Abstract
Chronic obstructive pulmonary disease (COPD) is one of the leading causes of death throughout the world and is largely associated with cigarette smoking. Despite the appreciation of the central role of smoking in the development of COPD, only a relatively small number of smokers (15%–20%) develop COPD. Recent studies depicting familial aggregation suggest that some subjects may have a genetic predisposition to developing COPD. In this respect, a number of single nucleotide polymorphisms have been reported in association with different COPD features (subphenotypes), although much of this data remains controversial. Classical genetic studies (including twin and family studies) assume an “equal-environment” scenario, but as gene-environment interactions occur in COPD, this assumption needs revision. Thus, new integrated models are needed to examine the major environmental factors associated with COPD which include smoking as well as air pollution, and respiratory infections, and not only genetic predisposition. Revisiting this area, may help answer the question of what has more bearing in the pathogenesis of COPD—the environment or the genomic sequence of the affected subjects. It is anticipated that an improved understanding of this interaction will both enable improved identification of individuals susceptible to developing this disease, as well as improved future treatments for this disease.
Keywords: chronic obstructive pulmonary disease, environment, genomics, pathogenesis
Introduction
The development of COPD is most likely a series of complex interactions between noxious airborne agents together with a genetic predisposition. Exposure to cigarette smoke and air pollution as well exposure to infectious agents are commonly associated with the development of COPD (Anthonisen et al 2005). Because COPD does not develop in all smokers, genetic predisposition such as protease-antiprotease imbalance is a plausible explanation for the development of airways inflammation and airflow obstruction (Molfino 2004a). Conversely, it is also possible that lungs adapt to environmental toxins, so that COPD usually does not develop in the majority of smokers (Molfino 2004b). While cigarette smoking is the major environmental factor associated with the development of COPD, some subjects continue to show decline in lung function after sustained smoking cessation. This suggests the presence or airways remodeling which persists and varies among subjects due to differences in the genetic architecture (Makris et al 2007; Pinho et al 2007). In addition, the role of air pollution and infections in the development and persistence of COPD even after sustained smoking cessation remain unclear, but their roles in provoking acute exacerbations are well established (Donaldson et al 2002; Lipton et al 2005; Naess et al 2007; Ko et al 2007).
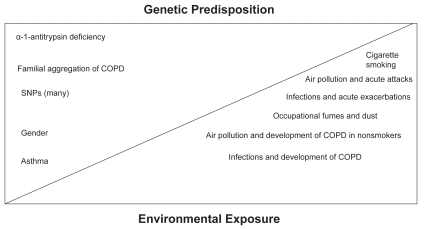
Figure 1 shows a simplified, classical, theoretical model of how genes and the environment may interact to induce and maintain the COPD phenotype. While these gene-environmental interactions are at present poorly understood, there may be a delicate balance between environmental stimuli and genes. In one extreme, such as that represented by PiZZ phenotype or indeed by severe smoking, either genetic predisposition or environmental factors may be sufficient to provoke COPD. It is more likely, however that in the COPD patient, the phenotype is under the influence of complex gene-gene and gene-environment interactions, as well as epigenetic modifications providing additional complexity to define a single etiopathogenic factor (Molfino 2007a). In this regard, although α1-antitrypsin deficiency results in COPD, it can also cause a variable phenotype because of the influence of environmental factors such as smoking (De Meo et al 2007). Moreover, although familial aggregation in COPD has been described, most of the studies have assumed an “equal-environment” model and while associations of single nucleotide polymorphisms (SNPs) with COPD phenotypes have been described, most remain controversial and unsubstantiated in other studies (Cookson 2006). Finally, it is also important to note that the influence of both gender as well as history of bronchial asthma may also contribute towards the COPD phenotype and are themselves subject to gene-gene interactions (Silverman et al 2000; Molfino 2007a).
Figure 1.
Interaction of environment and genes in chronic obstructive pulmonary disease (COPD).
