Abstract
Comparative genomics is a powerful tool to decipher gene and genome evolution. Placing multiple genome comparisons in a phylogenetic context improves the sensitivity of evolutionary inferences. In the genus Oryza, this comparative approach can be used to investigate gene function, genome evolution, domestication, polyploidy, and ecological adaptation. A large genomic region surrounding the MONOCULM1 (MOC1) locus was chosen for study in 14 Oryza species, including 10 diploids and 4 allotetraploids. Sequencing and annotation of 18 bacterial artificial chromosome clones for these species revealed highly conserved gene colinearity and structure in the MOC1 region. Since the Oryza radiation about 14 Mya, differences in transposon amplification appear to be responsible for the different current sizes of the Oryza genomes. In the MOC1 region, transposons were only conserved between genomes of the same type (e.g., AA or BB). In addition to the conserved gene content, several apparent genes have been generated de novo or uniquely retained in the AA lineage. Two different 3-gene segments have been inserted into the MOC1 region of O. coarctata (KK) or O. sativa by unknown mechanism(s). Large and apparently noncoding sequences flanking the MOC1 gene were observed to be under strong purifying selection. The allotetraploids Oryza alta and Oryza minuta were found to be products of recent polyploidization, less than 1.6 and 0.4 Mya, respectively. In allotetraploids, pseudogenization of duplicated genes was common, caused by large deletions, small frame-shifting insertions/deletions, or nonsense mutations.
Keywords: comparative genomics, genome evolution, microcolinearity, allotetraploid, conserved noncoding sequence
Comparative genomics has emerged as a powerful tool to decipher gene and genome evolution and improve genome annotation. Multiple species comparisons reveal novel insights into genome evolution (1), genome duplication (2), and new gene origination (3), and can also identify previously unknown or poorly characterized genome components, such as novel transposable elements (4) and novel functional elements (5).
Among plants, grasses have been the paradigm for comparative genomics for more than a decade (6). Comparative genetic and physical mapping revealed extensive genome colinearity among closely or distantly related grass species (7–9). Further fine-scale sequence comparisons have illustrated that the genes in grass genomes are generally colinear, with occasional small rearrangements (inversions, duplications, and deletions) that appear to be associated with unequal homologous or illegitimate recombination, and rarer gene movements from unlinked chromosomal sites without a known mechanism for mobility (10–16). Because only a few species were involved in these comparisons, instances of noncolinearity, such as predicted de novo gene origins, gene translocations, or pseudogenizations, could be identified but were not thoroughly investigated (1). To improve the precision and sensitivity of the evolutionary inferences drawn from the comparisons, the number of species compared had to be increased (5, 17). Because of its large number of species, history of genetic study, and well-characterized phylogeny and domestication, the Oryza genus is an ideal model system to study gene function, genome evolution, domestication, polyploidy, and ecological adaptation using a comparative approach (18).
The genus Oryza consists of 23 species with diverse ecological adaptations (19), including the Asian cultivated rice. Rice (Oryza sativa L.) is both an important food crop and a model plant for biological studies. Wild rice species have proven to be tremendous gene reservoirs to increase domesticated rice yield, quality, and resistance to diseases and insects. Wild rice species have furnished genes for the hybrid rice revolution, exhibit yield-enhancing traits, and have shown tolerance to biotic and abiotic stress (20, 21).
The Oryza species have 10 different genome types, including 6 diploid genome types (AA, BB, CC, EE, FF, and GG) and 4 allotetraploid genome types (BBCC, CCDD, HHKK, and HHJJ) (19). For the Oryza Map Alignment Project (OMAP) (18), representatives from each of these 10 genome types were selected for bacterial artificial chromosome (BAC) library construction, BAC end sequencing, and physical map construction (22, 23). From analysis of the OMAP data, Oryza comparative genomics has given novel insights into Oryza genome evolution (24–26) and genome size variation (27, 28). However, no systematic comparative sequence analysis has been performed across the Oryza. Moreover, the allotetraploidy in some Oryza species allows for study of the evolutionary dynamics of duplicated genes in polyploids.
To better understand Oryza genome evolution, MONOCULM1 (MOC1) genomic regions were sequenced and compared across 14 Oryza genomes. Located on the long arm of chromosome 6, MOC1 encodes a GRAS family nuclear protein that controls an important agronomic trait, the formation of tillers, in rice (29). This study revealed gene and transposable element (TE) dynamics during Oryza evolution.
