Abstract
The silencing mediator for retinoid and thyroid hormone receptors (SMRT) serves as a platform for transcriptional repression elicited by several steroid/nuclear receptors and transcription factors. SMRT exists in two major splicing isoforms, α and τ, with SMRTα containing only an extra 46-amino acid sequence inserted immediately downstream from the C-terminal corepressor motif. Little is known about potential functional differences between these two isoforms. Here we show that the pregnane X receptor (PXR) interacts more strongly with SMRTα than with SMRTτ both in vitro and in vivo. It is interesting that the PXR-SMRTα interaction is also resistant to PXR ligand-induced dissociation, in contrast to the PXR-SMRTτ interaction. SMRTα consistently inhibits PXR activity more efficiently than does SMRTτ in transfection assays, although they possess comparable intrinsic repression activity and association with histone deacetylase. We further show that the mechanism for the enhanced PXR-SMRTα interaction involves both the 46-amino acid insert and the C-terminal corepressor motif. In particular, the first five amino acids of the SMRTα insert are essential and sufficient for the enhanced binding of SMRTα to PXR. Furthermore, we demonstrate that Tyr2354 and Asp2355 residues of the SMRTα insert are most critical for the enhanced interaction. In addition, expression data show that SMRTα is more abundantly expressed in most human tissues and cancer cell lines, and together these data suggest that SMRTα may play a more important role than SMRTτ in the negative regulation of PXR.
Transcriptional regulation is a dynamic process involving both association and dissociation of the transcription factor with various coactivators and corepressors. One well investigated system is the prominent effects of coactivators and corepressors on the transcriptional activity of steroid/nuclear hormone receptors (NRs) (Westin et al., 2000). The silencing mediator of retinoid and thyroid hormone receptors (SMRT) (Chen and Evans, 1995; Ordentlich et al., 1999; Park et al., 1999) and the nuclear receptor corepressor (N-CoR) (Horlein et al., 1995) are two related corepressors known to mediate repression by several unliganded NRs through the recruitment of histone deacetylases (HDACs) (Nagy et al., 1997; Guenther et al., 2000). These corepressors are believed to act as protein platforms for the assembly of corepressor complexes necessary for transcriptional repression. Transcription repression is an important genomic event involved in many physiological processes such as development, homeostasis, cell growth, and differentiation (Privalsky, 2004).
In the absence of ligand, SMRT and N-CoR bind to the unliganded receptor through their NR-interacting domains (IDs) (Hu and Lazar, 1999; Ghosh et al., 2002). Upon ligand binding, the receptors undergo a conformational change, leading to the alteration of the corepressor-binding pocket that causes the release of corepressors and recruitment of coactivators. The human pregnane X receptor (PXR, also known as SXR and PAR) is a promiscuous sensor for several xenobiotic compounds (Watkins et al., 2001), and it binds to a diverse group of endogenous and exogenous ligands (Synold et al., 2001; Moore et al., 2003). PXR directly activates a subset of genes involved in drug metabolism (Xu et al., 2002). Therefore, drugs that activate PXR are likely to cause a higher risk of drug-drug interactions (Harmsen et al., 2007; Urquhart et al., 2007; Wipf et al., 2007).
The ability of PXR to regulate gene expression depends on its ability to form heterodimers with the retinoid X receptor (RXR) and bind to several PXR response elements. Several PXR response elements are found within the CYP3A promoters configured as direct repeats separated by three nucleotides (Kliewer et al., 1998), everted repeats separated by six nucleotides (Lehmann et al., 1998), or inverted repeats separated by eight nucleotides (Kast et al., 2002). In addition, the PXR-RXR heterodimers also bind tightly to several natural DR4 (direct repeats separated by four nucleotides)-type response elements. These include a DR4 motif in the intestinal multidrug resistance gene promoter responsible for its induction by rifampin (Geick et al., 2001) and a similar motif in the nitric-oxide synthase promoter responsible for its induction by clotrimazole (Toell et al., 2002), both through the PXR-RXR heterodimers. PXR-RXR heterodimers also consistently bind well to synthetic AG(G/T)TCA repeats of different spacing with a preferred affinity toward a DR4 element (Blumberg et al., 1998). Once activated, the PXR-RXR heterodimer recruits transcriptional coactivators such as the p160 proteins (Leo and Chen, 2000) to form a multiprotein complex to activate transcription (Kliewer et al., 1998). In addition, PXR can also cross-talk with other NR response elements, including those recognized by the constitutive androstane receptor (CAR) (Muangmoonchai et al., 2001; Kodama et al., 2004) and the antioxidant response element on the rat glutathione transferase A2 gene (Falkner et al., 2001).
SMRT is known to exist in cells as at least two major splicing isoforms: α and τ (Goodson et al., 2005). Compared with SMRTτ, SMRTα contains an extra small exon encoding a 46-amino acid sequence inserted after residue Gly2352, immediately downstream to the distal corepressor motif (ID2, residues 2342-2350). SMRTα and SMRTτ reportedly interact with thyroid hormone receptors (TRs) with different affinities (Goodson et al., 2005); however, the molecular mechanism of such a differential affinity remains unknown. SMRTτ has also been shown to interact directly with and regulate the transcriptional activity of PXR in a PXR ligand-sensitive manner (Johnson et al., 2006; Wang et al., 2006); however, it was unknown whether and how PXR might interact with SMRTα. In this study, we compared the binding affinities of SMRTα and SMRTτ toward several NRs, with a focus on PXR. We found that, in contrast to other NRs, PXR uniquely displayed a preferential binding toward SMRTα. It is interesting that this SMRTα interaction is resistant to PXR ligand-induced dissociation, and SMRTα elicited a greater inhibition on PXR activity than SMRTτ. It is noteworthy that we also uncovered critical residues in SMRTα that are responsible for its higher affinity toward PXR and showed that SMRTα is the dominant form expressed in most surveyed human tissues and cancer cells.
Materials and Methods
Chemicals. Rifampicin (Rif), clotrimazole (CTZ), and pregnenolone-16α-carbonitrile (PCN) were purchased from Sigma (St. Louis, MO). The rabbit anti-HA and mouse anti-FLAG antibodies were purchased from MBL International (Woburn, MA) and Stratagene (La Jolla, CA), respectively. All other reagents, including culture media for bacteria, yeast, and mammalian cells, were purchased from standard sources.
Plasmids. The expression vectors pGEX-SMRTτ S1/2 (aa 2077-2471) and pGEX-SMRTα S1/2 (aa 2077-2517) were as described previously (Goodson et al., 2005) and were kindly provided by Dr. Martin Privalsky. The pCMX-FLAG-cSMRTτ (2095-2471) and pCMX-FLAG-cSMRTα (2095-2517) were constructed by subcloning the Hind III to Nhe1 fragments of pGEX-SMRTτ S1/2 and pGEX-SMRTα S1/2 into the pCMX-FLAG vector, respectively. The SMRT ID1 (aa 2107-2187), SMRTτ ID2 (aa 2284-2379), SMRTα ID2 (aa 2284-2425), SMRTτ ID1-2 (aa 2107-2379), and SMRTα ID1-2 (aa 2107-2425) fragments were generated by PCR reactions with pfu polymerase (New England Biolabs, Ipswich, MA) and subcloned into various plasmid vectors. The full-length pCMX-F-SMRTτ and pCMX-F-SMRTα were constructed by assembling the Asp718 to Hind III fragment of pCMX-hSMRTe (Park et al., 1999) into the pCMX-FLAG-cSMRTτ and pCMX-FLAG-cSMRTα plasmids and then subcloned into pEGFP-C1 and pCMX-GAL4 plasmids at Asp718 and Nhe1 sites. The full-length human PXR (hPXR) and its ΔAF2 (aa 1-422) mutant in pGBT9, pCMXHA, and pCMX-GAL4 vectors were as described previously (Johnson et al., 2006). The point mutations mID1 (V2142A/I2143A), mID2 (I2345A/I2346A), mID1-2 (V2142A/I2143A, I2345A/I2346A), m3 (S2285E/K2286E/K2287E), and m4 (L2467A/I2468A, based on SMRTα sequence) were as described previously (Ghosh et al., 2002). These point mutants were regenerated in the SMRTα template by QuikChange site-directed mutagenesis (Stratagene). All constructs were double-confirmed by restriction enzyme digestion and DNA sequencing, and further information is available upon request.
