Summary
Skeletal muscle differentiation requires a cascade of transcriptional events to control the spatial and temporal expression of muscle-specific genes. Until recently, muscle-specific transcription was primarily attributed to prototypic enhancer-binding factors while the role of core promoter recognition complexes in directing myogenesis remained unknown. Here, we report the development of a purified reconstituted system to analyze the properties of a TAF3/TRF3 complex in directing transcription initiation at the Myogenin promoter. Importantly, this new complex is required to replace the canonical TFIID to recapitulate MyoD-dependent activation of Myogenin. In vitro and cell-based assays identify a domain of TAF3 that mediates co-activator functions targeted by MyoD. Our findings also suggest changes to CRSP/Mediator in terminally differentiated myotubes. This switching of the core promoter recognition complex during myogenesis allows a more balanced division of labor between activators and TAF coactivators thus providing another strategy to accommodate cell-specific regulation during metazoan development.
Introduction
Transcriptional activation is a major step in the regulation of gene expression during metazoan development. Formation of the pre-initiation complex (PIC), an obligate step leading to transcription, requires recruitment of the core promoter recognition complex, TFIID, to the DNA template. Promoter binding by the multi-subunit TFIID complex composed of the TATA binding protein (TBP) and 12–15 associated factors (TAFs) is followed by assembly of other basal factors including TFIIA, B, E, F, H and RNA Polymerase II to form the PIC (Albright and Tjian, 2000; Thomas and Chiang, 2006). Although essential for initiating transcription, the PIC is not generally thought to direct cell-specific functions. However, the discovery of cell-type specific TAF homologs within different tissues in flies, mice and humans raised the possibility that altered TFIID complexes may provide another mechanism to specify tissue selective programs of gene expression (Hiller et al., 2004; Hochheimer and Tjian, 2003).
Likewise, the seemingly ubiquitous and highly conserved TBP subunit of TFIID was initially presumed to be universally responsible for core promoter recognition. However, the identification of TBP related factors (TRFs) further challenged the view of a static or invariant core promoter recognition complex operating in metazoans (Hochheimer and Tjian, 2003). Among the diversified TRF family members, TRF3 and TBP are most closely related, although TRF3 is found in vertebrate genomes but not invertebrates or yeast (Persengiev et al., 2003). TBP and TRF3 may share some overlapping functions during embryonic development, but it also appears that these two factors perform non-redundant roles. Sequence analyses suggest that TRF3 shares a nearly identical C-terminal DNA-binding domain with TBP and in vitro studies confirmed similar DNA-binding activities for these two factors (Bartfai et al., 2004; Persengiev et al., 2003). Although TBP and TRF3 exhibit similar DNA binding properties, these two core promoter recognition proteins bear divergent N-terminal domains that may execute distinct gene selective functions by partnering with different associated subunits (TAFs) to acquire differential promoter selectivity. Indeed, a number of studies have found that TRFs likely carry out important developmental functions (Bartfai et al., 2004; Gazdag et al., 2007; Hart et al., 2007; Hochheimer and Tjian, 2003; Jallow et al., 2004). However, until recently, the TRFs had not been functionally linked to cell specific TAFs and such potentially new core promoter recognition complexes had not been shown to direct cellular differentiation.
We recently identified TAF3 as a subunit specifically associated with TRF3 to form a complex that is required for myogenic differentiation (Deato and Tjian, 2007). Skeletal myogenesis is a developmental process that involves a fine balance between continued cellular proliferation and terminal differentiation. Proliferating and differentiated skeletal muscle cells display distinct transcriptional programs wherein batteries of genes must be kept off while others must be activated. This carefully orchestrated transcriptional program has been, for the most part, attributed to the action of key myogenic enhancer binding factors (i.e. Myf5, MyoD, Myogenin and Mrf4) while potential contributions of the core promoter recognition apparatus remained largely unexplored (Tapscott, 2005). Recent experiments have unmasked a remarkable switching of the core promoter recognition complex from the prototypic TFIID in myoblasts to a TAF3/TRF3 complex in myotubes that opens potentially new vistas in the repertoire of mechanisms driving cellular differentiation (Deato and Tjian, 2007). These cell-based studies found that loss of TAF3 or TRF3 in myoblast cells blocks differentiation due in part to downregulation of a key myogenic factor, Myogenin. Interestingly, myoblast cells lacking TAF3 or TRF3 also induce ectopic retention of TFIID that correlates with a failure to direct proper expression of key myogenic markers during myoblast differentiation. Thus, the elimination of TFIID and concomitant engagement of the TAF3/TRF3 complex in myotubes appear to be necessary steps to regulate muscle specific gene expression during differentiation.
Unlike the bewildering and poorly understood array of functions attributed to the 12–15 TAFs in holo-TFIID, the single TAF3 subunit in complex with TRF3 offers a considerably simpler system to dissect its biochemical activities and potential coactivator function. We therefore developed an in vitro transcription system reconstituted with purified components of the pre-initiation complex to directly address core-promoter recognition properties of the TAF3/TRF3 complex. This stringent biochemical assay was also used to determine the minimum components necessary to support activator dependent transcription initiation mediated by the TAF3/TRF3 complex in conjunction with the in vivo regulator of myogenesis, MyoD. Next, we addressed the capacity, if any, of TFIID and TBP to drive in vitro transcription at the Myogenin promoter. Finally, we carried out an analysis in vitro and in vivo to probe the coactivator requirements and the function of TAF3 in regulating Myogenin expression. These studies provide evidence for coactivator properties of the TAF3/TRF3 complex and map a region of TAF3 that is directly targeted by the myogenic activator, MyoD. Our findings underscore the critically important integration between activator specificity and diversified core promoter recognition complexes in regulating cellular differentiation.
Results
TRF3 and TAF3/TRF3 can initiate Myogenin transcription in vitro
In skeletal muscle, the loss of the canonical holo-TFIID complex and replacement by a TAF3 /TRF3 complex appears to be required for turning on a select program of gene expression during myogenic differentiation (Deato and Tjian, 2007). However, the mechanism by which TAF3/TRF3 functions at target promoters remained unclear. In order to directly investigate the mechanism by which TAF3/TRF3 regulates transcriptional activation of Myogenin, we have developed a purified in vitro reconstituted system modified to test the transcription initiation properties of TRF3 and the TAF3/TRF3 complex.
First, to ensure that our chosen DNA template will accurately initiate from the Myogenin promoter in vitro, we mapped the in vivo transcription start site by primer extension. As expected, primer extension analysis identified a single major transcription start site for Myogenin (data not shown), consistent with previous promoter mapping studies (Edmondson et al., 1992). Importantly, the Myogenin transcript was detected in myotubes but not in myoblasts, reserve cells or mouse fibroblast (data not shown), consistent with the previously described expression profile of Myogenin in differentiated muscle cell types (Pownall et al., 2002).
