Abstract
Evolutionists have debated whether population-genetic parameters, such as effective population size and migration rate, differ between males and females. In humans, most analysis of this problem has focused on the Y chromosome and the mitochondrial genome, while the X chromosome has largely been omitted from the discussion. Past studies have compared FST values for the Y and mitochondrion under a model with migration rates that differ between the sexes but with equal male and female population sizes. In this study we investigate rates of coalescence for X-linked and autosomal lineages in an island model with different population sizes and migration rates for males and females, obtaining the mean time to coalescence for pairs of lineages from the same deme and for pairs of lineages from different demes. We apply our results to microsatellite data from the Human Genome Diversity Panel, and we examine the male and female migration rates implied by observed FST values.
Keywords: X chromosome, FST, separation of time scales, coalescent
Introduction
Evolutionists have long been interested in how demographic and population structural variables differ between males and females, and sex-biased dispersal processes are common in a variety of species (Lawson Handley and Perrin, 2007).
Differences between human males and females in parameters such as migration rate and effective population size have generally been investigated using the uniparentally-inherited Y chromosome and mitochondial genome. Past studies have observed differences in autosomal, Y-chromosomal and mitochondrial variation, and have typically explained these differences based on matrilocality or patrilocality (Wilkins and Marlowe, 2006; Wilkins, 2006).
In a patrilocal society, we expect to see more genetic differentiation across Y-chromosomal lineages than across mitochondrial lineages; such a pattern was observed using globally-distributed samples by Seielstad et al. (1998), while patterns consistent with matrilocality have been observed in Thailand (Oota et al., 2001) and Melanesia (Kayser et al., 2008). Recent studies have questioned the spatial scale at which one can expect to infer a genetic signature of patrilocality or matrilocality, arguing that this signal may be observable within geographic regions, but likely not at a global level (Wilder et al., 2004a; Wilkins and Marlowe, 2006).
The X chromosome has contributed comparatively little to the inference of sex-specific human migration rates. Garrigan et al. (2007) compared genetic variation using resequence data at 2 X-linked loci totaling 8486 bp, 6650 bp encompassing 13 Alu elements on the Y chromosome, and 780 bp of the cytochrome oxidase subunit III on the mitochondrion. Their inference of migration rates among 10 human populations did not produce a consistent pattern of sex-biased gene flow across all the loci investigated, though different rates of male and female migration were inferred for many pairs of populations.
Although variation in the Y chromosome and the mitochondrion has generally been used in studies of sex-specific differences in human dispersal, comparisons between variation observed on the X chromosome and on autosomes also have the potential to shed light on evolutionarily interesting differences between males and females (Schaffner, 2004). In contrast with the Y chromosome and the mitochondrial genome, each of which is effectively a single absolutely-linked locus, the X chromosome and autosomes offer numerous independent markers. The availability of multiple markers potentially adds power to the analysis, although recombination and the movement of the autosomes and X chromosome between males and females are expected to complicate the elucidation of sex-specific histories (Ramachandran et al., 2004; Wilkins and Marlowe, 2006).
Using 17 X-linked and 377 autosomal microsatellites genotyped in 52 globally-distributed populations in the Human Genome Diversity Panel (HGDP), Ramachandran et al. (2004) investigated differences in patterns of X-chromosomal and autosomal geographical variation around the world, as measured by FST among populations. These differences were studied by considering the different numbers of copies of X-linked and autosomal loci in a population, for a given female fraction of the total population size, and by deriving a formula for FST using a model of divergence from an ancestral population with subsequent isolation of descendant populations. Male and female effective population sizes were allowed to vary, but the model did not involve migration among subpopulations. Ramachandran et al. (2004) found that a ratio of the number of females to the total population size of 0.5 was sufficient to explain global differences in genetic variation between X-linked and autosomal microsatellites. However, the study could not explain differences in FST in some of the continental regions of the dataset where the divergence model might be less representative of population history (for example, Europe, where gene flow among populations post-divergence is likely to have been high).