Gene-environment interactions
Gene-environment interactions in COPD are suggested by the results of several studies. For example, in patients with the PiZZ phenotype who have low levels of circulating α1-antitrypsin (Stoller et al 2005), smoking is the single most important factor associated with a reduction in forced expiratory volume in 1 second (FEV1). Other factors associated with reduced FEV1 values in such phenotype include male gender, a diagnosis of asthma before age 16 years, and symptoms of asthma or chronic bronchitis (De Meo et al 2007). In subjects with less severe reduction of α1-antitrypsin, it is unclear whether the intermediate blood levels of α1-antitrypsin are sufficient to avert lung function decline, even if the individuals are exposed to noxious or toxic environments. In one study, lung function did not decline in subjects with less severe phenotypes (eg, PiMM, PiMS, or PiMZ), even if smokers were included in the analysis (Silva et al 2003). This finding contradicts previous reports in the literature (Dahl et al 2002). One possible explanation is that the PiMZ phenotype requires the presence of family history of COPD to exhibit a faster decline in FEV1. This suggests that other gene-gene interactions may be needed, in order to see a significant gene-environment interaction (Molfino 2007a).
Cigarette smoking and air pollution both affect the lung compartment, and their effects may be modulated by genes (perhaps even the whole genome sequence!) of exposed individuals (Conrad and Antonarakis 2007; Kehrer-Sawatzki 2007; Stranger et al 2007). In COPD, smoking cessation has been shown to be the only non-pharmacological intervention to slow the decline in FEV1. In this respect, the Lung Health Study studied 5,887 patients with mild to moderate COPD for 5 years and results of a 14-year follow-up were reported in 2005. Patients with sustained smoking cessation showed a significant increase in FEV1 during the first year after smoking cessation, followed by a steady decline thereafter. Patients with intermittent smoking cessation had a rate of decline in FEV1 between those with sustained cessation and those who continued to smoke. This shows how a major environmental change—apparently regardless of genetic architecture—can affect the initial progression of COPD (Anthonisen et al 2005). It may be that genes influence the continuous reduction in FEV1 in patients with sustained smoking cessation, but presently this is not well understood.
COPD can also develop in nonsmokers. Behrendt (2005) examined approximately 14000 subjects, of whom ~7500 were nonsmokers. In this group, 4.7% had mild COPD (>80% were women) and 1.9% had moderate-to-severe COPD (>80% men). Previous physician-diagnosed asthma and age were risk factors, whereas urban residence, occupation, and allergies were not. The study had several limitations since many factors were not collected or reported. These included gestational age, birth weight, lower respiratory tract infections in childhood, parental smoking during gestation and childhood, exposure to environmental tobacco smoke and other airway irritants throughout adulthood, and family history of COPD (Behrendt 2005).
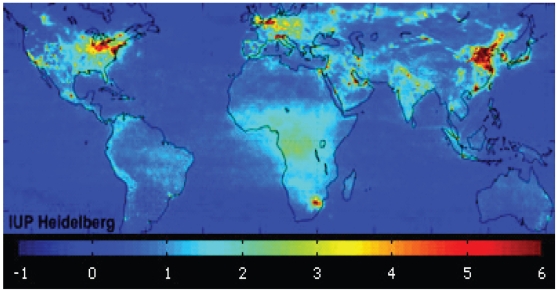
Air pollution is another major environmental factor per se involved in the development of COPD (Lipton et al 2005; Ko et al 2007; Naess et al 2007). Major outdoor pollutants include nitrogen dioxide (NO2), sulfur dioxide, ozone, and particulate matter (diesel exhaust). The major source of NO2 pollution is road traffic, and the highest concentrations are evident during the morning and evening rush hours. At a global level, the mean tropospheric NO2 corresponds to areas of major industrialization. Conversely, indoor pollution is a significant problem in sub-Saharan Africa (SSA), where it is caused by household energy use of biomass fuel, such as wood, charcoal, dung, and agricultural residues (Figure 2). Biomass burning alone causes ~400,000 annual deaths in SSA, both from respiratory infections in children and COPD in nonsmoking women (Bailis et al 2005).
Figure 2.
Global mean tropospheric nitrogen dioxide (NO2) vertical column density between January 2003 and June 2004. Concentrations increase with color intensity: blue, lowest NO2 concentrations; deep red, highest NO2 concentrations. Eighteen-month average taken from www.esa.int/images/pollution_global_L.
Once COPD is established, infections are the single best-characterized factor associated with acute exacerbations of COPD. The influence of infections in the development and persistence of COPD remains less clear. Patients with frequent exacerbations of COPD show a faster decline of FEV1 than patients with less frequent exacerbations even when smoking status is taken into consideration (Donaldson 2002). Interestingly, it has recently been reported that patients with frequent exacerbations of COPD have different genotypes in a particular allelic variant leading to a lack of the leukocyte chemotactic factor CCL1 (also known as i-309) which was associated with infection in some patients (Takabatake et al 2006).
