Abstract
Distinguishing self from non-self is a fundamental biological challenge. Many pathogens exploit the challenge of self discrimination by employing mimicry to subvert key cellular processes including the cell cycle, apoptosis, and cytoskeletal dynamics1-5. Other mimics interfere with immunity6, 7. Poxviruses encode K3L, a mimic of eIF2α, which is the substrate of Protein Kinase R (PKR), an important component of innate immunity in vertebrates8, 9. The PKR-K3L interaction exemplifies the conundrum imposed by viral mimicry. To be effective, PKR must recognize a conserved substrate (eIF2α) while avoiding rapidly evolving substrate mimics like K3L. Using the PKR-K3L system and a combination of phylogenetic and functional analyses, we uncover evolutionary strategies by which host proteins can overcome mimicry. We find that PKR has evolved under dramatic episodes of positive selection in primates. The ability of PKR to evade viral mimics is partly due to positive selection at sites most intimately involved in eIF2α recognition. We also find that adaptive changes on multiple surfaces of PKR produce combinations of substitutions that increase the odds of defeating mimicry. Thus, while it can appear that pathogens gain insurmountable advantages by mimicking cellular components, host factors like PKR can compete in molecular ‘arms races’ with mimics because of remarkable evolutionary flexibility at protein interaction interfaces challenged by mimicry.
List of key genes/proteins: Protein kinase R (PKR; EIF2AK2), eukaryotic translation initiation factor 2α (eIF2α; EIF2S1), K3L, PKR-like ER kinase (PERK; EIF2AK3), GCN2 (EIF2AK4), heme-regulated inhibitor (HRI; EIF2AK1)
To counteract viral infections, PKR phosphorylates the translation initiation factor eIF2α in the presence of double-stranded RNA (dsRNA) from viruses8, 9. This activity strongly inhibits protein synthesis and blocks the production of new virus particles. The crucial role for PKR in innate immunity is reflected by the evolution of numerous factors from diverse viruses that disable PKR to promote viral production10, including a poxvirus-encoded mimic of eIF2α called K3L (Figure S1). Host proteins like PKR that directly interact with viral antagonists like K3L can be subject to molecular ‘arms-races’ where amino acid substitutions that directly affect interactions can be rapidly fixed by positive selection11, 12.
To determine if PKR might be subject to positive selection, we cloned and sequenced cDNA of PKR from a panel of 20 primates representing over 30 million years of evolutionary divergence. By considering ratios of non-synonymous (dN) and synonymous (dS) substitutions, we found evidence for ancient, episodic positive selection in primate lineages (p<0.0003; Table S1, Figure 1a). In particular, one branch in Old World monkeys was calculated to have undergone 22 non-synonymous substitutions without any synonymous changes, one of the most intense episodes of positive selection reported for any primate gene (Supplementary data). Likelihood ratio tests13 using the entire phylogeny reveal that 17% of codons have evolved with an average dN/dS ratio of 3.7, strongly supporting a finding of positive selection (p<0.0001, Table S2 and S3), even after accounting for the potentially confounding effects of recombination and synonymous site variation14 (p<0.0001; Table S4 and S5). Positive selection is observed in each of the three domains of PKR: the dsRNA binding domain, the spacer region, and even the kinase domain, consistent with an extensive history of facing viral factors that directly bind PKR in these separate domains (Figure S1). Interestingly, several residues in the kinase domain, which make direct contacts with eIF2α15, are among the fastest evolving residues in PKR (Figure 1b and S1), suggesting that selective pressure to evade eIF2α mimics may have driven changes in these residues.