Yeast Two-Hybrid Assay. The GAL4 DBD fusion constructs (in pGBT or pAS vector) were cotransformed in combination with GAL4 activation domain (AD) fusion constructs (in pACT or pGAD vector) into yeast Y190 cells as indicated in individual experiments. Transformed cells were grown in synthetic complete media lacking tryptophan and leucine (-Trp-Leu) at 30°C for 24 h. Aliquots (100 μl) from individual cultures were added to 3 ml of fresh selection medium supplemented with solvent (DMSO) or indicated ligands. Cells were harvested 24 h later and analyzed by liquid β-galactosidase assay using o-nitrophenyl β-d-galactopyranoside as substrate. The average β-galactosidase units were calculated from three separate colonies.
GST Pull-Down Assay. GST and GST fusion proteins were expressed in bacteria BL21 cells and purified by glutathione agarose beads by standard procedure. Individual nuclear receptors were synthesized and labeled with [35S]methionine in rabbit reticulocyte lysate via TNT Quick-Coupled Transcription/Translation System (Promega, Madison, WI). Approximately 5 μg of purified GST and its fusion proteins coupled on agarose beads were mixed with 5 μl of in vitro translated probe with gentle rotation at 4°C overnight in a binding buffer (20 mM HEPES, pH 7.7, 75 mM KCl, 0.1 mM EDTA, 2.5 mM MgCl2, 0.05% Nonidet P-40, 1 mM dithiothreitol, and 0.1 mM methionine) supplemented with 10 mg/ml bovine serum albumin and 1% protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). The beads were washed three times with fresh binding buffer, collected by centrifugation at 3000 rpm for 5 min, and the bound probes were released by boiling in SDS sample buffer and analyzed by SDS-PAGE and autoradiography. For the effects of rifampicin on SMRT-PXR interaction, HA-PXR was overexpressed in HEK293 cells with or without rifampicin treatment. Total cell extracts were prepared and incubated with GST fusion proteins for 16 h at 4°C and analyzed as described above.
Coimmunoprecipitation. Coimmunoprecipitation was conducted according to a standard procedure using anti-FLAG (M2) agarose beads (Sigma). Whole-cell extracts were prepared from HEK293 cells transfected with indicated plasmids in a lysis buffer (20 mM HEPES, pH 7.9, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 1 mM dithiothreitol, and 0.5% Nonidet P-40). The cell extracts (100 μg each) were preabsorbed with protein A agarose beads for 1 h at room temperature before adding the anti-FLAG agarose (30 μl for each reaction). The binding reactions were incubated at 4°C overnight. The immunoprecipitates were then collected by centrifugation and washed extensively with phosphate-buffered saline containing 0.1% Nonidet P-40. The final precipitates were dissolved in SDS protein sample buffer and analyzed by SDS-PAGE and Western blot. Western blot was conducted using the enhanced chemiluminescence reagents according to the manufacturer's recommendations (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK).
Gel Electrophoresis Mobility Shift Assay. Gel electrophoresis mobility shift (gel-shift) assay was conducted as described previously (Chen and Evans, 1995; Blumberg et al., 1998). In brief, a double-stranded DNA of the DR4-type element of the following sequence: AGC TTA AGA GGT CAC GAA AGG TCA CTC GCA T (the underlined sequences are the two NR consensus half sites) was labeled with [32P]dCTP by standard Klenow fill-in reaction. The radioactive probe was purified using a spin column (Bio-Rad Laboratories, Hercules, CA). Approximately 5 × 104 dpm (approximately 1 ng) of the probe was incubated with 1 μl each of the in vitro translated hPXR442 and hRXRα443 in a binding buffer [7.5% glycerol, 20 mM HEPES, pH 7.5, 2 mM dithiothreitol, 0.1% Nonidet P-40, 1 μg of poly(dI-dC), and 100 mM KCl] for 20 min on ice. Approximately 6 μg of the purified GST or GST fusion proteins were added to the reaction for an additional 1 h at room temperature. The DNA-protein complexes were then separated on a 5% native polyacrylamide gel and analyzed by autoradiography.
Immunofluorescence Microscopy. COS-7 cells were plated on coverglasses in 24-well plates 1 day before transfection. Twenty-four hours after transfection, cells were fixed in a methanol/acetic acid [1:1 (v/v)] mixture and processed by indirect immunofluorescence staining as described previously (Li et al., 2000). After extensive washing, fluorescein isothiocyanate-conjugated goat anti-rabbit and rhodamine-conjugated goat anti-mouse antibodies were added. The cells were then stained with 4′, 6-diamidino-2-phenylindole dihydrochloride hydrate (Sigma Chemical Co.) and mounted on slides with ProLong Antifade reagents (Invitrogen, Carlsbad, CA). Standard epifluorescence microscopy was performed on a Zeiss inverted microscope Axiovert 200 equipped with a cool charge-coupled device camera (Axiocam; Carl Zeiss Inc., Thornwood, NY). The images were captured and analyzed by the Axiovision software (Zeiss).
Cell Culture and Transient Transfection. COS-7 cells were maintained in phenol red-free Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotic (Invitrogen). Transient transfection was performed by standard calcium phosphate precipitation method. Human liver HepG2 cells (5 × 105) were seeded in six-well plates and transfected with FuGENE-6 (Roche Diagnostics, Indianapolis, IN). After transfection, cells were washed with phosphate-buffered saline and refed with fresh medium containing vehicle (DMSO) or vehicle plus 10 μM rifampicin (where indicated). Luciferase activity was measured by plate luminometer and normalized with β-galactosidase activity as described previously (Zhang et al., 2004).
Real-Time PCR. Paired cDNAs from various human normal and tumor samples were purchased from BioChain Institute (Hayward, CA). Normal mouse and human liver cDNA libraries were purchased from Clontech (Mountain View, CA). Total RNA was also isolated from cell lines COS-7, HeLa, HepG2, A549, HEK293, and CV-1 cells using TRIzol reagents (Invitrogen). First-strand cDNA was synthesized from 2 μg of total RNA using poly(T) primers (200 ng) and Superscript III reverse transcriptase. Approximately 50 to 100 ng of cDNA template was mixed with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), and PCR reactions were performed in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). The following primers were used to detect SMRTα, SMRTτ, and endogenous control β-actin, respectively: 5′-CGG CTC ATG GGT GGC-3′ (upstream primer) and 5′-TCC GGC GGT TGC AGT CT-3′ (downstream primer); 5′-GCC TGC CCG CTG CTA TG-3′ (upstream primer) and 5′-TCC GGC GGT TGC AGT CT-3′ (downstream primer); 5′-ACG GCA TCG TCA CCA ACT G-3′ (upstream primer) and 5′-GGT TGG CCT TGG GGT TCA-3′ (downstream primer).
The amplification program was initiated with a heating step at 95°C for 10 min followed by 40 cycles of 95°C for 45 s, 54°C for 30 s, and 72°C for 1 min. The program was maintained at 72°C for another 10 min before proceeding to the dissociation step, which consisted of 95°C for 15 s, 60°C for 15 s, and 95°C again for 15 s. The threshold Ct data were determined with default setting using Applied Biosystems Sequence Detection Software version 2.2. Dissociation curve analysis and agarose gel electrophoresis were used to evaluate the specificity of PCR products. Relative quantification of SMRTα versus SMRTτ was normalized to the internal control β-actin and calculated based on the 2-ΔΔCt method. The amplification efficiencies of SMRTα, SMRTτ, and the reference β-actin were confirmed to be approximately equal.
