Abstract
Purpose
To provide a comprehensive, thorough analysis of somatic mutation and promoter hypermethylation of the von Hippel-Lindau VHL gene in the cancer genome, unique to clear cell renal cancer (ccRCC). Identify relationships between the prevalence of VHL gene alterations and alteration subtypes with patient and tumor characteristics.
Experimental Design
As part of a large kidney cancer case-control study conducted in Central Europe, we analyzed VHL mutations and promoter methylation in 205 well characterized, histologically-confirmed patient tumor biopsies utilizing a combination of sensitive, high-throughput methods (endonuclease scanning and Sanger sequencing), and analysis of 11CpG sites in the VHL promoter.
Results
We identified mutations in 82.4% of cases, the highest VHL gene mutation prevalence reported to date. Analysis of 11 VHL promoter CpG sites revealed that 8.3% of tumors were hypermethylated and all were mutation negative. In total, 91% of ccRCCs exhibited alteration of the gene through genetic or epigenetic mechanisms. Analysis of patient and tumor characteristics revealed that certain mutation subtypes were significantly associated with Fuhrman nuclear grade, metastasis, node positivity, and self-reported family history of RCC.
Conclusion
Detection of VHL gene alterations using these accurate, sensitive and practical methods provides evidence that the vast majority of histologically confirmed ccRCC tumors possess genetic or epigenetic alteration of the VHL gene and support the hypothesis that VHL alteration is an early event in ccRCC carcinogenesis. These findings also indicate that VHL molecular subtypes can provide a sensitive marker of tumor heterogeneity among histologically similar ccRCC cases for etiologic, prognostic, and translational studies.
Introduction
Considerable progress has been made understanding the genetic basis of kidney cancer (1, 2). The susceptibility genes associated with several forms of inherited renal cell cancer (RCC) have been identified by rigorous analysis of families using genetic linkage analysis and positional cloning (3-7). The most common subtype of RCC is the conventional, clear cell type (ccRCC), which accounts for approximately 75% of cases. In both familial and sporadic ccRCC, allelic inactivation of the VHL gene has been shown to occur through mutation, methylation, and/or chromosomal loss in the majority of ccRCCs analyzed (8-13).
Molecular studies examining tumor DNA from sporadic cases of ccRCC have provided strong evidence that VHL alteration is a common, early event in the carcinogenic process (12). In addition, specific types of VHL mutations may be associated with etiologic factors, disease progression, and prognosis (1). However, several studies have examined VHL alterations in multiple patient populations and have reported significant differences in the prevalence of mutations observed, ranging from 50−71% (8-10,12,13). Differences in mutation prevalence could be due to several factors, including: i) the patient population examined, ii) tumor histopathology, iii) the ratio of tumor to normal DNA in a sample, or iv) the method of mutation detection employed. For example, inclusion of non-ccRCC tumors in a molecular study would be expected to decrease the VHL mutation prevalence observed, as VHL mutations are rare in other types of RCC (14).
Accurate, sensitive, and practical high-throughput mutation detection methods must be used to analyze large numbers of well-characterized samples in order to correlate the prevalence, type, and location of VHL mutations with etiologic or prognostic risk factors. Previous studies have relied on scanning techniques, such as denaturing high performance liquid chromatography or single-strand conformational polymorphism, followed by annotation of variants using Sanger sequencing. Neither of these scanning techniques is amenable to a level of throughput that matches data generation by fluorescent Sanger sequencing on a capillary electrophoresis platform.
Recently, a new scanning technique has been developed that offers key advantages over previous approaches, especially when used in combination with Sanger sequencing. Endonuclease scanning utilizes enzymes purified from celery (Cel I and II) to detect mutations in amplified PCR products (15, 16, 17). The endonucleases cleave heteroduplexed DNA at mismatch sites and digests are separated by their size. Cleavage products and their sizes do not only indicate the presence of a variant, but also provide information of its location, allowing quick and easy identification of mutations when PCR products are sequenced. The primary objective of the current study was to apply this new mutation detection method to analyze VHL gene mutations in a set of tumor samples that were collected as part of a large, international molecular epidemiological study of kidney cancer etiology and survival, selected to represent a distribution by histopathological variables and known RCC risk factors. The second goal was to provide a comprehensive genetic and epigenetic analysis to elucidate the relationship between somatic mutations and promoter hypermethylation of the VHL gene in the cancer genome unique to ccRCC. This goal would be accomplished by using only DNA extracted from frozen tissue sections that were each histologically confirmed clear cell cases (ccRCC) by an NCI expert in renal tumor pathology (MM), by exhaustively searching for and confirming all mutations using a combination of analytical methods that could be practical, sensitive but suitable for analysis of a large number of cases, and lastly, by using Sanger sequencing evaluate 11 CpG sites in the VHL promoter in place of methylation specific PCR, which can result in both false positive and negative results.