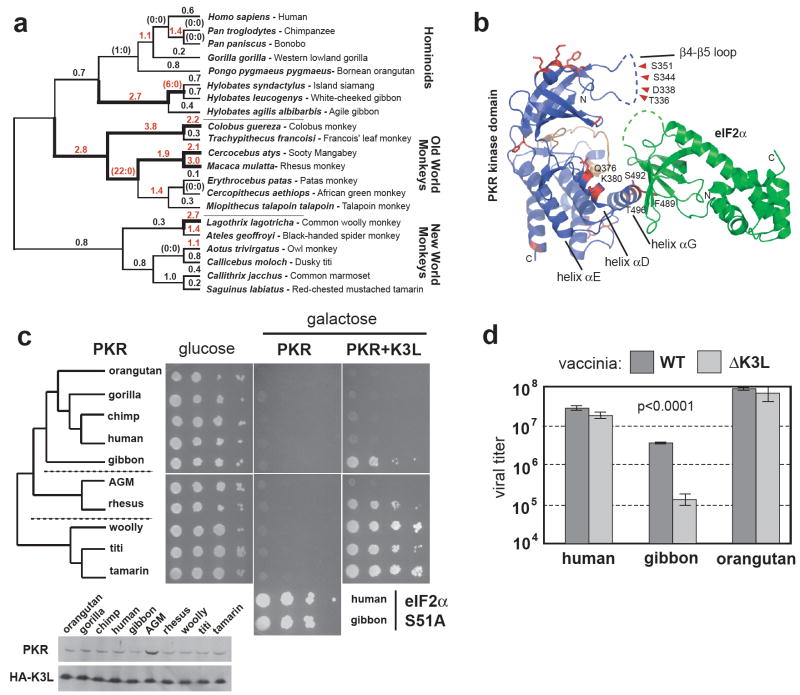
Figure 1. Widespread positive selection shaped PKR throughout primate evolution.
(a) PKR was sequenced from simian primates that together represent more than 30 million years of divergence. dN/dS values along each branch of the phylogeny are listed, and those with dN/dS>1 are highlighted in red. Branches with bold lines, overlapping the set in red, indicate lineages found to be under positive selection by complementary model fitting analysis (also see Table S6). Values in parentheses are shown for branches where no synonymous changes were observed (S=0) and indicate the number of non-synonymous changes (N).
(b) Sites under positive selection (red) are mapped onto a ribbons representation of the PKR kinase domain (blue) / eIF2α (green) complex (PDB code: 2A1A)15. The active site of PKR is shown in orange and a large portion of the β4-β5 loop (dashed blue line) is invisible from the structure deduced from the co-crystal for technical reasons15. Residues under positive selection near the interface of PKR with eIF2α and K3L are noted in the β4-β5 loop (Thr336, Asp338, Ser344, Ser351) and the αD (Gln376, Lys380) and αG (Phe489, Ser492, Thr496) helices.
(c) Plasmids encoding PKR variants from a panel of primates under pGal were introduced into yeast strains HM3 (eIF2α), HM2 (eIF2α and HA-vaccinia K3L), and J223 (eIF2α-S51A). Ten-fold serial dilutions of transformants were spotted on plates containing either glucose or galactose (see Full Methods). Immunoblot analysis of PKR (top panel) and HA-K3L (bottom panel) is also shown (see Full Methods). For AGM, resistance to K3L might reflect differences in PKR expression in yeast.
(d) Primary fibroblasts from the indicated primates were infected with WT or ΔK3L vaccinia virus in triplicate (moi=0.001). Virus production was assessed three days post infection by titering cell lysates. The significance of WT versus ΔK3L is indicated (Student’s t-test; bars show s.d.). Minor variations of this experiment (not shown) revealed that ΔK3L infections typically produced ~5-fold less virus than wildtype virus in gibbon cells.
Similarly, we find that positive selection has acted on the eIF2α mimic K3L (Figure S2). For instance, in a comparison of K3L from variola major (smallpox) and vaccinia viruses, we find a dN/dS of 2.80 (p<0.001), whereas fewer than 10% of orthologs in vaccinia and variola comparisons show any evidence of positive selection (average dN/dS = 0.10, Elde and Malik, unpublished data). This suggests that poxviral eIF2α mimics have also undergone positive selection and reflects the possibility that K3L has not achieved or maintained an optimal state of mimicry. Instead, K3L might continually evolve to counter adaptive changes in PKR.
