Abstract
The poor expression of Plasmodium falciparum proteins in heterologous systems and the difficulty in obtaining sufficient material directly from the parasite have limited the experimental characterization of many of the approximately 5200 proteins encoded by this organism. To improve the expression of P. falciparum proteins in the yeast Saccharomyces cerevisiae, we selected yeast ura3 mutants that acquired the ability to utilize the P. falciparum orthologue (PfOMPDC) of URA3 to grow on media lacking uracil. Two of these mutant strains, BY#29 and PJ#17, expressed up to 100-fold more of four P. falciparum proteins as a result of mutations in either HRP1 or KAP104, respectively. These mutations, as well as a temperature-sensitive rna15 mutation, likely decrease the efficiency of mRNA 3’ end formation and produce longer mRNAs of P. falciparum genes. These yeast strains may be useful for the analysis and purification of P. falciparum proteins.
Keywords: Plasmodium falciparum, protein expression, yeast, Saccharomyces cerevisiae
Plasmodium falciparum causes the most severe form of human malaria and is responsible for more than one million deaths each year [1]. Because no effective vaccine for malaria exists and resistance to currently used drugs is increasing, there is an urgent need for the identification of new vaccine and drug targets. Although the availability of the P. falciparum genome sequence [2] has been invaluable for malaria research, the majority of the parasite’s genes have insufficient homology to known proteins to predict their functions. At the experimental level, few genes have been characterized. A small number of P. falciparum proteins have been expressed and purified, but this approach has been hampered by the difficulties in expressing P. falciparum proteins in heterologous organisms or in vitro systems. These difficulties likely stem from unique features of P. falciparum genes, which are AT-rich (74% on average in protein coding regions), and which encode significantly longer proteins than do orthologous genes due to the presence of low complexity inserts consisting primarily of amino acids encoded by AU-rich codons. For E. coli expression, the overabundance of these AU-rich codons prompted the development of the pRIG plasmid, which boosts the levels of tRNAs corresponding to codons that are common in P. falciparum but rare in E. coli [3]. Although some P. falciparum proteins express well by this approach, many are still poorly expressed or insoluble [4, 5].
Alternative approaches to express P. falciparum proteins in heterologous systems have been investigated, with limited success, including the use of orthologous genes from other Plasmodium species or from Cryptosporidium parva [5]; expression in baculoviruses [4], the slime mold Dictyostelium discoideum [6], and wheat germ extracts [7]; and whole gene synthesis, in which the sequence of the gene is altered to match the AT content and codon usage of the heterologous host [4]. Another host cell that has been used to express several P. falciparum genes is the yeast Saccharomyces cerevisiae (for example, [8]. Despite some successes, however, many proteins fail to express in S. cerevisiae, primarily due to the high AT content of P. falciparum genes. In this study, we describe an unbiased selection of S. cerevisiae mutant strains that express the P. falciparum orotidine 5’-monophosphate decarboxylase (PfOMPDC, PF10_0225) protein.
Expression of P. falciparum genes in S. cerevisiae can result in mRNAs that are truncated after AT-rich sequences [8]. These AT-rich sequences resemble S. cerevisiae sequences that encode the positioning and efficiency elements in the 3’ untranslated regions of yeast RNAs that are responsible for specifying the site of polyadenylation [8]. Thus, mRNAs from P. falciparum genes expressed in yeast are often prematurely truncated and unable to synthesize full-length proteins [8]. Since the truncated mRNAs would encode partial open reading frames with no stop codons, we suspected that the yeast mRNA surveillance pathway specifically degrades these RNAs, preventing the accumulation of the encoded proteins [9]. If this interpretation is correct, then mutations in yeast genes required for degradation of aberrant RNAs would be predicted to increase production of truncated P. falciparum proteins. One such gene is SKI2, which encodes an RNA helicase that functions in conjunction with the exosome to degrade aberrant yeast mRNAs [9, 10]. We transformed ski2Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ski2∷KANMX4) and SKI+ yeast strains with P. falciparum protein expression constructs encoding the PfCRK1, PfMRK1 and PfPK6 genes as fusions to the bacterial lexA gene. Western blot analysis (Fig. 1A) demonstrated that P. falciparum fusion proteins were detectable in the ski2Δ mutant, but not in the SKI+ parent, and that the sizes of the proteins detected on the blot were much smaller than the predicted coding capacity of the genes (observed sizes of ~37, 34, and 30 kDa versus predicted sizes of 100, 58, and 59 kDa for the LexA-PfCRK1, LexA-PfMRK1, and LexA-PfPK6 fusion proteins, respectively), indicating that truncated proteins were produced in the ski2Δ strain. This result provides additional support for the hypothesis that premature transcription termination of P. falciparum mRNAs is the primary problem associated with expression of P. falciparum proteins in yeast [8].