Models to study gene-environment interactions
The hypothesis that subjects who develop COPD have genetic susceptibility to high-risk environments requires substantiation through an integrative model. The two most frequently used approaches in conditions in which gene-environment interactions are suspected include the “bottom-up” and “top-down” methods.
In the “bottom-up” approach, individual genetic and environmental factors are identified. The limitations of this approach are the difficulties in establishing statistical validity and in quantifying the gene-gene and gene-environment interactions. The “top-down” approach considers the risks at the population level, using twin and family association studies, as well as data on environmental factors in determining the trait. The major limitation of this approach is the impact of multiple genetic variants, as well as the whole genome sequence of each individual, on disease susceptibility, which leads to an increasingly intractable algebra (Wallace 2006).
In the “bottom-up” approach, genetic susceptibility studies have demonstrated that in addition to familial aggregation, TNF-α and IL-13 promoter polymorphisms, as well as TIMP-2 polymorphisms, are significantly associated with the presence of smoking-related COPD, whereas SNPs in MMP1 and MMP12, in the anti-oxidant genes GSTM1, GSTT1, GSTP1, HMOX-1, and mEPHX are associated with an accelerated decline of lung function in COPD (Molfino 2004a, 2004b, 2007a). More recently, SNPs have also been reported in relatives of patients with COPD, or associated with lung hyperinflation or other sub phenotypes (eg, signs, symptoms, or exercise tests in COPD).
Few of these results have been reproduced in other studies (Molfino 2007a). One potential explanation maybe the low frequency of SNPs with an occurrence of approximately one SNP in 300–500 base pairs in the human genome, it is possible that some of the associations were random. Therefore, a P value <0.005 and some plausible pathophysiological mechanism by which the SNP is relevant in COPD are highly desirable (Cookson 2006). Furthermore, in the “bottom-up” approach, rather than searching for direct main-effect associations between candidate genes and COPD, other experimental designs may identify how the genotype can moderate the response of a subject to environmental stimuli. In this regard, subjects carrying a variable number of tandem repeat polymorphisms in the D4 dopamine receptor gene experienced a craving significantly more often when in the presence of another smoker (Hutchinson et al 2002). Results of psychiatry studies will add complexity to the possible gene-environment interaction in COPD, since addiction is the main cause of cigarette smoking.
New approaches such as genome-wide association studies (GWAS) may be able to pinpoint genetic loci associated with COPD or its subphenotypes (Silverman 2006) This approach can be robust if one assumes the presence of non-confounder factors and no interaction with the environment. Advantages include reduced costs, since GWAS often follow a staged design, in which a proportion of samples are genotyped first and a proportion of the most promising markers are genotyped later in the remaining samples (Ye et al 2005). The standard approach is to focus on findings that are statistically significant when stage 2 data are analyzed; an alternative method is to analyze the results from different stages jointly (Skol et al 2007). In GWAS, the associations of the genome information (eg, SNPs) among different subjects, and the associations between different genetic loci and the phenotype (eg, FEV1 decline) can be expressed using probability functions. Based on these functions, reliable conclusions can be obtained about the associations between phenotype and genes using SNPs—called tagging SNPs (Molfino 2007a). Some limitations of GWAS include: the phenotype must be well characterized to enable selection of patients likely to share the genetic cause of COPD; thousands of patients and control subjects are needed to ensure the statistical reliability of the study; and bioinformatics challenges remain regarding identification of true positives. Another potential limitation involves the possibility of missing particular mutations in COPD, which may not be tagged by a specific SNP (Molfino 2007a). Moreover, the sequence of the human genome continues to uncover other forms of genetic variation (apparently normal) such as copy-number variation. Copy-number variations are frequent and inheritable, and include the deletion or duplication of DNA in ≥1 gene and the presence of ≥2 copy-number variations. They may influence susceptibility to the environment because they can change gene dose and expression (Conrad and Antonarakis 2007; Kehrer-Sawatzki 2007; Stranger et al 2007). Unless COPD is produced by a single gene disorder, it is possible that GWAS in COPD will only reveal genes that are associated with minimal or moderate susceptibility, as is the case with the already known environmental factors (Wallace 2006).