Materials and Methods
Tumor DNAs
A subset of patient tumor samples (N=205) from cases enrolled in a large case-control study of kidney cancer conducted in central and eastern Europe were selected to include a distribution of cases by tumor stage, grade, and known risk factors such as sex, body mass index (BMI), hypertension, smoking. We obtained informed consent from potential participants in accordance with the National Cancer Institute, International Agency for Research on Cancer, and local Institutional Review Boards. Tumor DNA extraction was performed following pathologic review, and manual macrodissection to remove non-tumor tissue. Sample areas that appeared to contain at least 70% tumor cells were used for DNA extraction. For each sample, 5 mm3 of tissue was sectioned and digested with 0.4 μg Proteinase K per μl of digestion buffer (500 mM KCl, 100 mM Tris-HCl, 15 mM MgCl2, 0.5% Tween 20) at 50°C overnight. A standard protocol1 was used to extract DNA from the digested samples. DNA was quantitated using a ND-1000 spectrophotometer (Nanodrop, Wilmington, DE).
PCR
Amplification of patient tumor DNAs was carried out in 50 μl reactions using 10−15 ng tumor DNA and 1.25 U of either HotMaster Taq DNA Polymerase (Eppendorf, Westbury, NY) or Hot-Start Taq DNA polymerase (Denville Scientific, Metuchen, NJ) with their respective 1x Reaction Buffers, and 0.2 mM of each dNTP, and 0.2 μM each of forward and reverse primer. Thermal cycling was accomplished using MJ Research (Bio-Rad, Waltham, MA) Dyad and Tetrad DNA Engines and a program of 95°C for 2 min, 10 cycles of touchdown PCR, and then 30 cycles of 95°C for 30 sec, 58°C for 30 sec, and 68°C for 30 sec; followed by a final 5 min extension at 68°C. PCR products were heteroduplexed using a program of 95°C for 5 minutes, cooling to 85°C at −2.0°C/sec., cooling to 25°C at −0.2°C/sec., then incubation at 4°C in a thermocycler. PCR products (5 μl) were analyzed by 2% agarose gel electrophoresis in 1x TAE and visualized with ethidium bromide.
Endonuclease Scanning
Heteroduplexed PCR samples were combined with 15 U of Surveyor Nuclease W and 1 μl Surveyor Enhancer W (Transgenomic, Omaha, NE) and incubated at 42°C for 20 min. Digestions were terminated by addition of 2 μl Stop Solution (0.5 M EDTA, pH 8.0) and analyzed on a WAVE® HPLC instrument equipped with a High Sensitivity Detection system and a DNASep® HT column (Transgenomic). Run parameters were a 12 μl injection volume using the double-strand sizing—multiple fragments application at a 50°C oven temperature. WAVE HPLC gradient parameters are described in Supplemental Methods. Detection was 260 nm for UV and 495 nm excitation/537 nm emission for fluorescence. A 100 bp DNA ladder (New England Biolabs, Ipswich, MA) was run as a size marker. Instrument control, data acquisition, and data analysis were performed using WAVE Navigator software. Positive and negative controls were included with each plate of PCR products to monitor endonuclease cleavage efficiency.
VHL Gene Sequencing
Excess PCR primers were removed from 10 μl of PCR product using the Ampure PCR Purification system (Agencourt Bioscience, Beverly, MA). Purified product was eluted in 30 μl of de-ionized water. Reaction chemistry using BigDye v. 3.1 (Applied Biosystems, Foster City, CA) and cycle sequencing were adapted from the manufacturer's recommendations. Cycle sequencing products were purified using CleanSeq reagents (Agencourt Bioscience Corp., Beverly, MA). Purified sequencing products were eluted in 40 μl of 0.01 μM EDTA and 30 μl was run on an ABI 3100 Genetic Analyzer. Sequence chromatograms were analyzed by several methods as described in the Suppl. Methods.
VHL Promoter Methylation
Standard methods were used for bisulfite modification of 100−500 ng tumor DNA (Zymo Research Labs, Orange, CA). Primers (Suppl. Methods) were designed to amplify both methylated and unmethylated alleles across 11 CpG dinucleotides of the VHL promoter. PCR was performed on 2 μl treated DNA in a MJ Research PTC200 thermal cycler: 5 min of denaturation at 95°C, 10 cycles of touchdown to a 50°C annealing temperature, then 35 cycles of 30 sec at 95°C, 30 sec at 50°C, and 60 sec at 72°C, followed by a final 5 min extension at 72°C. Nested PCR was performed on 1 μl of a 1:10 dilution of first round product using cycling conditions as described above. PCR products (5 μl) were visualized in 2.0% agarose and bi-directionally sequenced. Cytosine positions in CpGs were inspected for thymine or cytosine signal in chromatograms as follows: T only, not methylated; both cytosine and thymine, partially methylated; C only, fully methylated. Tumor samples containing at least 4 methylated CpGs analyzed (>36%) were considered methylated. All analyses were run in duplicate, blinded to VHL mutation status, and with positive (CpGenome Universal Methylated DNA, Chemicon/Millipore, Billerica, MA) and negative (K562 Human Genomic DNA, Promega, Madison, WI) controls.