Fig. 1.
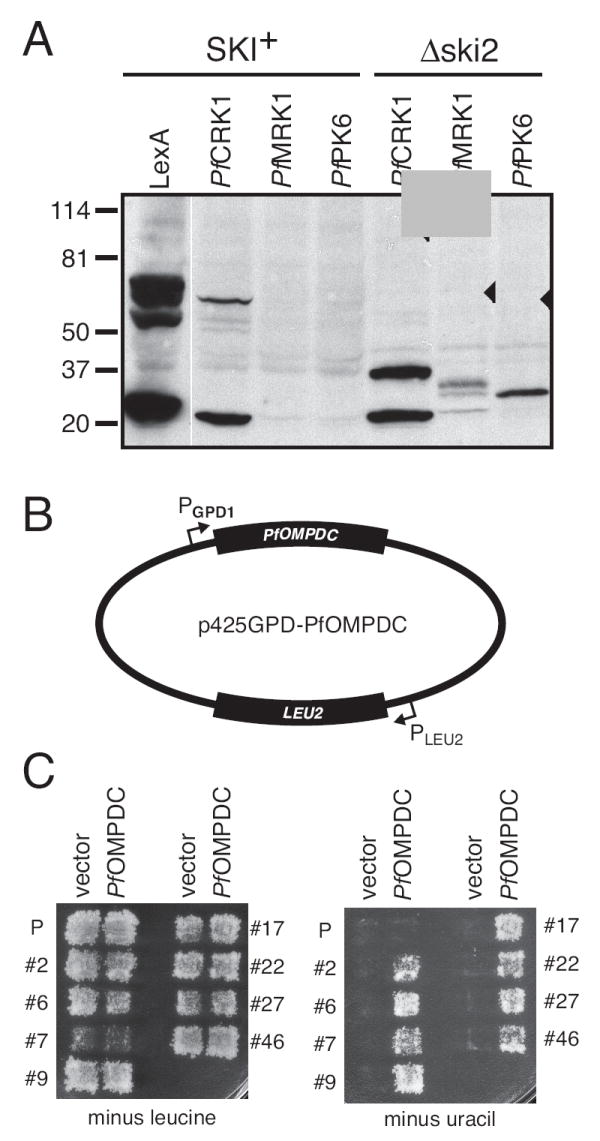
Expression of P. falciparum proteins in mutant yeast strains. A. An S. cerevisiae strain that lacks SKI2 expresses truncated P. falciparum proteins. Plasmids encoding the PfCRK1, PfMRK1, and PfPK6 genes in-frame with the E. coli lexA gene were transformed into BY4741 (SKI+) and ski2Δ yeast strains. Total protein lysates were separated by SDS-PAGE and subjected to immunoblot analysis with an anti-LexA antibody. Molecular weight markers are indicated at left. The predicted sizes of the LexA-PfCRK1, LexA-PfMRK1, and LexA-PfPK6 fusion proteins are 100, 58, and 59 kDa, respectively (indicated by black triangles). The ~26 kDa protein detected in lanes 1 and 4 is the same size as LexA protein produced by the parental LexA expression plasmid (not shown). B. Plasmid p425GPD-PfOMPDC, which encodes the P. falciparum orthologue of the S. cerevisiae URA3 gene. C. Selection of yeast strains that express functional PfOMPDC. The S. cerevisiae strains PJ69-4a [12] and BY4741 [11] were transformed with parental plasmid p425GPD or plasmid p425GPD-PfOMPDC. Yeast harboring either plasmid grew on media lacking leucine (due to expression of the S. cerevisiae LEU2 gene encoded by the plasmids), but did not grow on media lacking uracil. Spontaneous yeast mutants that acquired the ability to express PfOMPDC and grow on media lacking uracil were selected, cured of their plasmids by growth on 5-fluoroorotic acid (5-FOA), and transformed with either p425GPD or p425GPD-PfOMPDC. Growth of a subset of eight mutant strains on media lacking leucine or uracil is shown. “P” indicates the parental yeast strain PJ69-4a.
