Abstract
The central Extended Amygdala (EAc) is an ensemble of highly interconnected limbic structures of the anterior brain, and forms a cellular continuum including the Bed Nucleus of the Stria Terminalis (BNST), the central nucleus of the Amygdala (CeA) and the Nucleus Accumbens shell (AcbSh). This neural network is a key site for interactions between brain reward and stress systems, and has been implicated in several aspects of drug abuse. In order to increase our understanding of EAc function at the molecular level, we undertook a genome-wide screen (Affymetrix) to identify genes whose expression is enriched in the EAc. We focused on the less-well known BNST-CeA areas of the EAc, and identified 121 genes that exhibit more than 2-fold higher expression level in the EAc compared to whole brain. Among these, forty-three genes have never been described to be expressed in the EAc. We mapped these genes throughout the brain, using non-radioactive in situ hybridization, and identified eight genes with a unique and distinct rostro-caudal expression pattern along AcbSh, BNST and CeA. Q-PCR analysis performed in brain and peripheral organ tissues indicated that, with the exception of one (Spata13), all these genes are predominantly expressed in brain. These genes encode signaling proteins (Adora2, GPR88, Arpp21 and Rem2), a transcription factor (Limh6) or proteins of unknown function (Rik130, Spata13 and Wfs1). The identification of genes with enriched expression expands our knowledge of EAc at a molecular level, and provides useful information to towards genetic manipulations within the EAc.
Keywords: adult mouse, Affymetrix microarray, in-situ hybridization, central Extended Amygdala, gene, marker
Introduction
The central Extended Amygdala (EAc) is a network formed by basal forebrain structures that include the Bed Nucleus of the Stria Terminalis (BNST) and the central Amygdala (CeA) (Alheid and Heimer, 1988, Cassell et al., 1999, de Olmos and Heimer, 1999, Swanson, 2003). However, it has been proposed that the shell of the Nucleus Accumbens (AcbSh) would be part of this network (Koob, 2003). This particular division of the Extended Amygdala interfaces reward circuitry with stress systems, and is involved in both the acute reinforcing effects of drugs of abuse and the negative reinforcing effects of drug dependence. As such, the EAc may represent a common anatomical substrate for drug reward and stress-induced drug seeking and reinstatement (Koob, 2003, Shaham et al., 2003, Koob and Kreek, 2007). The EAc receives inputs mainly from limbic cortices, and projects to the ventral tegmental area, the lateral hypothalamus, the tegmental pedunculopontine nucleus and other various brain stem nuclei (in (Koob, 2003)). A number of transmitter systems that operate within the EAc have been described. GABA immunoreactive neurons have been characterized within the 3 components of the EAc (Sun and Cassell, 1993). Also BNST and CeA neurons express a number of neuropeptides that include CRF, NPY, vasopressin and galanin, and modulate EAc function (see (Koob, 2003, Kash and Winder, 2006)). However, our knowledge of phenotypic characteristics of EAc neurons remains limited.
Understanding the function of specific brain areas or circuits requires detailed information on molecules expressed by neuronals. Cellular composition and neuron function varies greatly across the brain, and large-scale gene expression studies are growing to explore gene patterning in the adult mammalian brain on a genome-wide basis. A number of transcriptome studies have examined large areas of the brain, such as the cortex, hippocampus and striatum (Bonaventure et al., 2002, de Chaldee et al., 2003, Ghate et al., 2007, Stansberg et al., 2007). Comparison of central and peripheral nervous systems has led to identify genes whose expression is restricted to either spinal cord or dorsal root ganglia (LeDoux et al., 2006). Transcriptional imprint of 24 neural brain tissues helped to construct a gene expression-based brain map in the adult mouse (Zapala et al., 2005). Further the exploration of specialized brain networks has provided regional-specific gene profiles using fine-dissection procedures. Genes restricted to the CA1, CA3 and DG hippocampal territories (Lein et al., 2004), or even along the dorso-ventral axis of the CA1 field has been identified (Leonardo et al., 2006). The analysis of subregions of the hypothalamus highlighted genes restricted to the ventromedial hypothalamus in the adult (Segal et al., 2005) or developing mouse brain (Kurrasch et al., 2007). The study of Amygdala revealed gene subsets whose expression matched anatomical boundaries of amygdaloid nuclei (Zirlinger, 2003). Recently Olsen and coll. compared gene profiles of the BNST to those of ventral and dorsal striatum, and identified distinct signaling and plasticity genes in these areas that all respond to dopamine and are involved in disorders ranging from Parkinson’s disease to drug addiction (Olsen et al., 2008).
The present study aimed at identifying genes with enriched expression in the EAc, in order to broaden our knowledge of genes operating within this brain network. In this study we have focused on the BNST/CeA components of the EAc. Using a micropunch-dissection procedure, we prepared a tissue sample from mouse brain CeA and BNST and compared the transcriptome in this sample to that of the whole brain. We identified 129 probe sets with a 2-fold enrichment or more in the EAc and mapped the expression pattern of 49 genes by in situ hybridization in the mouse brain. Eight genes showed an enriched expression pattern in the EAc, that we analyzed in greater details. These genes potentially influence some aspects of addictive behaviors, and may be useful for further genetic manipulations within the EAc.
Experimental procedures
Tissue dissection
Tissues were dissected from male 3 to 6 month old C57Bl/6J wild-type mice using a microdissection procedure as follows. Briefly, mice were killed by cervical dislocation. Brain was removed, washed in PBS buffer and placed into a matrix cooled on ice (ASI Instruments Inc, Warren, MI, USA) to obtain slices of 1 mm thickness. Accurate localization of brain structures was based on the stereotaxic atlas of mouse brain (Paxinos and Franklin, 2001) and areas corresponding to the Bed Nucleus of the Stria Terminalis (BNST, +0.5 to −0.5), Central nucleus of Amygdala (CeA, −0.5 to −1.5) were taken by bilateral punches (1.2 mm diameter). BNST and CeA samples were pooled to obtain the central Extended Amygdala (EAc) sample (see Figure 1A). Additionally, samples of the whole brain (WB), lateral hypothalamus (LH), spinal cord, thymus, lung, spleen, heart, liver, intestine, lung, stomach, kidney, testis, and lung were collected. All samples were stored at −80°C until use. All animal use procedures were in strict accordance with standard ethical guidelines (European Community Guidelines on the Care and Use of Laboratory Animals 86/609/EEC) and approved by the local ethical committee (Comité régional d’éthique en matière d’expérimentation animale de Strasbourg, CREMEAS, 2003-10-08-[1]-58).
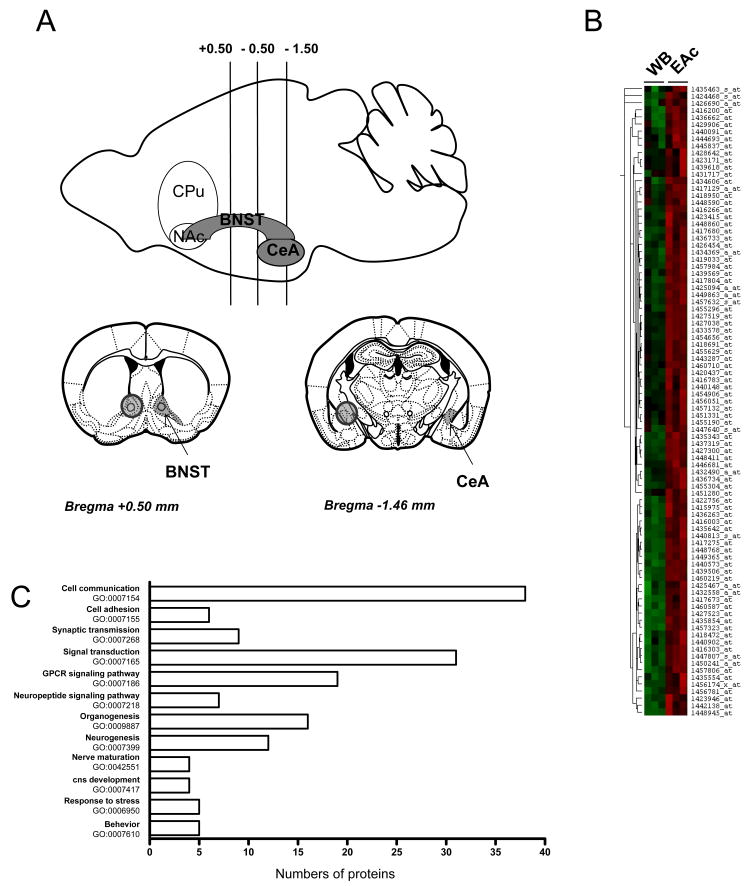
Figure 1. Affymetrix analysis for the identification of genes enriched in the EAc.
(A) This scheme shows brain areas under study: bilateral punches (1.2 mm diameter) were taken from mouse brain coronal slices (1 mm thick) to collect the bed nucleus of stria terminalis (BNST, +0.5 to −0.5) and the central nucleus of the Amygdala (CeA, −0.5 to −1.5) (see Methods for details). BNST and CeA punches were pooled and corresponded to central Extended Amygdala (EAc) samples. (B) This hierarchical cluster illustrates raw microarray data from three independent hybridizations for the 129 selected probe sets, and shows high expression in EAc (right columns, red) compared to whole brain (WB, left columns, left). The probe set selection was based on a standard statistical analysis by MAS 5.0 and a threshold of 2-fold change in EAc over WB was used (see Methods for details). Hierarchical cluster analysis was performed using the Cluster 3.0 and Treeview softwares. (C) Gene ontology analysis of the EAc enriched genes. Genes were categorized with the Biological Process domain and significantly enriched GO terms with a probability lower than 0.01 and including at least four proteins are represented.
RNA preparation and microarray hybridization
Total RNA was extracted from the different tissues using TRIzol reagent (Invitrogen, Cergy Pontoise, France) and following the manufacturer’s specifications. EAc samples were prepared with tissues pooled from 3 mice, WB samples were from 2 mice. The RNA quantity was measured using a spectrophotometer and quality was assessed by agarose gel electrophoresis. For the microarray experiments, cDNA synthesis, cRNA labeling, hybridization and scanning procedures were conducted according to standard Affymetrix’protocols (www.affymetrix.com) (Affymetrix Core Facility, IGBMC, Illkirch, France, http://www-microarrays.u-strasbg.fr). Three separate hybridizations were performed with independent pooled samples from both EAc and WB. In total, 6 Mouse Genome 430 2.0 oligonucleotide arrays representing 45101 transcripts or ESTs were used in this experiment.