Results
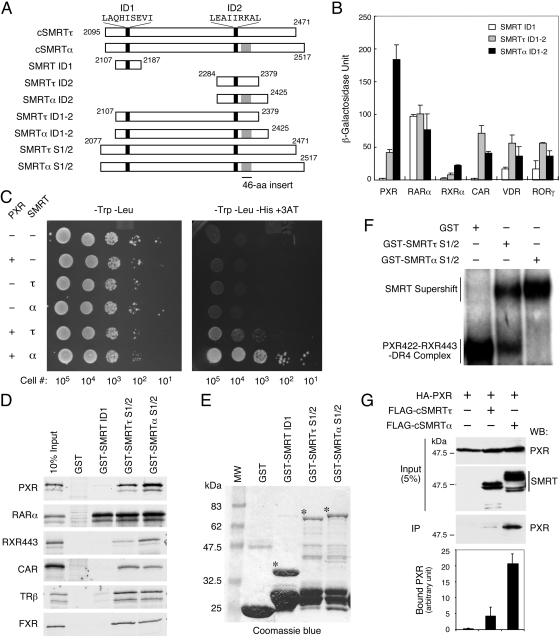
PXR Exhibits Preferential Interaction with SMRTα. The two major SMRT isoforms, τ and α, differ only by a 46-aa sequence inserted after the distal ID2 corepressor motif (Fig. 1A). To investigate the potential functional differences between them, we constructed corresponding pairs of their C-terminal NR-interacting domains (Fig. 1A) and first compared their interactions with various NRs. In a yeast two-hybrid assay (Fig. 1B), most NRs, including RARα, CAR, VDR, and RORγ interacted well with both SMRTτ ID1-2 (aa 2107-2379) and SMRTα ID1-2 (aa 2107-2425) with a slight preference toward SMRTτ. It is interesting that PXR displayed a clear preferential interaction with SMRTα ID1-2. Likewise, RXRα also exhibited an SMRTα preference, but its overall bindings to SMRT were much weaker. In addition, PXR displayed no interaction with SMRT ID1 (aa 2107-2187). In contrast, a strong association between RARα and SMRT ID1 was observed, consistent with our prior finding (Ghosh et al., 2002). These results suggest that different NRs may have different preferences toward these two SMRT isoforms and that PXR seems to have a unique preference toward SMRTα.
Fig. 1.
PXR interacts preferentially with SMRTα. A, schematic diagrams of various SMRTτ and SMRTα constructs used in this study. The proximal ID1 and distal ID2 corepressor motifs are indicated by black bars, with the motif core sequences shown at the top. The SMRTα-specific 46-aa insert is shown in gray. The amino acid positions of individual fragments are labeled numerically. B, PXR interacts preferentially with SMRTα in a yeast two-hybrid assay. The pGBT-hPXR, pAS-hRARα, pGBT-hRXRα, pGBT-hCAR, pGBT-hVDR, and pGBT-mRORγ were individually transformed into Y190 cells in combination with pACT-SMRT ID1 (aa 2107-2187), pACT-SMRTτ ID1-2 (aa 2107-2379), or pACT-SMRTα ID1-2 (aa 2107-2425). Three colonies from each plate were picked and grown in -Trp-Leu liquid media for 24 h. The expression of β-galactosidase was measured by liquid o-nitrophenyl β-d-galactopyranoside assay after normalization with cell numbers, and the average β-galactosidase units were calculated and plotted. C, survival assay of yeast cells cotransformed with pGBT-hPXR and pACT-SMRTτ ID1-2 or SMRTα ID1-2 constructs. Indicated numbers of transformed cells were spotted onto -Trp-Leu or -Trp-Leu-His + 3AT (50 mM) selection plates and incubated at 30°C for 2 days. The pGBT9 and pACT2 vectors were used as controls where indicated (-). D, interactions of SMRT isoforms with various NRs in GST pull-down assays. In vitro-translated 35S-labeled hPXR, hRARα, hRXRα443, hCAR, hTRβ, and hFXR were incubated with GST, GST-SMRT ID1, GST-SMRTτ S1/2 (aa 2077-2471), or GST-SMRTα S1/2 (aa 2077-2517) at 4°C overnight. After extensive washing with binding buffer, the bound proteins were collected by centrifugation and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. E, Coomassie blue staining of GST and GST-SMRT fusion proteins used in this study. The GST proteins were purified from bacteria BL21 cells and analyzed by SDS-PAGE and Coomassie staining. Notable size difference of the intact GST-SMRT ID1, GST-SMRTτ S1/2, and GST-SMRTα S1/2 are marked by asterisks (*). F, preferential interaction of PXR-RXR heterodimers with SMRTα on a DR4 PXR response element. Gel-shift assay was conducted with in vitro-translated hPXR422, hRXRα443, and a 32P-labeled DR4 element. Equal amounts (6 μg) of purified GST, GST-SMRTτ S1/2, or GST-SMRTα S1/2 were added to individual reaction. The DNA-protein complexes were separated onto a native polyacrylamide gel and detected by autoradiography. G, coimmunoprecipitation of PXR with cSMRTτ/α from mammalian cell extracts. Approximately 1 mg of protein extracts obtained from HEK293 cells coexpressing HA-PXR and FLAG-cSMRTτ (aa 2095-2471) or FLAG-cSMRTα (aa 2095-2517) were immunoprecipitated with monoclonal anti-FLAG antibody-conjugated agarose beads. Approximately 5% of the extract used in each immunoprecipitation reaction (input) was analyzed by Western blot to show the relative amount of HA-PXR and cSMRTτ/α in the cell extracts. The total amount of immunoprecipitates (IP) was analyzed by Western blot with rabbit anti-HA antibody to detect the coimmunoprecipitated HA-PXR. The precipitated HA-PXR in each reaction was quantitated by densitometry, and results are plotted and shown at the bottom.
To further investigate the preferential interaction of PXR with SMRTα, we analyzed the relative survivals of transformed Y190 cells on 3AT-containing plates to measure the activation of the second GAL4-dependent HIS3 reporter (Fig. 1C). Cells cotransformed with or without PXR and the indicated SMRT ID1-2 construct were serially diluted and spotted onto -Trp-Leu or -Trp-Leu-His + 3AT plates. On the -Trp-Leu plate, all transformed cells survived equally well, indicating comparable transformation efficiencies and growth rates. However, only cells that were cotransformed with PXR and SMRT showed clear survivals on the 3AT-containing plate, with SMRTα cotransformed cells displaying better survivals than SMRTτ-cotransformed cells. These results support the preferential interaction of PXR with SMRTα.
Next, we asked whether the SMRTα preference by PXR could be recapitulated by in vitro binding assay. GST pull-down assays were conducted with GST and GST-SMRT ID1, SMRTτ S1/2 (aa 2077-2471), and SMRTα S1/2 (aa 2077-2517) to pull down 35S-labeled PXR and other NRs (Fig. 1D). The amounts of intact GST-SMRTτ S1/2 and GST-SMRTα S1/2 fusion proteins were comparable as revealed by Coomassie blue staining (Fig. 1E), whereas the amount of GST-SMRT ID1 was slightly greater. In this assay, we found again that PXR bound better to SMRTα than SMRTτ, whereas it displayed no interaction with SMRT ID1. RARα consistently interacted equally well with SMRT ID1, SMRTτ S1/2, and SMRTα S1/2. Similar to PXR, RXR443 failed to bind SMRT ID1, but it interacted weakly with the S1/2 constructs with a preference toward SMRTα. In contrast, although CAR, TRβ, and FXR failed to interact with SMRT ID1, these receptors interacted approximately equally with the S1/2 fragments of both SMRTα and SMRTτ. These results indicate that the preferential interaction of PXR with SMRTα also occurs in vitro.