Here we investigate the rates of coalescence for X-linked and autosomal loci in an island migration model with sex-specific population sizes and migration rates. Past theoretical studies have examined the effect of sex-specific gene flow and genetic drift on times to coalescence and F-statistics (Wang, 1997; Rousset, 1999; Wang, 1999; Laporte and Charlesworth, 2002; Vitalis, 2002; Hedrick, 2007). We consider these issues from a coalescent perspective. We start with an exact discrete island model with migrating adults, and use a result due to Möhle (1998) to explicitly take the limit of the coalescent process as population size goes to infinity. We obtain simple expressions for FST at X-linked and autosomal loci in our model under the usual assumptions of the structured coalescent.
Applying the analytical results to the X-linked and autosomal microsatellite data from the HGDP (Cann et al., 2002; Ramachandran et al., 2004; Ramachandran et al., 2005; Rosenberg et al., 2005), we find that global patterns of population differentiation as measured by FST can be explained without requiring different migration rates for males and females. Within geographic regions, however, the inferred sex-specific migration rates differ substantially, although the direction of the deviation is not always the same.
The migration model
Consider an island model with D demes and four sex-specific parameters, each of which has the same value for all demes: fixed numbers of males and females (Nm and Nf, respectively), and fixed numbers of male and female migrants per generation (Mm and Mf, respectively). The total population size is DN = D(Nm + Nf) (each deme has the same number of individuals). Here we can write Nf = Nr, where r is the female fraction of the population size, assumed to be the same for each deme. It follows that Nm = N (1 − r). Denote by mf the backwards migration rate for females; that is, the probability that a female sampled from deme i has just migrated from some other deme in the generation during which sampling took place. The corresponding rate for males is mm. Since Mm and Mf are fixed, mf = Mf/Nf and mm = Mm/Nm. We shall assume throughout that mf and mm are of the order 1/N. Migration takes place after reproduction within demes, and the probability that a male (for example) migrates to a specific deme is mm/(D − 1).
We consider a single genetic locus. The resulting single-generation transition matrix for a sample of two autosomal lineages in this model has 10 states. For a sample of two X-linked lineages the model has 9 states, as listed in Table 1.
Table 1. States in the migration model.
Possible states in which two sampled lineages can be found in the island model with two sexes, and the columns of the autosomal and X-linked single-generation transition matrices that correspond to each state. Note that two sampled X-linked lineages cannot be found in the same male unless they have already coalesced.
| Columns | ||
|---|---|---|
| Autosomal | X-linked | Definition |
| 1 | 1 | In one female individual, not coalesced |
| 2 | In one male individual, not coalesced | |
| 3 | 2 | In two female individuals, same deme |
| 4 | 3 | In two male individuals, same deme |
| 5 | 4 | In one male and one female, same deme |
| 6 | 5 | In two female individuals, different demes |
| 7 | 6 | In two male individuals, different demes |
| 8 | 7 | In one male and one female, different demes |
| 9 | 8 | In one female, coalesced |
| 10 | 9 | In one male, coalesced |
Let PA be the 10 × 10 single-generation transition matrix for two lineages sampled from an autosomal locus, and let (PA)ij refer to the entry in the ith row and jth column of the matrix. Each matrix entry is the product of two terms: (a) a term involving migration or lack of migration among demes, and (b) a term describing inheritance.
For example, (PA)56, according to Table 1, is the entry describing the probability that two lineages sampled from one male and one female in the same deme came from female parents in different demes in the previous generation. (PA)56 is the product of (a) the probability that one male and one female lineage currently in the same deme were in different demes in the previous generation (either because one lineage was in a migrant or because both lineages were in migrants that arrived in the same deme), and (b) the probability that two autosomal lineages (one from a male and one from a female) both came from female parents. The latter probability is 1/4, since for each sampled individual we choose the maternal autosome with probability 1/2.