Results
Sequencing and Annotation of BAC Clones.
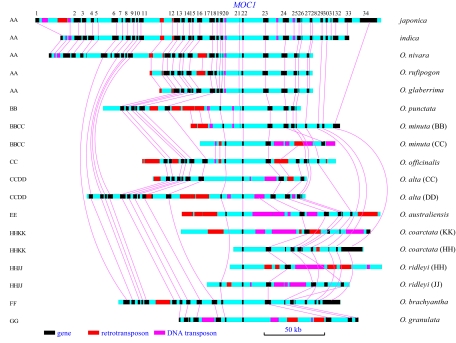
The 12 species in OMAP, plus indica rice (Oryza sativa L. ssp. indica cultivar 93–11) and japonica rice (Oryza sativa L. ssp. japonica cultivar Nipponbare), were included in the comparative study. Sixteen BAC clones from 8 diploid and 4 allotetraploid species were isolated from Oryza BAC libraries and sequenced (supporting information (SI) Table S1). About 2.4 Mb of data were generated in the MOC1 region across these 16 clones. Gene and transposable element (TE) annotation are shown in Fig. 1 and Table S2.
Fig. 1.
Genomic alignment of the MOC1 region in 18 Oryza genomes or subgenomes. The species are ordered by genome type. Horizontal light blue bars represent genomic sequence in the MOC1 region. Gene models are shown in black rectangles. Transposons (at least 1 kb) are shown in red and pink for retrotransposons and DNA transposons, respectively. Lines/curves connect orthologous genes with each other and orthologous transposons with each other.
In the preexisting japonica sequence (30), we refined the gene annotation of the MOC1 region. Of 47 initial gene models, 9 were removed because they were observed to be TE related. An additional 4 were removed because they do not have cDNA or EST support and were not found in all AA Oryza species, although some of these may represent de novo genes created in a specific AA lineage. Of the remaining 34 gene models that were further analyzed, 30 have cDNA or EST support. The remaining 4 gene models were found to be homologous to known proteins or were conserved in all Oryza species studied. Additionally, RT-PCR of gene 23 redefined its exon/intron structure (data not shown and Fig. S1).
To annotate the remaining non-japonica genomes, the FGENESH program (31) was used for gene prediction. From the 397 predicted gene models, 175 (44%) that are TE related or have no significant hits to cDNAs, ESTs, or known proteins and are not conserved in other studied Oryza genomes were removed. The remaining 256 gene models were annotated in the 14 Oryza genomes (Table S2). The exon/intron structures of 122 non-japonica genes were manually refined to match orthologous genes in japonica. In addition, the exon/intron structures of 18 genes were verified by RT-PCR (data not shown).
TEs were discovered and annotated using RepeatMasker and Repbase with subsequent manual validation (Table S2). Long terminal repeat (LTR) retrotransposons were the predominant class I TEs, and major class II TEs included Mutator, hAT, and CACTA elements. Although the extensive previous annotation of miniature inverted repeat transposable elements (MITEs) in O. sativa means that the use of Repbase should create a bias toward discovery of MITEs in AA genomes, these tiny elements were observed to have a higher density in the only studied FF genome, that of O. brachyantha (one MITE per 3.3 kb), than in AA genomes (averaging one MITE per 4.4 kb). All other genomes exhibited known MITE densities of less than one per 7 kb (Table S2).
In the tetraploid species, each subgenome identity was verified by phylogenetic analyses of gene models 22 (MOC1) and 23 (data not shown). Both of these genes have sequence information available for all Oryza species studied.
TEs Have Shaped Genome Architecture in the Genus Oryza.
The genome sizes of Oryza species range from ≈340 Mb (O. brachyantha) to ≈1,280 Mb (O. ridleyi) (22). In the MOC1 region, the TE content in diploid species was found to positively correlate with genome size (correlation coefficient = 0.89; P < 0.005). This correlation suggests that, in addition to polyploidization, TEs are the driving force behind Oryza genome expansion. In the MOC1 regions, detected DNA transposons and retrotransposons comprised an average of 29.5% of the sequence.