Earlier studies of the native Myogenin promoter have determined that sequences contained within 184 nucleotides upstream of the transcription start site are sufficient to confer muscle-specific expression, growth factor responsiveness and positive transcriptional autoregulation of Myogenin. This region consists of a TATA box flanked by two activator binding sites for MyoD (E-boxes) as well as a site for the serum-responsive activator, MEF2 (Edmondson et al., 1992). For the purposes of determining the transcription initiation properties of TRF3 and the TAF3/TRF3 complex this fragment of Myogenin should be sufficient to direct at least basal transcription and possibly activator dependent transcription associated with TRF3 or TAF3/TRF3 complex. To further enhance the sensitivity of our assay, two additional MyoD sites (E-boxes) have been appended to our in vitro transcription template (Figure 1C). Based on the mapping of our in vivo transcription start site accurate initiation in vitro from this Myogenin promoter should produce a primer extension product of 135 nucleotides.
Figure 1. Purified TRF3 and TAF3/TRF3 complex can substitute for TFIID for basal-level transcription of Myogenin in vitro.
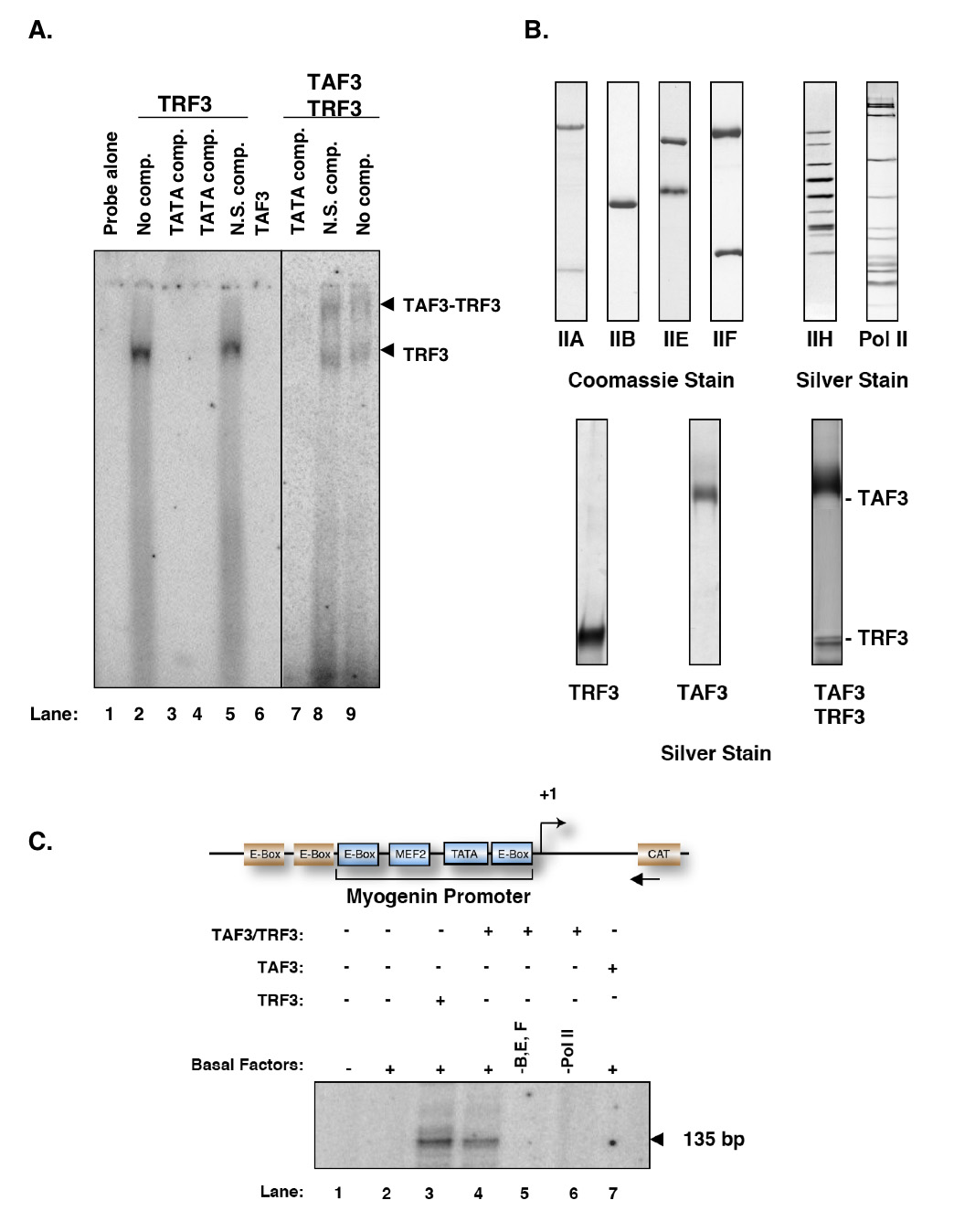
A. Purified TAF3 interacts with DNA-bound TRF3 in vitro. In electrophoretic mobility shift assay, the labeled 21bp fragment of Myogenin promoter containing the TATA box was used. Lane 1 contains the labelled probe alone; lane 2 contains TRF3 alone while lanes 3–4 include an unlabelled cold TATA competitor (TATA comp.). The binding reaction in lane 5 contains an unlabelled non-specific competitor (TATA mutant, N.S. comp.). The addition of purified TAF3 to reactions containing TRF3 produces a supershifted complex (lane 8 and lane 9 with N.S. comp.). In contrast, in reactions containing TAF3 alone has no DNA-binding activity (lanes 6–7) B. Silver stain of affinity purified basal factors (TFIIA, TFIIB, TFIIE, TFIIF, TFIIH and RNA Polymerase II) and TRF3, TAF3 and TAF3/TRF3 complex used to substitute for TFIID in our in vitro reconstituted transcription reaction. C. Factor requirements for basal transcription of Myogenin mediated by TRF3 or TAF3/TRF3 complex. In vitro transcription reactions contain general transcription factors (TFIIA, B, E, F and H) and RNA Polymerase II. In place of TFIID, purified TRF3 or TAF3/TRF3 complex was substituted to test for transcription initiation properties. In vitro transcription reactions were carried out on Myogenin template containing the endogenous promoter sequence elements (as marked) modified with two additional activator sites (E-boxes) and an upstream CAT gene sequence. Transcription products were analyzed by primer extension using specific primers directed at CAT gene. All transcription reactions unless otherwise indicated contains the Myogenin template, purified basal factors that excludes TFIID. Lane 1 contains DNA template alone while lane 2 only contains purified basal factors. Equal amounts of TRF3, TAF3 and TAF3/TRF3 complex were used in transcription reactions. The arrow indicates specific transcription product.