In an effort to obtain mutant yeast strains with an increased capacity to express full-length P. falciparum proteins, we began by transforming yeast strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) [11] and PJ69-4a (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ) [12] with a high copy number yeast expression construct based on the plasmid p425GPD [13] in which the strong yeast GPD1 promoter drives transcription of PfOMPDC (Fig. 1B). Both BY4741 and PJ69-4a contain a mutated URA3 gene, which renders them unable to grow on media lacking uracil unless a functional URA3 gene is supplied on a plasmid. PfOMPDC catalyzes the same reaction as S. cerevisiae Ura3, and is encoded by a 969 base pair ORF that is 76% AT, approximately the same as the average P. falciparum gene. Although the PJ69-4a parental strain carrying either the LEU2-containing vector or the PfOMPDC expression construct could grow on media lacking leucine, it could not use the PfOMPDC gene to grow on media lacking uracil (Fig. 1C); the same features were true for the BY4741 strain (data not shown). However, by plating a large number of yeast cells, we identified more than 50 spontaneously occurring mutants from each parental strain that grew on media lacking uracil. In general, these strains grew slowly, even on rich media, compared to the parental strains, with doubling times at least twice as long.
A subset of the mutant strains with the fastest growth rates was selected for further analysis. To confirm that the growth on media lacking uracil was due to mutations in the yeast genome and not to changes in the PfOMPDC expression plasmid, we cured the mutant strains of their plasmids by counterselection on 5’-fluoroorotic acid (5-FOA), which kills strains that have Ura3 activity. The resulting strains were re-transformed with empty expression plasmid or plasmid containing the PfOMPDC gene and replica-plated onto media lacking leucine or uracil. For 33 of 38 mutants tested, growth on media lacking uracil required the PfOMPDC expression plasmid (Fig. 1C, showing a representative subset of 8 strains). This result demonstrated that the yeast strains surviving the selection protocol had acquired the ability to transcribe full-length PfOMPDC mRNA and to translate that mRNA into a functional PfOMPDC protein capable of rescuing yeast growth on media lacking uracil.
To identify strains among these yeast mutants that were able to synthesize other P. falciparum proteins in addition to PfOMPDC, we transformed the two parental and 28 mutant strains with yeast expression plasmids for the P. falciparum proteins PfPPJ and PfPK6, each protein constructed as a fusion to the E. coli LexA protein. Immunoblotting with an antibody against LexA showed that neither parental strain expressed LexA-PfPPJ or LexA-PfPK6, whereas 20 mutant strains expressed full-length LexA-PfPPJ, 4 expressed full-length LexA-PK6, and 3 expressed both (Fig. 2A and data not shown). We further evaluated the two fastest growing of these strains, BY#29 and PJ#17, for their ability to express two additional P. falciparum proteins, PfMRK1 and PfMAP1, which could not be detected in parental strains, and found that both strains produced full-length LexA-PfMAP1, but only BY#29 synthesized detectable levels of LexA-PfMRK1 (Fig. 2A). Reprobing each blot with an antibody against S. cerevisiae glucose 6-phosphate dehydrogenase protein indicated equivalent or slight overloading of samples from the parental strains. By serial diluting total cell lysates from BY#29 and PJ#17, we used the blots to estimate that the mutant strains express at least 64- to 128-fold more LexA-PfPPJ than BY4741 and PJ6-4a, respectively (Fig. 2B). These experiments demonstrated that BY#29 and PJ#17 carry mutations that improve the expression of P. falciparum proteins in the absence of selection for the biochemical activity of these proteins.