PX denotes the 9 × 9 single-generation transition matrix for two lineages sampled from an X-linked locus. (PX)45 is the probability that two X-linked lineages sampled from one male and one female in the same deme came from female parents in different demes in the previous generation (Table 1). The probability (a) above, that the lineages were in different demes in the previous generation, will not differ between an X-linked and autosomal locus. However, the analog to (b) above, the probability that two X-linked lineages (one from a male and one from a female) came from two female parents is 1/2. This is because the male allele would have had to come from the female parent in the previous generation, while we choose the female’s allele from her maternal X with probability 1/2.
The matrices PA and PX are rather cumbersome due to their size. Since the terms describing migration among demes do not depend on whether the sampled locus is X-linked or autosomal, the matrices’ entries can be written more simply by using the notation for terms of type (a) above in the following manner. Let us denote the state in which two lineages, regardless of sex, are in the same deme as state I; state II represents two lineages being from different demes. Then is the probability that a sample of one male and one female now in state I was in state II in the previous generation, which corresponds to (a) in the previous paragraph. The probabilities for all types of samples are given in Appendix 1.
Using this notation, for example, (PA)39 is equal to the product of (a) (the probability two females currently in the same deme were in the same deme in the previous generation) and (b) 1/(8Nf) (the probability two sampled lineages, one from each sampled female, coalesce in a female in the previous generation). 1/(8Nf) is the probability that in both females the maternal autosome is selected (= 1/2 × 1/2) times the probability the loci were inherited from the same maternal chromosome (= 1/(2Nf)). (PX)62 is equal to (a) (the probability two sampled males are currently in different demes but were in the same deme in the previous generation) times (b) 1 − 1/Nf (the probability the sampled lineages come from two different females). Since a male’s X chromosome must come from his mother, the probability that two male X chromosomes are found in two different females is simply the probability the chromosomes do not come from the same female.
Suppose the sampled lineages are currently in the same individual but that the lineages have not coalesced (columns 1 and 2 in PA and column 1 in PX). Because migration occurs after reproduction within demes, the lineages had to be in a male and female (the individual’s parents) in the same deme in the previous generation, regardless of whether or not the individual from whom the lineages were sampled had migrated (see rows 1 and 2 of matrix (1) and row 1 of matrix (2)).
Thus we can write down both the autosomal and X-linked single generation transition matrices, PA and PX, as matrices (1) and (2). Above both matrices, we indicate the sex structure of the sample for each column (e.g., ℳ, ℳ denotes lineages sampled from two males), and the physical locations associated with states (e.g., in the same individual but not coalesced, or from different demes).
| (1) |
| (2) |
Results
We can rewrite both transition matrices in equations (1) and (2) in the form
| (3) |
Assuming that Mf and Mm do not depend on N (i.e., as N approaches infinity, the numbers of migrants per generation converge to some limiting constants, which are again denoted by Mf and Mm for convenience), then D = limN→∞ P and B = limN→∞ N(P - D) (which both do not depend on N). Note that, in equation (3), EN = P - D - B/N denotes some error matrix with terms of the order of m2, 1/N2, and m/N. See Appendix 1 for an example of this decomposition.
The entries in D represent a fast process, namely the movement of lineages between males and females according to Mendelian inheritance, while the entries in B represent rare processes of migration and coalescence which are assumed to occur once over a period on the order of N generations. Möhle’s theorem (1998) states that if R = limt→∞ Dt exists (letting the fast process run to its conclusion), then the rates of coalescence and migration among demes when time is scaled by N generations are given by the product matrix G = RBR. Specifically, limN→∞ PNt = RetG (Möhle, 1998).