Using the Artemis Comparison Tool (32), clear divisions between conserved genic and variable intergenic regions were found to be primarily the result of different TE insertions in intergenic regions (Fig. S2). TEs in genic regions were composed mainly of short MITEs, such as Tourist, Stowaway, Explorer, Crackle, and Gaijin elements. In contrast, TEs in intergenic regions were often long transposons, including LTR retrotransposons, plus LINE, hAT, CACTA, and Mutator elements, which can extend up to 10 kb. In some cases, these intergenic TEs were nested within clusters. In intergenic regions, besides intact gypsy and copia LTR retrotransposons, many solo LTRs were observed (Table S2). However, no Helitron was detected. In genomes of the same type, many TEs were located in orthologous positions. In contrast, few orthologous TEs were identified between different genome types (Fig. S2). For example, only 8 of 107 (7.5%) TEs were orthologous between japonica (AA) and O. punctata (BB). In contrast, ≈95% of the TEs were found to be orthologous between 2 AA genomes (japonica and O. glaberrima). Similarly, ≈98% of the TEs were observed to be orthologous between 2 BB genomes (O. punctata and the BB subgenome of O. minuta).
High Gene Colinearity and Rare Exceptions in the MOC1 Region.
Although TEs resulted in highly variable intergenic regions, gene content, gene order, and transcriptional orientation are highly conserved in the MOC1 regions (Fig. 1). Of 222 genes, 217 (97.7%) in non-japonica species have japonica orthologs located in colinear positions.
Three exceptions to strict colinearity were identified. First, in the O. coarctata KK subgenome, a unique 60-kb fragment with 3 genes was detected (Fig. 1). Based on a BLAST search, this fragment was homologous to a japonica region 400 kb upstream of MOC1, indicating a DNA rearrangement in that lineage that gave rise to this part of the O. coarctata genome. Second, a 3-gene segment was found in the japonica and indica MOC1 regions but absent in the other genome types studied (genes 31–33; Fig. 1). These 3 genes are homologous to 3 adjacent genes on japonica chromosome 1 (Fig. S3). The segment was not introduced by retrotransposition because the genes retain introns. Additionally, although fragments of Mutator elements were found in the intergenic regions of the 3 genes, no terminal inverted repeat or target site duplication was identified. Third, in O. granulata, a tandem duplication of gene 23 was observed (Fig. S3). Both duplicated genes are separated by a 50-kb repeat region, comprised of Mutator elements and LTR retrotransposons. One copy appears to be a pseudogene caused by a 28-bp deletion in the ninth exon that led to a frameshift mutation.
Conserved Exon/Intron Structure of Orthologous Genes.
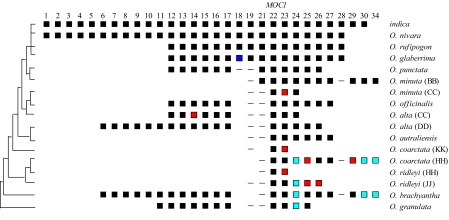
Besides extensive gene colinearity across the genus Oryza, exon/intron structures of orthologous genes are also highly conserved. Of 217 non-japonica genes, 192 (88.5%) had identical exon/intron structures to their japonica orthologs (Fig. 2). Most introns had the canonical GT/AG splice site. One exception was that the second intron splicing site of gene 12 was changed to GC/AG in O. officinalis and the O. alta CC subgenome. Despite a 4-kb retrotransposon insertion in the third intron of gene 14 in the O. alta DD subgenome, the exon/intron structure remained unchanged. The FGENESH program predicted that the TE-related domains would fuse with gene 14 exons to form a new gene. However, RT-PCR experiments revealed that the retrotransposon was spliced out of the transcript (data not shown).
Fig. 2.
Gene evolution in the MOC1 region in Oryza. Numbers on the top represent genes in japonica. Each row represents an Oryza genome or subgenome. Black rectangles indicate that the genome has an ortholog with identical exon/intron structure to japonica. Blue rectangles indicate that the genome has an ortholog with different exon/intron structure from the japonica gene. Red rectangles indicate pseudogenes. Light blue rectangles indicate genes with ambiguous structure or functionality. Short lines indicate the presence of partial homology to candidate genes (18, 19, 21, and 28) that are predicted to have been created de novo in the AA lineage leading to japonica. Candidate genes 20, 31, 32, and 33 are not shown because their predicted exon/intron structures have not been confirmed experimentally in japonica rice. The Oryza phylogenetic tree is shown on the left.
Possible de Novo Gene Creation in the MOC1 Region.