PXR needs to form heterodimers with RXR on DNA elements to regulate target gene expression. If SMRTα plays a more important role than SMRTτ in regulating PXR activity, we anticipated that SMRTα should also interact with PXR-RXR-DNA complexes better than SMRTτ. This was tested by gel-shift assay using in vitro translated receptors, 32P-labeled DR4 element, and purified GST-SMRT S1/2 fusion proteins (Fig. 1F). Because the receptor's AF2-helix is known to reduce corepressor binding (Liu et al., 2004), AF2-deletion mutants of both PXR (PXR422) and RXR (RXR443) were used in these binding reactions. We found that the S1/2 fragments of both SMRTτ and SMRTα were capable of interacting with the PXR-RXR-DR4 complex in the gel-shift assay. It is interesting that SMRTα again showed a more efficient binding to the PXR-RXR-DNA complex than SMRTτ, suggesting that the SMRTα preference also occurs at the level of receptor-DNA complex.
Finally, we tested whether the SMRTα preference for PXR also occurs in mammalian cells. Coimmunoprecipitation assay was conducted using HEK293 cell extracts transfected with HA-PXR and FLAG-cSMRTτ (aa 2095-2471) or FLAG-cSMRTα (aa 2095-2517) (Fig. 1G). Cell extracts were immunoprecipitated by anti-FLAG agarose beads, followed by Western blot detection of PXR. Both PXR and the two SMRT isoforms were expressed at similar levels as shown in the inputs. Remarkably, we found that PXR was preferentially immunoprecipitated with SMRTα by approximately 4-fold higher than with SMRTτ (bottom), suggesting that PXR preferentially interacts with SMRTα also in mammalian cells. Together, these results strongly suggest that PXR interacts preferentially with SMRTα both in vitro and in vivo.
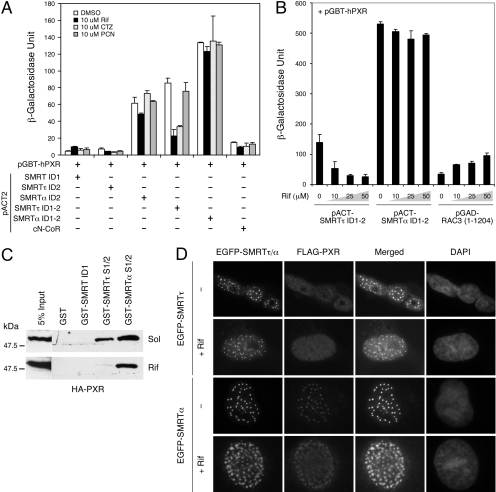
SMRTα-PXR Interaction Is Resistant to Ligand-Induced Dissociation. We have shown previously that the interaction of PXR with SMRTτ could be disrupted by PXR agonists (Johnson et al., 2006); thus, it is interesting to test whether the interaction of PXR with SMRTα is also sensitive to PXR ligands. A series of corresponding SMRTτ/α constructs were compared for their interactions with PXR and their responses to PXR ligands by yeast two-hybrid assay (Fig. 2A). Under conditions in which PXR had little interaction with SMRT ID1, SMRTτ ID2 (aa 2284-2379), or cN-CoR (aa 1929-2440), strong associations of PXR with SMRTα ID2 (aa 2284-2425), SMRTτ ID1-2, and SMRTα ID1-2 were observed in the absence of ligand (Fig. 2A). This suggests that SMRTα ID2 is the preferred binding site for PXR, consistent with our prior observation (Johnson et al., 2006). Also consistently, the human PXR-specific ligands Rif and CTZ diminished PXR's interaction with SMRTτ ID1-2, whereas the mouse PXR-specific ligand PCN had little or no effect. It is remarkable that none of these PXR ligands was capable of diminishing PXR's association with SMRTα ID2 or SMRTα ID1-2. These results suggest that SMRTα also differs from SMRTτ in a way that its interaction with PXR is resistant to ligand-induced dissociation.
Fig. 2.
SMRTα-PXR interaction is resistant to ligand-induced dissociation. A, effects of PXR ligands on SMRT-PXR interactions in a yeast two-hybrid assay. Yeast colonies cotransformed with pGBT-hPXR and the indicated pACT-SMRT constructs or pACT-cN-CoR were treated with the hPXR-specific ligands Rif (10 μM) and CTZ (10 μM), or with the mPXR-specific ligand PCN (10 μM) for 48 h. The empty pACT2 vector was used as a control where indicated (-). The SMRTα ID2 and SMRTα ID1-2 show stronger, ligand-resistant interactions with PXR in comparison with SMRTτ. B, rifampicin concentration-dependent dissociation of SMRTτ/α-PXR interaction in a yeast two-hybrid assay. Yeast transformants containing pGBT-hPXR and pACT-SMRTτ ID1-2, pACT-SMRTα ID1-2, or pGAD-RAC3 (1-1204) were grown in -Trp-Leu media for 24 h. Aliquots of each sample were treated with increasing concentrations of Rif (10, 25, and 50 μM) and incubated for another 36 h. Rifampicin had little effect on the interaction of PXR with SMRTα ID1-2, whereas it reduced its interaction with SMRTτ ID1-2 and enhanced interaction with the coactivator RAC3. C, rifampicin had little effect on the formation of SMRTα-PXR complex. HA-PXR was overexpressed in HEK293 cells in the absence (Sol) or presence of Rif (10 μM). Approximately 50 μg of cell extracts was incubated with 5 μg of purified GST, GST-SMRT ID1, GST-SMRTτ S1/2, or GST-SMRTα S1/2 for 16 h at 4°C. The bound HA-PXR proteins were collected by centrifugation and analyzed by SDS-PAGE and Western blot using anti-HA antibody. D, SMRTα colocalizes with PXR in mammalian cells in the presence of rifampicin. COS-7 cells were transfected with pEGFP-hSMRTτ (full-length) or pEGFP-hSMRTα (full-length) together with FLAG-hPXR. Cells were recovered in Dulbecco's modified Eagle's medium media containing 10 μM rifampicin or DMSO for 12 h. Cells were fixed, and colocalization between SMRT and PXR was detected by immunostaining with anti-FLAG antibody and the EGFP signals. Rifampicin causes clear dissociation of PXR from SMRTτ nuclear foci, whereas it has little effect or no impact on the SMRTα-PXR colocalization.
To investigate the resistance of SMRTα-PXR complex to ligand-induced dissociation in greater detail, we compared the effects of rifampicin on PXR's interactions with SMRTτ ID1-2, SMRTα ID1-2, and RAC3 (aa 1-1204) in a ligand concentration-dependent manner (Fig. 2B). Consistent with a prior finding (Johnson et al., 2006), rifampicin disrupted the SMRTτ-PXR interaction, although it concomitantly enhanced the RAC3-PXR interaction. In contrast, the SMRTα-PXR interaction remained strong at all concentrations of rifampicin, suggesting that the SMRTα-PXR complex is indeed resistant to rifampicin.
To recapitulate the ligand resistance of SMRTα-PXR interaction in vitro, we conducted a GST pull-down assay using HA-PXR expressed in mammalian cells and treated with rifampicin. Purified GST and GST fusions of SMRT ID1, SMRTτ S1/2, and SMRTα S1/2 were mixed with cell extracts containing unliganded or rifampicin-bound HA-PXR, and the bound PXR was detected by Western blot (Fig. 2C). With solvent alone, we found that both SMRTτ S1/2 and SMRTα S1/2 pulled down significant amounts of HA-PXR, whereas GST and GST-SMRT ID1 could not. SMRTα consistently pulled down more PXR than SMRTτ. It is interesting that rifampicin disrupted PXR's interaction with SMRTτ, whereas it had only a minimal effect on the interaction with SMRTα. These results suggest that, in contrast to SMRTτ, SMRTα is capable of binding with PXR in the presence of rifampicin.