We show DX (= limN→∞ PX) and RX in (4) and (5) below while the detailed derivations of the corresponding autosomal matrices and of GX and GA appear in Appendix 2. In the case of PX given by matrix (2), DX = limN→∞ PX is
| (4) |
The columns in matrix (4) can be interpreted using the definitions in Table 1. The terms in DX are familiar terms based on the inheritance of X chromosomes, as are the entries of RX = limt→∞(DX)t:
| (5) |
When applying Möhle’s result to PA and PX, a block structure emerges in the R and G matrices for both X-linked and autosomal loci, exemplified by the blocks seen in matrix (5). We can collapse some states together by summing the entries in their columns and by collapsing some rows, reducing the analysis to 3 × 3 matrices. For example, we can sum the entries (see Appendix 2) and get a single rate of staying in the same deme for two lineages sampled in the same female individual, but not coalesced (the state described by row and column 1 of PX). The sum has the same value for each i = 1, 2, 3, 4. This is because, in the fast process occurring according to DX and DA, lineages move quickly between males and females, so the current sex structure of the sample becomes unimportant and instead we need only follow whether sampled lineages are in the same deme or in diffrent demes. Thus, the product matrices GX and GA for X-linked and autosomal lineages in this process simplify to ℊX and ℊA (equations (6) and (7), respectively; see Appendix 2 for derivation).
| (6) |
| (7) |
Using first-step analysis, we can calculate the expected times to coalescence for a pair of lineages, sampled from the same deme (E[Tsame]) or sampled from different demes (E[Tdiff]). In the discrete time processes studied here, the expected time to arrive in state j given that the current state is i equals the time to make the jump from state i to another state plus the expected time it takes to reach state j after the jump is made. We are interested in time to coalescence, so we need to solve the following equations to get, for example, and :
Solving these and the analogous equations for X-linked loci gives equations (8 – 11), measured in units of N generations.
| (8) |
| (9) |
| (10) |
| (11) |
Using our notation, Slatkin’s (1991) formulation of FST at an autosomal locus in a set of D demes is . The relationship between coalescence times and FST in this formulation depends on the mutation rate being very small. As D approaches infinity, we get
| (12) |
| (13) |
Given estimates of FST at X-linked and autosomal loci, and assuming some value on the interval (0,1) for r, we can estimate Mf and Mm from (12) and (13) as:
| (14) |
| (15) |
Application to HGDP-CEPH data
A total of 783 autosomal microsatellites from Marshfield Screening Sets #10 and #52 have been reported in the HDGP individuals from 52 populations. Screening Set #10 also contained the 17 non-pseudoautosomal X-linked microsatellites studied by Ramachandran et al. (2004), and Screening Set #52 provided 19 additional non-pseudoautosomal X-linked microsatellites studied here. The data files used in this analysis are available from the authors.
We inferred the sex of individuals from their X-chromosomal genotypes at the 36 loci examined, and verified the inferences against the corresponding inferences made using the X-chromosomal data of Conrad et al. (2006). With one exception, individuals treated as males in our analysis all had <15% heterozygous loci and females all had >19% loci on the X chromosome, among loci with no missing data. The exception, individual #139, was verified to be male on the basis of the data of Conrad et al. (2006), which included a larger number of X-chromosomal loci. Males were treated as hemizygous for calculations. Some males were reported as heterozygous at non-pseudoautosomal X-linked loci; in such cases males were coded as having missing data at these loci.
We calculated FST based on the 36 X-linked and 783 autosomal microsatellites typed in the Human Genome Diversity Panel, using Weir’s estimator (Weir, 1996) for the proportion of genetic variation distributed among populations. FST was calculated among all populations, as well as among populations within the same continental region, as defined previously by Rosenberg et al. (2002); the estimator was obtained separately for X-linked loci and for autosomal loci, following equation (5.3) on page 174 of Weir (1996). For the computation we grouped all Bantu individuals into one population with a sample size of 20 individuals. We obtained confidence intervals for X-linked and autosomal FST values by bootstrapping separately over each set of loci 1000 times (see intervals in Tables 2 and 3).
Table 2. Estimated ratio of Mf/Mm, using data from 1048 individuals.