To carry out our in vitro reconstituted transcription reactions, recombinant TRF3 or the TAF3/TRF3 complex was first affinity-purified from insect cells (Figure 1B). Next, we carried out electrophoretic mobility shift assays to ascertain that the purified factors can bind to TATA box containing promoter fragments (Figure 1A). As expected, TRF3 binds to a Myogenin promoter DNA fragment as efficiently as TBP (Figure 1A, lanes 2–5) consistent with previous reports (Bartfai et al., 2004; Jallow et al., 2004). By contrast, TAF3 failed to bind DNA alone but was able to “supershift” the probe DNA when in complex with TRF3 (Figure 1A, lanes 6, 8–9). Having established that these purified factors are able to recognize and bind Myogenin core promoter template, we next use them to supplement our in vitro reconstituted transcription system consisting of purified RNA Polymerase II and the general transcription factors TFIIA, B, E, F and H but lacking TBP and the prototypic TFIID complex (Figure 1B). In vitro transcription assays carried out with the Myogenin template revealed that TRF3 and the TAF3/TRF3 complex can substitute for TBP and TFIID in directing accurate basal level transcription from the Myogenin promoter (Figure 1C, lanes 3–4). We did not detect significant differences between TRF3 and the TAF3/TRF3 complex in directing basal transcription from the Myogenin promoter. As expected, omission of purified general transcription factors (TFIIB, E and F) as well as RNA Polymerase II abolishes transcriptional activity (Figure 1C, lanes 5–6). Likewise, reactions reconstituted with general transcription factors and TAF3 but no TRF3 also failed to initiate transcription from the Myogenin promoter (Figure 1C, lane 7). Taken together, these initial observations suggest that either TRF3 or the TAF3/TRF3 complex is sufficient to replace TBP/ TFIID for basal level transcription in our in vitro experiments and direct accurate initiation from the Myogenin promoter in vitro.
TAF3 is required for activator-dependent transcription of Myogenin
Myogenin is a key regulator of myogenic differentiation and during mouse development, abundant levels of Myogenin transcript are detected coincident with terminal differentiation and myotube formation (Pownall et al., 2002). The timing and cell-type specific expression of Myogenin in vivo is achieved, in part, through action of the muscle-specific activator, MyoD along with various cofactors (Berkes et al., 2004; Tapscott, 2005). Although selective interactions between key myogenic activators and the transcription initiation machinery have been proposed to play an important role in the transmission of activator signals (Black et al., 1998), to date, the molecular mechanism and key cofactors required for Myogenin expression at the level of transcription initiation remained uncharacterized. Thus, to determine the potential direct contribution of TAF3/TRF3 in the MyoD-dependent activation of Myogenin transcription, we turned to our purified reconstituted in vitro transcription system. However, before embarking on a detailed analysis of activator dependent transcription reactions, we first needed to test the ability of our Myogenin promoter fragment (184 bp) to direct activator responsiveness using a cell-based luciferase reporter assay. In this assay, we transiently co-transfect the activator (FLAG-MyoD) together with our reporter construct containing either control vector sequences or the native Myogenin promoter into mouse fibroblast cells that do not express endogenous MyoD (Figure 2A) but do express significant levels of TAF3/TRF3. The resulting luciferase activities indicate that the short Myogenin template can efficiently respond to co-transfected muscle-specific activator, MyoD. A control promoter lacking MyoD binding sites failed to support activated transcription (Figure 2A). To confirm that the observed activation is indeed due to MyoD, we analyzed the transfected fibroblast cell lysates for MyoD expression and confirmed that activation of Myogenin is only observed when MyoD is expressed (Figure 2A). These cell-based assays thus confirm that our truncated 184bp fragment bearing the native Myogenin promoter to be used in our in vitro reconstituted transcription reactions contains cis-regulatory elements capable of directing MyoD dependent transcriptional activation.
Figure 2. TAF3 is required for MyoD-dependent activated transcription of Myogenin.

A. Luciferase reporter assay to determine the Myogenin promoter response to the activator MyoD. Diagram of the reporter constructs transfected into 3T3 fibroblast cells. For this reporter assay, pGL3 vector control sequences alone or pGL3 vector containing the endogenous Myogenin promoter sequences (184 bp fragment) were used to drive the expression of luciferase. Constructs were introduced into fibroblast cells in the presence or absence of exogenous FLAG-MyoD. Luciferase expression was determined and plotted as fraction relative to total protein (luciferase/ug protein). Each bar represents the mean of triplicate samples per condition. The error bars represent the standard deviation. Protein immunoblot analyses were performed to determine relative levels of transfected FLAG-MyoD in fibroblast cells. B. TAF3 is required to potentiate MyoD-dependent transcription activation of Myogenin in vitro. Silver stain of the myogenic activator FLAG-MyoD co-expressed with its heterodimer E47 were affinity purified from insect cells using anti-FLAG antibody. This purified factor was included in our purified in vitro reconstituted transcription system to test the effects of activator in TRF3- or TAF3/TRF3-mediated transcription reactions. Using conditions described in Figure 1C, TRF3- or TAF3/TRF3-mediated transcription reactions were supplemented with the purified activator MyoD/E47 in vitro. The arrow indicates specific transcription products analyzed by primer extension.
To directly test the role of TRF3 or the TAF3/TRF3 complex in mediating activated transcription, we affinity-purified MyoD and its well-established in vivo E-protein heterodimer partner E47 (Lassar et al., 1991) from insect cells expressing both recombinant products (Figure 2B). The addition of purified activators (MyoD/E47) to reconstituted reactions containing TAF3/TRF3 (but no added CRSP/MED) activated transcription from the Myogenin promoter 3–5 fold (Figure 2B, lane 6). By contrast, addition of MyoD/E47 activators to reconstituted reactions containing TRF3 but no TAF3 failed to support activator dependent transcription (Figure 2B, lane 3). As observed previously, basal level transcription using TRF3 and TAF3/TRF3 containing reactions were indistinguishable (Figure 2B, lanes 2 and 5). Taken together, these observations suggest that TRF3 may be sufficient to support basal transcription but not MyoD/E47 directed activation. Importantly, TAF3 appears to be specifically required to support activated transcription of Myogenin – a critical finding that we could not easily discern from our in vivo studies. These studies suggest that the muscle-specific activator MyoD may require a novel core promoter recognition complex and coactivator target (TAF3/TRF3) in place of TFIID to support activated transcription of Myogenin. These studies thus provide strong evidence that TAF3 but not TRF3 can serve as an important coactivator in mediating transcription regulation during myogenesis. These experiments may also suggest that MyoD activation of Myogenin transcription is CRSP/MED independent.