Subcloning
Cloning of PCR products utilized topoisomerase-activated vector (18) and a TOPO cloning kit (Invitrogen, Carlsbad, CA). Amplified inserts were prepared by colony PCR (19), and sequenced as described above before quantifying the number of clones containing mutant alleles.
Statistical Analysis
VHL mutation and promoter methylation were considered as dichotomous variables per case (no/yes). Tumor and subject characteristics such as clinical stage, grade, node stage (N0, N1, N2), body mass index (BMI) (<25, 25−35, 35+), pack-years of smoking (0, 1−20, 20+), and smoking status (never, former, current), were considered as categorical variables. Other variables such as metastasis (M0, M1) self-reported hypertension (no/yes), family history of cancer (no/yes-kidney and no/yes-any), sex, and age at diagnosis (<50, ≥50 years) were considered as dichotomous variables. Prevalences of VHL alterations were calculated by dividing the number of cases with an alteration by the total number of cases analyzed. Cases with missing information were excluded from analyses. Chi-square tests were applied to contingency table (2×2) analysis to test for differences between the number of cases with or without an alteration in each group. Ordered logistic regression was used to analyze associations between categorical variables and cases with particular VHL alterations. All analyses were conducted using STATA 9.0 (Stata Corporation, College Station, TX) and all statistical tests were two-sided.
Results
Pilot Study
DNA extracted from 22 RCC patient tumors with previously-characterized mutations in the VHL gene was used to compare the specificity and sensitivity of endonuclease scanning relative to DHPLC. All mutations were detected using both techniques. To assess the sensitivity of endonuclease scanning, serial dilutions of PCR products from six tumor DNA samples with known mutations were combined with normal amplicons and analyzed. Both techniques successfully detected mutations in DNA dilutions that contained 3−5% mutant/normal DNA (data not shown).
Patient and Tumor Characteristics
Cases were selected to include a distribution by sex, age at diagnosis, histopathological parameters such as tumor stage and grade, and other kidney cancer risk factors that were prevalent in this population, such as hypertension, high BMI, tobacco smoking, and family history of cancer. Table 1 provides a frequency distribution of patient and tumor characteristics for cases that were included in this study. Most cases (63%) were from the Czech Republic. Sex, smoking habits, BMI category, and prevalence of hypertension were similar in those in the entire study population.
Table 1.
Distribution of patient and tumor characteristics among cases in this study
| N | % | |
|---|---|---|
| Total Cases | 205 | 100% |
| Center | ||
| Romania | 36 | 18% |
| Poland | 21 | 10% |
| Russia | 19 | 9% |
| Czech Republic* | 129 | 63% |
| Sex | ||
| Male | 122 | 60% |
| Female | 83 | 40% |
| Smoking Status | ||
| Never | 89 | 45% |
| Ever | 110 | 55% |
| BMI† | ||
| <25 | 54 | 26% |
| 25−35 | 147 | 72% |
| >35 | 4 | 2% |
| Self-Reported Hypertension | ||
| No | 108 | 53% |
| Yes | 97 | 47% |
| Tumor Stage | ||
| T | 42 | 20% |
| T2 | 92 | 45% |
| T3/T4 | 54 | 26% |
| missing | 17 | 8% |
| Metastasis | ||
| Mx | 37 | 18% |
| M0 | 148 | 72% |
| M1 | 9 | 4% |
| Missing | 11 | 5% |
| Fuhrman Nuclear Grade | ||
| I | 2 | 1% |
| II | 154 | 75% |
| III | 39 | 19% |
| IV | 3 | 1% |
| Missing | 7 | 3% |
Czech centers include Brno, Olomouc, Prague, Ceske Budejovice
Body Mass Index (BMI) at time of interview
Analysis of VHL Mutations
Detailed annotation of all mutations and SNPs can be found in Supplementary Table 1. Representative results from analysis of endonuclease digests are shown in Supplementary Figures 1 and 2a-f. Endonuclease cleavage product sizes were determined by overlay of a digest profile with a sizing ladder using Navigator software. This allowed rapid location of variants in sequence chromatograms. We observed 100% concordance between mutations identified using endonuclease, and those detected in forward and reverse sequencing chromatograms. Overall, VHL was mutated in 82.4% (169/205) of cases (Table 2). Double mutations were found in 3.4% (7/205) of cases. The P25L variant that has been previously described (10, 20) was present in eight patients (4%). As in previous studies, this variant was not considered a mutation, however, six of these cases (75%) also possessed another VHL alteration. A total of 3% (N=7) of tumors exhibited coding SNPs and 2% (N=4) possessed an IVS 2+43 intronic SNP. A total of 176 VHL mutations were identified, including deletions (34%, N=59), insertions (24%, n = 42), missense (24%, N=42), nonsense (10%, N=18), and splice junction alterations (9%, N=15). Ten of 15 splice junction mutations occurred in the first base of an intron and all were observed within 3 bases of an exon. The prevalence of mutations by exon was: exon 1, 37% (N=65); exon 2, 34% (N=59); and exon 3, 30% (N=52). We found a high (6%, N=11) prevalence of alterations that appear to affect exon 2/3 splicing and may truncate the protein downstream of codon 155. Figure 1A shows that alterations of the VHL gene were distributed from codon 52 (a deletion) to 213 (an insertion), excluding the P25L variant (10, 20). Subtypes of mutations were distributed as previously reported (8-10, 13).