Some lineages contained apparent novel genes in the MOC1 region. To determine if these genes represented gene gain or gene loss, we examined their distribution in a phylogenetic context. Gene 18 was identified only in AA genomes. In O. punctata (BB), partial homology to gene 18 was observed in the first exon and first intron that contains a fragmented LINE (Fig. S4), but no homology to gene 18 was detected outside of genome type BB. To distinguish whether this gene originated in the AA lineage de novo or as a duplicated locus that was later deleted from other genomes, gel blot hybridization analysis was performed on japonica, O. rufipogon, O. punctata, O. brachyantha, and 3 additional AA species not included in OMAP (O. glumaepatula, O. longistaminata, and O. meridionalis). The hybridized blot showed that gene 18 was present as a single copy in japonica, O. rufipogon, and O. glumaepatula, but no bands were observed for the other Oryza species (data not shown). In O. glaberrima, a 7-bp insertion in the first exon of gene 18 led to a new exon (Fig. S4), suggesting that the structure of gene 18 is unstable in AA genomes.
Not all candidate de novo genes originated by such major changes. For example, gene 21 has a single exon of 180 bp. The ORF of gene 21 was identified only in AA and BB genomes (Fig. 2). However, DNA sequence with more than 90% identity was found in the syntenic regions of japonica (AA) and O. granulata (GG) (Fig. S4). Thus, gene 21 may have originated de novo by cumulative point mutations and small indels in an existing sequence. Alternatively, gene 21 might actually be a conserved noncoding sequence (CNS). Similar cases were observed for gene 19 and gene 28 (Fig. 2). In contrast to gene 21, these genes show weak sequence homology to orthologous regions of distant species (data not shown).
Timing the Radiation of the Genus Oryza and the Origins of Allotetraploids.
Although a robust phylogeny of the Oryza genus has been reconstructed (19, 33), the dates of the ancestral divergences of Oryza lineages remain controversial. The divergence time of Oryza species was estimated with 4 genes that were identified in all diploid species (Table S3). To limit the estimates to genes that evolve at similar rates, O. brachyantha, which evolves faster than other Oryza species (33), and allotetraploids, which have duplicated genes that may have relaxed selection constraints, were excluded from this analysis. The earliest split within Oryza was estimated at about 14 Mya; the AA and BB lineages are predicted to have diverged from each other about 7.5 Mya.
Estimation of when the tetraploid species originated required estimates for the divergence times of both “progenitor” lineages. Of the 4 tetraploid species in our study, only O. minuta had both putative maternal (O. punctata) and paternal (O. officinalis) diploid progenitors identified (19). Thus, the tetraploid origin for O. minuta (BBCC) could be calculated on the basis of the divergence times of the diploid genome and the corresponding subgenome in the allotetraploid. Six gene pairs in O. punctata (BB) and the O. minuta BB subgenome and 3 gene pairs in the O. minuta CC subgenome and O. officinalis (CC) were combined to estimate that the O. minuta parental lineages diverged ≈0.4 Mya, and thus the polyploidy event must have occurred within the last 400,000 years.
Because O. officinalis (CC) is proposed to be a close relative of the maternal progenitor of O. alta (CCDD) (19), when the tetraploid genome formed in O. alta could also be calculated. Based on the substitution patterns in 9 orthologous gene pairs in O. officinalis and the CC subgenome of O. alta, the tetraploid formation event of CCDD was estimated as less than 1.6 Mya.
Pseudogenization of Duplicated Genes in Allotetraploids.
In the allotetraploid Oryza genomes, pseudogenization of duplicated genes resulted from nonsense mutations, frameshift mutations, and sequence deletions (Fig. 2). In the O. minuta CC subgenome, gene 23 was pseudogenized by a nonsense mutation. In the O. alta CC subgenome, gene 14 was pseudogenized by 3 frameshift mutations, leading to a premature stop codon. In O. coarctata and O. ridleyi, 6 of 18 genes were pseudogenized. In the O. ridleyi JJ subgenome, 3 kb was deleted between gene 25 and gene 26, which eliminated exons in both genes and led to a double pseudogenization. In all cases, a maximum of one copy of each pair of duplicated genes was pseudogenized.
Evolutionary Dynamics of Duplicated Genes in O. minuta (BBCC).
To reveal the evolutionary dynamics of duplicated genes in recently formed allotetraploids, we calculated nonsynonymous substitution rates (KA) and synonymous substitution rates (KS) between the tetraploid genes and their orthologs in the diploid “progenitor” lineage. In O. minuta, 4 of 8 genes have KA/KS > 1.