In contrast to the rapid evolution of PKR, its substrate, eIF2α, is essentially unchanged in simian primates at the amino acid level (dN/dS = 0 comparing human and rhesus). Thus, PKR must recognize an unchanging substrate while evolving to discriminate against mimics like K3L to be effective. Considering that most viruses, including poxviruses16, 17, evolve at faster rates than primates, such challenges by mimics are daunting for hosts. Nevertheless, PKR can inhibit viruses encoding eIF2α mimics10, suggesting that adaptive changes in PKR might help overcome mimicry by these factors.
We investigated whether primate PKR orthologs differ in ability to discriminate against K3L from vaccinia, the model poxvirus. Because the entire clade of extant poxviruses is very young relative to the divergence between primates16, 17, we could not investigate strict co-evolutionary dynamics between PKR and K3L. Instead, we used vaccinia K3L as a means to study the evolutionary strategies afforded PKR for counteracting substrate mimics faced over the course of primate evolution, which could leave PKR alleles either susceptible or resistant to vaccinia K3L. Even though primate PKR alleles did not necessarily evolve against vaccinia K3L, our approach allowed us to identify the mechanisms by which host proteins might defeat mimicry more generally. Examining host-virus evolution from a similar perspective led to the identification of a region in the restriction factor Trim5α that confers specificity against ancient, extinct retroviruses, but fails to protect humans from HIV18, 19.
A growth assay in yeast has provided a facile test of PKR function20. Human PKR recognizes and phosphorylates yeast eIF2α, owing to its high level of similarity to primate eIF2α, to cause growth arrest15, 21. We expressed 10 divergent primate PKR cDNAs in yeast to determine if they differed in ability to phosphorylate eIF2α. All primate PKR genes tested caused consistent levels of growth arrest, which specifically depended on phosphorylation of eIF2α22 (Figure 1c, middle panel). However, co-expression with vaccinia virus K3L uncovered dramatic differences in K3L inhibition of primate PKR orthologs, which leads to a rescue of growth23 (Figure 1c, right panel). PKR alleles from Old and New World monkeys, and from white-cheeked gibbon were generally quite susceptible to suppression of growth arrest by K3L from vaccinia and variola, whereas other hominoid PKR alleles showed only modest suppression by K3L (Figure 1c and S3). Thus, rapid evolution of primate PKR did not appear to significantly alter eIF2α recognition, but resulted in considerable differences in susceptibility to K3L. In particular, we find in the hominoid lineage that human, chimp, gorilla, and orangutan PKR orthologs are 1000-fold more resistant to growth rescue by K3L than gibbon PKR.
We further corroborated the large differences in K3L susceptibility uncovered by the yeast assay by infecting human, orangutan and gibbon fibroblast cell lines with either wildtype vaccinia virus or a strain with a K3L gene deletion (ΔK3L). Consistent with our yeast assays and previous reports in human cells24, we found that ΔK3L virus had no significant effect on viral titer in human or orangutan cells but led to a substantial drop in titer in gibbon cells (Figure 1d). Vaccinia virus therefore depends on K3L for full infectivity in gibbon cells, where PKR is susceptible to K3L.
We wished to map critical genetic differences between ‘resistant’ and ‘susceptible’ PKR alleles to understand the basis of K3L resistance. We first investigated helix αG of the kinase domain because residues 489, 492, and 496 play key roles in recognition of eIF2α15, yet have evolved under recurrent positive selection (Figure 1b and 2). While gibbon PKR (helix αG: Tyr489-Ala492-Thr496 or Y-A-T) is susceptible to K3L in growth assays, the human αG configuration (F-S-T) in an otherwise gibbon PKR backbone increases gibbon PKR resistance to vaccinia K3L (Figure 2a, rows 1 and 2). In fact, the A492S substitution (Y-S-T) alone confers to gibbon PKR greatly increased resistance to K3L (Figure 2a, row 3). These findings reveal that even a single change in PKR at the common interface with substrate and mimic has the capacity to reverse a ‘susceptibility’ phenotype.