Last, we analyzed the effects of rifampicin on colocalization of PXR with SMRT in mammalian cells (Fig. 2D). Full-length SMRTτ is known to accumulate at nuclear foci (Park et al., 1999). EGFP-SMRTτ also formed nuclear foci and displayed colocalization with PXR at those foci, similar to a previous finding (Johnson et al., 2006). It is interesting that rifampicin treatment caused a clear dissociation of PXR from SMRTτ nuclear foci, whereas SMRT foci themselves were not affected. Likewise, EGFP-SMRTα also formed nuclear foci and colocalized efficiently with PXR in the absence of ligand. We were surprised to find that rifampicin was unable to release PXR from these SMRTα foci, suggesting that the association of PXR with SMRTα in mammalian cells is also resistant to ligand-induced dissociation. Taken together, these results strongly suggest that the PXR-SMRTα complex is resistant to ligand-induced dissociation both in vitro and in vivo.
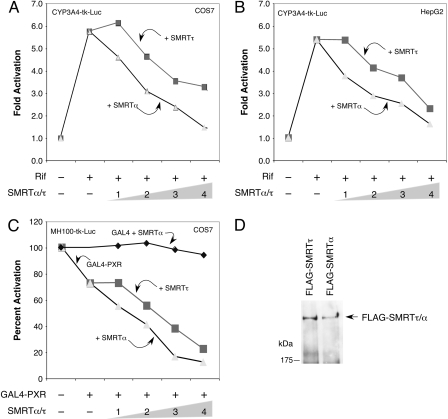
PXR Transcriptional Activity Is Preferentially Inhibited by SMRTα. Because SMRTα displays distinct properties from SMRTτ in terms of interacting with PXR and its ligand sensitivity, it was of interest to compare their abilities in suppressing PXR transcriptional activity. To do so, we used a cell-based assay with a CYP3A4 promoter-driven luciferase reporter activated by rifampicin in the presence of PXR. We found that both SMRTτ and SMRTα were capable of suppressing reporter gene activity in a concentration-dependent manner in COS-7 (Fig. 3A) and HepG2 cells (Fig. 3B). It is interesting that SMRTα exhibited a stronger inhibitory effect on PXR's transcriptional activity than SMRTτ at all concentrations in both cell types. Furthermore, SMRTα also exhibited a stronger corepressor activity than SMRTτ on a GAL4-dependent luciferase reporter MH100-tk-luc in a GAL4-PXR-dependent manner (Fig. 3C). As a control, we found that SMRTα had little effect on the expression of MH100-tk-luc reporter in the presence of GAL4 DBD alone. Both full-length SMRTα and SMRTτ were expressed at similar levels in the transfected cells as detected by Western blot (Fig. 3D). Together, these results indicate that SMRTα has a stronger inhibitory effect than SMRTτ on PXR-mediated transcriptional activity.
Fig. 3.
Transcriptional activity of PXR is preferentially suppressed by SMRTα. COS-7 (A) and human liver HepG2 (B) cells were transiently transfected with pCMXHA-hPXR and pCYP3A4-tk-luc reporter together with a β-galactosidase expression vector as an internal control. Increasing amounts of pCMX-FLAG-SMRTα (full-length) or pCMX-FLAG-SMRTτ (full-length) were cotransfected as indicated (in micrograms). After transfection, cells were refed with fresh media containing 10 μM Rif where indicated (+) and recovered for 16 h. Relative -fold activations of the reporter in comparison with the control sample without treatment or SMRT cotransfection were determined from three independent experiments. SMRTα inhibits PXR transactivation stronger than SMRTτ in both cell types in a dose-dependent manner. C, transcriptional repression by GAL4-PXR is preferentially enhanced by SMRTα. COS-7 cells were transfected with GAL4-hPXR with increasing amounts (in micrograms) of pCMX-FLAG-SMRTτ/α (full-length) construct along with the GAL4-dependent MH100-tk-luc reporter and a β-galactosidase control vector. SMRTα did not affect GAL4 activity, whereas it preferentially enhanced the transcriptional repression activity of GAL-PXR. D, Western blot analysis showing comparable expression levels of FLAG-SMRTτ/α full-length proteins in the transfected cells.
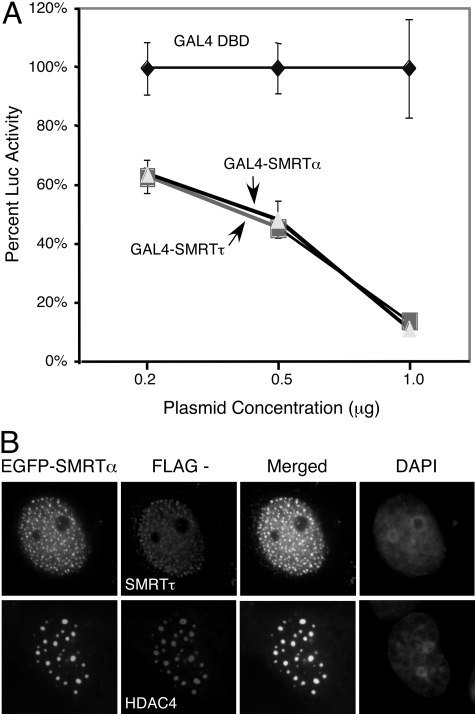
SMRTα and SMRTτ Possess Comparable Intrinsic Repression Activity. The higher corepressor activity of SMRTα over SMRTτ on PXR is consistent with its higher PXR binding affinity and the resistance to ligand-induced dissociation. However, the corepressor function could also be affected by intrinsic basal transcription repression activity. To address this possibility, we compared the basal transcriptional potentials between SMRTα and SMRTτ using GAL4 DBD fusion proteins on the GAL4-dependent MH100-tk-luc reporter. In this assay, we found that GAL4-SMRTτ (full-length) and GAL4-SMRTα (full-length) exhibited strong repression activity at similar levels (Fig. 4A), suggesting that these SMRT isoforms have similar potentials in repressing basal transcription. Furthermore, SMRTτ is known to accumulate at discrete nuclear foci, where it colocalizes with HDACs (Privalsky, 2001; Wu et al., 2001). Immunostaining of coexpressed EGFP-SMRTα (full-length) and FLAG-SMRTτ shows that these two proteins colocalized at discrete nuclear foci (Fig. 4B), suggesting that SMRTα has a distribution similar to that of SMRTτ. In addition, we found that EGFP-SMRTα also efficiently recruited HDAC4 to these nuclear foci, suggesting that SMRTα also interacts with HDACs. These results suggest that SMRTα and SMRTτ have comparable intrinsic repression activities; therefore, the differences in their abilities to inhibit PXR cannot be attributed to differences in their repression potentials.
Fig. 4.
SMRTα and SMRTτ possess comparable intrinsic repression activities. A, transcriptional repressions by GAL4-SMRTα (full-length) and GAL4-SMRTτ (full-length) are comparable. HEK293 cells were transfected with increasing amounts of GAL4 DBD or GAL4 DBD fusions of either full-length SMRTα (GAL4-SMRTα) or full-length SMRTτ (GAL4-SMRTτ), together with a GAL4-dependent MH100-tk-luc reporter. The relative percentages of luciferase activity in comparison with GAL4 DBD alone (set as 100%) were determined from three independent experiments. B, SMRTα colocalizes with SMRTτ and HDAC4. COS-7 cells were transiently transfected with EGFP-SMRTα (full-length) in combination with FLAG-SMRTτ (full-length) or FLAG-HDAC4. Transfected cells were fixed and immunostained with anti-FLAG monoclonal antibody and rhodamine-conjugated secondary antibody and visualized in comparison with the EGFP-SMRTα signals. Cell nuclei were stained with 4,6-diamidino-2-phenylindole.