Since our in vitro experiments were reconstituted in the absence of the prototypic TFIID complex, we next wanted to test the specificity of the Myogenin promoter by adding back native purified TFIID. In particular, we wanted to test whether TFIID would fail to substitute for TAF3/TRF3 in mediating activator dependent transcription from the Myogenin promoter in vitro as predicted from our previous in vivo studies. To address the possibility that the coactivator function of TAF3 within the TAF3/TRF3 complex is non-redundant with coactivation functions of TFIID, we substituted purified TBP or TFIID for TRF3 and TAF3/TRF3 in our reconstituted reactions and then assayed for activated transcription of Myogenin (Figure 3A). First, we compared basal transcription levels in reactions reconstituted with TRF3, TAF3/TRF3, TBP or TFIID. Consistent with previous studies where we found no significant differences in DNA-binding activity associated with TBP versus TRF3, we now observed comparable levels of basal activity initiated by these factors (Figure 3B, lanes 3,4,6 and 7). These findings indicate that TBP and TFIID can bind and accurately initiate basal levels of Myogenin transcription in vitro as efficiently as TRF3 and TAF3/TRF3. We next tested these reconstituted systems for their ability to support activated transcription upon addition of MyoD/E47 in reactions containing TBP but no TRF3 or the TAF3/TRF3 complex. Addition of TBP to these reactions failed to support activated transcription consistent with our previous in vitro findings (Figure 3C lanes 8–9) (Dynlacht et al., 1991). Likewise, transcription reactions supplemented with activators and TRF3 but no TAF3 (Figure 3C, lanes 4–5) or TAF3 alone (Figure 3C, lanes 2–3) also failed to potentiate Myogenin transcription. Adding MyoD/E47 to transcription reactions containing TFIID, which contains substoichiometric amounts of TAF3, failed to support activated transcription of Myogenin (Figure 3C, lanes 10–11). In contrast, addition of the TAF3/TRF3 complex under the same reaction conditions efficiently activated Myogenin (Figure 3C, lanes 6–7). As expected, a similar amount of TFIID added to a set of reactions programmed with a non-muscle specific promoter template containing E1B TATA box and activator binding sites for Sp1 (or G3BCAT) produced strong activator dependent transcription in the presence of Sp1confirming that our purified TFIID is fully functional (Figure 3C). These in vitro studies are consistent with our previous in vivo analyses of differentiation-deficient TAF3 or TRF3 depleted myoblasts, wherein TFIID was unable to substitute for the TAF3/TRF3 complex in regulating Myogenin expression (Deato and Tjian, 2007). Taken together, these observations suggest that selective use of TAF containing coactivator complexes (in this case, TAF3/TRF3) is required to support MyoD activation of Myogenin transcription. Moreover, distinct activator interactions with the core-promoter recognition complex may contribute an important mechanism for differential coactivator recruitment to cell type specific promoters.
Figure 3. Selective use of TAF3/TRF3 complex is required to support MyoD-dependent activation of Myogenin transcription.

A. Inclusion of purified TBP from insect cells and immunopurified TFIID complex isolated from HeLa cells in our in vitro reconstituted transcription reactions (shown here in silver stain gels). B. TBP and TFIID can initiate basal transcription of Myogenin in vitro. Transcription reactions contain basal transcription factors (TFIIA, B, E, F and H) and RNA Polymerase II. Reactions initiated by purified TRF3 or TAF3/TRF3 were compared to reactions containing TBP or TFIID or TAF3 alone complex to assess the basal level of Myogenin transcription. C. TFIID complex cannot substitute for TAF3/TRF3 function in mediating MyoD- dependent activation of Myogenin. Using conditions described in B, MyoD/E47 were included in reconstituted transcription reactions initiated by TRF3, TAF3/TRF3, TBP or TFIID. As a positive control, a similar amount of TFIID used for Myogenin transcription reaction was used on the TFIID-dependent DNA template G3BCAT in the presence of Sp1. The numerical values represent the fold activation. The fold of MyoD-dependent activation of Myogenin was calculated by dividing the scanned transcriptional signal value generated from reaction done with MyoD over the value obtained from transcription reactions done without the activator. Arrowheads indicate transcription product analyzed by primer extension.
TAF3 is a direct coactivator target for MyoD
Previous studies have found that TAFs can serve as binding partners/targets for activators and these interactions have functional consequences in potentiating transcription (Albright and Tjian, 2000). We therefore set out to determine whether TAF3 may likewise serve as a direct target for MyoD. To test the coactivator contribution of TAF3 in mediating activator-dependent Myogenin transcription, we utilized a truncated version of TAF3 that lacks the N-terminal 1–516 amino acid residues of the protein (TAF3ΔN) but retains its C-terminal TRF3 interaction domain (data not shown). To begin our analysis, recombinant TAF3ΔN/TRF3 complex was affinity-purified from insect cells (Figure 4A). Next, we compared transcription levels of Myogenin in reactions initiated by either TAF3/TRF3 or TAF3ΔN/TRF3 and as expected, we observed comparable levels of accurate basal transcription initiation (Figure 4B, lanes 3–4). Next, we tested the ability of the TAF3ΔN/TRF3 complex to support activator-dependent transcription of Myogenin. TAF3ΔN/TRF3, in contrast to the wild-type TAF3/TRF3 complex failed to support MyoD dependent activation of Myogenin transcription (Figure 4C, lanes 5–6 and 7–8). These findings suggest that the N-terminal region of TAF3 contains a domain responsible for the coactivator function of TAF3.
Figure 4. N-terminal region of TAF3 is important for MyoD-dependent activation of Myogenin.

A. Interaction of TAF3ΔN with TRF3 is comparable to wildtype TAF3/TRF3 complex. The FLAG-TAF3ΔN/TRF3 complex was purified from insect cells (silver stain). B. TAF3ΔN/TRF3 complex can initiate basal transcription of Myogenin in vitro. Using conditions described in Figure 1C, TAF3ΔN/TRF3 complex was added to our purified transcription system to test the capacity of this complex to initiate transcription of Myogenin. For comparison, an equal amount of TAF3/TRF3 complex was used as a positive control. C. TAF3ΔN/TRF3 complex cannot support activated transcription of Myogenin in vitro. Using similar transcription conditions as in part B, the capacity of TAF3ΔN/TRF3 complex to potentiate activated transcription of Myogenin in the presence of MyoD/E47 was tested. Fold activation was calculated as in Figure 3. Arrowheads indicate transcription product analyzed by primer extension.