Estimates of the among-population component of genetic variation based on 783 autosomal and 36 X-linked microsatellites in the HGDP-CEPH individuals, for global and various regional subsets of the data. Also reported are the estimated ratios of Mf/Mm when r = 0.5, calculated using 1048 individuals (Rosenberg, 2006). “C/S Asia” refers to Central/South Asian populations from the panel (see Rosenberg et al., 2002). Confidence intervals for FST were obtained by bootstrapping over loci 1000 times. The number of values out of 106 used to generate intervals for Mf/Mm, after the exclusion of of negative estimates, is also given.
| Sample | Number of populations | FST, autosomal (95% C.I.) | FST, X-linked (95% C.I.) | Mf/Mm at r = 0.5 (95% C.I.) | Number of ratios ≥ 0 | |||
|---|---|---|---|---|---|---|---|---|
| World | 52 | 0.0561 | (0.0543, 0.0579) | 0.0718 | (0.0620, 0.0839) | 1.1524 | (0.4149, 4.4257) | 999535 |
| Africa | 6 | 0.0300 | (0.0286, 0.0314) | 0.0539 | (0.0401, 0.0691) | 0.0936 | (0.0078, 1.1324) | 716775 |
| Eurasia | 21 | 0.0158 | (0.0152, 0.0165) | 0.0226 | (0.0183, 0.0276) | 0.6345 | (0.1519, 2.7184) | 998081 |
| Europe | 8 | 0.0079 | (0.0071, 0.0087) | 0.0122 | (0.0069, 0.0180) | 0.4127 | (0.0208, 9.1672) | 818855 |
| Middle East | 4 | 0.0137 | (0.0130, 0.0145) | 0.0162 | (0.0121, 0.0208) | 2.1807 | (0.4004, 32.2857) | 852251 |
| C/S Asia | 9 | 0.0137 | (0.0130, 0.0145) | 0.0149 | (0.0095, 0.0208) | 4.9120 | (0.3772, 59.5790) | 636089 |
| East Asia | 18 | 0.0125 | (0.0117, 0.0134) | 0.0156 | (0.0102, 0.0215) | 1.4720 | (0.1563, 23.0857) | 865762 |
| Oceania | 2 | 0.0635 | (0.0577, 0.0692) | 0.0847 | (0.0544, 0.1220) | 0.8746 | (0.05421, 18.3075) | 863700 |
| America | 5 | 0.1174 | (0.1127, 0.1219) | 0.1367 | (0.1166, 0.1567) | 2.1349 | (0.7401, 19.5850) | 964990 |
Table 3. Estimated ratio of Mf/Mm, using data from 952 individuals.
This table reports a similar analysis to that reported in Table 2, but using 952 individuals (Rosenberg, 2000). This set of HGDP individuals contains no two individuals with a second-degree relationship (half siblings, avuncular, or grandparent/grandchild). Confidence intervals for FST were obtained by bootstrapping over loci 1000 times. The number of values out of 106 used to generate intervals for Mf/Mm, after the exclusion of negative estimates, is also given.
| Sample | Number of populations | FST, autosomal (95% C.I.) | FST, X-linked (95% C.I.) | Mf/Mm at r = 0.5 (95% C.I.) | Number of ratios ≥ 0 | |||
|---|---|---|---|---|---|---|---|---|
| World | 52 | 0.0455 | (0.0438, 0.0472) | 0.0586 | (0.0491, 0.0702) | 1.1354 | (0.3530, 5.2649) | 992554 |
| Africa | 6 | 0.0260 | (0.0245, 0.0274) | 0.0465 | (0.0338, 0.0611) | 0.1026 | (0.0087, 1.2628) | 720323 |
| Eurasia | 21 | 0.0150 | (0.0144, 0.0156) | 0.0218 | (0.0172, 0.0266) | 0.5896 | (0.1265, 2.8729) | 994778 |
| Europe | 8 | 0.0076 | (0.0069, 0.0084) | 0.0112 | (0.0061, 0.0167) | 0.5722 | (0.0262, 15.7893) | 808661 |
| Middle East | 4 | 0.0130 | (0.0121, 0.0137) | 0.0150 | (0.0111, 0.0194) | 2.5593 | (0.4367, 40.5666) | 817864 |
| C/S Asia | 9 | 0.0127 | (0.0119, 0.0134) | 0.0146 | (0.0089, 0.0205) | 2.7676 | (0.2370, 34.5307) | 719969 |
| East Asia | 18 | 0.0113 | (0.0105, 0.0121) | 0.0134 | (0.0081, 0.0190) | 2.1811 | (0.1713, 35.2105) | 745435 |
| Oceania | 2 | 0.0552 | (0.0493, 0.0616) | 0.0753 | (0.0410, 0.1155) | 0.7702 | (0.0320, 19.1807) | 766059 |
| America | 5 | 0.0836 | (0.0799, 0.0876) | 0.0942 | (0.0789, 0.1087) | 3.0884 | (0.9013, 32.3565) | 909730 |
We use equations (14) and (15) to estimate the ratio of female migrants to male migrants using observed FST values from the data, for a given assumed proportion of females in the population. Note that in order for Mf and Mm to be interpretable they must be positive, which may not be the case for certain combinations of FST and r values. In order for both Mf and Mm to be greater than zero, the condition 2FST,A(2 − r)/[3 + FST,A(1 − 2r)] < FST,X < 4FST,A(2 − r)/[3 + FST,A(5 − r)] must be satisfied. The region in which Mf/Mm is positive for various fixed values of r, as FST,X and FST,A vary on the interval [0,1], is shown in Figure 1.