Table 2.
Subtypes of von Hippel-Lindau (VHL) gene mutations observed among 205 histologically confirmed clear cell renal tumors
|
Number of Cases (N = 205) |
Number of Mutations (N = 176) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VHL status |
N |
% |
Type |
N |
% |
Location |
N |
% |
RSI *Bin |
N |
% |
Database |
N |
% |
| Mutated | 169 | 82 | Deletion | 59 | 34 | Exon 1 | 62 | 35 | Low | 80 | 45 | Yes | 124 | 70 |
| 1 Mutation | 162 | 79 | Insertion | 42 | 24 | Intron 1 | 4 | 2 | Medium | 92 | 52 | No | 52 | 30 |
| 2 Mutations | 7 | 3 | Missense | 42 | 24 | Exon 2 | 50 | 28 | High | 4 | 2 | |||
| No mutation | 36 | 18 | Nonsense | 18 | 10 | Intron 2 | 11 | 6 | ||||||
| P25L‡ | 8 | 4 | Splicing† | 15 | 9 | Exon 3 | 49 | 28 | ||||||
| Silent‡ | 7 | 3 | ||||||||||||
Relative Signal Intensity (RSI) of mutant allele compared to wild type
Splicing mutations include intronic nucleotide changes within 3 bases of the intron-exon boundry.
P25L and silent mutations are shown for reference and were not classified as functional mutations in future calculations.
Figure 1.
Distribution of VHL mutations by codon. A, all mutations; B, mutations with a low RSI value (5−30%).
Confirmation of VHL Mutant Status
Analysis of 20 mutation positive cases from half of the VHL mutant cases (N=20) and a subset that were wild-type (N=19) was repeated and scored in a blinded manner. All but one mutation was confirmed (95% agreement, 38/39), and this false positive was excluded from subsequent calculations. The second half of cases were re-amplified (268/273 amplicons) using a high fidelity polymerase (different from the first round analyses) and VHL mutation or wild-type status was confirmed in all cases. Five samples failed to amplify and were not repeated due to limited DNA quantities.
Relative Signal Intensity (RSI)
Examination of mutant and wild type peak heights averaged from forward and reverse sequencing chromatograms revealed that the fluorescence signal of mutant nucleotides in DNA amplified from many tumors were dramatically less than those from wild-type nucleotide peaks. Figure 1B presents the distribution of mutations with a low RSI. To calculate RSI of mutant peak heights, we averaged visual estimates of peak heights from forward and reverse sequencing traces, divided by total signal (mutant + wild-type peaks) present at a single nucleotide position. These RSI estimates were grouped into high (>60%), medium (31−60%), and low (5−30%) bins to estimate the proportion of mutations that might be difficult to analyze using sequencing software. For example, mutations in the low RSI bin exhibited less than 30% signal compared to the total fluorescence, whereas greater than 70% of signal was observed from wild-type nucleotides. As shown in Table 2, 46% of variants exhibited a low RSI value. These samples represent cases whose mutant signal could be easily masked by wild-type sequence, especially in the presence of high background. Automated detection of low RSI mutations was unreliable by commercially available sequencing software.
Confirmation of Allele Frequencies by Subcloning
To determine whether visual estimates of RSI reflected the proportion of mutant allele in the original tumor DNA or whether PCR and sequencing were introducing a bias towards wild type, PCR products from 10 tumors containing mutations with RSI values between 10−50% were amplified and subcloned (Table 3). Mutant and wild type alleles were counted in an average of 48 subclones per tumor, each of which was sequenced on both strands. Generally, mutant allele frequency determined by subcloning agreed to within ± 12% of the visual RSI estimate (7/10 cases). Five tumors with deletions each possessed a RSI value that was lower than the mutant allele frequency determined from subcloning. This may indicate that either visual estimation of RSI for deletions was inaccurate or alternatively, that deletions actually show lower fluorescent signal compared to that of wild type alleles in sequence chromatograms.
Table 3.