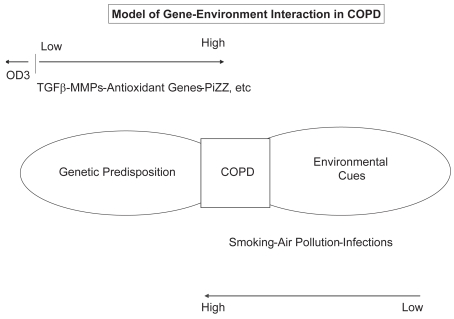
Since the development of COPD appears to require that a highly predisposed individual be present in a high-risk environment, a 4-category model may also be useful as a starting point for “top-down” analysis (Figure 3) (Molfino 2007b). This model, developed by Khoury and Wagener (1995), abandons the classical assumptions of twins studies, as well as the Fisher (1918) assumption, which suggests that genes are risk factors for common traits in a manner dominated by an additive polygenic term. This model has been adapted and shown that there may be limited potential for reducing the incidence of common diseases, such as COPD, through environmental interventions targeted by genotype (Wallace 2006).
Figure 3.
Four-category model of gene-environment interaction in chronic obstructive pulmonary disease (COPD).
It appears that the individual genotype that determines the susceptibility to complex disease tends to be exaggerated by the classical models due to the assumption of “equal environment” in twin studies or no gene-environment interactions at all. The model proposed by Wallace reveals that inherited genetic variants may be important in determining susceptibility only for the relatively rare familial forms of disease (Wallace 2006). Thus, the studies of familial aggregation may be incorrect, and the search for additional susceptibility genes may turn out to be largely fruitless.
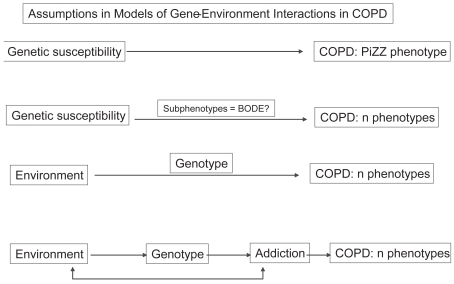
It appears that there is a need to revise our assumptions on how COPD develops (Caspi and Moffitt 2006; Molfino 2007b) (Figure 4). The gene-to-COPD approach assumes direct linear relations between genes and COPD (eg, PiZZ with severe phenotype). This model has been used to study a number of COPD phenotypes (eg, in broader COPD populations than the PiZZ homozygous). Subphenotypes are thought to have simpler genetic underpinnings than COPD itself. Thus, this assumption pursues the hypothesis that it will be easier to identify genes associated with subphenotypes than with the whole constellation of COPD signs and symptoms. This approach adds an intermediate measure to the final diagnosis of COPD (eg, the BODE index: body mass index, FEV1, dyspnea, and exercise capacity index; Figure 4) (Celli et al 2004).
Figure 4.
Assumptions in models of gene-environment interactions in chronic obstructive pulmonary disease (COPD). BODE, multidimensional index including body mass index, airflow obstruction, dyspnea, and exercise capacity index. Adapted from Caspi and Moffitt (2006).
A third approach (Figure 4) seeks to incorporate information about the environment (eg, pollutants). This approach of environment-gene interaction differs fundamentally from the “main-effects” approach, which assumes that genes cause COPD, an approach used in single-gene diseases (Fisher 1918). The environment-gene approach assumes that the environment causes the disorder and that genes cause the susceptibility to pathogens (eg, women and children in SSA). There is no expectation of an association between gene and COPD in this model in the absence of environmental pollutants or pathogens. The environment-gene approach in COPD is based on the fact that COPD is due to a major environmental cause and that subjects show a heterogeneous response to that cause. The fourth approach is more complex because it includes an additional co-morbidity such as nicotine addiction (Caspi and Moffit 2006). Therefore, it may be applicable to the majority of patients with COPD in the industrialized world.
Molecular mechanisms in gene-environment interactions
We are just beginning to understand how the environment can interact with the genome. The genetic response to the environment can be in the form of large-scale recombination, chromosomal breakage-fusion-bridge cycles, and loss of chromosomes or chromosome fragments. Such changes may be induced or facilitated by transposition of DNA (Madlung and Comai 2004). In addition, chemical modifications of histones, around which DNA is coiled and coincide with modifications in transcriptional activity, can be inherited. The modifications that alter DNA activity without altering its basic nucleotide structure are referred to as epigenetic changes and include the chemical modifications of DNA or histones, usually with methyl or acetyl groups, which have the potential to change gene expression and bring the phenotype into being (Wolffe and Matzke 1999). Methylation of DNA functions as gene silencing, and is vital to the maintenance of proper cell division. Acetylation of histones leads to gene activity, and gene expression is controlled by packing and unpacking DNA from inaccessible to accessible nucleosomes. Packing and unpacking is achieved by acetylation and deacetylation of the histones of the nucleosome core (Segal 2006). In COPD, it has been shown that deacetylation is needed to repress the expression of pro-inflammatory cytokines (which is accompanied by hyper-acetylation) (Barnes et al 2005) and to enable the response to corticosteroids (Barnes 2006) It is possible that other molecular mechanisms exist and remain to be uncovered.