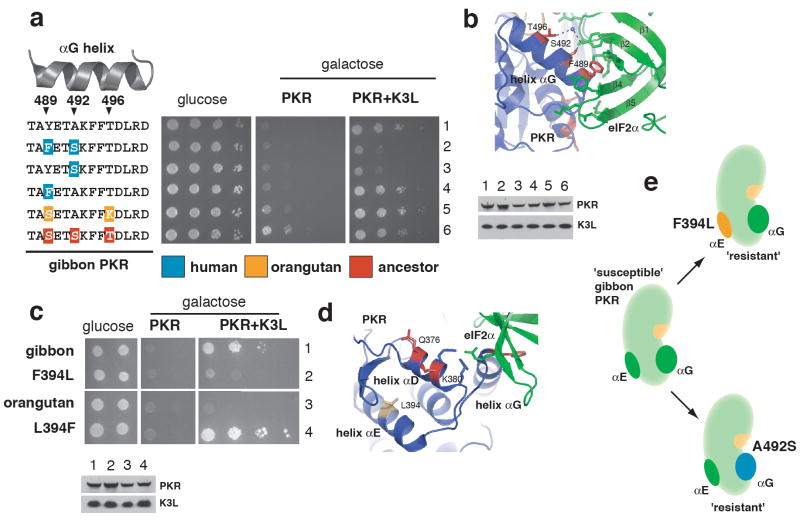
Figure 2. Distinct surfaces of the PKR kinase domain are critical to K3L resistance.
(a) Plasmids encoding gibbon PKR alleles with substitutions in the αG helix were introduced into yeast strains HM3 (eIF2α alone) and HM1 (eIF2α and K3L). Ten-fold serial dilutions of transformants are shown. Corresponding immunoblot analysis is also shown using antibodies against PKR (top panel) and K3L (bottom panel).
(b) A ribbon representation of the PKR/eIF2α complex highlighting the association of side chains of residues under positive selection with side chains of eIF2α. Phe489, Ser492, and Thr496 form a face of the αG helix directly interacting with eIF2α15.
(c) Plasmids encoding gibbon and orangutan PKR alleles with substitutions in the αE helix were introduced into yeast strains HM3 and HM1. Ten-fold serial dilutions of transformants are shown along with corresponding immunoblot analysis.
(d) Residues under positive selection (Gln376 and Lys380) and residue Leu394 from a ribbon representation of human PKR and eIF2α are shown15.
(e) A schematic depicting that single substitutions in either the αE and αG helices can confer resistance against vaccinia K3L to gibbon PKR.
Surprisingly, a second determinant, not in helix αG, explains the resistance of orangutan PKR to K3L. When we tested the αG configuration (S-A-K) of the ‘resistant’ orangutan PKR allele (Figure 1c) in the gibbon backbone, this S-A-K allele was still quite susceptible (Figure 2a, row 5). To identify the source of orangutan PKR resistance, we tested chimeras between orangutan and gibbon PKR and found that a region in the kinase domain containing helices αD and αE from orangutan PKR greatly increased the resistance of gibbon PKR to K3L (data not shown). When we tested individual substitutions in this region, we found that the F394L substitution of the αE helix was responsible for conferring resistance to gibbon K3L (Figure 2c). Importantly, the opposite L394F substitution greatly reduced resistance in orangutan PKR (Figure 2c). Unlike helix αG, helix αE discrimination appears independent of PKR contact with its substrate because it is positioned away from the eIF2α interface (Figure 2d)15. In addition, positive selection in helix αD suggested that this region could contribute to escaping mimicry, either directly or by virtue of co-evolution between helix αD and αG (Table S9)25. However, we did not find functional evidence for a role for αD over the evolutionary timeframe we examined for this particular mimic (Figure S4). Therefore, susceptible gibbon PKR alleles can gain resistance to vaccinia K3L by single substitutions in either the αG or αE helices (Figure 2e), increasing the chances of escaping mimicry.