To identify the substitutions that took place in each lineage, we compared the sequence in the allotetraploids and the parental diploid progenitors with japonica as the outgroup (Table S4). Taking all of the genes together, both nonsynonymous (P < 0.05) and synonymous (P < 0.01) substitutions were in excess in O. minuta, indicating that the duplicated genes in this allo tet raploid are evolving under relaxed constraint.
Conserved Noncoding Sequences in the Vicinity of MOC1.
Intergenic regions were highly variable and nonhomologous between distant Oryza genomes due to TE insertions and deletions (Fig. S2). However, 3 CNS regions flanking MOC1 with a sequence identity of up to 96% were identified by comparing japonica and O. granulata (Fig. S5). These 3 CNS regions were 3.8, 3, and 2 kb long. They were conserved in all Oryza genomes studied. To investigate whether these CNS regions evolve neutrally or under selective constraints, we analyzed the substitutions of TE and intron sequences between japonica and O. granulata to get a distribution of neutral substitutions (Fig. S6). The substitution rates of orthologous genes between japonica and O. granulata were compared with those of the CNS regions. The 3 CNS regions evolved similarly to coding sequences and had significantly fewer substitutions than expected under the neutral model (Fig. S5), suggesting that purifying selection plays an important role in these CNS regions. These CNS regions were also found to be conserved in orthologous positions in sorghum and maize genomes (Fig. S5).
Discussion
With the completion of the rice genome sequence (30), the Oryza genus is becoming an ideal system for comparative genomics (18, 22, 23, 27). Though previous comparative studies in grasses revealed some genome evolution patterns (1), these comparisons involved only a few species (often distantly related) and thus were unlikely to uncover mechanisms involved in recent rearrangements. Instead, to uncover these mechanisms, we compared 14 species/subspecies and 18 genomes/subgenomes across the Oryza genus. This study provides insight into plant genome evolution, new gene origination, conserved noncoding regions, and the evolution of genes and noncoding sequences in polyploids.
Comparative Genomics Improves Genome Annotation.
Gene annotation is an imperfect process, especially in complex genomes rich in transposable and retrotransposable elements (34). Comparative genomics can dramatically improve gene and genome annotation (14, 17). In this study, more than 40% of the gene models predicted by FGENESH were found to be TE related. If these gene models remained in the annotation, many lineage-specific genes would have been erroneously predicted in Oryza. Despite extensive studies of rice, nearly 30% of gene models in the MOC1 region appear to have been misannotated, including gene models with “full-length” cDNA support (Fig. S1).
Except candidate novel genes, orthologs of most rice genes can be recognized easily in other Oryza species, including distant Oryza species such as O. brachyantha and O. granulata. In all Oryza species, most genes have identical gene structure to their japonica ortholog; the exon sequences are highly conserved, whereas introns have limited sequence identity. Potentially functional CNS elements embedded in variable intergenic regions were identified as well. Thus, full genome sequencing of a distant Oryza species could dramatically improve gene annotation of rice. O. brachyantha is a good candidate for this comprehensive sequence analysis because of its small genome size (≈360 Mb) (22) and phylogenetic position (19).
De Novo Origination of New Genes.
New genes can originate by gene duplication, retroposition, exon shuffling, gene fusion, and gene fission using preexisting genes as raw materials (35). Only recently have de novo genes originating from noncoding DNA sequences been investigated (36, 37), but no example of this type of gene origin has been proven in plants.
In the Oryza genus, novel genes in the AA genomes can be easily identified because multiple AA genomes have been included in OMAP. In the AA genomes, 4 putative protein-coding genes may have originated de novo from noncoding sequence. The predicted proteins were not found to be homologous to any known proteins. Perhaps these candidate new genes originated by dramatic structural rearrangements of preexisting repetitive sequences or by the gradual accumulation of mutations in previously unselected sequence. Alternatively, these apparent de novo genes could be remnants of still-unidentified TEs that have retained, for reasons unknown, perhaps by simple chance, an ORF status across several AA genome species despite the rapid degradation and deletion of nonfunctional sequences in rice (38). These possibly TE-derived sequences may have acquired some new function that qualifies them as truly new genes. Little is known about the process of transposon domestication in plants (39), but it is a field that is ripe for further enquiry. Part of the reason that the exact evolutionary mechanisms for predicted de novo gene origin remains unknown is because the current OMAP lacks intermediate species between the AA and BB genomes. The study of more divergent AA species, such as those in O. longistaminata or O. meridionalis, could reveal the evolutionary dynamics of de novo gene origination in the MOC1 region.