Conclusions
Chronic obstructive pulmonary disease is a complex condition mainly associated with cigarette smoking, as well as exposure to air pollution and respiratory pathogens. It is evident that only a relatively small number of exposed individuals develop COPD, and a fraction suffer exacerbations when exposed to noxious agents or pathogens. The classical approach to understanding the pathogenesis of the disease is to accept that different genetic loci determine the phenotype in COPD and to continue the search for such predisposing loci.
Models of gene-environment interaction suggest that either it may be difficult to identify predisposing loci or, if identified, they will be found to have minimal effects on the development of COPD. Thus, new approaches to the study of COPD are needed. It may be appropriate to avoid the equal-environment assumption and to consider haplotype or even whole genome sequences to determine susceptibility to COPD.
Future study designs should characterize the phenotype and history of potential exposures of study subjects. The latter category should include stratification of the intensity of smoking, determination of pollutants, type of respiratory infections not only during the subject’s recent adult years, but also during childhood, and occupational exposures.
Footnotes
Disclosure
Dr Molfino and Coyle are employees of MedImmune, Gaithersburg, MD. Presented in part at the 2007 European Society of Clinical Investigation Scientific meeting in Uppsala, Sweden.
References
- Anthonisen NR, Skeans MA, Wise RA, et al. The effects of a smoking cessation intervention on 14.5-year mortality: a randomized clinical trial. Ann Intern Med. 2005;142:233–9. doi: 10.7326/0003-4819-142-4-200502150-00005. [DOI] [PubMed] [Google Scholar]
- Bailis R, Ezzati M, Kammen DM. Mortality and greenhouse gas impacts of biomass and petroleum energy futures in Africa. Science. 2005;308:98–103. doi: 10.1126/science.1106881. [DOI] [PubMed] [Google Scholar]
- Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Respir J. 2005;25:552–63. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- Barnes PJ. How corticosteroids control inflammation: Quintiles Prize Lecture 2005. Br J Pharmacol. 2006;148:245–54. doi: 10.1038/sj.bjp.0706736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt CE. Mild and moderate-to-severe COPD in nonsmokers: distinct demographic profiles. Chest. 2005;128:1239–44. doi: 10.1378/chest.128.3.1239. [DOI] [PubMed] [Google Scholar]
- Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 2006;7:583–90. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- Celli BR, Cote CG, Marin JM, et al. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:1005–12. doi: 10.1056/NEJMoa021322. [DOI] [PubMed] [Google Scholar]
- Conrad B, Antonarakis SE. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- Cookson WO. State of the art. Genetics and genomics of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3:473–5. doi: 10.1513/pats.200603-036MS. [DOI] [PubMed] [Google Scholar]
- Dahl M, Tybjaerg-Hansen A, Lange P, et al. Change in lung function and morbidity from chronic obstructive pulmonary disease in alpha1-antitrypsin MZ heterozygotes: A longitudinal study of the general population. Ann Intern Med. 2002;136:270–9. doi: 10.7326/0003-4819-136-4-200202190-00006. [DOI] [PubMed] [Google Scholar]
- DeMeo DL, Sandhaus RA, Barker AF, et al. Determinants of airflow obstruction in severe alpha 1-antitrypsin deficiency. Thorax. 2007;62:806–13. doi: 10.1136/thx.2006.075846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GC, Seemungal TA, Bhowmik A, et al. Relationship between exacerbation frequency and lung function decline in chronic obstructive pulmonary disease. Thorax. 2002;57:847–52. doi: 10.1136/thorax.57.10.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Philosophical Transactions of the Royal Society of Edinburgh. 1918;52:399–433. [Google Scholar]