Microarray data analysis
Microarrays were scanned using Affymetrix GeneChip Scanner 3000. Quantification and initial analysis of the microarrays were done using Gene Chip operating Software (GCOS v1, Affymetrix UK Ltd). The comparisons of gene expression profiles between EAc and WB samples were done using a standard significance analysis (MAS 5.0 software, Affymetrix UK Ltd). For selecting differentially expressed genes, we first searched for probes considered as detected. Probe sets were eliminated in the analysis when described as absent more than 4 times among 6, or when having all signal values bellow 25 (corresponding to the median value of signals for all samples). This step led to the selection of 25174 probe sets. We then used p-values from the MAS 5.0 comparative analysis to find probe sets, which led to acceptable False Discovery Rate (FDR). A statistical significance of 0.0025 corresponded to a FDR value of 2.5%. This threshold selected 2799 probe sets. To limit the number of genes for further analysis, we then applied more stringent criteria. We re-analyzed each hybridization set separately. We eliminated probe sets with signals under 100 in the EAc sample. We then selected probe sets with a “signal log2 ratio” of EAc/WB equal or superior to 1, corresponding to a difference in expression level of at least 2 fold. Finally, lists obtained from each hybridization set were combined and probe sets that were differentially expressed in at least 2 out of the 3 hybridizations were selected. Finally, this led to a group of 129 probe sets that we defined as enriched in the EAc. A student t-test was then performed on this subgroup to confirm that average signals from triplicate hybridizations from the EAc differed from those measured in the whole brain. Hierarchical clustering was performed on probe sets selected as enriched in the EAc using the Cluster 3.0 and Treeview software (Eisen et al., 1998, de Hoon et al., 2004). Genes enriched in the EAc were annotated for association with biological processes using an optimized Gene Ontology (GO) analysis (as described in (Chalmel et al., 2005, Abou-Sleymane et al., 2006, Befort et al., 2008b) and with biological functions using Ingenuity Pathway Analysis (IPA) network. For GO analysis, over-represented GO terms with a probability lower than 0.01 and including at least 4 proteins were selected. In IPA analysis, biological functions and/or diseases that are most significant to the dataset were identified. The p-value for a given annotation is calculated by considering the number of “focus genes” that participate in that function and the total number of genes that are known to be associated with that process in Ingenuity’s knowledge base (http://www.ingenuity.com). In our analysis, we show only the functions with the six highest p-values.
In situ hybridization
Plasmids containing candidate genes were obtained from the Deutsches Ressourcenzentrum für Genomforschung (RZPD, Berlin, Germany). Clone inserts were amplified by PCR (100 μl), using vector-specific primers and 0.25 μl of bacterial glycerol stock as template material. PCR reactions were purified using Millipore’s (Millipore Corporation, Bedford, USA) Montage 96 and amplicons used as template for in-vitro transcription of sense and anti-sense Dig-labeled riboprobes. To this aim 1μg linearized DNA was transcribed using T7, T3 or Sp6 polymerases and the 10x DIG RNA labeling mix (Roche Diagnostics, Meylan, France) according to the manufacturer’s instructions. Probes were quantified by spectrophotometry and quality assessed by agarose gel electrophoresis. Adult mice were killed by cervical dislocation and brains were rapidly extracted and fresh frozen in OCT. The OCT-embedded brain blocks were stored at −20°C until use. Brain sections 25 μm thick were processed for in situ hybridization using Genepaint robotic equipment and procedures (www.genepaint.org; (Carson et al., 2002) as previously described (Ghate et al., 2007). Briefly, 600 ng of probe at a concentration of 20 ng/μl was hybridized onto the sections at 64°C for 5.5 hours. The dig-label was detected using anti-Dig-POD antibody (Roche, 1:500 dilutions in Tris-NaCl pH 7.5 solution containing Perkin Elmer blocking reagent and 0.1% Tween). The signal was amplified and revealed using tyramide amplification process-kit (Perkin Elmer, Waltham, USA) and BCIP (0.15mg/ml)/NBT (0.4 mg/ml, Roche) color substrates. Images were recorded using a CCD camera (Leica Instruments, Rueil-Malmaison, France). In each in situ hybridization set, neuropeptide Y and preproenkephalin were used as positive controls and a blank hybridization (no probe) as the negative control. For ISH analysis, we used criteria of classification adapted from the GenePaint annotation procedures (http://www.genepaint.org/) as described in (Gofflot et al., 2007). Three different levels of expression were defined: 0: no color precipitate detected; 1: weak expression, a few particles of color precipitate per cell; 2: strong expression, color precipitate completely filling cells. For those, we distinguished two different types of expression patterns: ubiquitous distribution throughout the brain or restricted distribution to specific regions of the brain, including EAc.
Quantitative RT-PCR
Total RNA was extracted from the different tissues using TRIzol reagent (Invitrogen, Cergy Pontoise, France) with EAc and LH samples, prepared with tissues pooled from 3 mice, whole brain samples from 2 mice and all the other tissues from individual mouse. Total RNA (2.5 μg) from each pool (n=2) was treated for 30 min at 37°C by DNase I RNase free (4 U, Invitrogen, Cergy Pontoise, France) in First strand Superscript buffer (Invitrogen, Cergy Pontoise, France) and reaction was stopped by incubating the mix 5 min at 75°C. RNA was then pre-incubated with oligodT primer (8 μM), random Hexamer (16 μM) and dNTPs (500 μM each) in a volume of 30 μl for 5 min at 65°C. Finally, First strand Superscript buffer, DTT (0,01M) and Superscript II (200U, Invitrogen, Cergy Pontoise, France) were added in final volume of 46 μl for 50 min at 42°C. Reaction was stopped by 15 min incubation at 70°C. Real-time PCR was performed in triplicate on a MyIQ BioRad instrument using iQ SYBR Green supermix, cDNA (0.5 μl) and gene-specific primers (200 nM) in a 25 μl reaction as recommended by the manufacturer (Bio-Rad, Marnes-la-Coquette, France). Gene-specific primers were designed using primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to obtain a 75–150 bp product (see supplemental Table S1). Relative quantification for a given gene in any area was normalized to its level in whole brain (ΔCt) and expressed as fold change (WB level being equal to 1).
Results
Identification of genes enriched in the central Extended Amygdala
To identify genes whose expression is enriched in the EAc, we searched for genes whose expression level is higher in EAc compared to WB. We used the Genechip Mouse Genome 430 2.0 oligonucleotide arrays (Affymetrix) and compared hybridization signals of RNA transcripts in the EAc to those obtained in WB. Our gene selection procedure involved a multi-step procedure based on the FDR, the gene expression level and the fold change EAc/WB (see methods). Using this approach, we identified 129 probe sets showing statistical differential expression with a fold-change of 2 at least, that we categorized as enriched in EAc (Table 1, Figure 1B). A statistical analysis further performed directly on mean signals of each probe set (see Methods) showed that 95.3 % of the EAc probe sets were significantly enriched over whole brain (p value<0.05). Gene annotation from Affymetrix identified distinct probe sets corresponding to identical genes, with 9 genes represented by two different probe sets. In the end we identified 121 genes enriched in the EAc. Figure 1B presents a hierarchical cluster analysis of the enriched EAc transcripts with the corresponding signals in all 6 arrays.
Table 1. EAc-enriched genes.
This table shows the list of 129 probe sets selected from the Affymetrix analysis, with high expression in EAc compared to whole brain (WB). Data are expressed as fold-change of EAc over WB, with corresponding p values from the MAS5.0 analysis (Affy p-value). A FDR of 2.5% was used for the initial selection of probe sets (see details in Methods). Student t-test was further performed to compare mean signals for EAc versus WB data and validate the selection. Probe sets whose expression was investigated in this study by in situ hybridization (ISH) on sagittal (S) or coronal (C) sections are indicated (see Figures 2, 3 and Supplemental Figure 1). Gene encoding a glial protein (Gli) or previously described in EAc using ISH analysis (Litt) have been identified from literature mining and are also indicated in the table. Twins indicate probe sets corresponding to the same gene.