Comparison of mutant allele frequencies estimated by subcloning and RSI*
|
Mutant Allele Frequency |
||||||
|---|---|---|---|---|---|---|
| Tumor ID | Exon | Sequencing of Tumor DNA† | Sequencing of Clones | RSI (%)* | Cloning‡ | (%) |
| 4_1 | 3 | 763 deletion | 763 delCTCTACG | 20 | 18/56 | 32 |
| 4_6 | 2 | 613 G>T | 613 G>T | 40 | 17/48 | 35 |
| 4_8 | 2 | 652 A>G | 652 A>G | 50 | 21/38 | 55 |
| 4_12 | 1 | 469 C>T | 469 C>T | 40 | 9/31 | 29 |
| 4_16 | 2 | 581 deletion | 581 deletion | 30 | 21/58 | 36 |
| 4_31 | 3 | IVS2−1 G>A | IVS2−1 G>A | 25 | 3/40 | 8 |
| 4_47 | 3 | 752 deletion | 752 delTCGTCA | 30 | 18/54 | 33 |
| 4_53 | 2 | 574 insertion | 574 delGA | 10 | 20/57 | 35 |
| 4_65 | 2 | 570 deletion | 570 delCAGAGAT | 20 | 24/55 | 44 |
| 4_70 | 2 | 665 T>C | 665 T>C | 30 | 21/50 | 42 |
Relative Signal Intensity (RSI) of mutant allele compare with total fluorescence
Insertions and deletions were not characterized in tumor DNA sequencing
Number of positive transformants
VHL Promoter Methylation
Another mechanism by which genes can be inactivated is through promoter hypermethylation. Most studies have used methylation-specific PCR (MSP), which relies on methylation of a few CpGs to determine the methylation status of the promoter (8, 10, 13, 21). As with MSP, we analyzed bisulfite-treated DNA, but primers were designed to equally amplify both methylated and unmethylated alleles. To provide a comprehensive analysis, after bisulfite treatment we used Sanger sequencing to evaluate cytosine positions in CpGs. Cytosine (methylated):thymine (unmethylated) ratios in 11 sequential CpGs of the VHL promoter. Mutation negative (94%, 34/36) and positive (12%, 21/169) tumors were selected for analysis. Seventeen (8.3%, 17/205) cases possessed at least 4 methylated CpGs and were considered hypermethylated and potentially silenced. All methylated cases were negative for VHL mutation (Figure 2). In summary, 91% (186/205) of cases demonstrated potential VHL inactivation through mutation (82.4%, N=169) or hypermethylation (8.3%, N=17).
Figure 2.
Analysis of VHL promoter methylation across 11 CpGs among mutation negative (N=16) and mutation positive (N=10) ccRCC DNAs. Methylation status of each CpG is indicated as follows: Yellow=unmethylated, blue=partial methylation, red=fully methylated, and the hatched box indicates that the CpG was uninformative. Positive (methylated) and negative (unmethylated) controls included in each analysis are shown at the bottom and VHL mutation status is shown on the left. Fully and partially methylated CpG sites are summed in the far right column. Cases that had at least 4 methylated CpG sites in the VHL promoter were considered methylation positive.
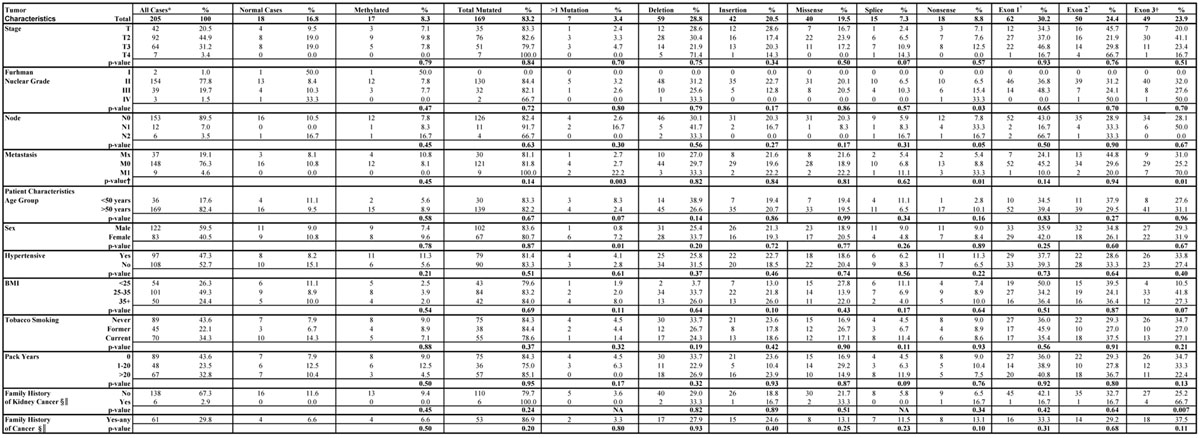
VHL Status by Patient and Tumor Characteristics
VHL alterations were subsequently stratified by tumor histopathologic and patient characteristics as summarized in Table 4. The overall prevalence of mutated cases was not associated with any of the tumor or patient characteristics examined, however, the prevalence of certain subtypes of mutations was. For example, tumors with distant metastases (M1) had significantly more double mutations than those without metastases (M0) (22.2% vs. 2.7%; P=0.003). Nonsense mutations were associated with Fuhrman nuclear grade (P=0.03) and lymph node positivity (P=0.05), and were more prevalent among M1 than M0 cases (P=0.01). The location of alterations also appeared to differ between groups. For example, exon 3 mutations were more prevalent in M1 than M0 cases (70.0% versus 25.2%; P=0.01), and among cases with a family history of kidney cancer (66.7% versus 25.2%; P=0.007). Of note, all six cases with a positive family history of kidney cancer possessed tumor DNA harboring a VHL mutation and 4/6 mutations (66.7%) were located in Exon 3. Lastly, the prevalence of VHL alterations located in Exon 3 increased from 8.3% among subjects with low BMI (<25) to 25.5% among those with high BMI (>35) (Ptrend=0.07).