Fig. 2.
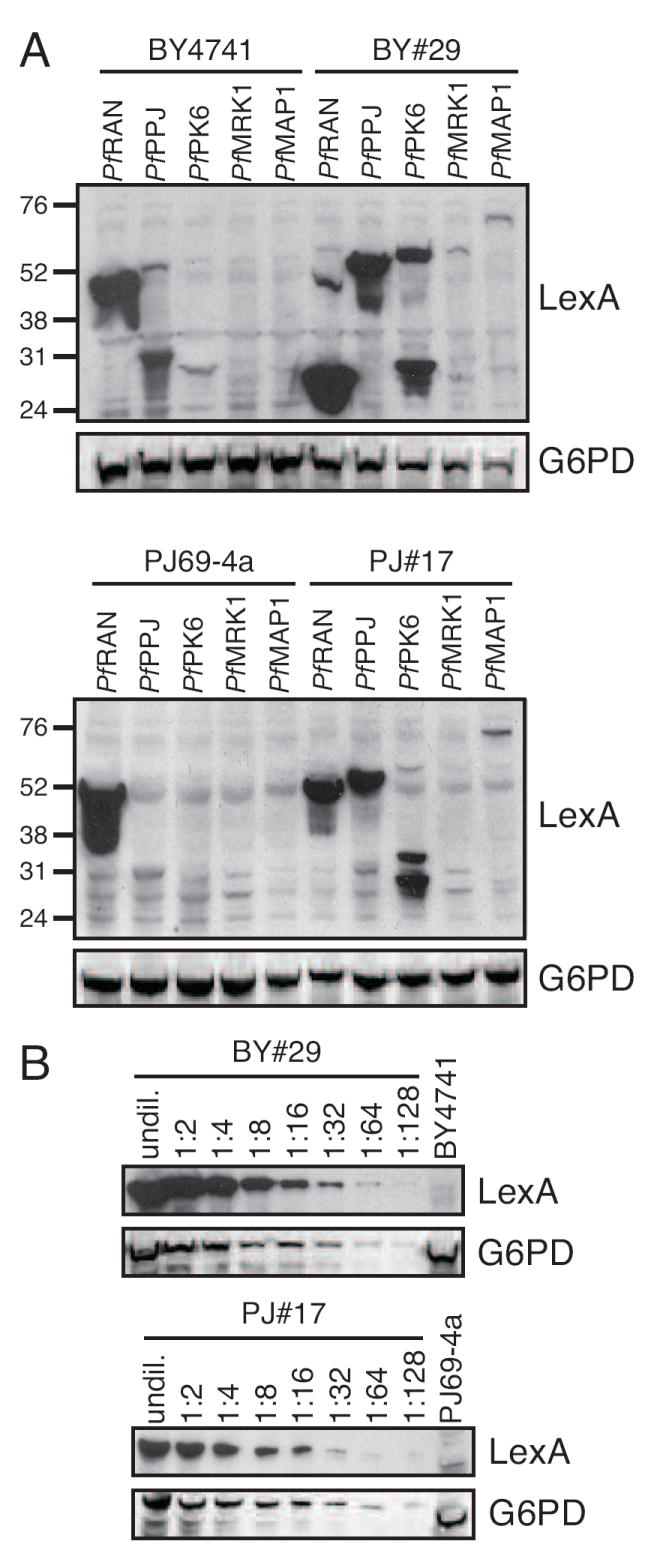
A. Expression of P. falciparum proteins in mutant yeast strains that express PfOMPDC. Total protein lysates were prepared from the parental yeast strains BY4741 and PJ69-4a, and from PfOMPDC-expressing strains BY#29 and PJ#17, transformed with the lexA expression plasmid or plasmids encoding the P. falciparum genes PfRAN, PfPPJ, PfPK6, PfMRK1, and PfMAP1 as fusions with the lexA gene. Lysates were separated by SDS-PAGE and subjected to immunoblot analysis with an anti-LexA antibody. The blots were then reprobed with an antibody against the yeast glucose-6-phosphate dehydrogenase as a loading control. Molecular weight markers are indicated at left. The predicted sizes of the LexA-PfRan, LexA-PfPK6, LexA-PfPPJ, LexA-PfMRK1, and LexA-PfMap1 fusion proteins are 46, 58, 59, 59, and 114 kDa, respectively. B. Estimate of the fold increase in LexA-PfPPJ expression in BY#29 and PJ#17 compared to parental strains. Undiluted and two-fold serial dilutions of total protein lysates from BY#29 and PJ#17 and undiluted total lysate from BY4741 and PJ69-4a expressing PfPPJ were subjected to SDS-PAGE and probed with anti-LexA antibody. Blots were reprobed with an antibody against the yeast glucose-6-phosphate dehydrogenase (G6PD) as a loading control.
To identify the mutations responsible for the improved expression of P. falciparum genes, we rescued the slow-growth phenotype of strains BY#29 and PJ#17 by transforming them with a yeast genomic DNA library [14]. By testing individual genes encoded on inserts of plasmids that restored wildtype growth, we determined that the mutated gene responsible for the slow growth phenotype and the ability to use the PfOMPDC gene to grow on media lacking uracil was HRP1 in strain BY#29, and KAP104 in strain PJ#17 (Fig. 3A and data not shown). Sequencing of the 1602 bp ORF of the HRP1 gene from BY#29 revealed a C to A mutation at position 1592 that converts the Pro at position 531 to Glu. Analysis of the 2754 bp KAP104 ORF from PJ#17 revealed a G to T mutation at position 1777 that results in a stop codon at position 593. We also identified a 114 base pair deletion of nucleotides 1042–1155 in KAP104 in another mutant strain (PJ#27) that generates an in-frame deletion of amino acids 348 to 385.