Our analyses suggested that human PKR contained residues associated with increased resistance to K3L from both αG and αE helices. Indeed, we found that a human PKR allele carrying ‘susceptible’ mutations in both its αE (L394F) and αG (F489Y/S492A) helices loses wildtype resistance to K3L (Figure 3a, row 5). We tested all combinations of resistant and susceptible substitutions at positions 394 (helix αE), 489 and 492 (helix αG) in human PKR and found that six out of eight combinations of human PKR alleles resist K3L. The two exceptions are F-Y-A (described above) and F-F-A (Figure 3a, row 4), which is only slightly more resistant to K3L than F-Y-A, revealing a weak effect associated with the positively selected residue at position 489. While the human and gibbon PKR backbones bear similar outcomes at all positions (Figures 3b), the ‘susceptible’ human alleles still appear more resistant to vaccinia K3L than the ‘susceptible’ gibbon alleles, hinting at an additional K3L resistance determinant in the human PKR sequence (data not shown).
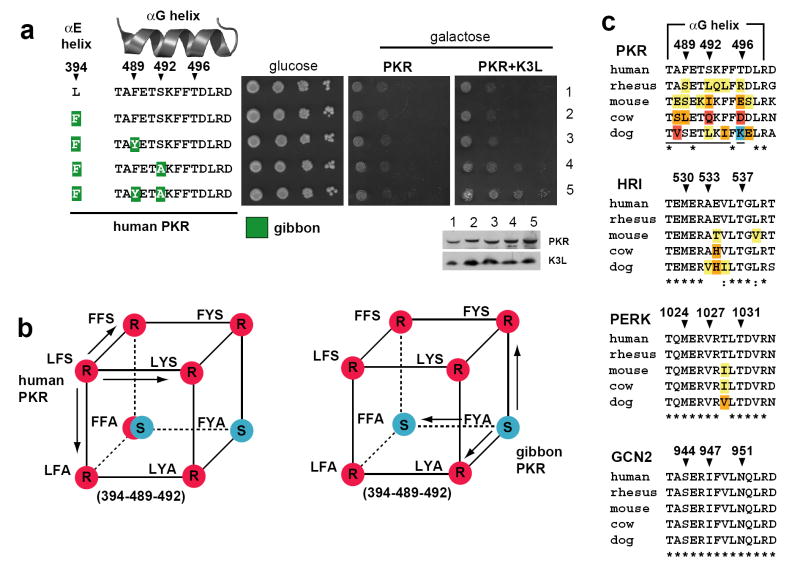
Figure 3. PKR chimeras reveal masking of K3L sensitivity by Leu394.
(a) Ten-fold serial dilutions of transformants expressing alleles of human PKR with combinations of substitutions in the αE and αG helices are shown along with corresponding immunoblot analysis.
(b) Phenotype ‘cubes’ summarizing the K3L susceptibility of alleles with all combinations of substitutions between human and gibbon PKR at positions 394, 489, and 492 from Figures 2a, 3a and S5. Red and blue dots indicate resistance and sensitivity to K3L respectively. With the exception of F-F-A, which shows some measure of resistance to K3L in the human background (indicated by the red crescent), each set of substitutions have similar phenotypes in the human and gibbon backgrounds. Each single substitution in wildtype human PKR results in a variant still resistant to K3L, while in two of three cases gibbon PKR becomes resistant (indicated by arrows).
(c) Sequence alignments of the αG helix for each member of the eIF2α kinase family from several mammals highlights the conservation of this region compared to rapid evolution of PKR (black arrowheads indicate residues of the αG helix under positive selection in PKR). The frequency of substitutions among the panel at each position is indicated by a color code (yellow for a single substitution, orange for a second, red for a third, and blue for a fourth) with human sequence as a reference. Residues making contacts with eIF2α are indicated with lines below the PKR alignment.