| Enriched EA markers
| ||||||||
|---|---|---|---|---|---|---|---|---|
| ProbeSet | RefSeq ID | Gene Name | Symbol | ISH | Fold Change EAc | Affy p-values | t-test EA vs WB | twins |
| 1427300_at | NM_010713 | LIM homeobox protein 8 | Limh8 | S | 11.05 | 2E-05 | 0.006 | |
| 1450723_at | NM_021459 | ISL1 transcription factor, LIM/homeodomain (islet 1) | Isl1 | 6.73 | 2.7E-05 | 0.014 | ||
| 1419411_at | NM_009312 | tachykinin 2 | Tac2 | Litt | 6.35 | 2E-05 | 0.084 | |
| 1432558_a_at | NM_013779 | melanoma antigen, family L, 2 | Magel2 | S | 4.84 | 2E-05 | 0.003 | 1417217_at |
| 1431717_at | AK014386 | RIKEN full-length enriched library, clone:3526401B18 | Rik352 | 4.44 | 2E-05 | 0.221 | ||
| 1441382_at | NM_001033360 | Mus musculus G protein-coupled receptor 101 (Gpr101), | Gpr101 | 4.21 | 5.2E-05 | 0.041 | ||
| 1422586_at | NM_021306 | endothelin converting enzyme-like 1 | Xce | Litt | 4.21 | 2E-05 | 0.034 | |
| 1421978_at | NM_008078 | glutamic acid decarboxylase 2 | GAD65 | Litt | 3.94 | 4E-05 | 0.001 | |
| 1456781_at | AK043872 | RIKEN full-length enriched library, clone:A830044L07 | RikA83 | 3.70 | 4.6E-05 | 0.014 | ||
| 1444693_at | NM_023116 | Mus musculus calcium channel, voltage-dependent, beta 2 subunit (Cacnb2), | Cacnb2 | 3.64 | 6.8E-05 | 0.031 | ||
| 1417997_at | NM_022414 | neuroglobin | Ngb | 3.59 | 2.7E-05 | 0.011 | 1417996_at | |
| 1439569_at | NM_010287 | G protein-coupled receptor 83 | GPR83 | S | 3.52 | 2E-05 | 0.026 | 1423415_at |
| 1427509_at | AK122358 | Mus musculus similar to mKIAA0734 protein (LOC381075), mRNA | KIAA073 | 3.50 | 2.3E-05 | 0.023 | ||
| 1460668_at | NM_010253 | galanin | galn | Litt | 3.47 | 2E-05 | 0.042 | |
| 1420437_at | NM_008324 | indoleamine-pyrrole 2,3 dioxygenase | Indo | Gli | 3.45 | 1.3E-04 | 0.037 | |
| 1441429_at | NM_010572 | insulin receptor substrate 4 | Irs4 | S | 3.41 | 2E-05 | 0.025 | |
| 1427523_at | NM_011381 | sine oculis-related homeobox 3 homolog (Drosophila) | Six3 | 3.39 | 1.1E-04 | 0.003 | ||
| 1448411_at | NM_011716 | Wolfram syndrome 1 homolog (human) | Wfs1 | SC - Litt | 3.38 | 2E-05 | 0.002 | |
| 1422860_at | NM_024435 | RIKEN cDNA 5033428E16 gene | Nts | Litt | 3.31 | 2E-05 | 0.068 | |
| 1425094_a_at | NM_008500 | LIM homeobox protein 6 | Limh6 | SC | 3.27 | 5.2E-05 | 0.012 | |
| 1446681_at | Mm.184283 | Mus musculus transcribed sequences | 3.24 | 3E-05 | 0.037 | |||
| 1451280_at | NM_028755 | Mus musculus cyclic AMP-regulated phosphoprotein, 21 (Arpp21) | Arpp21 | SC | 3.15 | 2E-05 | 0.013 | |
| 1457806_at | NM_001033420 | dedicator of cyto-kinesis 1 | Dock1 | 3.12 | 2E-05 | 0.007 | ||
| 1419033_at | NM_001037750 | Mus musculus transcribed sequence with weak similarity to protein pir:I58401 | PirI584 | 3.12 | 2E-05 | 0.028 | ||
| 1416266_at | NM_018863 | prodynorphin | Pdyn | Litt | 3.06 | 7.8E-05 | 0.006 | |
| 1440148_at | NM_199058 | G protein-coupled receptor 6 | GPR6 | 3.06 | 3E-05 | 0.070 | ||
| 1417680_at | NM_145983 | potassium voltage-gated channel, shaker-related subfamily, member 5 | Kcna5 | Gli | 3.05 | 5.2E-05 | 0.018 | |
| 1425467_a_at | NM_011123 | proteolipid protein (myelin) | Myelin | Gli | 2.97 | 2E-05 | 0.059 | |
| 1433652_at | NM_183336 | immunoglobulin superfamily, member 1 | IGSF1 | Litt | 2.96 | 2E-05 | 0.007 | |
| 1454906_at | NM_011243 | retinoic acid receptor, beta | Rarb | Litt | 2.96 | 2E-05 | 0.067 | |
| 1440091_at | NP_034955 | myeloid ecotropic viral integration site-related gene 1 | Mrg1 | S | 2.96 | 2.1E-04 | 0.057 | |
| 1455304_at | XM_146948 | Mus musculus similar to Munc13-3 (LOC235480), mRNA | Munc13-3 | 2.94 | 2E-05 | 0.003 | 1437319_at | |
| 1435854_at | NM_153520 | transmembrane protein 10 | Tmem10 | 2.88 | 2E-05 | 0.014 | ||
| 1433578_at | NM_173403 | hypothetical protein E130304D01 | E130 | S | 2.83 | 2E-05 | 0.002 | |
| 1427038_at | NM_001002927 | preproenkephalin 1 | PENK1 | S - Litt | 2.81 | 2E-05 | 0.003 | |
| 1455296_at | NM_001012765 | adenylate cyclase 5 | Adcy5 | S | 2.80 | 2E-05 | 0.009 | |
| 1423415_at | NM_010287 | G protein-coupled receptor 83 | GPR83 | S | 2.78 | 2E-05 | 0.019 | 1439569_at |
| 1440570_at | AK045773 | RIKEN full-length enriched library, clone:B230310I20 | Rik230 | 2.77 | 2.3E-05 | 0.022 | ||
| 1460587_at | AK131933 | Gabrg1 | Sox2 | 2.77 | 1.7E-04 | 0.039 | ||
| 1433434_at | NM_178737 | expressed sequence AW551984 | AW55 | 2.77 | 2E-05 | 0.035 | ||
| 1442561_at | NM_207010 | MAM domain containing 1 | MAM1 | 2.76 | 6.8E-05 | 0.026 | ||
| 1427227_at | NM_010252 | gamma-aminobutyric acid (GABA-A) receptor, subunit gamma 1 | GABA-A | Litt | 2.75 | 4.6E-05 | 0.066 | 1460408_at |
| 1443287_at | XM_917427 | RIKEN full-length enriched library, clone:E330020H17 | Rik3300 | 2.73 | 2E-05 | 0.001 | ||
| 1436733_at | NM_178756 | RIKEN cDNA E130309F12 gene | Rik130 | SC | 2.72 | 2E-05 | 0.063 | 1436734_at |
| 1439618_at | NM_011866 | phosphodiesterase 10A | Pde10A | 2.72 | 5.2E-05 | 0.031 | 1432490_a_at | |
| 1423946_at | NM_145978 | PDZ and LIM domain 2 | PDLIM2 | S | 2.71 | 7.8E-05 | 0.051 | |
| 1445837_at | NM_023872 | Mus musculus potassium voltage-gated channel, subfamily Q, member 5 (Kcnq5), | Kcnq5 | 2.70 | 2E-05 | 0.021 | ||
| 1417217_at | NM_013779 | melanoma antigen, family L, 2 | Magel2 | S | 2.69 | 4.6E-05 | 0.019 | 1432558_a_at |
| 1447640_s_at | NM_016768 | Mus musculus pre B-cell leukemia transcription factor 3 (Pbx3), | Pbx3 | S | 2.69 | 2E-05 | 0.038 | |
| 1434394_at | NM_001024917 | FLJ10680 protein | FLJ106 | S | 2.69 | 1.7E-04 | 0.019 | |
| 1454656_at | XM_147847 | spermatogenesis associated 13 | Spata13 | SC | 2.68 | 1E-04 | 0.003 | |
| 1441629_at | NM_028736 | Mus musculus glutamate receptor interacting protein 1 (Grip1) | Grip1 | 2.65 | 3E-05 | 0.070 | ||
| 1416783_at | NM_009311 | tachykinin 1 | Tac1 | Litt | 2.65 | 2E-05 | 0.050 | |
| 1460408_at | NM_010252 | gamma-aminobutyric acid (GABA-A) receptor, subunit gamma 1 | GABA-A | 2.63 | 2E-05 | 0.043 | 1427227_at | |
| 1450975_at | NM_019431 | calcium channel, voltage-dependent, gamma subunit 4 | Cacng4 | 2.62 | 3E-05 | 0.595 | ||
| 1436263_at | NM_008614 | myelin-associated oligodendrocytic basic protein | Mobp | Gli | 2.62 | 3E-05 | 0.001 | |
| 1434594_at | NM_177336 | RIKEN cDNA B230373P09 gene | Dho6 | S | 2.61 | 3.5E-4 | 0.008 | |
| 1448590_at | NM_009933 | procollagen, type VI, alpha 1 | Col6a1 | Gli | 2.61 | 2E-05 | 0.038 | |
| 1416997_a_at | NM_010404 | huntingtin-associated protein 1 | Hap1 | Litt | 2.61 | 1.3E-04 | 0.091 | |
| 1448860_at | NM_080726 | rad and gem related GTP binding protein 2 | Rem2 | SC | 2.60 | 1.3E-04 | 0.051 | |
| 1417996_at | NM_022414 | neuroglobin | Ngb | Litt | 2.60 | 7.8E-05 | 0.050 | 1417997_at |
| 1429906_at | AK032623 | RIKEN full-length enriched library, clone:6430710A17 | Rik643 | 2.59 | 4.6E-05 | 0.169 | ||
| 1435343_at | XM_976471 | dedicator of cytokinesis 10 | Dock10 | S | 2.58 | 2E-05 | 0.005 | |
| 1436734_at | NM_178756 | RIKEN cDNA E130309F12 gene | Rik130 | SC | 2.54 | 1.9E-04 | 0.002 | 1436733_at |
| 1455629_at | NM_010076 | dopamine receptor D1A | DRD1a | Litt | 2.54 | 2.3E-05 | 0.004 | 1456051_at |
| 1456051_at | NM_010076 | dopamine receptor D1A | DRD1a | Litt | 2.54 | 2.7E-05 | 0.042 | 1455629_at |
| 1423171_at | NM_022427 | G-protein coupled receptor 88 | GPR88 | SC | 2.54 | 2E-05 | 0.157 | |
| 1450241_a_at | NM_010161 | ecotropic viral integration site 2a | Evi2a | S | 2.54 | 2E-05 | 0.006 | |
| 1455190_at | NM_010319 | guanine nucleotide binding protein (G protein), gamma 7 subunit | Gng7 | Litt | 2.53 | 2E-05 | 0.023 | |
| 1428642_at | NM_029529 | frc, fringe-like 1 (Drosophila) | Slc35d3 | S | 2.53 | 4.6E-05 | 0.049 | |
| 1440573_at | NM_021563 | Mus musculus Erbb2 interacting protein (Erbb2ip), transcript variant 2, mRNA | Erbb2ip | S | 2.53 | 3.5E-05 | 0.012 | |
| 1445148_at | AK049014 | RIKEN full-length enriched library, clone:C230091K16 | RikC23 | 2.52 | 2E-05 | 0.078 | ||
| 1447807_s_at | XM_126961 | pleckstrin homology domain containing, family H (with MyTH4domain) member 1 | Max-1 | S | 2.49 | 2E-05 | 0.021 | |
| 1448945_at | NM_026385 | transmembrane 4 superfamily member 11 | Trn4sf11 | Gli | 2.47 | 2E-05 | 0.039 | |
| 1429589_at | AK018118 | RIKEN full-length enriched library, clone:6330404F12 | Rik633 | 2.46 | 2E-05 | 0.035 | ||
| 1427519_at | NM_009630 | adenosine A2a receptor | Adora2 | SC | 2.45 | 2E-05 | 0.001 | 1460710_at |
| 1433551_at | NM_173016 | hypothetical protein 9430073I07 | Vat-1 | S | 2.45 | 2E-05 | 0.023 | |
| 1418950_at | NM_010077 | dopamine receptor 2 | DRD2 | Litt | 2.44 | 5.2E-05 | 0.050 | |