Table 4.
Von Hippel-Lindau alteration subtypes associated with patient descriptive and clinical characteristics
‡Total number of mutations
Subgroups that do not add to 205 are due to missing clinical or risk factor information
p-value for comparison of M0 vs M1 only
both groups are compared to cases without family history of cancer
first degree relative with cancer or kidney cancer
Discussion
In this study, DNA from 205 histologically confirmed, macrodissected ccRCC tumors were analyzed for VHL gene alterations using a novel combination of endonuclease scanning and fluorescent Sanger sequencing. When applied in parallel, mutations were detected in 82.4% (169/205) of ccRCC cases. This is the highest prevalence of VHL mutations thus far reported and the largest number of histologically confirmed ccRCC tumors analyzed in a single study. Detailed analysis of the VHL promoter identified an additional 8.3% of tumors that were hypermethylated and potentially silenced. Notably, in this study methylation and mutation were mutually exclusive. Together, a total of 91% of cases exhibited genetic and epigenetic alteration of the VHL gene. Analysis of patient and tumor characteristics revealed that the overall prevalence of mutations was not associated with clinical parameters normally associated with disease progress, but particular mutation subtypes were associated with Fuhrman nuclear grade, metastasis, node positivity, and a family history of kidney cancer. Sensitive and accurate methods of mutation detection such as those described here will be an advantage to future studies by minimizing misclassification of cases by VHL mutation/promoter methylation status, and reducing the risk of biasing associations towards the null.
Some of the largest studies (between 93−205 RCC cases) that had previously examined VHL mutation and methylation have reported fewer VHL gene mutations (between 42 (ref 13) and 71% (ref 10)). These studies utilized single strand conformational polymorphism (SSCP) and denaturing high performance liquid chromatography (DHPLC) (8-10, 13). As observed previously (10), we document a high prevalence of alterations at codons 65, 114, 147, and 155, though only 6% of mutations occurred in codons 147/148 (13). Mutations were distributed evenly across exons whereas other studies reported higher frequencies in exons 1 (8, 10) and 2 (13). Lastly, 3.4% (7/205) of tumors in this study displayed two VHL alterations, Banks et al. (10) reported a single case (1%, 1/93), while most other studies reported none (8, 13). Hypermethylation of the VHL promoter was observed exclusively in mutation negative tumors and the prevalence observed is within the range previously reported (5 and 17%, respectively, refs. 8 and 13). In contrast, one earlier study reported that 21% of tumors were hypermethylated and half of these (11/19, 58%) possessed VHL mutations (10). This discrepancy may result from different approaches used to assess promoter methylation status or from differences between the histological RCC subtypes examined. In this study, cases were exhaustively reviewed by one expert to ensure reliability that all cases included in this study were histologically confirmed ccRCC.
Once mutations were identified using endonuclease scanning and sequencing, the RSI value was estimated to determine the proportion that would be difficult or impossible to detect using automated sequence anlalysis. To avoid misclassification of low RSI VHL mutants, PCR product from a subset of mutant cases were subcloned to confirm their identity. Mutation status was confirmed in 10/10 cases and in 7 of 10 tumors, the mutant allele frequency calculated from the percentage of mutation positive transformants agreed with the RSI visual estimate.