Duplicated Gene Evolution in Allotetraploids.
In allotetraploids, each gene has 2 copies, one from each subgenome. Based on the classical gene duplication model (40), one gene copy remains functional through purifying selection, and the other copy usually accumulates deleterious mutations due to relaxation of selection constraints. An alternative gene duplication model is the degeneration divergence complementation (DDC) model, which emphasizes that degenerative mutations facilitate preservation of duplicate genes (41).
Some Oryza allotetraploidizations support the DDC model, and others support the classical Ohno model (40). In O. minuta and O. alta, which appear to have arisen as polyploids less than 2 Mya, only ≈5% (2 of 38) of duplicated genes were identified as pseudogenes. Similarly, in Gossypium hirsutum, whose allotetraploid was predicted to have formed ≈1.5 Mya, 3% of duplicated genes have been pseudogenized (42). In contrast, one-third of O. coarctata- and O. ridleyi-duplicated genes were observed to be pseudogenized.
Although it is clear that no current diploid is a true progenitor of any current polyploid, surrogates for diploid progenitors can sometimes be identified as existing diploids that are closely related to an existing polyploid. For O. minuta, both maternal and paternal diploid progenitor surrogates were analyzed (19), such that nucleotide differences that arose in the allotetraploid could be identified. In O. minuta, more nonsynonymous substitutions were observed in the duplicated genes than in their diploid “progenitor” orthologs. Furthermore, half of the gene pairs had a KA/KS > 1, suggesting relaxed selective constraint or positive selection for duplicated genes in O. minuta. Notably, compared with its diploid progenitors, the O. minuta synonymous substitution rate is also accelerated. Because most synonymous substitutions are neutral, this rate increase cannot be explained by changes in selective constraint and might be related to the small population size at the early stage of O. minuta speciation (43).
O. coarctata Has a Unique Genome Type.
Although most Oryza genome types were determined by traditional genome or molecular analysis (44, 45), O. coarctata was designated as an HHKK genome type based solely on its phylogenetic position (19). When the HH subgenomes in O. coarctata (HHKK) and O. ridleyi (HHJJ) were compared, no homology was observed in the intergenic regions. These findings contrast with other subgenome comparisons that show homologous sequences and shared TE elements in intergenic regions, such as the BB and CC genome types (Fig. S2). Moreover, the gene sequence differences between the predicted HH subgenome types in O. coarctata and O. ridleyi were more different from between AA and BB genome types. Both of these subgenomes were estimated to have diverged from each other ≈11 Mya. Hence, “HH” subgenomes in O. coarctata and O. ridleyi are likely to belong to different genome types. To avoid confusion in future research, we suggest O. coarctata should be designated as KKLL.
Conserved Noncoding Sequences.
CNSs are highly conserved sequences that are not known to be transcribed or translated. CNSs comprise ≈1%–2% of the human genome (46). CNSs are usually shorter and less conserved in plants than in animals (47, 48).
The major CNSs observed in the vicinity of MOC1 are very large (2–3.8 kb). Exhaustive searches identified no long ORFs, RNA genes, or protein homology. These CNSs appear to have evolved under strong purifying selection. CNSs in the Vgt1 locus of maize have been shown to be associated with flowering time variations (49). A CNS cluster in a knotted1 transcription factor gene intron may serve as a site of negative regulation (50). The functional significance of CNSs flanking the MOC1 gene remains to be investigated, but possible roles in regulating MOC1 or serving as an unknown class of gene in their own right seem likely.
Genome Structure, Function, and Evolution.
Comparative sequence analysis revealed extensive gene colinearity across the genus Oryza. This gene colinearity unambiguously revealed orthologous genes. However, repetitive sequences, such as retrotransposons and DNA transposons, have also shaped the Oryza genome landscape dramatically. Some orthologous genes reside in highly repetitive DNA blocks in some species, such as gene 23 in O. ridleyi. The effects, if any, of the surrounding repetitive DNA blocks on gene expression and function remain to be determined. Furthermore, polyploidy and the evolution of duplicated genes following polyploidization can have profound impacts on genome function and evolution (51). New gene origination adds a new dimension to genome evolution and the adaptation of organisms (35). Future comparative genomic studies should include gene expression and functional characterization to improve gene annotation, detect subtle changes in orthologous gene structure that might affect gene function, and identify functional modifications and innovations in different organisms. In this regard, whole-genome sequencing of multiple Oryza species would open a new era in plant biology, especially in comparative and functional genomics.