| 1417804_at | NM_011242 | RAS, guanyl releasing protein 2 | CDC25L | SC | 2.42 | 2E-05 | 0.006 | |
| 1442138_at | XM_488192 | Mus musculus G protein-coupled receptor 62 (Gpr62) | Gpr62 | 2.42 | 2.3E-05 | 0.058 | ||
| 1449863_a_at | NM_010056 | distal-less homeobox 5 | Dlx5 | S | 2.42 | 1.3E-04 | 0.004 | |
| 1437319_at | XM_146948 | Mus musculus similar to Munc13-3 (LOC235480), mRNA | Munc13-3 | 2.41 | 2E-05 | 0.005 | 1455304_at | |
| 1449682_s_at | NM_023716 | TUBULIN, BETA 5 homolog | Tubbh | S | 2.39 | 2E-05 | 0.045 | |
| 1418472_at | NM_023113 | aspartoacylase (aminoacylase) 2 | Aspa | Gli | 2.39 | 2E-05 | 0.029 | |
| 1436662_at | NM_021377 | Mus musculus VPS10 domain receptor protein SORCS 1 (Sorcs1), | Sorcs1 | S | 2.38 | 3.5E-04 | 0.067 | |
| 1435463_s_at | NM_177390 | Mus musculus myosin ID (Myo1d), mRNA | MyosinID | 2.38 | 2.1E-04 | 0.050 | ||
| 1457323_at | XM_905537 | hypothetical protein LOC632191 | LOC632191 | 2.37 | 1.3E-04 | 0.006 | ||
| 1457984_at | NM_205769 | Mus musculus transcribed sequence with strong similarity to protein sp:P00722 | CRF | Litt | 2.36 | 1E-04 | 0.014 | |
| 1451331_at | NM_144828 | protein phosphatase 1, regulatory (inhibitor) subunit 1B | DARPP32 | Litt | 2.35 | 2E-05 | 0.013 | |
| 1425263_a_at | NM_001025245 | myelin basic protein | Mbp | Gli | 2.35 | 6.1E-04 | 0.166 | |
| 1435642_at | NM_178774 | RIKEN cDNA 9630019K15 gene | Rik963 | S | 2.33 | 2E-05 | 0.017 | |
| 1428434_at | NM_028325 | RIKEN cDNA 2810028A01 gene | Sizn | S | 2.32 | 2E-05 | 0.006 | |
| 1418691_at | NM_011268 | regulator of G-protein signaling 9 | RGS9 | Litt | 2.31 | 2E-05 | 0.009 | |
| 1440803_x_at | NM_021382 | tachykinin receptor 3 | Tacr3 | Litt | 2.31 | 6.8E-05 | 0.005 | |
| 1434098_at | NM_183427 | glycine receptor, alpha 2 subunit | Glra2 | S | 2.31 | 3E-05 | 0.064 | |
| 1417129_a_at | NM_010825 | myeloid ecotropic viral integration site-related gene 1 | Mrg1 | S | 2.30 | 2E-05 | 0.007 | 1457632_s_at |
| 1436449_at | AK142834 | Mus musculus transcribed sequences | Pcdhx | 2.28 | 0.000147 | 0.010 | ||
| 1417090_at | NM_009037 | reticulocalbin | Rcn1 | S | 2.28 | 4.6E-05 | 0.002 | |
| 1423506_a_at | NM_010923 | neuronatin | Nnat | S | 2.28 | 2E-05 | 0.078 | |
| 1432490_a_at | NM_011866 | phosphodiesterase 10A | Pde10A | S | 2.27 | 1.8E-04 | 0.011 | 1439618_at |
| 1417275_at | NM_010762 | myelin and lymphocyte protein, T-cell differentiation protein | Mal | Gli | 2.27 | 2E-05 | 0.013 | |
| 1460710_at | NM_009630 | adenosine A2a receptor | Adora2 | SC | 2.24 | 2E-05 | 0.002 | 1427519_at |
| 1448768_at | NM_010814 | myelin oligodendrocyte glycoprotein | Mog | Gli | 2.24 | 2E-05 | 0.005 | |
| 1437418_at | Mm.11171 | Mus musculus transcribed sequence with weak similarity to protein ref:NP_061720.1 | NP061720 | 2.24 | 2.4E-04 | 0.124 | ||
| 1417673_at | NM_016719 | growth factor receptor bound protein 14 | Grb14 ppGANT ase | S | 2.23 | 8.9E-05 | 0.029 | |
| 1440902_at | NM_029972 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 | 5 | S | 2.23 | 2E-05 | 0.071 | |
| 1460219_at | NM_010758 | myelin-associated glycoprotein | Mag | S | 2.22 | 2E-05 | 0.008 | |
| 1422756_at | NM_009508 | vesicular inhibitory amino acid transporter | Slc32a1 | Litt | 2.22 | 2E-05 | 0.006 | |
| 1449365_at | NM_053190 | endothelial differentiation, sphingolipid G-protein-coupled receptor, 8 | Edg8 | Gli | 2.21 | 4E-05 | 0.002 | |
| 1456174_x_at | NM_010884 | N-myc downstream regulated 1 | Ndrg1 | S | 2.20 | 2E-05 | 0.081 | |
| 1457132_at | Mm.26805 | Mus musculus transcribed sequences | 2.20 | 2.7E-05 | 0.140 | |||
| 1439506_at | XM_983842 | Mus musculus gene model 98, (NCBI) (Gm98) | C11orf9 | 2.19 | 7.8E-05 | 0.004 | ||
| 1457632_s_at | NM_010825 | myeloid ecotropic viral integration site-related gene 1 | Mrg1 | S | 2.19 | 2E-05 | 0.001 | 1417129_a_at |
| 1425892_a_at | NM_010932 | Prepronociceptin | Pnoc | Litt | 2.18 | 2.3E-05 | 0.012 | |
| 1444345_at | AW123227 | Mus musculus transcribed sequences | 2.16 | 3E-04 | 0.359 | |||
| 1440813_s_at | XM_621025 | plexin B3 | PLNX6 | Gli | 2.16 | 2E-05 | 0.020 | |
| 1416200_at | NM_133775 | RIKEN cDNA 9230117N10 gene | Rik923 | S | 2.16 | 2.3E-05 | 0.023 | |
| 1434369_a_at | NM_009964 | crystallin, alpha B | Crya2 | Gli | 2.15 | 2E-05 | 0.033 | |
| 1424468_s_at | NM_153537 | RIKEN cDNA D330037A14 gene | Rik330 | S | 2.15 | 1.7E-04 | 0.042 | |
| 1416003_at | NM_008770 | claudin 11 | Cldn11 | S | 2.14 | 3E-05 | 0.008 | |
| 1435554_at | NM_172051 | Transmembrane and coiled coil domains 3 (Tmcc3), mRNA | Tmcc3 | 2.13 | 2E-05 | 0.073 | ||
| 1415975_at | NM_025821 | calcium regulated heat stable protein 1 | Chrsp-24 | 2.11 | 7.8E-05 | 0.012 | ||
| 1434606_at | NM_010153 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | Erbb3 | Gli | 2.09 | 1E-04 | 0.021 | |
| 1449106_at | NM_008161 | glutathione peroxidase 3 | Gpx3 | 2.09 | 8.6E-04 | 0.142 | ||
| 1426690_a_at | NM_011480 | sterol regulatory element binding factor 1 | Sebf1 | Gli | 2.06 | 1.1E-04 | 0.073 | |
| 1424843_a_at | NR_002840 | growth arrest specific 5 | Gas5 | S | 2.06 | 4.6E-05 | 0.008 | |
| 1426454_at | NM_007486 | Rho, GDP dissociation inhibitor (GDI) beta | Arhgdib | S | 2.05 | 6.2E-04 | 0.045 | |
| 1416303_at | NM_019980 | LPS-induced TN factor | LITAF | S | 2.04 | 2E-05 | 0.009 | |
| 1426545_at | NM_144812 | trinucleotide repeat containing 6b | Tnrc6b | S | 2.04 | 3.5E-04 | 0.832 | |
To characterize the enriched transcriptome in the Extended Amygdala, we used a GO database and annotated the 121 genes selected, as previously described (Chalmel et al., 2005, Befort et al., 2008b). We found 98 genes associated with proteins, among which 82 were associated with GO terms. These genes were then grouped into categories of biological processes, in which their encoded proteins are involved (GOBP) and these categories are illustrated in Figure 1C. This analysis showed that some GO categories are enriched, particularly in the response to stress (GO:0006950), behavior (GO:0007610), cell communication (GO:0007154), signal transduction (GO:0007165) and synaptic transmission (GO:0007268). Interestingly, a large group included neuropeptide signaling pathway (GO:0007218) and GPCR signaling pathway (GO:0007186) and some transcripts were also associated with neurogenesis (GO:0007399), organogenesis (GO: 00099887), central nervous system development (GO:0007417) and nerve maturation (GO:00042551). To interrogate potential functional interactions among EAc enriched genes, we also used the Ingenuity Pathways Analysis (Table 2). Our results showed that, among the top biological functions, potential interactions were revealed by putative protein networks involved in Cell Morphology, Gene Expression, Cell Signaling and Nervous System Development and Function. Interestingly, these two bioinformatics analysis were consistent and revealed that the genes we have identified in our study participate in functions described in the literature as involving the EAc and suggest new functions for this network.
Table 2.
Top gene networks identified in Ingenuity Pathway analysis. Number of genes and corresponding top functions for which expression is higher in the Extended Amygdala as compared to the whole brain.
| Top biological functions | Score | Focus genes | genes |
|---|---|---|---|
| Behavior, Digestive System Development and Function, Nervous System Development and Function | 52 | 24 | Adcy5, Adora2, Rik352, CRF, Dlx5, Drd1a, Drd2, Galn, Gpx3, Grb14 Grp1, Rik923, Kcnq5, Mrg1, Nts, Pdyn, Penk, Pnoc, PirI584, Sebf1,, Tac1, Tac2, Tacr3 |
| Neurological Disease, Cell Morphology, Nervous System Development and Function | 28 | 15 | Akt, Cacnb2, Col6a1, Erbb3, Gad2, Gas5, Indo, Irs4, Isl1, Litaf, Mag, Mbp, Mog, Slc12A7, SlcA1 |
| Cancer, Neurological Disease, Cell Death | 17 | 10 | Arpp21, Chrsp-24, Dock10, Glra2, MyosinID, Nnat, Rik330, Plnx6, Tubbh, Wfs1 |
| Reproductive System Development and Function, Gene Expression, Amino Acid Metabolism | 16 | 10 | Cacng4, Dock1, Edg8, Gaba-A, Igsf1, Kcna5A, Mal, Pbx3, Pdlim2, Rgs9 |
| Cell Death, Hepatic System Disease, Liver Necrosis/Cell Death | 15 | 9 | Cldn11, Gng7, Hap1, Limh6, Limh8, Ngb, Pde10A, Cdc25L, Six3 |
| Cell Signaling, Molecular Transport, Vitamin and Mineral Metabolism | 14 | 9 | Arhgdib, Aspa, ppGantase5, GPR6, GPR83, Flj106, Ndrg1, Rcn1, Sorcs1 |
Altogether, this microarray screen identifies 23 genes that were already reported by others in EAc (see Table 1 column ISH label Litt.), some of which are widely studied (for example tachykinin 1 and 2, prodynorphin or dopamine D1 receptor). Importantly, the data also highlight a large number of genes whose expression in EAc is reported here for the first time.