Fig. 3.
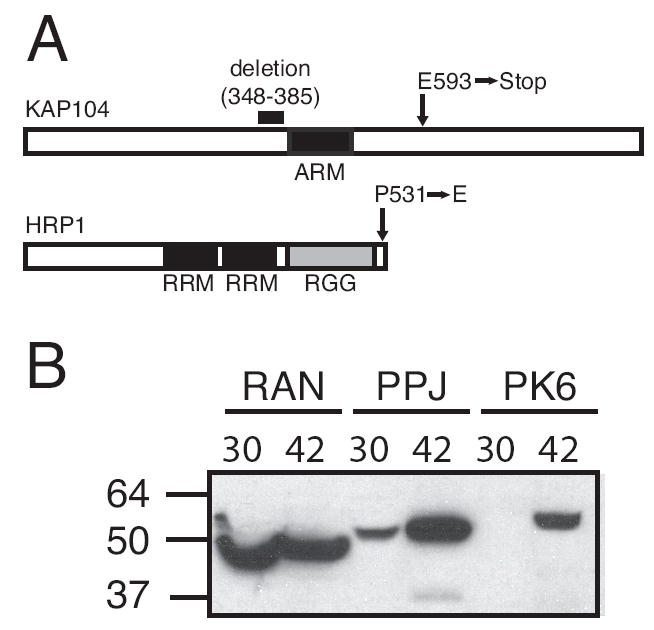
A. S. cerevisiae strains that express functional PfOMPDC have mutations in Hrp1 or Kap104 proteins. To identify the genes responsible for the increased expression of PfOMPDC, BY#29 and PJ#17, which grow slowly, were transformed with a high copy number yeast genomic library [14] and plated on media lacking uracil. Plasmids from large, fast-growing colonies were isolated and sequenced from both ends, which narrowed the chromosomal region harboring the mutated gene to approximately 10 kb on chromosome 15 of BY#29 and to approximately 5.2 kb on chromosome 2 of PJ#17. Candidate genes encoded on the genomic DNA library plasmids were PCR-amplified, cloned into plasmid p424GPD, and transformed into the mutant strains harboring p425GPD-PfOMPDC to determine if expression of the candidate gene restored wildtype growth rate and reversed the ability of the mutant strains to express PfOMPDC. This analysis identified HRP1 and KAP104 as the genes that were mutated in BY#29 and PJ#17, respectively. This figure represents the Kap104 (918 amino acids) and Hrp1 (534 amino acids) proteins as white bars. Black and gray regions indicate protein domains. The positions of mutations in these proteins that confer enhanced expression of P. falciparum genes are indicated by arrows (point mutations) or a horizontal bar (deletion). B. A yeast strain with a temperature-sensitive mutation in RNA15 expresses P. falciparum proteins. The yeast strain CMHy58.1A, which encodes a temperature-sensitive mutation in RNA15 [18], was transformed with parental lexA expression plasmid or plasmids encoding the P. falciparum genes PfRAN, PfPPJ, or PfPK6 as fusions with the LexA gene. Yeast were grown at 25° C to an OD600 of 1.0, and shifted to either 30° or 42° C. Total protein lysates were prepared after 4 h at the elevated temperatures, separated by SDS-PAGE and subjected to immunoblot analysis with an anti-LexA antibody. Molecular weight markers are indicated at left.