To further address this possibility, we determined the potential direct binding interaction between TAF3 and MyoD in gst-pulldown experiments. In this analysis, whole cell lysates were prepared from insect cells expressing either Flag-TAF3 or Flag-TAF3ΔN (Figure 5A). Lysates from these cells were then incubated with glutathione-bound recombinant Gst-MyoD or control Gst (Figure 5A). Following extensive bead washes, immunoblot analyses of bound samples revealed that TAF3 can bind directly to MyoD while the TAF3ΔN mutant failed to bind (Figure 5A). This TAF3- MyoD interaction does not appear to be DNA dependent as the binding remained unaffected upon treatment with DNAse I nor is it dependent on the C-terminal region Plant Homeobox Domain (PHD) of TAF3 (data not shown). These protein-protein binding studies suggest that most likely, a motif within the N-terminal region of TAF3 is required for mediating a direct interaction with the muscle-specific activator MyoD at least in vitro.
Figure 5. TAF3 is a direct target for MyoD.
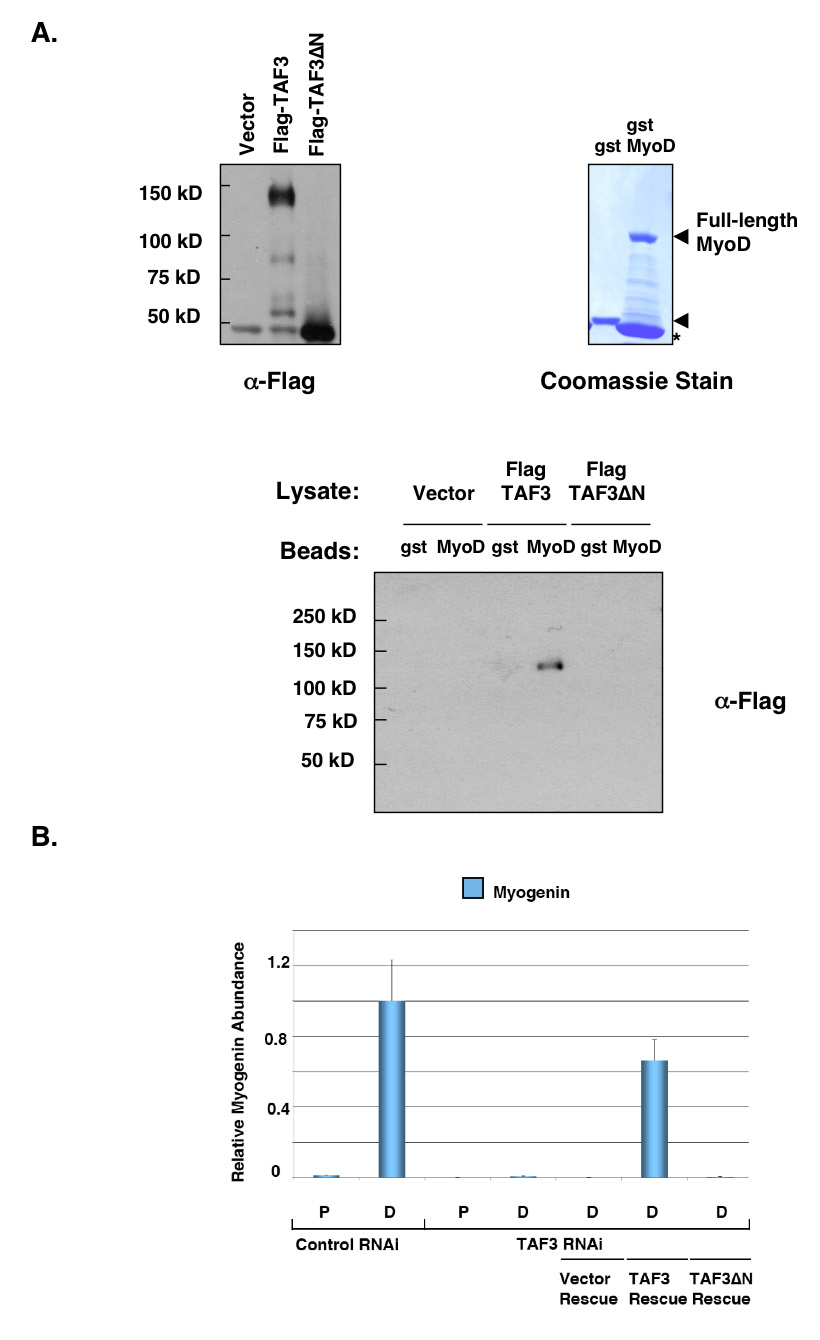
A. TAF3 directly interacts with MyoD. FLAG-TAF3 and FLAG-TAF3ΔN were expressed in insect cells. Protein lysates were prepared and incubated with glutathione-bound Gst-MyoD or Gst control. Asterisk (*) on Coomassie stain gel indicates degradation band. The samples bound on beads were analyzed by protein immunoblot using anti-FLAG antibody. B. Recovery of Myogenin expression in rescued-TAF3 RNAi cells. C2C12 cells depleted for endogenous TAF3 were grown in both proliferating (P) and differentiation (D) conditions. These cells were then rescued by introducing an RNAi-resistant FLAG-TAF3, FLAG-TAF3ΔN or vector control expression construct. Total RNA was isolated from these TAF3-rescued RNAi cell lines to analyze the recovery of Myogenin expression by quantitative PCR (Q-PCR). Samples were normalized to U6 RNA levels, and represented as mean from four reactions. The error bars represent the standard deviation.
Having identified a potentially important N-terminal domain for interaction between TAF3 and MyoD in vitro, we next tested a functional requirement for this putative interaction domain during myogenic differentiation. For these studies, we utilized our previously described myoblast C2C12 TAF3-RNAi cell line that is depleted of endogenous TAF3. Our published analysis of this TAF3 deficient cell line showed dramatic defects in myoblast to myotube differentiation. Moreover, these cells were severely downregulated for Myogenin expression (Deato and Tjian, 2007). Thus, these TAF3-depleted cells provide a useful tool for testing the relevance of our newly identified TAF3-MyoD interaction in vivo. In particular, we can test the ability of wildtype TAF3 and TAF3ΔN to rescue the capacity of these cells to re-express Myogenin. By quantitative-PCR analysis, we found that full-length TAF3 restored Myogenin expression upon differentiation (Figure 5B). In contrast, re-expressing TAF3ΔN failed to restore Myogenin expression (Figure 5B). Taken together, these observations suggest that the N-terminal region of TAF3 likely bears an important target for the muscle-specific activator MyoD, both in vitro and in myoblast cells. An interaction between TAF3 and MyoD is apparently required for MyoD/E47 activated transcription of Myogenin. Thus, as expected for this class of coactivators, TAF3 may be an important target for transmitting the activator signals of MyoD to a TAF3/TRF3 directed transcription complex that presumably enhances active PIC formation at the Myogenin promoter.