One of the most notable findings from testing the ‘susceptible’ and ‘resistant’ PKR variants was that helices αE and αG had distinct roles in defeating K3L. Leu394 resisted K3L regardless of whether human, gibbon, or orangutan PKR had a ‘susceptible’ αG helix (Figure S5 and 2c). Thus, the mutational profile of the αE and αG helices is not strictly independent, because helix αE masks αG in terms of K3L resistance. Only in cases where helix αE is ‘susceptible’ does the configuration of αG matter. Interestingly, residue 394 of helix αE toggles exclusively between leucine and phenylalanine at a much slower rate than residues of helix αG, not only in primates, but also among mammals in general (Figure S6). Our finding that Leu394 confers overriding resistance to vaccinia K3L strongly suggests that toggling can unmask potentially adaptive substitutions in the rapidly evolving αG helix. The fact that Phe394 is fixed in numerous species, including the new world monkeys we sampled, suggests that phenylalanine rather than leucine might confer resistance against substrate mimics different from the two we tested in this study. Therefore, toggling at position 394 reveals how a single substitution, in combination with positive selection in helix αG, might effectively increase the adaptive space PKR can explore, greatly increasing the odds of defeating substrate mimics.
Positive selection appears to be a major evolutionary driver of many host-pathogen interactions11, 18, 26. Dramatic positive selection seen in both primate PKR and poxvirus K3L, and the presence of substrate mimics in unrelated viruses27, clearly points to the fact that both host and viral genomes have been under intense pressure to gain advantages in these ancient and ongoing evolutionary battles. The positive selection we observed in primate PKR is likely to reflect selection driven by ancient viruses with K3L-like factors that strongly influenced susceptibility to present-day mimics. For example, positive selection in the gibbon lineage driven by ancient mimics may have left gibbon PKR susceptible to vaccinia K3L. Similar trade-offs have been observed for variants of antiviral proteins under strong positive selection that might have defeated ancient retroviruses, but are currently susceptible to HIV-119.
Mimicry adds a layer of complexity to host-pathogen interfaces. Because PKR must distinguish an essentially unchanging substrate from rapidly evolving mimics like K3L, it is surprising that most present-day hominoid species are resistant to vaccinia K3L (Figure 1c). Our studies reveal evolutionary mechanisms that might allow host genes like PKR to stave off mimicry. This strategy involves not only positive selection, but also multiple discrimination interfaces (αE and αG helices) and a combinatorial outcome of resistance or susceptibility based on these surfaces, which together can increase discrimination against rapidly evolving mimics.
PKR appears well suited for molecular arms races against mimics due to a striking level of evolutionary flexibility. Because the biochemical activity of PKR depends on recognition of an unchanging substrate, strong purifying selection at the interaction interface would be expected. Indeed, other members of the eIF2α kinase family, which do not primarily serve antiviral roles and are not known to directly encounter viral mimicry, have highly conserved αG helices (Figure 3c) and evolve under purifying selection (dN<dS, Figure S1). Despite extensive amino acid diversity in helix αG, variants of PKR retain the ability to recognize eIF2α. The contrasting evolutionary trajectories of helix αG in the family of eIF2α kinases suggests that host factors challenged by mimics, like PKR, rely on a high degree of flexibility to escape mimicry. We speculate that substantial selective pressures for distinguishing substrate mimics may even result in substitutions causing a reduction in substrate recognition until potential compensatory mutations might arise. Consistent with this scenario, introducing an ancestral helix αG or one from orangutan into PKR from gibbon results in slightly compromised substrate recognition (Figure 2a, rows 5 and 6, middle panel; also see Figure S7, middle panel), yet full substrate recognition is restored for helix αG from orangutan in the context of the whole protein (Figure 1c, orangutan, middle panel). Compromising one function to explore a greater adaptive landscape for another function is likely a theme for genetic gains of functional novelty28, 29. Because contending with viral mimicry can be essential for combating infectious disease, compromises to components of key cellular processes targeted by mimics1-5, 30 might be a ‘hidden’ evolutionary cost of such high-stakes genetic conflicts.