The Hrp1 and Kap104 proteins are both involved in mRNA 3’ end processing and transport of polyadenylated RNA out of the nucleus. Hrp1 is both an RNA-binding protein that binds to and shuttles polyA+ RNA out of the nucleus [15] and a component of cleavage stimulation factor 1 (CSF1), a complex that binds to immature mRNAs and specifies the cleavage site at which the polyA tail will be added [16]. Mutations in HRP1 result in defects in mRNA 3’ end formation and accumulation of polyA+ RNA in the nucleus. Kap104 is a nuclear importin β subunit protein whose role is to shuttle polyA+ RNA-binding proteins, including Hrp1, into the nucleus [17]. Mutations in KAP104 disrupt nuclear export of polyA+ RNA by blocking re-import of polyA+ shuttling proteins into the nucleus, which leads to defects in 3’ end formation that result in aberrantly long mRNAs [17, 18]. Both KAP104 and HRP1 are essential for yeast viability, which likely accounts for the slow growth of the mutant strains.
To determine if other strains with mutation in genes required for 3’end formation also improved expression of P. falciparum genes in yeast, we assayed a strain (CMHy58.1A, MATa ura3–52 leu2Δ1 his3DΔ200 rna15–58) with a temperature-sensitive mutation in RNA15 [18]. Rna15 is also a component of CSF1 [19], but does not shuttle in and out of the nucleus and has no known role in RNA transport [18]. CMHy58.1A cells have severe defects in 3’ end formation at both 37° and 42° C [18]. Immunoblot analysis of the expression of three LexA-P. falciparum fusion proteins in CMHy58.1A revealed that expression of LexA-PfRAN was not affected at the nonpermissive temperature, whereas the levels of full-length LexA-PfPPJ and LexA-PfPK6 were significantly increased (Fig. 3B). LexA-PfPPJ was also detected in CMHy58.1A at 30° C (Fig. 3B), suggesting that 3’end formation is also affected at temperatures that are permissive for growth. Consistent with this observation, CMHy58.1A transformed with the PfOMPDC expression plasmid was able to grow slowly on media lacking uracil. These results suggests that defects in 3’ end formation, as opposed to the ability to shuttle in and out of the nucleus, are primarily responsible for the improved expression of P. falciparum genes in BY#29 and PJ#17.