Loss of Mediator Subunits in Differentiated Myotubes
It has been well-established that in addition to TFIID, the multi-subunit complex CRSP/MED can function to mediate activator dependent transcription. Similar to TFIID, this co-factor complex can be targeted by diverse arrays of sequence-specific DNA-binding activators. In previous in vitro transcription reactions, most promoters required both TFIID and CRSP/MED in order to reconstitute an activator dependent transcription (Taatjes et al., 2004). It was therefore surprising to find that in our fully reconstituted transcription system responsive to MyoD/E47, no added CRSP/MED components appeared to be necessary. We next set out to track the fate of CRSP/MED subunits during myotube differentiation in C2C12 cells. Just as we previously tracked TBP and TFIID subunits in myotubes, we performed immunoblot analyses to examine the expression profile of CRSP/MED subunits in various cell types including myoblast, reserve cells and myotubes isolated from C2C12 cells (Figure 6). As an additional control, lysates from fibroblast cells were also included as our non-muscle cell-type control. Our immunoblot results revealed a surprising reduction and near complete loss of CRSP/MED subunits in differentiated myotubes reminiscent of the fate of canonical holo-TFIID complex in these cells (Figure 6). In contrast, myoblasts and reserve cells contain a normal complement of most CRSP/MED subunits. Interestingly, MED1 expression was only detected in fibroblasts while none of the C2C12 derived skeletal muscle cell types expressed detectable levels of this largest subunit of the CRSP/MED complex (Figure 6). As expected, TAF3 and TRF3 are both highly expressed in myotubes consistent with the requirement for Myogenin expression. To directly test the requirement for CRSP/MED complex in our reconstituted transcription reactions, we have incubated our Myogenin template with TAF3/TRF3, MyoD/E47 and CRSP/MED complex. In this assay, we find no significant contribution of CRSP/MED on activated Myogenin transcription (Figure 6B). Although these observations suggest that the CRSP/MED complex may also be altered or possibly discarded in differentiated myotubes, more work is required to dissect the fate of CRSP/MED. It remains possible that smaller sub-complexes or an alternative co-factor complex not related to CRSP/MED may functionally replace this co-factor in myotubes.
Figure 6. Loss of CRSP/Mediator subunits in differentiated myotubes.

A. Representative subunits of CRSP/MED complex are dramatically altered in differentiated myotubes. Cell types derived from C2C12 and fibroblast cells were analyzed by protein immunoblots using various antibodies against CRSP/MED subunits. As a positive control, the same set of protein lysates were also analyzed for TAF3, TRF3 and Myogenin expression. Coomassie staining of the gel loaded with equal amounts of cell lysate was used as loading control. B. Purified CRSP/MED complex is included in reconstituted in vitro transcription of Myogenin. Fold activation is calculated as described in Figure 3. Arrowheads indicate the expected primer extension product.
Discussion
Enhancer-binding activators such as MyoD and related basic helix-loop-helix (bHLH) transcription factors are well- established drivers of skeletal muscle differentiation (Tapscott, 2005). Although binding of MyoD to its cognate recognition sites at muscle genes is a required step, occupancy at target promoters is apparently not sufficient to mediate transcriptional activation. Instead, this key regulator of myogenesis along with some of its associated proteins are subject to post-translational modifications that can lead to either activation (promote differentiation) or repression (block differentiation) of MyoD dependent promoters (Sartorelli and Caretti, 2005; Tapscott, 2005). However, dramatic change in the composition of the core transcription apparatus was not a mechanism anticipated for directing muscle differentiation. The recent in vivo finding that holo-TFIID is functionally replaced by a TAF3/TRF3 complex during myoblast to myotube differentiation highlights an unexpected and possibly integral regulatory function of the PIC during cell lineage specific transcription (Deato and Tjian, 2007). Here we report direct biochemical evidence for the differential utilization of diversified core promoter recognition complexes to accommodate tissue-specific and gene-selective regulation during metazoan development.
We have used in vitro reconstituted assays to determine the biochemical properties of the TAF3/TRF3 complex that direct muscle-specific promoter recognition and coactivator selectivity. We establish that TAF3/TRF3 is a bona fide core promoter recognition complex capable of initiating both basal and activator-dependent transcription at the Myogenin promoter. In stark contrast, the canonical TFIID can mediate basal levels of transcription but failed to support MyoD dependent transcription. These in vitro studies have also allowed us to manipulate and dissect the cofactor requirements needed to mediate activator-specific transcription initiation at the Myogenin promoter. Importantly, these in vitro studies provide direct evidence for TAF3 as a “muscle-gene” coactivator directly targeted by the myogenic activator, MyoD. Thus, TAF3/TRF3 can serve as a bona fide alternative core promoter recognition complex that operates both in vitro and in vivo to drive cell-type selective gene regulation.
Our studies indicate that like the prototypic TAF subunits of TFIID, TAF3 mediates its coactivator functions, at least in part, by interacting directly with a muscle specific activator (MyoD). Intriguingly, although a sub-stoichiometric amount of TAF3 can at times be found associated with purified TFIID, only TAF3/TRF3 was able to support MyoD-dependent activation of Myogenin in our in vitro reconstituted reactions. This inability of TAF3-containing TFIID to potentiate activator–dependent transcription from the Myogenin promoter suggests that the larger TFIID complex may arrange the target protein-protein interface in a manner that is incompatible for productive transactions with MyoD. By contrast, MyoD-TAF3 interactions appear well suited to direct specific recruitment of the TAF3/TRF3 complex at the Myogenin promoter. Thus, the TAF3/TRF3 complex behaves as a bona fide coactivator wherein an activator signal is transmitted by direct interaction with components of the promoter recognition complex, underscoring the role of TAFs as molecular adaptors that connect activators to the core transcription machinery.