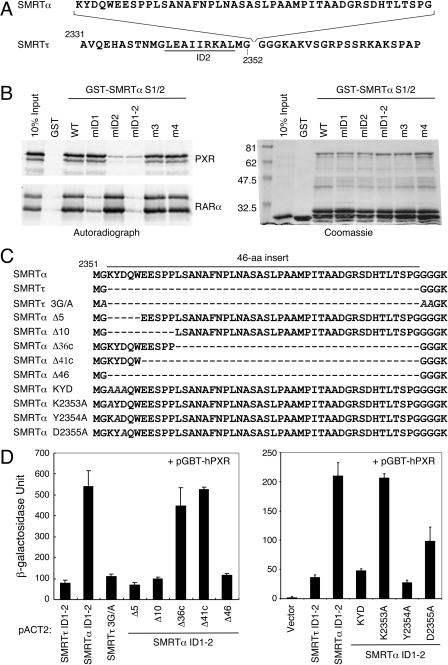
Determinants of SMRTα Preferential Binding by PXR. We have shown previously that SMRTτ interacts with PXR through its ID2 domain (Johnson et al., 2006; Wang et al., 2006). Likewise, SMRTα seems to use the ID2 domain for PXR interaction as well, because the ID1 domain alone does not interact with PXR, whereas the ID1-2 fragment of SMRTα does (Fig. 1 and 2). The only difference between SMRTα and SMRTτ is the α-specific 46-aa insert after residue Gly2352, immediately downstream from the ID2's “LEAIIRKAL” core motif (Fig. 5A). Therefore, this 46-aa sequence must be involved in the enhanced binding of SMRTα by PXR. To investigate how this 46-aa sequence enhances interaction with PXR, we first hypothesized that this 46-aa insert might enable the ID1 domain to interact with PXR, thus creating two binding sites. This was tested by measuring PXR's interactions with a series of point mutations on GST-SMRTα S1/2 by GST pull-down assay (Fig. 5B). The wild-type GST-SMRTα S1/2 and its mutants were expressed, purified, and confirmed by Coomassie blue staining (Fig. 5B, right). It is interesting that we found that only the ID2 mutation (mID2, or I2345A/I2346A), but not the ID1 mutation (mID1, or V2142A/I2143A) or the two non-ID mutations (m3 and m4), disrupted SMRTα's interaction with PXR. The ID1 and ID2 double mutation (mID1-2) also disrupted PXR binding. In contrast, mID1 but not mID2 mutation diminished SMRTα's interaction with RARα. These results suggest that PXR still interacts with SMRTα through its ID2 domain; thus, the 46-aa insert does not enable ID1 to interact with PXR. Therefore, the enhanced interaction of SMRTα is probably mediated directly through the ID2 motif and the 46-aa sequence.
Fig. 5.
Molecular determinants of SMRTα preference for PXR binding. A, sequence of the SMRTα-specific 46 amino acids is shown at the top with the flanking SMRTτ sequences shown at the bottom. This SMRTα-specific sequence is inserted after glycine at position 2352 after the distal ID2 corepressor motif (underlined). The numbers indicate amino acid positions. B, the ID2 motif mutation in SMRTα disrupts PXR interaction. The wild-type (WT) GST-SMRTα S1/2 and its site-directed mutants, mID1 (V2142A/I2143A), mID2 (I2345A/I2346A), mID1-2 (V2142A/I2143A, I2345A/I2346A), m3 (S2285E/K2286E/K2287E), and m4 (L2467A/I2468A), were tested for binding with 35S-labeled hPXR and hRARα in a GST pull-down assay. Right, Coomassie blue-stained GST proteins. The mID2-containing mutants (mID2 and mID1-2) show decreased binding to PXR, whereas only mID1-containing mutations (mID1 and mID1-2) affect their interactions with RARα. C, sequence comparison of the SMRTα-specific 46-aa insert and its mutants. The solid line on top marks the 46-aa sequence. Dashed lines represent deletions. The alanine substitutions in the SMRTτ 3G/A and the KYD, K2353A, Y2354A, and D2355A mutants of SMRTα are italicized. The plasmid pACT-SMRTα ID1-2 was used as a template to construct these mutants used in the following assays. D, yeast two-hybrid assays showing interactions of PXR with SMRTα-specific 46-aa-related mutants. Yeast Y190 cells were cotransformed individually with pGBT-hPXR and indicated pACT-SMRTτ/α ID1-2 wild-type and indicated mutant constructs.
In the SMRTτ sequence, a methionine and four consecutive glycine residues follow the ID2 core motif immediately. Glycine has no side chain and therefore can adopt different conformations. It frequently occurs in turns of proteins and is sometimes known as a “helix breaker.” Therefore, we replaced the first three glycines in the SMRTτ sequence with alanines to extend, theoretically, the length of the ID2 corepressor helix (Fig. 5C, SMRTτ 3G/A mutant). However, this mutation was ineffective in enhancing PXR's interaction with SMRTτ ID1-2 (Fig. 5D), suggesting that a mere extension of the ID2 helix is not sufficient to enhance PXR interaction. Hence, we hypothesized that the 46-aa sequence might participate directly in stabilizing the ID2-PXR interaction or by providing an additional binding surface for PXR. The potential contribution of these 46 amino acids to PXR's interaction was analyzed first by deletion analysis. First, we confirmed that deletion of the entire 46-aa insert from SMRTα (Δ46 mutant) converted its PXR binding efficiency to the level of SMRTτ (Fig. 5D, left). We were surprised to find that deletion of the first 5 (Δ5) or 10 (Δ10) amino acids of the 46-aa insert was each sufficient to reduce SMRTα's interaction to the level of SMRTτ. On the other hand, deletion of the C-terminal 36 (Δ36c) or 41 amino acids (Δ41c) had little effect on SMRTα's interaction with PXR. These results clearly suggest that the first five amino acids (KYDQW) of the 46-aa insert are necessary and sufficient for the preferential interaction of SMRTα with PXR.
To further pinpoint the exact residues that are responsible for the enhanced SMRTα interaction with PXR, we conducted site-directed mutagenesis on the above five amino acids. It is interesting that we found that replacement of the first three residues from KYD to AAA was sufficient to reduce the SMRTα-PXR interaction to the level of SMRTτ (Fig. 5D, right). Additional mutational analysis showed that the Lys2353 to alanine mutation (K2353A) had no effect, whereas the Tyr2354 to alanine mutation (Y2354A) completely abolished the enhanced interaction. In contrast, mutation of Asp2355 to alanine (D2355A) caused a partial decline in the SMRTα-PXR interaction. These results indicate that amino acids Tyr2354 and Asp2355 are both involved and are critical for the preferential association of SMRTα by PXR.