To reduce risk of false negative results, prior to DNA extraction all frozen tumor biopsies were macrodissected to remove normal tissue based on a pathology review of one H&E stained slide to obtain at least 70% tumor tissue per sample. For this reason, the observed high percentage of variants (46%) with low RSI values was unexpected. Tumors in general contain various amounts of normal tissue and cells (22, 23). Kidney tumors in particular can be highly vascularized (24). Tumor infiltration by normal tissue or blood cells such as lymphocytes, can reduce the proportion of tumor to normal DNA in a sample. Linehan et al. (25) examined seven RCC tumors that were free of any visible normal tissue. After enzymatic dispersion of RCC tissue, they observed that only 20−50% were tumor cells (mean 26 ± 15%). Similarly, Belldegrun et al. (26) examined 37 tumors and found that 6−75% of cells were tumor cells (mean of 39 ± 3%). It is plausible that the low amount of mutant allele estimated by RSI in approximately half of the tumors examined here resulted from a low ratio of tumor to normal tissue that was not removed by macrodissection. Lymphocyte infiltration, vascular architecture, and surrounding normal tissue appear to be a significantly greater component of kidney tumors compared to other types of tumor tissues. Contamination of tumor tissue with normal tissue in patient samples may also explain discrepancies in the reported VHL mutation frequency in ccRCC. The presence of significant amounts of normal cells in tumor tissue strongly argues for application of highly sensitive and accurate mutation detection methods such as those presented here to ensure that all mutations are detected. This conclusion is based on the observation that the VHL mutation frequency we detected with the methods described surpasses the renal cancer VHL somatic mutation frequency detected in all previous studies (14).
After we completed this comprehensive analysis of somatic mutations and methylation in the cancer genome of ccRCC, we correlated the genetic findings with patient clinical and descriptive characteristics. Analysis of mutations prevalence among patient and tumor subgroups revealed that total mutation prevalence was not associated with any of the parameters examined. This finding is similar to other studies and supports the hypothesis that VHL gene inactivation is likely an early event in the ccRCC carcinogenic process (8-10, 27-29). Unlike some previous studies, we did not observe a higher prevalence of VHL mutation/hypermethylation among high stage tumors (13) nor did we did not observe differences in the prevalence of hypermethylation or double mutations by sex (10). We did find that late stage metastatic lesions had more double mutations, nonsense mutations, and mutations located in exon 3 than M0 or Mx cases. We also observed that the prevalence of nonsense mutations was significantly associated with grade and node positivity and that cases with a family history of kidney cancer had significantly more mutations located in Exon 3. Survival studies will be necessary to determine whether these molecular subtypes have any prognostic relevance.
A goal of personalized medicine is to identify tumors with particular alterations that can be used to predict a clinical response to treatment in the patient. A recent study demonstrated that patients with metastatic RCC that possessed VHL methylation or truncating mutations had prolonged time to tumor progression during treatment with anti-angiogenic therapy (21). We show that segregation of tumors by VHL alteration subtype could be achieved in 91% of ccRCC cases. These findings indicate that VHL molecular subtypes could provide a sensitive biomarker of ccRCC tumor heterogeneity among histologically similar cases for etiologic, prognostic, and translational studies (1, 2, 21,30,31).
Supplementary Material
Suppl. Table 1. Summary of von Hippel-Lindau (VHL) gene alterations in 205 clear cell RCC patient samples
Acknowledgements
We thank Stan Lilleberg, Kathryn Walters, Marjorie Smithhisler, John Troy, Mike Geimer, Tammy Putmon, Heather Marshall, Rathi Thiagarajan, and Phyllis Nimeroff for excellent technical assistance. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research with federal funds from contracts N01-C0-12400 (LS) and CA102600 (FW), and by European Commission INCO-COPERNICUS Grant IC15-CT96-0313. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- VHL
von Hippel-Lindau
- RCC
renal cell carcinoma
- ccRCC
clear cell RCC
- DHPLC
denaturing high performance liquid chromatography
- RSI
relative signal intensity
- PCR
polymerase chain reaction
- MSP
methylation-specific PCR
- BMI
body mass index
Footnotes
References
- 1.Linehan WM, Walther MM, Zbar B. The genetic basis of cancer of the kidney. J Urol. 2003;170:2163–72. doi: 10.1097/01.ju.0000096060.92397.ed. [DOI] [PubMed] [Google Scholar]