The coactivator domain of TAF3 that interacts with MyoD maps to a N-terminal region that also contains the histone fold domain implicated in various TAF-TAF and TAF-partner interactions (Gangloff et al., 2001). In myotubes and in vitro, this TAF3 domain is required to mediate MyoD-dependent transcriptional activation of Myogenin. In contrast, the C-terminal region of TAF3 interacts with TRF3 and does not appear to mediate interactions with MyoD (data not shown). Interestingly, this C-terminal region contains a Plant Homeobox Domain (PHD) commonly found in chromatin modifying cofactors that was recently shown to bind trimethylated lysine 4 on histone H3, a modification that marks active promoters in human cells (Bienz, 2006; Vermeulen et al., 2007). One study also suggested that when TAF3 is a component of TFIID it mediates its coactivation function through the PHD domain (Vermeulen et al., 2007). Here we find that in the context of myogenesis the coactivation domain of TAF3 targeted by MyoD is independent of the PHD domain. However, it is possible, even likely, that TAF3 contains multiple coactivator domains that can operate in different contexts depending on the composition of its other partner subunits (i.e. TAF3/TRF3 versus TFIID) in the complex. Thus, the N-terminal coactivator domain of TAF3 might direct interactions with MyoD while the PHD domain could serve to read nucleosome modifications on target gene promoters. Since our in vitro reconstituted reactions were reformed with naked DNA template, this latter function of TAF3 would not be manifested in our system but may be important for transcription from chromatin templates. Indeed, both coactivation functions may be important to distinguish transcriptional programs in differentiated cells.
Like TFIID, the CRSP/MED coactivator complex is another key component of the core transcription machinery that can be targeted by activators to form the pre-initiation complex (PIC). A recent in vivo study of Drosophila transcription initiation uncovered an unexpected functional crosstalk between TFIID and CRSP/MED in regulating the transcriptional output of inducible genes (Marr et al., 2006). In addition, genetic loss of function analyses have suggested the differential requirements for CRSP/MED subunits during cellular differentiation (Ge et al., 2002; Urahama et al., 2005). In conventional in vitro transcription reactions, TFIID and CRSP/MED are thought to perform non-overlapping functions in order to potentiate activator-dependent transcription (Taatjes et al., 2004). We were therefore curious to determine the functional requirements for CRSP/MED in myoblast to myotube differentiation wherein TFIID has been replaced by TAF3/TRF3. Remarkably, the absence of CRSP/MED had no measurable effect in our TAF3/TRF3 reconstituted system that supports MyoD-dependent activation of Myogenin template. Although, we cannot rule out the possibility that other muscle specific promoters or chromatinized Myogenin template may require a Mediator-like activity, it appears that at least for the non-chromatinized Myogenin promoter, CRSP/MED appears largely dispensable for MyoD-dependent transcription activation in vitro. Consistent with this in vitro result, we found that many of the CRSP/MED subunits are indeed severely reduced or absent from myotubes while most subunits were abundantly present in myoblasts with the exception of MED 1. This intriguing finding raises the possibility that the canonical CRSP/MED complex may also be dramatically altered and possibly disposable during terminal differentiation. It remains possible that other Mediator-like complexes or sub-complexes may substitute for the canonical complex in differentiated skeletal muscle. Paradoxically, myoblast to myotube differentiation involves the activation of genes by nuclear receptors (VDR, RAR and TR), a class of activators thought to target CRSP/MED (Halevy and Lerman, 1993). It will be of interest for future studies to further explore the existence and function of CRSP/MED or other muscle-specific Mediator-like complexes in myotubes.
The replacement of TFIID with a TAF3/TRF3 complex in differentiated skeletal muscles has revealed a potentially new transcriptional mechanism that involves diversified core promoter recognition complexes interacting with muscle-specific activators to orchestrate spatial and temporal patterns of gene expression. Such a more equitable parsing of duties between muscle specific activators working in concert with cell selective coactivators may represent an important mechanism evolved to regulate metazoan gene expression; one that allows for a simple yet efficient way to permanently turn off a majority of genes while selectively turning on transcription patterns in differentiated cells.
Experimental Procedures
Plasmids and Insect Cell Expression
TAF3 was cloned as described in (Deato and Tjian, 2007) and the TAF3ΔN truncation was generated by PCR-amplification of DNA that corresponds to residues 517 to 990 of TAF3. TRF3 was cloned as described in (Deato and Tjian, 2007). E2A gene was PCR-amplified from mouse cDNA obtained from Open Biosystems. MyoD fragment was subcloned from pCMV-FLAG-MyoD construct. The amplified DNA fragments were cloned into pIEX-2 vector (Novagen) and transfected into SF9 insect cells with Insect Gene Juice Reagent (Novagen) following manufacturer’s conditions.
Antibodies
TAF3 and TRF3 antibodies were previously described (Deato and Tjian, 2007). Monoclonal antibodies against tubulin and Myogenin were purchased from Developmental Studies Hybridoma Bank (University of Iowa). Antibodies used in immunoblots against MyoD, TFIIH and Cdk8 were purchased from Santa Cruz Biotechnology. The following antibodies against Mediator subunits were purchased: MED1 antibody was obtained from Bethyl Labs, MED12 and MED18 from Novus Biologicals, MED14 from Santa Cruz Biotechnology. The MED26 rabbit polyclonal antibody was raised against the full-length mouse MED26. Anti-FLAG antibodies were purchased from Sigma.
Cell Culture
C2C12 rescue cell lines were maintained as described in (Deato and Tjian, 2007). SF9 cells were maintained in Grace’s Media (Invitrogen). When used for transfection, SF9 cells were maintained in Bacvector Media (Novagen). NIH 3T3 cells (ATCC) were maintained in DME media containing 10% FBS, penicillin and streptomycin and glutamine.
Quantitative PCR Analyses
Total RNA was isolated from various cell lines mentioned in this study as described in (Deato and Tjian, 2007). Data shown in this study is from four independent sets of experiments.
Electrophoretic Mobility Shift Assay
This assay was performed based on conditions previously reported for TBP (Lescure et al., 1994). Briefly, purified TRF3(20 ng) and the separately purified TAF3 (20ng) were incubated with labeled probe in a 20ul reaction 12mM Hepes 8.0, 60mM KCl, 5mM MgCl2, 1mM DTT, 0.5mM EDTA, 0.05% NP-40, 10% glycerol and 500ng of poly (dG-dC)/(dG-dC) (GE Biosciences). Gel mobility shift reactions were incubated for 20 minutes at room temperature and separated on 5% nondenaturing gel containing 3mM MgCl2 followed by autoradiography. For these sets of reactions, the Myogenin TATA sequence (GAGGGTTTAAATGGCACCCAG) was used as gel mobility shift probe and specific (TATA) competitor while (GAGGGTCCCGCCGGCACCCAG) was used as non-specific competitor. The unlabelled competitors were used at 50–100 fold excess.
Luciferase Reporter Assay
The endogenous Myogenin promoter was PCR amplified from mouse genomic DNA (BD Bioscience) using specific primers directed at −184 and +46 region of Myogenin promoter. The DNA fragment was cloned into pGL3-Basic vector (Promega). The constructs (pGL3-Basic vector control and pGL3-Myogenin) were co-expressed with pCMV-FLAG-MyoD in 3T3 fibroblast cells following manufacturer’s instructions for Lipofectamine 2000 (Invitrogen) transfections. Luciferase expression from these reporter constructs was analyzed following the instructions for Dual Luciferase Assay System (Promega). Data shown in this study is an average from three independent experiments.