In summary, we used the inability of S. cerevisiae ura3 strains to express the P. falciparum orthologue of the yeast URA3 gene and to grow in the absence of uracil as a selection for yeast mutants with increased expression of the parasite’s proteins. Yeast hrp1 and kap104 mutants produced sufficient PfOMPDC to survive the selection, and also showed increased expression of four other P. falciparum proteins for which no selection was imposed. In addition, an existing temperature-sensitive rna15 mutant with defects in 3’ end processing activity expressed full-length P. falciparum proteins, with the additional feature that it can be grown and transformed at the permissive temperature and shifted to the restrictive temperature when P. falciparum protein production is required.
These data provide additional support for the hypothesis that misinterpretation of AU-rich P. falciparum sequences as mRNA 3’ end processing signals is the primary impediment to expression of P. falciparum genes in S. cerevisiae [8]. In addition, these strains should prove valuable as a heterologous system for P. falciparum protein expression. First, the parasite’s proteins may be produced for purification and biochemical analysis or crystallization. Second, other yeast mutations can be placed into these backgrounds for complementation by P. falciparum genes, enabling subsequent analysis of the encoded proteins. These studies can take advantage of the extensive array of yeast functional genomics tools, as has been done for bacterial proteins (for example, see ref. [20]). Third, yeast can be an effective platform for small molecule screens. For example, pairs of yeast strains expressing a Plasmodium gene and its human orthologue can be screened for small molecules that preferentially inhibit the Plasmodium protein. Finally, PJ#17 is a yeast two-hybrid strain and should improve the results of yeast two-hybrid screens with P. falciparum proteins by enabling the expression of larger fragments from a wider variety of genes.
Acknowledgments
We thank M. Vignali and C. Zhang for assistance with characterization of BY#29, and C. Sibley, W. Van Voorhis, M. Vignali and Y. Zhuang for comments on the manuscript. This work was supported by grant P50 GM64655 from the NIH. S.F. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner MJ, Hall N, Fung E, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baca AM, Hol WG. Overcoming codon bias: a method for high-level overexpression of Plasmodium and other AT-rich parasite genes in Escherichia coli. Int J Parasitol. 2000;30:113–118. doi: 10.1016/s0020-7519(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 4.Mehlin C, Boni E, Buckner FS, et al. Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol Biochem Parasitol. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Vedadi M, Lew J, Artz J, et al. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol Biochem Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 6.van Bemmelen MX, Beghdadi-Rais C, Desponds C, et al. Expression and one-step purification of Plasmodium proteins in dictyostelium. Mol Biochem Parasitol. 2000;111:377–390. doi: 10.1016/s0166-6851(00)00330-3. [DOI] [PubMed] [Google Scholar]
- 7.Mudeppa DG, Pang CK, Tsuboi T, et al. Cell-free production of functional Plasmodium falciparum dihydrofolate reductase-thymidylate synthase. Mol Biochem Parasitol. 2007;151:216–219. doi: 10.1016/j.molbiopara.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Sibley CH, Brophy VH, Cheesman S, et al. Yeast as a model system to study drugs effective against apicomplexan proteins. Methods. 1997;13:190–207. doi: 10.1006/meth.1997.0511. [DOI] [PubMed] [Google Scholar]
- 9.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 10.Anderson JS, Parker RP. The 3’ to 5’ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3’ to 5’ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 14.Shimogawa MM, Graczyk B, Gardner MK, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry M, Borland CZ, Bossie M, Silver PA. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessler MM, Henry MF, Shen E, et al. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3’-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 18.Hammell CM, Gross S, Zenklusen D, et al. Coupling of termination, 3’ processing, and mRNA export. Mol Cell Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minvielle-Sebastia L, Preker PJ, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3’-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 20.Alto NM, Shao F, Lazar CS, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]


