms following alloactivation of human T cells. Immunology. 1997;91:204–211. doi: 10.1046/j.1365-2567.1997.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coggeshall KM. Regulation of signal transduction by the Fc γ receptor family members and their involvement in autoimmunity. Curr Dir Autoimmun. 2002;5:1–29. doi: 10.1159/000060554. [DOI] [PubMed] [Google Scholar]
- 70.Ma BY, Mikolajczak SA, Danesh A, Hosiawa KA, Cameron CM, Takaori-Kondo A, Uchiyama T, Kelvin DJ, Ochi A. The expression and the regulatory role of OX40 and 4-1BB heterodimer in activated human T cells. Blood. 2005;106:2002–2010. doi: 10.1182/blood-2004-04-1622. [DOI] [PubMed] [Google Scholar]
- 71.Cooper D, Bansal-Pakala P, Croft M. 4-1BB (CD137) controls the clonal expansion and survival of CD8 T cells in vivo but does not contribute to the development of cytotoxicity. Eur J Immunol. 2002;32:521–529. doi: 10.1002/1521-4141(200202)32:2<521::AID-IMMU521>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 72.Saoulli K, Lee SY, Cannons JL, Yeh WC, Santana A, Goldstein MD, Bangia N, DeBenedette MA, Mak TW, Choi Y, Watts TH. CD28-independent, TRAF2-dependent costimulation of resting T cells by 4-1BB ligand. J Exp Med. 1998;187:1849–1862. doi: 10.1084/jem.187.11.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cannons JL, Chamberlain G, Howson J, Smink LJ, Todd JA, Peterson LB, Wicker LS, Watts TH. Genetic and functional association of the immune signaling molecule 4-1BB (CD137/TNFRSF9) with type 1 diabetes. J Autoimmun. 2005;25:13–20. doi: 10.1016/j.jaut.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 74.Ilan N, Madri JA. PECAM-1: old friend, new partners. Curr Opin Cell Biol. 2003;15:515–524. doi: 10.1016/s0955-0674(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 75.Prager E, Staffler G, Majdic O, Saemann M, Godar S, Zlabinger G, Stockinger H. Induction of hyporesponsiveness and impaired T lymphocyte activation by the CD31 receptor:ligand pathway in T cells. J Immunol. 2001;166:2364–2371. doi: 10.4049/jimmunol.166.4.2364. [DOI] [PubMed] [Google Scholar]
- 76.Reedquist KA, Ross E, Koop EA, Wolthuis RM, Zwartkruis FJ, van Kooyk Y, Salmon M, Buckley CD, Bos JL. The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J Cell Biol. 2000;148:1151–1158. doi: 10.1083/jcb.148.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zehnder JL, Shatsky M, Leung LL, Butcher EC, McGregor JL, Levitt LJ. Involvement of CD31 in lymphocyte-mediated immune responses: importance of the membrane-proximal immunoglobulin domain and identification of an inhibiting CD31 peptide. Blood. 1995;85:1282–1288. [PubMed] [Google Scholar]
- 78.Michels J, O’Neill JW, Dallman CL, Mouzakiti A, Habens F, Brimmell M, Zhang KY, Craig RW, Marcusson EG, Johnson PW, Packham G. Mcl-1 is required for Akata6 B-lymphoma cell survival and is converted to a cell death molecule by efficient caspase-mediated cleavage. Oncogene. 2004;23:4818–4827. doi: 10.1038/sj.onc.1207648. [DOI] [PubMed] [Google Scholar]
- 79.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.