- 2.Mathew A, Devesa SS, Fraumeni JF, Jr, Chow WH. Global increases in kidney cancer incidence, 1973−1992. Eur J Cancer Prevent. 2002;11:171–8. doi: 10.1097/00008469-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Latif F, Tory K, Gnarra J, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–20. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt LS, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–10. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 6.Toro JR, Nickerson ML, Wei MH, et al. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet. 2002;73:95–106. doi: 10.1086/376435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nickerson ML, Warren MB, Toro JR, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2:157–64. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 8.Kondo K, Yao M, Yoshida M, et al. Somatic von Hippel-Lindau disease gene mutation in clear-cell renal carcinomas associated with end-stage renal disease/acquired cystic disease of the kidney. Genes Chromosomes Cancer. 2002;34:58–68. doi: 10.1002/gcc.10123. [DOI] [PubMed] [Google Scholar]
- 9.Van Houwelingen KP, van Dijk BAC, Hulsbergen-van de Kaa CA, et al. Prevalence of von Hippel-Lindau gene mutations in sporadic renal cell carcinoma: results from The Netherlands cohort study. BMC Cancer. 2005;5:57–67. doi: 10.1186/1471-2407-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banks RE, Tirukonda P, Taylor C, et al. Genetic and epigenetic analysis of von Hippel-Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer Res. 2006;66:2000–11. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- 11.Stolle C, Glenn G, Zbar B, et al. Improved detection of germline mutations in the von Hippel-Lindau disease tumor suppressor gene. Hum Mutat. 1998;12:417–23. doi: 10.1002/(SICI)1098-1004(1998)12:6<417::AID-HUMU8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 12.Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 13.Brauch H, Weirich G, Brieger J, et al. VHL alterations in human clear cell renal cell carcinoma: association with advanced tumor stage and a novel hot spot mutation. Cancer Res. 2000;60:1942–8. [PubMed] [Google Scholar]
- 14.Kim WY, Kaelin WG. The role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 15.Gerard GF, Shandilya H, Qiu P, Shi Y, Lo J. In: Genetic Variance Detection Technologies for Pharmacogenomics. Hecker KH, editor. DNA Press; Eagleville (PA): pp. 95–129. [Google Scholar]
- 16.Yang B, Wen X, Kodali NS, et al. Purification, cloning, and characterization of CEL I nuclease. Biochemistry. 2000;39:3533–41. doi: 10.1021/bi992376z. [DOI] [PubMed] [Google Scholar]
- 17.Qiu P, Shandilya H, D'Alessio JM, O'Connor K, Durocher J, Gerard GF. Mutation detection using Surveyor nuclease. BioTechniques. 2004;36:702–7. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 18.Shuman S. Novel approach to molecular cloning and polynucleotide synthesis using Vaccinia DNA topoisomerase. J Biol Chem. 1994;269:32678–84. [PubMed] [Google Scholar]
- 19.Nickerson ML, Warren MB, Zbar B, Schmidt LS. Random mutagenesis-PCR to introduce alterations into defined DNA sequences for validation of SNP and mutation detection methods. Hum Mutat. 2001;17:210–19. doi: 10.1002/humu.6. [DOI] [PubMed] [Google Scholar]
- 20.Rothberg PG, Bradley JF, Baker DW, Huelsman KM. Is P25L a “Real” VHL mutation? Molec Diag. 2001;6:49–54. doi: 10.1054/modi.2001.21637. [DOI] [PubMed] [Google Scholar]
- 21.Rini BI, Jaeger E, Weinberg V, et al. Clinical response to therapy targeted at vascular endothelial growth factor in metastatic renal cell carcinoma: impact of patient characteristics and Von Hippel-Lindau gene status. BJU Intl. 2006;98:756–62. doi: 10.1111/j.1464-410X.2006.06376.x. [DOI] [PubMed] [Google Scholar]
- 22.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. New Eng J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 23.Asano H, Toyooka S, Tokumo M, et al. Detection of EGFR gene mutation in lung cancer by mutant-enriched polymerase chain reaction assay. Clin Cancer Res. 2006;12:43–8. doi: 10.1158/1078-0432.CCR-05-0934. [DOI] [PubMed] [Google Scholar]
- 24.Hutson TE. Targeted therapy for renal cell carcinoma: a new treatment paradigm. Proc Bayl Univ Med Cent. 2007;20:244–8. doi: 10.1080/08998280.2007.11928297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linehan M, Miller E, Anglard P, Merino M, Zbar B. Improved detection of allele loss in renal cell carcinomas after removal of leukocytes by immunologic selection. J Natl Cancer Inst. 1989;81:287–90. doi: 10.1093/jnci/81.4.287. [DOI] [PubMed] [Google Scholar]
- 26.Belldegrun A, Muul LM, Rosenberg SA. Interleukin 2 expanded tumor-infiltrating lymphocytes in human renal cell cancer: isolation, characterization, and antitumor activity. Cancer Res. 1988;48:206–14. [PubMed] [Google Scholar]
- 27.Suzuki H, Ueda T, Komiya A, et al. Mutational state of von Hippel-Lindau and adenomatous polyposis coli genes in renal tumors. Oncology. 1997;54:252–7. doi: 10.1159/000227697. [DOI] [PubMed] [Google Scholar]
- 28.Hamono K, Esumi M, Igarashi H, et al. Biallelic inactivation of the von Hippel-Lindau tumor suppressor gene in sporadic renal cell carcinoma. J Urol. 2002;167:713–7. doi: 10.1016/S0022-5347(01)69132-8. [DOI] [PubMed] [Google Scholar]
- 29.Schraml P, Struckmann K, Hatz F, et al. VHL mutations and their correlation with tumor cell proliferation, microvessel density, and patient prognosis in clear cell renal cell carcinoma. J Pathol. 2002;196:186–93. doi: 10.1002/path.1034. [DOI] [PubMed] [Google Scholar]
- 30.Motzer RJ, Hutson TE, Tomczak P, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–24. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 31.Escudier B, Eisen T, Stadler WM, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–34. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Table 1. Summary of von Hippel-Lindau (VHL) gene alterations in 205 clear cell RCC patient samples