Methods
BAC Clone Identification, Sequencing, and Annotation.
BAC clone identification, sequencing, and gene and TE annotation are described in SI Text.
Phylogenetic Analysis and Chronology.
A neighbor-joining phylogeny based on the Kimura 2-parameter model (52) was created in MEGA4 (53). Robustness was evaluated with 1,000 bootstrap replicates.
Synonymous and nonsynonymous substitutions were calculated based on Nei and Gojobori's model (54) using the PAML toolkit (55). A synonymous substitution rate of 6.5 × 10−9 synonymous base substitutions per site per year (56) was used to date lineage separation events.
Simulations to Detect Purifying Selection in CNS Regions.
TE and intron sequences were used to estimate neutral substitution (d) in these genomes. Because orthologous TEs and introns were difficult to identify between japonica and O. granulata, orthologous TEs and introns between japonica and O. glaberrima were combined to estimate d using the Jukes-Cantor model (57). Sequences of 6 orthologous genes (gene 14, 15, 16, 17, 22, and 24) were merged to estimate the KS value between japonica, O. glaberrima, and O. granulata. Based on the assumption that d is positively correlated with KS, we estimated dTE and dintron between japonica and O. granulata as 0.39 and 0.19, respectively.
Second, the theoretical distribution of observed neutral substitutions was simulated. Employing the Jukes-Cantor model (57), p = 3 × (1 –e−4d/3)/4, where p is the ratio of different base pairs of 2 aligned sequences, pTE and pintron were estimated as 0.31 and 0.17, respectively. To generate the theoretical distribution of observed neutral substitutions, 20,000 replicates of 50-bp-long sequences under either pTE or pintron were generated (Fig. S6). Based on these distributions, 50-bp windows for TEs with ≤10 substitutions or for introns with ≤5 substitutions between japonica and O. granulata were judged as evolving significantly more slowly than TEs or introns.
Supplementary Material
Acknowledgments.
We thank Dr. Song Ge (Institute of Botany, Chinese Academy of Sciences) for his critical reading of the manuscript. This work was supported by the Chinese Academy of Sciences (Grants KSCX2-YW-N-028 and CXTD-S2005–2) and the National Natural Science Foundation of China (Grants 30600034, 30621001, 30623011, and 30770143).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ032625–FJ032640).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812798106/DCSupplemental.
References
- 1.Bennetzen JL. Patterns in grass genome evolution. Curr Opin Plant Biol. 2007;10:176–181. doi: 10.1016/j.pbi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Ilic K, SanMiguel PJ, Bennetzen JL. A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc Natl Acad Sci USA. 2003;100:12265–12270. doi: 10.1073/pnas.1434476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang S, et al. Repetitive element-mediated recombination as a mechanism for new gene origination in Drosophila. PLoS Genet. 2008;4:e3. doi: 10.1371/journal.pgen.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai J, Li Y, Messing J, Dooner HK. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennetzen JL, Freeling M. Grasses as a single genetic system: Genome composition, collinearity and compatibility. Trends Genet. 1993;9:259–261. doi: 10.1016/0168-9525(93)90001-x. [DOI] [PubMed] [Google Scholar]
- 7.Ahn S, Tanksley SD. Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hulbert SH, Richter TE, Axtell JD, Bennetzen JL. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc Natl Acad Sci USA. 1990;87:4251–4255. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JS, et al. Comprehensive molecular cytogenetic analysis of sorghum genome architecture: Distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics. 2005;171:1963–1976. doi: 10.1534/genetics.105.048215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M, SanMiguel P, Bennetzen JL. Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics. 1998;148:435–443. doi: 10.1093/genetics/148.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feuillet C, Keller B. High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han F, et al. Sequence analysis of a rice BAC covering the syntenous barley Rpg1 region. Genome. 1999;42:1071–1076. doi: 10.1139/g99-060. [DOI] [PubMed] [Google Scholar]
- 13.Tikhonov AP, et al. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubcovsky J, et al. Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol. 2001;125:1342–1353. doi: 10.1104/pp.125.3.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chantret N, et al. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops) Plant Cell. 2005;17:1033–1045. doi: 10.1105/tpc.104.029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chantret N, et al. Contrasted microcolinearity and gene evolution within a homoeologous region of wheat and barley species. J Mol Evol. 2008;66:138–150. doi: 10.1007/s00239-008-9066-8. [DOI] [PubMed] [Google Scholar]