Expression pattern by in situ hybridization
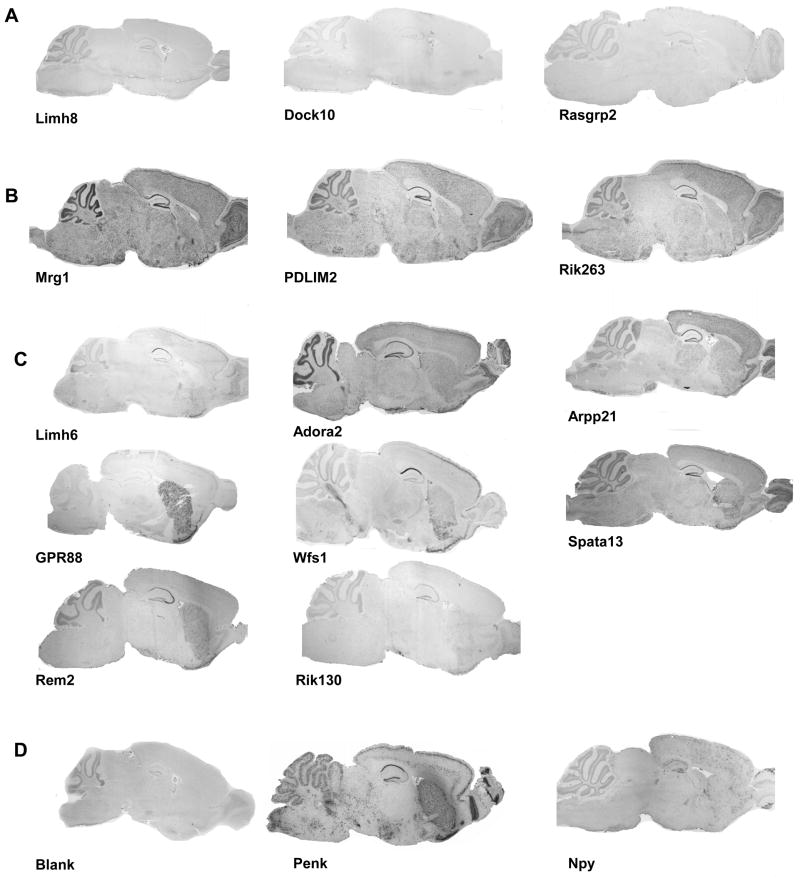
We further performed in situ hybridization (ISH) throughout the brain to examine expression patterns for a sub-selection of genes. We focused on 49 genes whose expression pattern in BNST and/or CeA was not studied in the mouse previously (Table 1 column ISH label S or SC). We first performed non isotopic ISH on sagittal sections of mouse brain. Our results show that a signal was detectable in the EAc for all tested probes confirming our microarray analysis (see Supplemental Figure S2). Expression patterns could be described as (i) low expression, (ii) strong and fairly ubiquitous expression or (iii) significant expression restricted to specific brain regions including the EAc (see Figure 2 and Methods). Altogether, 17 genes (see supplementary Figure S2A) showed a weak signal under our hybridization conditions and those were not further investigated. Figure 2A shows sagittal ISH for three of these genes, Limh8, Dock10 and Rasgrp2 whose expression was close to the detection limit. Twenty-four other genes showed a hybridization signal widely spread along the brain (see supplementary Figure S2B). Figure 2B shows an example for three of these genes, namely Mrg1, Rik263, PDLIM2 with staining throughout the brain. Lastly, 8 genes were clearly detected in EAc, with weak or restricted expression in other brain regions. Figure 2C shows the sagittal expression pattern for these genes namely Limh6, Adora2, Arpp21, GPR88, Wfs1, Spata13, Rem2 and Rik130.
Figure 2. Expression pattern of EAc-enriched genes by in situ hybridization on sagittal brain sections.
Dig-labeled RNA in situ hybridization (ISH) was performed on 25μm sagittal adult mouse brain sections 49 genes whose specific expression pattern in EAc network has not been reported earlier. In this figure, examples of low expressed genes (A), strong and ubiquitously expressed genes (B) and potential novel EAc markers (C) are shown. Neuropeptide Y (Npy) was used as a low intensity positive control, preproenkephalin (Penk) was used as a high intensity positive control and a blank hybridization was used as the negative control (D). Representative images are shown, and the complete ISH analysis on sagittal sections for all 49 genes is shown in supplemental Figure S2. Expression patterns were classified as detailed in methods. Gene symbols are indicated as in Table 1.
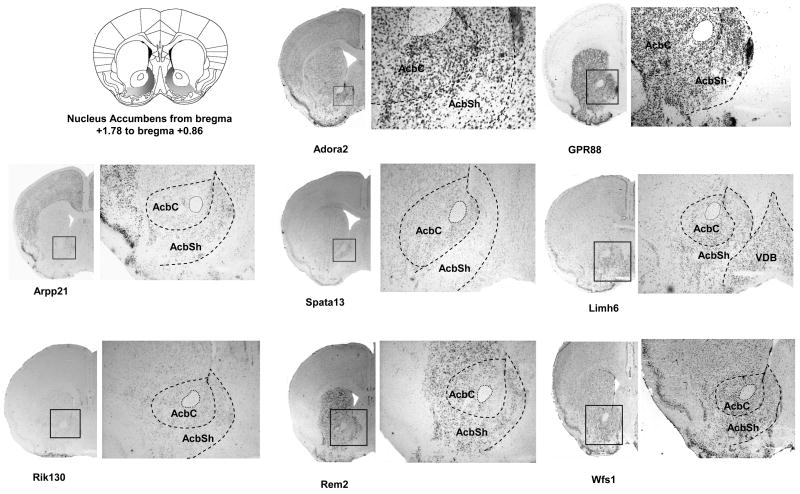
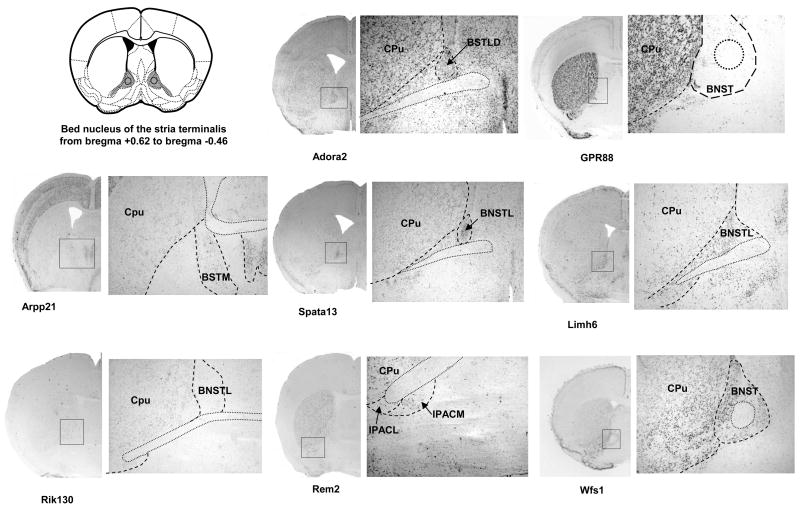
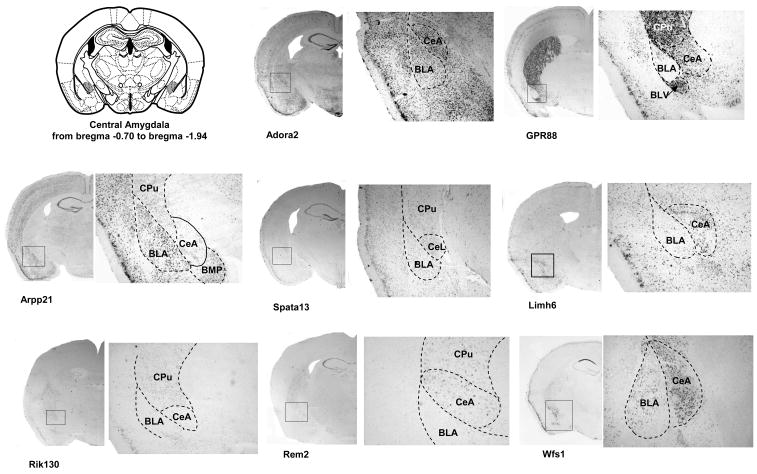
We next performed ISH on coronal sections for the last 8 genes (Figure 3). We focused our analysis on sections corresponding to the EAc, at the levels of BNST (Figure 3B) and CeA (Figure 3C). As the AcbSh was proposed to be part of the Extended Amygdala (Koob, 2003), we also included analysis of the staining observed at the level of this structure (Figure 3A). The expressed sequence tag RIKEN E130309F12 probe (Rik130) presented a weak expression in the AcbSh that diminished gradually rostro-caudally and no signal could be detected in the CeA. The Rad and gem related GTP binding protein 2 transcript (Rem2) was strongly expressed in the Nucleus Accumbens including shell and core, and in the interstitial nucleus of the posterior limb of the anterior commissure (IPACL), which is part of BNST. A patchy distribution in the CeA was observed and Rem2 was also clearly expressed in the dorsal striatum. Concordant with our sagittal screen, the adenosine A2 receptor gene (Adora2) showed a strong and consistent expression in all areas of the EAc network. In the BNST, the lateral division was particularly labeled (Figure 3B). Adenosine A2 receptor mRNA expression was also present in several brain areas such as the striatum (Cpu and core of the Nucleus Accumbens), the cortex, the hippocampus and the piriform cortex. The cyclic AMP-regulated phosphoprotein 21 gene (Arpp21) presented an interesting expression pattern, with specific mRNA expression in the AcbSh, without signal in adjacent structures like the core of the Nucleus Accumbens and the caudate putamen (Figure 3B). At the level of the BNST, a specific signal was detected in the medial division. No detectable staining was obtained in the CeA while the Arpp21 gene was highly expressed in the basolateral and basomedial amygdaloid nucleus. Specific signal was also measured in the medial preoptic nucleus, the cortex and the hippocampus. GPR88 gene showed strong expression throughout the striatum, including the AcbSh (Figure 3A). GPR88 mRNA expression was undetectable in the BNST (Figure 3B). A strong staining was observed in the medial division of the CeA with no signal in the basolateral Amygdala. The GPR88 mRNA was also clearly present in the piriform cortex. For the LIM homeobox 6 (Limh6) mRNA, we obtained a strong and homogenous expression throughout areas of the Extended Amygdala including AcbSh, the lateral division of the BNST and the CeA. We also noticed expression of this gene on the nucleus of the vertical limb of the diagonal band (VDB) (Figure 3A). A strong signal was measured for the spermatogenesis associated 13 (Spata 13) gene in the dorsal part of the lateral division of BNST compared to other BNST nuclei, as well as in the AcbSh and CeA. Additionally, a low Spata 13 staining was observed in the CA1 field of the hippocampus. Finally, the wolframin gene (Wfs1) showed strong expression throughout the Extended Amygdala. This gene was strongly expressed in the AcbSh and BNST, with a low staining in the caudate putamen. (Figure 3A and 3B). Interestingly, expression of this gene was strong in the CeA, and extremely low in the BLA clearly defining the boundaries between these two amygdaloid nuclei (Figure 3C). Wolframin mRNA was also strongly expressed specifically in the CA1 field of the hippocampus and the piriform cortex.
Figure 3. Expression analysis of EAc-enriched genes in AcbSh, BNST and CeA on coronal brain sections.
Dig-labeled RNA in situ hybridization (ISH) was performed for a selection of 8 potential EAc markers using 25μm coronal sections of mouse brain. A scheme from the mouse brain atlas with the coordinates (Paxinos and Franklin, 2001) shows location of (A) the shell of the Nucleus Accumbens (AcbSh), (B) the bed nucleus of stria terminalis (BNST) and (C) the central nucleus of the Amygdala. Representative ISH images are shown for each candidate gene with an enlargement (zoom) for specific areas of interest. Abbreviations: AcbSh, accumbens nucleus, shell; AcbC, accumbens nucleus, core ; VDB, nucleus of the vertical limb of the diagonal band; CPu, caudate putamen; IPACL, interstitial nucleus of the posterior limb of the anterior commissure, lateral part; IPACM, interstitial nucleus of the posterior limb of the anterior commissure, medial part BSTLD, Bed Nucleus of the Stria Terminalis, lateral division, dorsal part; BSTL, Bed Nucleus of the Stria Terminalis, lateral division; BSTM, Bed Nucleus of the Stria Terminalis, medial division; BLA, basolateral amygdaloid nucleus, anterior part ; BLV, basolateral amygdaloid nucleus, ventral part CeA central amygdaloid nucleus; CeL, central amygdaloid nucleus, lateral division; BMP, basomedial amygdaloid nucleus, posterior part (Paxinos and Franklin, 2001)
In conclusion, these 8 genes show a specific and distinct expression pattern within the EAc. They are expressed throughout the AcbSh and CeA, and show locally restricted expression in the subdivisions of the BNST structure.