Methods Summary
Details of phylogenetic and other evolutionary analyses18, vaccinia infection experiments24, genotypes of yeast strains and yeast growth assays15, 21 are presented in the Full Methods accompanying this paper.
Supplementary Material
Supplementary information accompanies this paper.
Acknowledgments
We thank Tom Dever (NIH) for providing yeast strains and advice on the yeast growth assays, Jim Tartaglia (Sanofi Pasteur) for K3L antibody, Bertram Jacobs (Arizona State Univ.) for wildtype and ΔK3L viruses, and Sue Biggins and Suzanne Furuyama for yeast expression plasmids and advice. We are also grateful to Michael Emerman, Steve Henikoff, Sue Biggins, Aaron Turkewitz, Dan Gottschling, Doug Koshland, Eric Smith, Julie Kerns, Sara Sawyer, and Danielle Vermaak for their comments and suggestions. We are supported by NIH grant AI026672 (A.P.G.) and a Searle Scholar and Burroughs Wellcome Investigator Award (H.S.M.). N.C.E. is an Ellison Medical Foundation Fellow of the Life Sciences Research Foundation.
Footnotes
Sequences of PKR have been deposited in Genbank under accession numbers EU733254-EU733271. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to H.S.M. (hsmalik@fhcrc.org).
References
- 1.Murphy PM. Molecular mimicry and the generation of host defense protein diversity. Cell. 1993;72:823–6. doi: 10.1016/0092-8674(93)90571-7. [DOI] [PubMed] [Google Scholar]
- 2.Angot A, Vergunst A, Genin S, Peeters N. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 2007;3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benedict CA, Norris PS, Ware CF. To kill or be killed: viral evasion of apoptosis. Nat Immunol. 2002;3:1013–8. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- 4.Izard T, Tran Van Nhieu G, Bois PR. Shigella applies molecular mimicry to subvert vinculin and invade host cells. J Cell Biol. 2006;175:465–75. doi: 10.1083/jcb.200605091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stebbins CE, Galan JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–5. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 6.Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 7.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–82. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 8.Meurs E, et al. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–90. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 9.Dever TE, Dar AC, Sicheri F. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2007. pp. 319–344. [Google Scholar]
- 10.Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–10. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 14.Scheffler K, Martin DP, Seoighe C. Robust inference of positive selection from recombining coding sequences. Bioinformatics. 2006;22:2493–9. doi: 10.1093/bioinformatics/btl427. [DOI] [PubMed] [Google Scholar]
- 15.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, et al. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci U S A. 2007;104:15787–92. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babkin IV, Shchelkunov SN. The time scale in poxvirus evolution. Mol Biol (Mosk) 2006;40:20–4. doi: 10.1134/s0026893306010043. [DOI] [PubMed] [Google Scholar]
- 18.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102:2832–7. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316:1756–8. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 20.Chong KL, et al. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. Embo J. 1992;11:1553–62. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey M, et al. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–13. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 22.Dever TE, et al. Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci U S A. 1993;90:4616–20. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2alpha. Mol Cell Biol. 1997;17:4146–58. doi: 10.1128/mcb.17.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langland JO, Jacobs BL. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology. 2002;299:133–41. doi: 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- 25.Poon AF, Lewis FI, Pond SL, Frost SD. An evolutionary-network model reveals stratified interactions in the V3 loop of the HIV-1 envelope. PLoS Comput Biol. 2007;3:e231. doi: 10.1371/journal.pcbi.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Essbauer S, Bremont M, Ahne W. Comparison of the eIF-2alpha homologous proteins of seven ranaviruses (Iridoviridae) Virus Genes. 2001;23:347–59. doi: 10.1023/a:1012533625571. [DOI] [PubMed] [Google Scholar]
- 28.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–8. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–88. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawyer SL, Malik HS. Positive selection of yeast nonhomologous end-joining genes and a retrotransposon conflict hypothesis. Proc Natl Acad Sci U S A. 2006;103:17614–9. doi: 10.1073/pnas.0605468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information accompanies this paper.