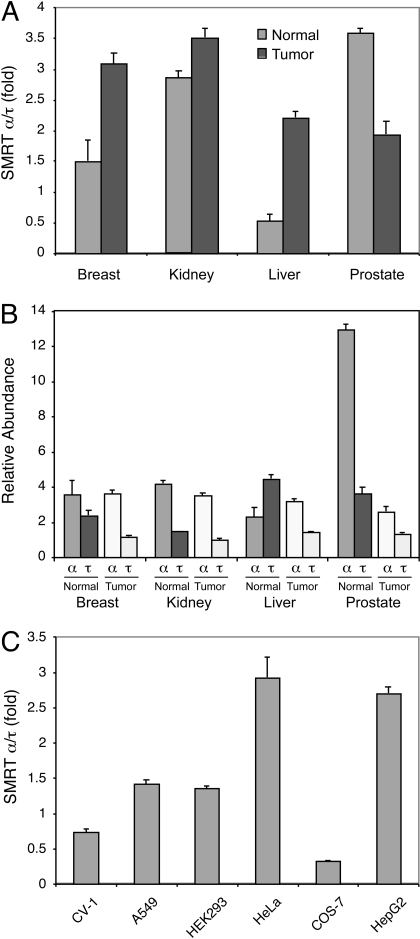
Expression of SMRT Isoforms in Human Tissues and Cancer Cells. To shed light onto the potential physiological relevance of the SMRT isoforms, we decided to compare the relative expression levels of SMRTα versus SMRTτ in various human tissues and cancer cell lines. Paired normal versus tumor cDNAs from various human tissues and cDNAs generated from established cell lines were amplified by real-time PCR using a set of primers that specifically amplify either SMRTα or SMRTτ. Dissociation curve analysis showed a specific peak at the predicted melting temperature of each amplified PCR product. Electrophoresis on a 1.8% agarose gel confirmed the specificity of the same PCR product with one single band at the expected size. In this experiment, we found that SMRTα is the predominant form expressed in both normal and tumor tissues of the breast, kidney, and prostate (Fig. 6, A and B). In particular, the normal prostate seems to express the highest level of SMRTα (Fig. 6B). It is interesting that although SMRTα remained the dominant form in the tumor samples of liver, SMRTτ was more abundant in normal liver tissue (Fig. 6B). In addition, SMRTα is also the major form expressed in several established cell lines (Fig. 6C).
Fig. 6.
Expression of SMRT isoforms in human tissues and cancer cells. cDNAs from normal (N) and tumor (T) tissues of the breast, kidney, liver, and prostate and from various established cell lines were amplified using primer sets specific for SMRTα or SMRTτ as described under Materials and Methods. Real-time PCR reactions were performed in an ABI PRISM 7900HT Sequence Detection System. Relative quantification of SMRTα versus SMRTτ was normalized to the internal control β-actin and calculated according to the 2-ΔΔCt method. A, SMRTα/τ expression ratios in human normal and tumor tissues. B, relative abundance of SMRTα and SMRTτ among tested human normal and tumor tissues. C, SMRTα/τ expression ratios in established cell lines.
Discussion
The xenobiotic receptor PXR plays an important role in the metabolism of many prescribed drugs by controlling the expression of many drug-metabolizing enzymes in liver. In this study, we compared the roles of two different SMRT isoforms, α and τ, in regulating PXR activity. We found that PXR preferentially interacts with the α isoform in a ligand-resistant manner and that SMRTα represses PXR activity more efficiently compared with SMRTτ. We further uncovered the amino acid residues that are responsible for PXR preferential binding to SMRTα. In addition, we found that SMRTα is the dominant isoform expressed in several human tissues and cancer cell lines, suggesting an important role for SMRTα in regulating PXR activation.
There are two distinct corepressor motifs in the SMRT sequence that exhibit different binding affinities toward different NRs. For instance, TR interacts with both the upstream ID1 motif and the downstream ID2, whereas RAR interacts primarily with ID1, although PXR and RXR apparently prefer the ID2 (Ghosh et al., 2002; Johnson et al., 2006). Because the SMRTα-specific 46-aa sequence is located immediately after the ID2 LxxIIxxxL core motif, it is reasonable to speculate that this 46-aa sequence might affect the binding of ID2 to other proteins. Our data suggest that the 46-aa sequence does not contain additional interacting surface for PXR and does not enable PXR-ID1 interaction (Fig. 5). It is interesting that SMRTα reportedly interacted better with TRs on DNA in vitro (Goodson et al., 2005). Although this difference was not seen in our assay (Fig. 1), it remains possible that the presence of DNA might influence SMRT isoform preference. However, if TR does possess differential affinity toward SMRT isoforms, the difference may not be as profound as that for PXR. Our previous structural modeling of the PXR LBD-SMRT ID2 complex (Wang et al., 2006) was unable to address the involvement of the 46-aa insert because of its length and distance from the ID2 core motif. Future improvement in our modeling system will be informative to predict any molecular interaction with PXR involving the 46-aa sequence.
In contrast to ligand-reversible association of SMRTτ with PXR, SMRTα retains a strong interaction with PXR in the presence of PXR ligands (Fig. 2). It is known that certain NR variants possess ligand-irreversible association with SMRT (Tagami et al., 1998). Our current data further suggest that different SMRT isoforms may have different affinity toward the same receptor. In addition to the ligand-irreversible effect, several other possibilities may also explain the preferential inhibition of PXR by SMRTα. For example, in the presence of SMRTα, rifampicin might be unable to produce a conformational change in PXR that is required to release the corepressor. On the other hand, SMRTα might prevent rifampicin from binding to PXR. It is equally possible that the presence of SMRTα might interfere with the ability of PXR to recruit coactivators.
To shed light into the relative importance of SMRT isoforms on regulating PXR activity, we compared the expression of SMRTα versus SMRTτ in various human tissues and cancer cell lines (Fig. 6). We were surprised to find that SMRTα was found at higher levels than SMRTτ in most tissues and cell lines, especially in the normal prostate. It is remarkable that SMRTτ was found in higher levels in the normal liver sample (Fig. 6, A and B). Although both SMRTα and SMRTτ are ubiquitously expressed and the amount of SMRTα in normal liver tissue is slightly lower than in other tested tissues, the amount of SMRTτ in normal liver tissue is much higher compared with other tissues. It is important to note that human PXR is most abundantly expressed in liver and intestine (Blumberg et al., 1998; Lehmann et al., 1998). Because the interaction between PXR and SMRTτ is sensitive to PXR ligand-induced dissociation, and SMRTτ is more abundant than SMRTα in normal liver tissue, it is possible that the PXR-SMRTτ interaction may be more relevant for the inductive response of PXR activation by ligands in the liver. Furthermore, we speculate that the relatively more abundant SMRTα in other tissues and cancerous samples and cell lines might play a role in limiting PXR activation in these other tissues, because the PXR-SMRTα interaction is stronger and more resistant to ligand-induced dissociation.
PXR coordinately regulates drug clearance in response to a wide variety of xenobiotic compounds; thus, reducing PXR activity may diminish drug clearance and increase the potency of therapeutic drugs, causing dangerous drug-drug interaction. There has been a great amount of interest in drug discovery with an emphasis on understanding structure-function relationship for attenuating drug-mediated PXR activation (Gao et al., 2007; Ung et al., 2007). The fact that the key amino acids of the SMRTα-specific 46-aa insert is located within the first 5 amino acids, which is only 2 residues away from the ID2 core sequence, may provide a novel therapeutic target using an extended ID2 motif via a peptide interference mechanism. Indeed, the SMRT corepressor motif has been designed as a peptide to occupy the lateral groove of BCL6 and compete with corepressor binding (Polo et al., 2004). These peptides not only attenuate BCL6-mediated transcriptional repression but also reactivate the expression of BCL6 target genes, resulting in the disruption of endogenous BCL6 repression complexes. Thus, this current study may provide a molecular basis for rational drug designs aimed at enhancing the efficacy of therapeutic drugs by inhibiting PXR-mediated drug metabolism.
Acknowledgments
We thank Martin L. Privalsky at University of California-Davis, Davis, California, for providing the pGEX-SMRTτ S1/2 (aa 2077-2471) and pGEX-SMRTα S1/2 (aa 2077-2517) plasmid vectors.
This work was supported by the National Institutes of Health [Grant DK52542].