Distribution of EAc-enriched genes in the central nervous system and peripheral tissues by qPCR
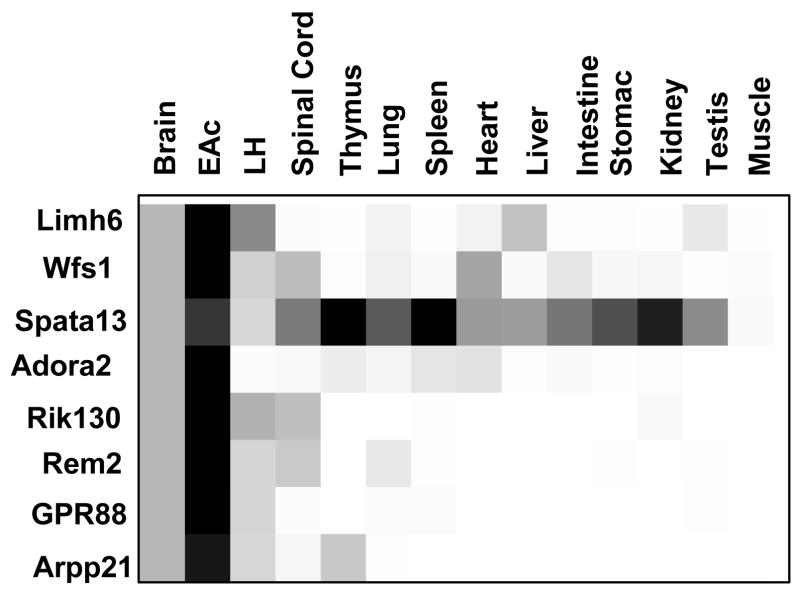
To examine the general expression pattern of these 8 genes throughout the central nervous system and in peripheral organs, we performed quantitative PCR on cDNA samples from WB, EAc and LH (a brain region that we have investigated in a separate study, see Befort et al (Befort et al., 2008a), as well as spinal cord, thymus, lung, spleen, heart, liver, intestine, stomach, kidney, testis, and muscle (Figure 4). The results confirmed high expression in the EAc for all the tested genes compared to whole brain. The Spata-13 gene showed the less restricted pattern of expression, with high expression levels in thymus, kidney and spleen (4.7-, 3.1- and 1.7-fold higher than in the EAc, respectively). In contrast, the GPR88 gene was detected only in the brain, with its highest level of expression in the EAc. Expression of the Rik130 transcript was restricted to the nervous system, including the spinal cord. Finally the Wfs1, Limh6, Rem2, Arpp21 and Adora2 genes showed detectable expression in several peripheral tissues, and none of the transcripts tested could be detected in muscle.
Figure 4. Expression of EAc-enriched genes in the central nervous system and peripheral tissues by qPCR.
Quantitative PCR reactions were performed in triplicate on 2 independent samples (3 mice pooled for EAc, 2 mice pooled for WB and individual n=2 mice all other tissues, see Methods for details) and data are expressed as a fold-change over WB considered as the reference sample. Each gene is represented by a single box row, with expression levels illustrated using a grey scale from white (low level) to black (high level). All the genes are mainly expressed in the central nervous system (CNS), with the exception of Spata-13 expressed in most tested tissues at levels similar or higher to CNS.
Discussion
Using Affymetrix microarrays, we have identified 129 probe-sets corresponding to 121 genes with prominent expression in the EAc. We used a strategy where gene expression in a sample from the region of interest (CeA and BNST) is compared with expression in a sample from the whole brain. Others have used a similar approach in the past, and successfully identified neuronal markers (Cahoy et al., 2008), subregional-specific genes within mouse Amygdala nuclei (Zirlinger et al., 2001, Zirlinger and Anderson, 2003), or regional gene markers within the several hippocampal fields (Lein et al., 2004, Leonardo et al., 2006).
Twenty-three genes enriched in EAc have been reported earlier as being expressed in the rodent EAc (Tables 1). For example, we detected enriched expression of neuropeptide precursor genes such as preprodynorphin (3.06-fold in EAc over WB), preproenkephalin (2.81-fold in EAc over WB) or preprotachykinin 2(6.35-fold in EAc over WB) genes, whose expression patterns were described previously in specific areas of the Extended Amygdala (Harlan et al., 1987, Iadarola et al., 1989, Song and Harlan, 1994). We also identified genes encoding myosin D (2.38-fold in EAc over WB), neuronatin (2.28-fold in EAc over WB) and myelin associated glycoprotein (2.22-fold in EAc over WB). These genes were described as specific markers of particular Amygdala nuclei in a microarray screen analysis from Zirlinger and coll (Zirlinger, 2003, Zirlinger and Anderson, 2003). Among the top enriched genes, we finally identified the wolframin gene (3.38-fold in EAc over WB). This transcript was previously described as specifically expressed in the CA1 region of the hippocampus, and central Amygdala (Takeda et al., 2001, Leonardo et al., 2006). Altogether, our identification of previously reported Amygdalar genes validates our gene selection strategy.
Importantly, literature mining indicated that expression in the EAc was unknown for 49 other genes. Our further ISH mapping analysis revealed that 8 of these genes show an expression pattern of particular interest within the EAc. We compared the expression patterns of these 8 genes with the Allen Brain Atlas (Lein et al., 2007) and, with the exception of Arpp21 and Wfs1, our analysis showed a similar pattern in the three focused areas of interest. The two expression patterns for Arpp21 transcript presented in the Allen Brain Atlas showed a more widespread expression than our ISH data. However, our ISH results were consistent with Arpp21 protein distribution in rat brain (Ouimet et al., 1989). For the Wfs1 transcript, the pattern in the Allen Brain Atlas appeared ubiquitous, under our criteria (see methods), compared with the expression profile in the rat brain (Takeda et al., 2001) or shown here.
Altogether, with the exception of Rik130, all these genes encode a protein with either fully identified or potential function. For each of these genes, the unique and distinct expression pattern suggests a potential role in regulating the EAc network.
Spata13 showed moderate enrichment in EAc (ISH and qPCR), and was the only gene with strong expression in peripheral organs (Q-PCR) including thymus, lung or kidney. This gene (also called Asef2) was identified as a guanine-nucleotide exchange factor for Rac1 and Cdc42 (Hamann et al., 2007, Kawasaki et al., 2007). Rik130 was specifically expressed in the brain compared to peripheral tissues, but the expression in the EAc was quite low. No information is available on Rik130 distribution or function. A blast search in the Refprot database indicates that the Rik130 protein shows 74% homology with phosphatidic acid phosphatase type 2B (ppap2B), a gene ubiquitously and strongly expressed throughout the brain (http://brain-map.org/).
Limh6 encodes a protein belonging to the LIM homeodomain proteins, a family of transcription factors. Expression of this gene was high in brain, particularly in EA and barely detectable in the periphery. The LIM protein family is involved in many processes of CNS development, from cell fate specification to the establishment of neuronal connectivity. During embryogenesis Limh6 was found involved in the migration of cortical GABAergic interneurons (Cobos et al., 2006, Cuzon et al., 2008). Noticeably, several authors have proposed a role for Limh6 in the development of the Extended Amygdala in mice (Choi et al., 2005, Garcia-Lopez et al., 2008) or zebra fish (Mueller et al., 2008). Our Affymetrix analysis also identified Limh8, another member of the family, as a gene enriched in the EAc (Table 1), and the expression of Limh8 was also described in the adult mouse Amygdala in another report (Zirlinger and Anderson, 2003). These findings suggest that LIM genes, expressed in the adult brain, may contribute to remodeling of the EAc network (Grueter and Winder, 2005, Samson et al., 2005).
Genes with a noticeable expression pattern within the EAc include four genes involved in cell signaling, namely Adora2a, the orphan GPR88, Arpp21 and Rem2. Adora2a encodes the adenosine 2A receptor, a Gs-coupled G protein coupled receptor known to be expressed in the striatum and olfactory tubercules (Santicioli et al., 1993) and involved in many brain functions (for review see (Yaar et al., 2005)). Relevant to our finding of Adora2 expression in the EAc, previous evidence have suggested a role for Adora2 in Amygdala function. The Adora2 agonist CGS21680 was associated with an apoptosis regression in the Amygdala following myocardial infarction (Boucher et al., 2006) and a polymorphism in the human Adora2a gene was associated with panic disorders (Yamada et al., 2001, Hamilton et al., 2004, Lam et al., 2005). GPR88 is an orphan G-protein coupled receptor previously reported as a striatal transcript (Mizushima et al., 2000, Ghate et al., 2007). In accordance, our mapping data confirm prominent expression of this gene in both EAc and the caudate putamen. GPR88 function is unknown, but the alteration of GPR88 expression was reported under several experimental conditions. GPR88 mRNA was up-regulated in the rat arcuate ventromedial nucleus during lactation (Xiao et al., 2005), in the prefrontal cortex following metamphetamine or valpoate exposure (Ogden et al., 2004), and down-regulated in the striatum of human patients with Huntington’s disease (Hodges et al., 2006). Arpp-21 is a cyclic AMP-regulated phosphoprotein of 21 kDa whose expression was reported in the caudate putamen and substantia nigra (Girault et al., 1990). Arpp-21 has been associated to several aspects of dopamine signaling (Tsou et al., 1993, Ivkovic and Ehrlich, 1999), and is down-regulated in substantia nigra of sporadic parkinsonian patients (Grunblatt et al., 2007). Finally, Rem2 encodes a Ras-like GTPAse (Finlin et al., 2000) initially described as a modulator of N-type current in primary neuron cultures (Chen et al., 2005). Recently, this protein was shown to play a key role in the development of glutamatergic and GABAergic synapses in rat hippocampus primary cultures (Paradis et al., 2007).
Most remarkable was the expression of Wfs1 gene, which showed best EAc enrichment among the final eight-gene subset. This gene appeared strongly expressed throughout the AcbSh, BNST and CeA, and weak -if no- expression was detected in most other brain regions. ISH indicated scarce expression in the caudate, olfactory tubercle, locus coeruleus, cerebellar cortex (Takeda et al., 2001). Our mapping study also highlighted the intriguing hippocampal expression of Wfs1, uniquely restricted to the CA1 field (see Figure 3C) as was previously described by two groups (Takeda et al., 2001, Leonardo et al., 2006). Expression of Wfs1 was also detectable in the spinal cord, heart and intestine. Mutations in the Wfs1 gene are responsible for the Wolfram syndrome, an autonomous recessive disorder which is highly variable in its clinical manifestations and includes diabetes mellitus, diabetes insipidus, deafness, optic atrophy and psychiatric abnormalities (Cryns et al., 2003). The Wfs1gene was originally characterized by positional cloning and encodes a transmembrane protein, wolframin, (Inoue et al., 1998) whose precise role in cell physiology remains to be determined. Knockout of the Wfs1 gene in mice leads to a loss of pancreatic beta-cell, suggesting a role in maintaining some populations of endocrine cells (Ishihara et al., 2004) (Riggs et al., 2005), however the potential neurological and behavioral phenotypes of Wfs1 null mutants have not been reported as yet. Interestingly the Wfs1 messenger was up-regulated in rat Amygdala (Koks et al., 2002) and mouse pre-frontal cortex (Raud et al., 2007) when animals were exposed to cat odor, suggesting a implication of wolframin in stress responses. Together, both anatomical and functional data suggest a prominent implication of Wfs1 in emotionally related behaviors.