- 17.Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 18.Wing RA, et al. The Oryza Map Alignment Project: The golden path to unlocking the genetic potential of wild rice species. Plant Mol Biol. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- 19.Ge S, Sang T, Lu BR, Hong DY. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA. 1999;96:14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brar DS, Khush GS. Alien introgression in rice. Plant Mol Biol. 1997;35:35–47. [PubMed] [Google Scholar]
- 21.Xiao J, et al. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics. 1998;150:899–909. doi: 10.1093/genetics/150.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ammiraju JS, et al. The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 2006;16:140–147. doi: 10.1101/gr.3766306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H, et al. Construction, alignment and analysis of 12 framework physical maps that represent the 10 genome types of the genus Oryza. Genome Biol. 2008;9:R45. doi: 10.1186/gb-2008-9-2-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, et al. New insights into Oryza genome evolution: High gene colinearity and differential retrotransposon amplification. Plant Mol Biol. 2007;64:589–600. doi: 10.1007/s11103-007-9178-3. [DOI] [PubMed] [Google Scholar]
- 25.Kim H, et al. Comparative physical mapping between Oryza sativa (AA genome type) and O. punctata (BB genome type) Genetics. 2007;176:379–390. doi: 10.1534/genetics.106.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J, Wing RA, Bennetzen JL, Jackson SA. Evolutionary history and positional shift of a rice centromere. Genetics. 2007;177:1217–1220. doi: 10.1534/genetics.107.078709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piegu B, et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammiraju JS, et al. Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 2007;52:342–351. doi: 10.1111/j.1365-313X.2007.03242.x. [DOI] [PubMed] [Google Scholar]
- 29.Li X, et al. Control of tillering in rice. Nature. 2003;422:618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- 30.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 31.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carver TJ, et al. ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 33.Zou XH, et al. Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol. 2008;9:R49. doi: 10.1186/gb-2008-9-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennetzen JL, et al. Consistent over-estimation of gene number in complex plant genomes. Curr Opin Plant Biol. 2004;7:732–736. doi: 10.1016/j.pbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Long M, Betran E, Thornton K, Wang W. The origin of new genes: Glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 36.Levine MT, et al. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci USA. 2006;103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Q, et al. On the origin of new genes in Drosophila. Genome Res. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson ME, Lisch DR, Quail PH. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003;34:453–471. doi: 10.1046/j.1365-313x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 40.Ohno S. Evolution by Gene Duplication. London: Springer; 1970. [Google Scholar]
- 41.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cronn RC, Small RL, Wendel JF. Duplicated genes evolve independently after polyploid formation in cotton. Proc Natl Acad Sci USA. 1999;96:14406–14411. doi: 10.1073/pnas.96.25.14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bromham L, Penny D. The modern molecular clock. Nat Rev Genet. 2003;4:216–224. doi: 10.1038/nrg1020. [DOI] [PubMed] [Google Scholar]
- 44.Aggarwal RK, Brar DS, Khush GS. Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet. 1997;254:1–12. doi: 10.1007/s004380050384. [DOI] [PubMed] [Google Scholar]
- 45.Li HW, Chen CC, Wu HK, Lu KCL. In: Rice Genetics and Cytogenetics. Tsunoda S, Takahashi N, editors. Amsterdam: Elsevier; 1964. pp. 118–131. [Google Scholar]
- 46.Dermitzakis ET, Reymond A, Antonarakis SE. Conserved non-genic sequences—an unexpected feature of mammalian genomes. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 47.Guo H, Moose SP. Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell. 2003;15:1143–1158. doi: 10.1105/tpc.010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaplinsky NJ, et al. Utility and distribution of conserved noncoding sequences in the grasses. Proc Natl Acad Sci USA. 2002;99:6147–6151. doi: 10.1073/pnas.052139599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salvi S, et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA. 2007;104:11376–11381. doi: 10.1073/pnas.0704145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inada DC, et al. Conserved noncoding sequences in the grasses. Genome Res. 2003;13:2030–2041. doi: 10.1101/gr.1280703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 53.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 54.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 55.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 56.Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, Allison JB, editors. Mammalian Protein Metabolism. New York: Academic; 1969. pp. 21–123. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.