- Hutchinson KE, La Chance H, Naiura R, et al. The DRD4 VNTR polymorphism influences reactivity to smoking cues. J Abnorm Psychol. 2002;111:134–43. doi: 10.1037//0021-843x.111.1.134. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H. What a difference copy number variation makes. Bioessays. 2007;29:311–13. doi: 10.1002/bies.20554. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Wagener DK. Epidemiological evaluation of the use of genetics to improve the predictive value of disease risk factors. Am J Hum Genet. 1995;56:835–44. [PMC free article] [PubMed] [Google Scholar]
- Ko FW, Tam WW, Chan DP, et al. The temporal relationship between air pollutants and hospital admissions for chronic obstructive pulmonary disease in Hong Kong. Thorax. 2007;62:780–5. doi: 10.1136/thx.2006.076166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton R, Banerjee A, Dowling KC, et al. The geography of COPD hospitalization in California. COPD. 2005;2:435–44. doi: 10.1080/15412550500346543. [DOI] [PubMed] [Google Scholar]
- Madlung A, Comai L. The effect of stress on genome regulation and structure. Ann Bot (Lond) 2004;94:481–95. doi: 10.1093/aob/mch172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris D, Moschandreas J, Damianaki A, et al. Exacerbations and lung function decline in COPD: New insights in current and ex-smokers. Respir Med. 2007;101:1305–12. doi: 10.1016/j.rmed.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Molfino NA. Genetics of COPD. Chest. 2004a;125:1929–40. doi: 10.1378/chest.125.5.1929. [DOI] [PubMed] [Google Scholar]
- Molfino NA. Lung function evolution and respiratory symptoms. Arch Bronconeumol. 2004b;40:429–30. doi: 10.1016/s1579-2129(06)60350-9. [DOI] [PubMed] [Google Scholar]
- Molfino NA. Current thinking on genetics of chronic obstructive pulmonary disease. Curr Opin Pulm Med. 2007a;13:107–13. doi: 10.1097/MCP.0b013e328013e97d. [DOI] [PubMed] [Google Scholar]
- Molfino NA. COPD: what is genes and what is environment? Eur J Clin Inv. 2007b;37:37. [Google Scholar]
- Naess O, Nafstad P, Aamodt G, et al. Relation between concentration of air pollution and cause-specific mortality: four-year exposures to nitrogen dioxide and particulate matter pollutants in 470 neighborhoods in Oslo, Norway. Am J Epidemiol. 2007;165:435–43. doi: 10.1093/aje/kwk016. [DOI] [PubMed] [Google Scholar]
- Pinho RA, Chiesa D, Mezzomo KM, et al. Oxidative stress in chronic obstructive pulmonary disease patients submitted to a rehabilitation program. Respir Med. 2007;101:1830–5. doi: 10.1016/j.rmed.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GE, Sherrill DL, Guerra S, et al. A longitudinal study of alpha1- antitrypsin phenotypes and decline in FEV1 in a community population. Chest. 2003;123:1435–40. doi: 10.1378/chest.123.5.1435. [DOI] [PubMed] [Google Scholar]
- Silverman EK, Weiss ST, Drazen JM, et al. Gender-related differences in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2000;162:2152–8. doi: 10.1164/ajrccm.162.6.2003112. [DOI] [PubMed] [Google Scholar]
- Silverman EK. Progress in chronic obstructive pulmonary disease genetics. Proc Am Thorac Soc. 2006;3:405–8. doi: 10.1513/pats.200603-092AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Stoller JK, Snider GL, Brantly ML, et al. American Thoracic Society/European Respiratory Society Statement: Standards for the diagnosis and management of individuals with alpha-1 antitrypsin deficiency. Pneumologie. 2005;59:36–68. doi: 10.1055/s-2004-830176. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabatake N, Shibata Y, Abe S, et al. A single nucleotide polymorphism in the CCL1 gene predicts acute exacerbations in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;174:875–85. doi: 10.1164/rccm.200603-443OC. [DOI] [PubMed] [Google Scholar]
- Wallace HM. A model of gene-gene and gene-environment interactions and its implications for targeting environmental interventions by genotype. Theor Biol Med Model. 2006;3:35. doi: 10.1186/1742-4682-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–6. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- Ye Y, Zhong X, Zhang H. A genome-wide tree- and forest-based association analysis of comorbidity of alcoholism and smoking. BMC Genet. 2005;6(suppl 1):S135. doi: 10.1186/1471-2156-6-S1-S135. [DOI] [PMC free article] [PubMed] [Google Scholar]