In conclusion, we have identified a set of genes with enriched expression in the EAc, and examined the expression pattern for a number of these genes particularly in the EAc. This study extends our knowledge of genes expressed within a brain network, which is central in reward dysregulation and stress disorders. It will be interesting to further characterize neural populations expressing these genes, and particularly to examine whether these transcripts are present in GABAergic neurons that are best characterized within the EAc (Sun and Cassell, 1993). Additionally, BAC transgenic approaches has been used to label (Gong et al., 2003) or to isolate specific cells population in the brain (Nielsen et al., 2006, Cahoy et al., 2008) (Cahoy et al., 2008) (Lovatt et al., 2007). The generation of transgenic mice expressing eGFP under the control of EAc specific genes could be useful to label EAc neurons and study their function and plasticity under pathological stimulations.
Supplementary Material
Probe sets ID are indicated, with the corresponding name and Refseq information from Affymetrix. Forward and reverse primer sequences used for qPCR analysis of 8 candidate EAc marker genes are indicated.
Dig-labeled RNA in situ hybridization (ISH) was performed on 25μm sagittal adult mouse brain sections 49 genes whose expression pattern in EAc network has not been reported earlier. This figure shows the 17 genes with low expression (A) and the 24 genes with strong and ubiquitously expressed genes (B). Expression patterns were classified as detailed in methods. Sagittal ISH of the 8 potential EAc markers are shown on Figure 2. Gene symbols are indicated as in Table 1.
Acknowledgments
We thank Pascal Dollé, C. Thaller and G. Eichele for their advice about in situ hybridization and Julie Le Merrer for careful reading of the manuscript. We are thankful to IGBMC Core facilities. This work was supported by Centre National de la Recherche Scientifique, Institut de la Santé Et de la Recherche Médicale, Université Louis Pasteur. The authors wish to thank the National Institute of Health (NIH-NIAAA AA016658, NIDA 016768), and the European Union (GENADDICT/FP6 005166; EURExpress # LSHG-CT-2004-512003) for financial support. AG was supported by an IBRO PhD fellowship. JM was supported by grants from the National Research Fund and the Ministère de la Culture, de l’Enseignement Supérieur et de la Recherche of Luxembourg.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abou-Sleymane G, Chalmel F, Helmlinger D, Lardenois A, Thibault C, Weber C, Merienne K, Mandel JL, Poch O, Devys D, Trottier Y. Polyglutamine expansion causes neurodegeneration by altering the neuronal differentiation program. Hum Mol Genet. 2006;15:691–703. doi: 10.1093/hmg/ddi483. [DOI] [PubMed] [Google Scholar]
- Alheid GF, Heimer L. New perspectives in basal forebrain organization of special relevance for neuropsychiatric disorders: the striatopallidal, amygdaloid, and corticopetal components of substantia innominata. Neuroscience. 1988;27:1–39. doi: 10.1016/0306-4522(88)90217-5. [DOI] [PubMed] [Google Scholar]
- Befort K, Filliol D, Darcq E, Ghate A, Matifas A, Lardenois A, Muller J, Thibault C, Dembele D, Poch O, Kieffer BL. Gene expression is altered in the lateral hypothalamus upon activation of the mu opioid receptor. Ann N Y Acad Sci. 2008a;1129:175–184. doi: 10.1196/annals.1417.028. [DOI] [PubMed] [Google Scholar]
- Befort K, Filliol D, Ghate A, Darcq E, Matifas A, Muller J, Lardenois A, Thibault C, Dembele D, Le Merrer J, Becker JAJ, Poch O, Kieffer BL. Mu-opioid receptor activation induces transcriptional plasticity in the central extended amygdala. J neurosci. 2008b;27:2973–2984. doi: 10.1111/j.1460-9568.2008.06273.x. [DOI] [PubMed] [Google Scholar]
- Bonaventure P, Guo H, Tian B, Liu X, Bittner A, Roland B, Salunga R, Ma XJ, Kamme F, Meurers B, Bakker M, Jurzak M, Leysen JE, Erlander MG. Nuclei and subnuclei gene expression profiling in mammalian brain. Brain Res. 2002;943:38–47. doi: 10.1016/s0006-8993(02)02504-0. [DOI] [PubMed] [Google Scholar]
- Boucher M, Wann BP, Kaloustian S, Cardinal R, Godbout R, Rousseau G. Reduction of apoptosis in the amygdala by an A2A adenosine receptor agonist following myocardial infarction. Apoptosis. 2006;11:1067–1074. doi: 10.1007/s10495-006-6313-6. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JP, Thaller C, Eichele G. A transcriptome atlas of the mouse brain at cellular resolution. Curr Opin Neurobiol. 2002;12:562–565. doi: 10.1016/s0959-4388(02)00356-2. [DOI] [PubMed] [Google Scholar]
- Cassell MD, Freedman LJ, Shi C. The intrinsic organization of the central extended amygdala. Ann N Y Acad Sci. 1999;877:217–241. doi: 10.1111/j.1749-6632.1999.tb09270.x. [DOI] [PubMed] [Google Scholar]
- Chalmel F, Lardenois A, Thompson JD, Muller J, Sahel JA, Leveillard T, Poch O. GOAnno: GO annotation based on multiple alignment. Bioinformatics. 2005;21:2095–2096. doi: 10.1093/bioinformatics/bti252. [DOI] [PubMed] [Google Scholar]
- Chen H, Puhl HL, 3rd, Niu SL, Mitchell DC, Ikeda SR. Expression of Rem2, an RGK family small GTPase, reduces N-type calcium current without affecting channel surface density. J Neurosci. 2005;25:9762–9772. doi: 10.1523/JNEUROSCI.3111-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Dong HW, Murphy AJ, Valenzuela DM, Yancopoulos GD, Swanson LW, Anderson DJ. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46:647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;16(Suppl 1):i82–88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Cryns K, Sivakumaran TA, Van den Ouweland JM, Pennings RJ, Cremers CW, Flothmann K, Young TL, Smith RJ, Lesperance MM, Van Camp G. Mutational spectrum of the WFS1 gene in Wolfram syndrome, nonsyndromic hearing impairment, diabetes mellitus, and psychiatric disease. Hum Mutat. 2003;22:275–287. doi: 10.1002/humu.10258. [DOI] [PubMed] [Google Scholar]
- Cuzon VC, Yeh PW, Yanagawa Y, Obata K, Yeh HH. Ethanol consumption during early pregnancy alters the disposition of tangentially migrating GABAergic interneurons in the fetal cortex. J Neurosci. 2008;28:1854–1864. doi: 10.1523/JNEUROSCI.5110-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chaldee M, Gaillard MC, Bizat N, Buhler JM, Manzoni O, Bockaert J, Hantraye P, Brouillet E, Elalouf JM. Quantitative assessment of transcriptome differences between brain territories. Genome Res. 2003;13:1646–1653. doi: 10.1101/gr.1173403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- de Olmos JS, Heimer L. The concepts of the ventral striatopallidal system and extended amygdala. Ann N Y Acad Sci. 1999;877:1–32. doi: 10.1111/j.1749-6632.1999.tb09258.x. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin BS, Shao H, Kadono-Okuda K, Guo N, Andres DA. Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem J. 2000;347(Pt 1):223–231. [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez M, Abellan A, Legaz I, Rubenstein JL, Puelles L, Medina L. Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development. J Comp Neurol. 2008;506:46–74. doi: 10.1002/cne.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghate A, Befort K, Becker JA, Filliol D, Bole-Feysot C, Demebele D, Jost B, Koch M, Kieffer BL. Identification of novel striatal genes by expression profiling in adult mouse brain. Neuroscience. 2007;146:1182–1192. doi: 10.1016/j.neuroscience.2007.02.040. [DOI] [PubMed] [Google Scholar]
- Girault JA, Walaas SI, Hemmings HC, Jr, Greengard P. ARPP-21, a cAMP-regulated phosphoprotein enriched in dopamine-innervated brain regions: tissue distribution and regulation of phosphorylation in rat brain. Neuroscience. 1990;37:317–325. doi: 10.1016/0306-4522(90)90402-p. [DOI] [PubMed] [Google Scholar]
- Gofflot F, Chartoire N, Vasseur L, Heikkinen S, Dembele D, Le Merrer J, Auwerx J. Systematic gene expression mapping clusters nuclear receptors according to their function in the brain. Cell. 2007;131:405–418. doi: 10.1016/j.cell.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Grueter BA, Winder DG. Group II and III metabotropic glutamate receptors suppress excitatory synaptic transmission in the dorsolateral bed nucleus of the stria terminalis. Neuropsychopharmacology. 2005;30:1302–1311. doi: 10.1038/sj.npp.1300672. [DOI] [PubMed] [Google Scholar]
- Grunblatt E, Zander N, Bartl J, Jie L, Monoranu CM, Arzberger T, Ravid R, Roggendorf W, Gerlach M, Riederer P. Comparison analysis of gene expression patterns between sporadic Alzheimer’s and Parkinson’s disease. J Alzheimers Dis. 2007;12:291–311. doi: 10.3233/jad-2007-12402. [DOI] [PubMed] [Google Scholar]
- Hamann MJ, Lubking CM, Luchini DN, Billadeau DD. Asef2 functions as a Cdc42 exchange factor and is stimulated by the release of an autoinhibitory module from a concealed C-terminal activation element. Mol Cell Biol. 2007;27:1380–1393. doi: 10.1128/MCB.01608-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SP, Slager SL, De Leon AB, Heiman GA, Klein DF, Hodge SE, Weissman MM, Fyer AJ, Knowles JA. Evidence for genetic linkage between a polymorphism in the adenosine 2A receptor and panic disorder. Neuropsychopharmacology. 2004;29:558–565. doi: 10.1038/sj.npp.1300311. [DOI] [PubMed] [Google Scholar]
- Harlan RE, Shivers BD, Romano GJ, Howells RD, Pfaff DW. Localization of preproenkephalin mRNA in the rat brain and spinal cord by in situ hybridization. J Comp Neurol. 1987;258:159–184. doi: 10.1002/cne.902580202. [DOI] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, Olson JM, Jones L, Luthi-Carter R. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Iadarola MJ, Naranjo JR, Duchemin AM, Quach TT. Expression of cholecystokinin and enkephalin mRNA in discrete brain regions. Peptides. 1989;10:687–692. doi: 10.1016/0196-9781(89)90160-5. [DOI] [PubMed] [Google Scholar]
- Inoue H, Tanizawa Y, Wasson J, Behn P, Kalidas K, Bernal-Mizrachi E, Mueckler M, Marshall H, Donis-Keller H, Crock P, Rogers D, Mikuni M, Kumashiro H, Higashi K, Sobue G, Oka Y, Permutt MA. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome) Nat Genet. 1998;20:143–148. doi: 10.1038/2441. [DOI] [PubMed] [Google Scholar]