ABBREVIATIONS: NR, nuclear receptor; PXR, pregnane X receptor; SMRT, silencing mediator for retinoid and thyroid hormone receptors; N-CoR, nuclear receptor corepressor; HDAC, histone deacetylase; ID, interacting domain; RXR, retinoid X receptor; CAR, constitutive androstane receptor; TR, thyroid hormone receptor; PCN, pregnenolone-16α-carbonitrile; GST, glutathione transferase; Rif, rifampicin; CTZ, clotrimazole; HA, hemagglutinin; aa, amino acid(s); PCR, polymerase chain reaction; AD, activation domain; DMSO, dimethyl sulfoxide; PAGE, polyacrylamide gel electrophoresis; HEK, human embryonic kidney; RAR, retinoic acid receptor; VDR, vitamin D receptor; ROR, retinoid-related orphan receptor; EGFP, enhanced green fluorescent protein.
References
- Blumberg B, Sabbagh W Jr, Juguilon H, Bolado J Jr, van Meter CM, Ong ES, and Evans RM (1998) SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev 12 3195-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD and Evans RM (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377 454-457. [DOI] [PubMed] [Google Scholar]
- Falkner KC, Pinaire JA, Xiao GH, Geoghegan TE, and Prough RA (2001) Regulation of the rat glutathione S-transferase A2 gene by glucocorticoids: involvement of both the glucocorticoid and pregnane X receptors. Mol Pharmacol 60 611-619. [PubMed] [Google Scholar]
- Gao YD, Olson SH, Balkovec JM, Zhu Y, Royo I, Yabut J, Evers R, Tan EY, Tang W, Hartley DP, et al. (2007) Attenuating pregnane X receptor (PXR) activation: a molecular modelling approach. Xenobiotica 37 124-138. [DOI] [PubMed] [Google Scholar]
- Geick A, Eichelbaum M, and Burk O (2001) Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem 276 14581-14587. [DOI] [PubMed] [Google Scholar]
- Ghosh JC, Yang X, Zhang A, Lambert MH, Li H, Xu HE, and Chen JD (2002) Interactions that determine the assembly of a retinoid X receptor/corepressor complex. Proc Natl Acad Sci U S A 99 5842-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson ML, Jonas BA, and Privalsky ML (2005) Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors. J Biol Chem 280 7493-7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, and Shiekhattar R (2000) A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev 14 1048-1057. [PMC free article] [PubMed] [Google Scholar]
- Harmsen S, Meijerman I, Beijnen JH, and Schellens JH (2007) The role of nuclear receptors in pharmacokinetic drug-drug interactions in oncology. Cancer Treat Rev 33 369-380. [DOI] [PubMed] [Google Scholar]
- Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, and Glass CK (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377 397-404. [DOI] [PubMed] [Google Scholar]
- Hu X and Lazar MA (1999) The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402 93-96. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Li CW, Chen LY, Ghosh JC, and Chen JD (2006) Regulation and binding of pregnane X receptor by nuclear receptor corepressor silencing mediator of retinoid and thyroid hormone receptors (SMRT). Mol Pharmacol 69 99-108. [DOI] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, and Edwards PA (2002) Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem 277 2908-2915. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92 73-82. [DOI] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, and Yamamoto Y (2004) Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 24 7931-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, and Kliewer SA (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102 1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo C and Chen JD (2000) The SRC family of nuclear receptor coactivators. Gene 245 1-11. [DOI] [PubMed] [Google Scholar]
- Li H, Leo C, Zhu J, Wu X, O'Neil J, Park EJ, and Chen JD (2000) Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol 20 1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Shaw CK, Reineke EL, Liu Y, and Kao HY (2004) Retinoid X receptor α (RXRα) helix 12 plays an inhibitory role in the recruitment of the p160 coactivators by unliganded RXRα/retinoic acid receptor α heterodimers. J Biol Chem 279 45208-45218. [DOI] [PubMed] [Google Scholar]
- Moore JT, Moore LB, Maglich JM, and Kliewer SA (2003) Functional and structural comparison of PXR and CAR. Biochim Biophys Acta 1619 235-238. [DOI] [PubMed] [Google Scholar]
- Muangmoonchai R, Smirlis D, Wong SC, Edwards M, Phillips IR, and Shephard EA (2001) Xenobiotic induction of cytochrome P450 2B1 (CYP2B1) is mediated by the orphan nuclear receptor constitutive androstane receptor (CAR) and requires steroid co-activator 1 (SRC-1) and the transcription factor Sp1. Biochem J 355 71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, and Evans RM (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89 373-380. [DOI] [PubMed] [Google Scholar]
- Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, and Evans RM (1999) Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci U S A 96 2639-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Schroen DJ, Yang M, Li H, Li L, and Chen JD (1999) SMRTe, a silencing mediator for retinoid and thyroid hormone receptors-extended isoform that is more related to the nuclear receptor corepressor. Proc Natl Acad Sci U S A 96 3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Dell'Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, Prive GG, Licht JD, and Melnick A (2004) Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med 10 1329-1335. [DOI] [PubMed] [Google Scholar]
- Privalsky ML (2001) Regulation of SMRT and N-CoR corepressor function. Curr Top Microbiol Immunol 254 117-136. [DOI] [PubMed] [Google Scholar]
- Privalsky ML (2004) The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol 66 315-360. [DOI] [PubMed] [Google Scholar]
- Synold TW, Dussault I, and Forman BM (2001) The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med 7 584-590. [DOI] [PubMed] [Google Scholar]
- Tagami T, Kopp P, Johnson W, Arseven OK, and Jameson JL (1998) The thyroid hormone receptor variant alpha2 is a weak antagonist because it is deficient in interactions with nuclear receptor corepressors. Endocrinology 139 2535-2544. [DOI] [PubMed] [Google Scholar]
- Toell A, Kröncke KD, Kleinert H, and Carlberg C (2002) Orphan nuclear receptor binding site in the human inducible nitric oxide synthase promoter mediates responsiveness to steroid and xenobiotic ligands. J Cell Biochem 85 72-82. [PubMed] [Google Scholar]
- Ung CY, Li H, Yap CW, and Chen YZ (2007) In silico prediction of pregnane X receptor activators by machine learning approaches. Mol Pharmacol 71 158-168. [DOI] [PubMed] [Google Scholar]
- Urquhart BL, Tirona RG, and Kim RB (2007) Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol 47 566-578. [DOI] [PubMed] [Google Scholar]
- Wang CY, Li CW, Chen JD, and Welsh WJ (2006) Structural model reveals key interactions in the assembly of the pregnane X receptor/corepressor complex. Mol Pharmacol 69 1513-1517. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, and Redinbo MR (2001) The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292 2329-2333. [DOI] [PubMed] [Google Scholar]
- Westin S, Rosenfeld MG, and Glass CK (2000) Nuclear receptor coactivators. Adv Pharmacol 47 89-112. [DOI] [PubMed] [Google Scholar]
- Wipf P, Gong H, Janjic JM, Li S, Day BW, and Xie W (2007) New opportunities for pregnane X receptor (PXR) targeting in drug development. lessons from enantio- and species-specific PXR ligands identified from a discovery library of amino acid analogues. Mini Rev Med Chem 7 617-625. [DOI] [PubMed] [Google Scholar]
- Wu X, Li H, and Chen JD (2001) The human homologue of the yeast DNA repair and TFIIH regulator MMS19 is an AF-1-specific coactivator of estrogen receptor. J Biol Chem 276 23962-23968. [DOI] [PubMed] [Google Scholar]
- Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, et al. (2002) Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature 415 813-817. [DOI] [PubMed] [Google Scholar]
- Zhang A, Yeung PL, Li CW, Tsai SC, Dinh GK, Wu X, Li H, and Chen JD (2004) Identification of a novel family of ankyrin repeats containing cofactors for p160 nuclear receptor coactivators. J Biol Chem 279 33799-33805. [DOI] [PubMed] [Google Scholar]