In vitro gst-pulldown / binding assay
Recombinant Gst and Gst-MyoD proteins were expressed and purified from E. coli and subsequently bound on Glutathione Sepharose 4B beads (GE Bioscience). Bound proteins were washed in Gst wash buffer 1(100 mM Tris 8.0, 120 mM NaCl and 0.1% Triton X-100) and Gst wash buffer 2 (Gst wash buffer 1 containing 500 mM NaCl). The full-length Flag-TAF3 or truncated version, Flag-TAF3ΔN was expressed in SF9 cells. Transfected SF9 cells were lysed in 50 mM Tris 7.4, 150 mM NaCl, 10% glycerol and 0.5% Tween-20. To set-up the binding reaction, equal amounts of bound Gst and Gst-MyoD beads were incubated with 200 ul of Flag-TAF3 transfected SF9 lysate. The samples were incubated for 4 hrs at 4°C. Samples were washed using (50 mM Tris 7.4, 250 mM NaCl, 10% glycerol and 0.5% Tween 20).
Protein Purification
Recombinant FLAG- tagged proteins TRF3, TBP and TAF3 were purified from transfected SF9 cells. Insect cells were transfected with pIEX-2 constructs for TRF3, TBP and TAF3 following Insect Gene Juice (Novagen) transfection conditions. SF9 cells were lysed in buffer containing 50 mM Tris 7.4, 150 mM NaCl, 10% glycerol and 0.5% Tween 20. The protein lysates from transfected cells were incubated with FLAG-affinity resin M2 (Sigma) for 4 hrs at 4°C. Next, M2 beads were first washed in 50 mM Tris 7.4, 250 mM NaCl, 0.5% Tween 20 for 1 hr at 4°C with buffer changes every 20 minutes. Following this step, the beads were subsequently washed in 50 mM Tris 7.4, 500 mM NaCl, 0.5% Tween 20 for an additional 2 hrs at 4°C. M2-bound proteins were eluted using 3X-FLAG peptide in elution buffer containing 50 mM Tris 7.4, 150 mM NaCl and 10% glycerol. Similar purification steps and wash conditions were also followed to co-purify FLAG-TAF3/TRF3 complex, FLAG-TAF3ΔN/TRF3 complex and FLAG-MyoD/ E47 heterodimer complex. All eluted proteins were dialyzed against 0.1 M HEMG buffer (25 mM HEPES 7.9, 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol and 100 mM KCl).
In vitro transcription assay
The modified Myogenin promoter (as described in the text) was used as DNA template in our in vitro transcription reactions. Briefly, the Myogenin core promoter (−184 to +46) with two additional activator sites was cloned into modified pGL3-Basic vector with CAT sequences inserted at NcoI site. The transcription template (200 ng/ul) was incubated with either purified TBP, TRF3, TAF3 (or TAF3ΔN)/TRF3 complex or TFIID (0.2ng/ul) at room temperature. After 5 min., a mix of all purified basal factors [TFIIA (50ng/ul), TFIIB (5ng/ul), TFIIE (50ng/ul), TFIIF (80ng/ul), TFIIH (1ng/ul), RNA Polymerase II (2.5ng/ul)] were added into the 25ul reaction and incubated at 30°C for 30 minutes. For activator dependent reactions, samples were first incubated with basal factor mix for 5 minutes at 30°C then purified MyoD/E47 (20ng) was added to the reactions and incubated at 30°C for 25 minutes. Next, rNTPs were added at a final concentration of 0.63 mM and the transcription reaction was allowed to proceed for an additional 30 minutes. The transcription reaction was subsequently terminated by adding 100 ul of stop solution (20 mM EDTA, 1% SDS, 200 mM NaCl) with 0.2 mg/ml glycogen (Ambion) per reaction. RNA was isolated and transcription products were analyzed by primer extension using end-labeled CAT oligonucleotide. For primer extension, the precipitated RNA was dissolved in hybridization buffer (10 mM Tris 7.5, 1 mM EDTA and 250 mM KCl). Samples were incubated at 72°C for 2 minutes and transferred to 55°C block and incubated for 1hr. Reverse transcription reaction was performed using MLV-RT (Ambion) for 30 minutes at 37°C. Samples were subsequently loaded on 6% denaturing acrylamide gel followed by autoradiography. Fold activation was calculated as the ratio of activated transcription and basal transcription values obtained as average signal from Phosphorimager scan (Molecular Dynamics).
Acknowledgements
The authors would like to thank K.Wright, Y. Fong, A. Levitzki and D. Rio for critical comments on the manuscript, and for helpful discussions. We thank Y. Fong for providing us with purified CRSP/MED complex. We would also like to thank R. Bell and E. Borbon for preparation of Hela nuclei. M.Deato was a predoctoral fellow of the California Institute for Regenerative Medicine (CIRM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Bartfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orban L, Muller F. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Black BL, Molkentin JD, Olson EN. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol Cell Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht BD, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Cheng TC, Cserjesi P, Chakraborty T, Olson EN. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol. 1992;12:3665–3677. doi: 10.1128/mcb.12.9.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Romier C, Thuault S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci. 2001;26:250–257. doi: 10.1016/s0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- Gazdag E, Rajkovic A, Torres-Padilla ME, Tora L. Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggests different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction. 2007;134:51–62. doi: 10.1530/REP-06-0337. [DOI] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- Halevy O, Lerman O. Retinoic acid induces adult muscle cell differentiation mediated by the retinoic acid receptor-alpha. J Cell Physiol. 1993;154:566–572. doi: 10.1002/jcp.1041540315. [DOI] [PubMed] [Google Scholar]
- Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci U S A. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Davi RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lescure A, Lutz Y, Eberhard D, Jacq X, Krol A, Grummt I, Davidson I, Chambon P, Tora L. The N-terminal domain of the human TATA-binding protein plays a role in transcription from TATA-containing RNA polymerase II and III promoters. Embo J. 1994;13:1166–1175. doi: 10.1002/j.1460-2075.1994.tb06366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Dixit BL, Maston GA, Kittler EL, Green MR. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci U S A. 2003;100:14887–14891. doi: 10.1073/pnas.2036440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat Rev Mol Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Urahama N, Ito M, Sada A, Yakushijin K, Yamamoto K, Okamura A, Minagawa K, Hato A, Chihara K, Roeder RG, Matsui T. The role of transcriptional coactivator TRAP220 in myelomonocytic differentiation. Genes Cells. 2005;10:1127–1137. doi: 10.1111/j.1365-2443.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]