- Ishihara H, Takeda S, Tamura A, Takahashi R, Yamaguchi S, Takei D, Yamada T, Inoue H, Soga H, Katagiri H, Tanizawa Y, Oka Y. Disruption of the WFS1 gene in mice causes progressive beta-cell loss and impaired stimulus-secretion coupling in insulin secretion. Hum Mol Genet. 2004;13:1159–1170. doi: 10.1093/hmg/ddh125. [DOI] [PubMed] [Google Scholar]
- Ivkovic S, Ehrlich ME. Expression of the striatal DARPP-32/ARPP-21 phenotype in GABAergic neurons requires neurotrophins in vivo and in vitro. J Neurosci. 1999;19:5409–5419. doi: 10.1523/JNEUROSCI.19-13-05409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash TL, Winder DG. Neuropeptide Y and corticotropin-releasing factor bi-directionally modulate inhibitory synaptic transmission in the bed nucleus of the stria terminalis. Neuropharmacology. 2006;51:1013–1022. doi: 10.1016/j.neuropharm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y, Sagara M, Shibata Y, Shirouzu M, Yokoyama S, Akiyama T. Identification and characterization of Asef2, a guanine-nucleotide exchange factor specific for Rac1 and Cdc42. Oncogene. 2007;26:7620–7267. doi: 10.1038/sj.onc.1210574. [DOI] [PubMed] [Google Scholar]
- Koks S, Planken A, Luuk H, Vasar E. Cat odour exposure increases the expression of wolframin gene in the amygdaloid area of rat. Neurosci Lett. 2002;322:116–120. doi: 10.1016/s0304-3940(02)00110-6. [DOI] [PubMed] [Google Scholar]
- Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. Am J Psychiatry. 2007;164:1149–1159. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. Neuroadaptive mechanisms of addiction: studies on the extended amygdala. Eur Neuropsychopharmacol. 2003;13:442–452. doi: 10.1016/j.euroneuro.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Kurrasch DM, Cheung CC, Lee FY, Tran PV, Hata K, Ingraham HA. The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning. J Neurosci. 2007;27:13624–13634. doi: 10.1523/JNEUROSCI.2858-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Hong CJ, Tsai SJ. Association study of A2a adenosine receptor genetic polymorphism in panic disorder. Neurosci Lett. 2005;378:98–101. doi: 10.1016/j.neulet.2004.12.012. [DOI] [PubMed] [Google Scholar]
- LeDoux MS, Xu L, Xiao J, Ferrell B, Menkes DL, Homayouni R. Murine central and peripheral nervous system transcriptomes: comparative gene expression. Brain Res. 2006;1107:24–41. doi: 10.1016/j.brainres.2006.05.101. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci. 2004;24:3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo ED, Richardson-Jones JW, Sibille E, Kottman A, Hen R. Molecular heterogeneity along the dorsal-ventral axis of the murine hippocampal CA1 field: a microarray analysis of gene expression. Neuroscience. 2006;137:177–186. doi: 10.1016/j.neuroscience.2005.08.082. [DOI] [PubMed] [Google Scholar]
- Lovatt D, Sonnewald U, Waagepetersen HS, Schousboe A, He W, Lin JH, Han X, Takano T, Wang S, Sim FJ, Goldman SA, Nedergaard M. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J Neurosci. 2007;27:12255–12266. doi: 10.1523/JNEUROSCI.3404-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima K, Miyamoto Y, Tsukahara F, Hirai M, Sakaki Y, Ito T. A novel G-protein-coupled receptor gene expressed in striatum. Genomics. 2000;69:314–321. doi: 10.1006/geno.2000.6340. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF, Guo S. Early teleostean basal ganglia development visualized by zebrafish Dlx2a, Lhx6, Lhx7, Tbr2 (eomesa), and GAD67 gene expression. J Comp Neurol. 2008;507:1245–1257. doi: 10.1002/cne.21604. [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J Neurosci. 2006;26:9881–9891. doi: 10.1523/JNEUROSCI.2246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, Kuczenski R, Niculescu AB. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9:1007–1029. doi: 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- Olsen CM, Huang Y, Goodwin S, Ciobanu DC, Lu L, Sutter TR, Winder DG. Microarray analysis reveals distinctive signaling between the bed nucleus of the stria terminalis, nucleus accumbens, and dorsal striatum. Physiol Genomics. 2008;32:283–298. doi: 10.1152/physiolgenomics.00224.2006. [DOI] [PubMed] [Google Scholar]
- Ouimet CC, Hemmings HC, Jr, Greengard P. ARPP-21, a cyclic AMP-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Immunocytochemical localization in rat brain. J Neurosci. 1989;9:865–875. doi: 10.1523/JNEUROSCI.09-03-00865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C, Greenberg ME. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin K. The mouse brain in stereotaxic coordinates. Academic Press; San Diego: 2001. [Google Scholar]
- Raud S, Sutt S, Plaas M, Luuk H, Innos J, Philips MA, Koks S, Vasar E. Cat odor exposure induces distinct changes in the exploratory behavior and Wfs1 gene expression in C57Bl/6 and 129Sv mice. Neurosci Lett. 2007;426:87–90. doi: 10.1016/j.neulet.2007.08.052. [DOI] [PubMed] [Google Scholar]
- Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, Welling C, Murray J, Schmidt RE, Herrera PL, Permutt MA. Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia. 2005;48:2313–2321. doi: 10.1007/s00125-005-1947-4. [DOI] [PubMed] [Google Scholar]
- Samson RD, Duvarci S, Pare D. Synaptic plasticity in the central nucleus of the amygdala. Rev Neurosci. 2005;16:287–302. doi: 10.1515/revneuro.2005.16.4.287. [DOI] [PubMed] [Google Scholar]
- Santicioli P, Del Bianco E, Maggi CA. Adenosine A1 receptors mediate the presynaptic inhibition of calcitonin gene-related peptide release by adenosine in the rat spinal cord. Eur J Pharmacol. 1993;231:139–142. doi: 10.1016/0014-2999(93)90695-e. [DOI] [PubMed] [Google Scholar]
- Segal JP, Stallings NR, Lee CE, Zhao L, Socci N, Viale A, Harris TM, Soares MB, Childs G, Elmquist JK, Parker KL, Friedman JM. Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J Neurosci. 2005;25:4181–4188. doi: 10.1523/JNEUROSCI.0158-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham Y, Shalev U, Lu L, De Wit H, Stewart J. The reinstatement model of drug relapse: history, methodology and major findings. Psychopharmacology (Berl) 2003;168:3–20. doi: 10.1007/s00213-002-1224-x. [DOI] [PubMed] [Google Scholar]
- Song DD, Harlan RE. The development of enkephalin and substance P neurons in the basal ganglia: insights into neostriatal compartments and the extended amygdala. Brain Res Dev Brain Res. 1994;83:247–261. doi: 10.1016/0165-3806(94)00145-6. [DOI] [PubMed] [Google Scholar]
- Stansberg C, Vik-Mo AO, Holdhus R, Breilid H, Srebro B, Petersen K, Jorgensen HA, Jonassen I, Steen VM. Gene expression profiles in rat brain disclose CNS signature genes and regional patterns of functional specialisation. BMC Genomics. 2007;8:94. doi: 10.1186/1471-2164-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Cassell MD. Intrinsic GABAergic neurons in the rat central extended amygdala. J Comp Neurol. 1993;330:381–404. doi: 10.1002/cne.903300308. [DOI] [PubMed] [Google Scholar]
- Swanson LW. The amygdala and its place in the cerebral hemisphere. Ann N Y Acad Sci. 2003;985:174–184. doi: 10.1111/j.1749-6632.2003.tb07081.x. [DOI] [PubMed] [Google Scholar]
- Takeda K, Inoue H, Tanizawa Y, Matsuzaki Y, Oba J, Watanabe Y, Shinoda K, Oka Y. WFS1 (Wolfram syndrome 1) gene product: predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum Mol Genet. 2001;10:477–484. doi: 10.1093/hmg/10.5.477. [DOI] [PubMed] [Google Scholar]
- Tsou K, Girault JA, Greengard P. Dopamine D1 agonist SKF 38393 increases the state of phosphorylation of ARPP-21 in substantia nigra. J Neurochem. 1993;60:1043–1046. doi: 10.1111/j.1471-4159.1993.tb03252.x. [DOI] [PubMed] [Google Scholar]
- Xiao XQ, Grove KL, Lau SY, McWeeney S, Smith MS. Deoxyribonucleic acid microarray analysis of gene expression pattern in the arcuate nucleus/ventromedial nucleus of hypothalamus during lactation. Endocrinology. 2005;146:4391–4398. doi: 10.1210/en.2005-0561. [DOI] [PubMed] [Google Scholar]
- Yaar R, Jones MR, Chen JF, Ravid K. Animal models for the study of adenosine receptor function. J Cell Physiol. 2005;202:9–20. doi: 10.1002/jcp.20138. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hattori E, Shimizu M, Sugaya A, Shibuya H, Yoshikawa T. Association studies of the cholecystokinin B receptor and A2a adenosine receptor genes in panic disorder. J Neural Transm. 2001;108:837–848. doi: 10.1007/s007020170033. [DOI] [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, Winrow C, Lockhart DJ, Reilly JF, Young WG, Bloom FE, Lockhart DJ, Barlow C. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirlinger M. Selection and validation of microarray candidate genes from subregions and subnuclei of the brain. Methods. 2003;31:290–300. doi: 10.1016/s1046-2023(03)00158-0. [DOI] [PubMed] [Google Scholar]
- Zirlinger M, Anderson D. Molecular dissection of the amygdala and its relevance to autism. Genes Brain Behav. 2003;2:282–294. doi: 10.1034/j.1601-183x.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- Zirlinger M, Kreiman G, Anderson DJ. Amygdala-enriched genes identified by microarray technology are restricted to specific amygdaloid subnuclei. Proc Natl Acad Sci U S A. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Probe sets ID are indicated, with the corresponding name and Refseq information from Affymetrix. Forward and reverse primer sequences used for qPCR analysis of 8 candidate EAc marker genes are indicated.
Dig-labeled RNA in situ hybridization (ISH) was performed on 25μm sagittal adult mouse brain sections 49 genes whose expression pattern in EAc network has not been reported earlier. This figure shows the 17 genes with low expression (A) and the 24 genes with strong and ubiquitously expressed genes (B). Expression patterns were classified as detailed in methods. Sagittal ISH of the 8 potential EAc markers are shown on Figure 2. Gene symbols are indicated as in Table 1.