Figure 1.
The region in which the ratio Mf/Mm is positive, as computed from equations (14) and (15) for fixed values of r, with FST,X and FST,A varying on the interval [0,1]. The region is shaded in grey. The solid line is 2FST,A(2 − r)/[3+ FST,A(1 − 2r)], which FST,X must be greater than for Mm to be greater than zero. The dashed line is 4FST,A(2 − r)/[3 + FST,A(5 − r)], which FST,X must be less than for Mf to be greater than zero.
We obtained intervals for Mf/Mm (Tables 2 and 3) by taking the 1000 bootstrapped FST,X and 1000 boostrapped FST,A values, and computing Mf/Mm for all 106 possible pairs of boostrapped FST values. We disregarded those estimates of Mf/Mm which were negative, choosing to interpret negative estimates of Mf and Mm as providing little support for r = 0.5 or for our migration model. The number of values used to generate the intervals in Tables 2 and 3 after the exclusion of negative estimates is also given.
Since the initial announcement of the Human Genome Diversity Panel (Cann et al., 2002), subsequent analyses have called attention to individuals who appear to be duplicated or closely related. Here we calculate FST for two sets of HGDP individuals (Tables 2 and 3): 1048 individuals, where one individual from each pair of putatively duplicated individuals (Mountain and Ramakrishnan, 2005; Rosenberg, 2006) is excluded; and 952 individuals, a proper subset of the set of 1048, where individuals with first- and second-degree relationships are excluded (Rosenberg, 2006).
Discussion
In this paper, we applyMöhle’s theorem (1998) to transition matrices for X-linked and autosomal loci sampled in an island model of D demes with sex-specific population sizes and migration rates, and we obtain simple expressions under the model for expected times to coalescence for two sampled alleles and for FST at X-linked and autosomal loci. Möhle’s result is useful because it gives us a continuous-time limit of a discrete-time process where events are occurring on two time scales: in this case, the fast process of movement of lineages between males and females, and the slow processes of movement of individuals among demes and of coalescence.
The entries in matrices (6) and (7) give us the rates at which, when time is measured in units of N generations, two sampled lineages move among three states: being in the same deme, being in different demes, or being “coalesced”. ( A)12 gives the rate (over N generations) at which autosomal lineages move out of the same deme into different demes, (
A)12 gives the rate (over N generations) at which autosomal lineages move out of the same deme into different demes, ( A)21 gives the rate of movements of lineages into the same deme from different demes, and (
A)21 gives the rate of movements of lineages into the same deme from different demes, and ( A)13 gives the rate of coalescence, which can only happen in the model when lineages are in the same deme. For both
A)13 gives the rate of coalescence, which can only happen in the model when lineages are in the same deme. For both  A and
A and  X the last row contains only zeros because coalescence is an absorbing state.
X the last row contains only zeros because coalescence is an absorbing state.
The rates of coalescence given by ( A)13 and (
A)13 and ( X)13 are familiar: they are half the reciprocals of the variance effective population size of autosomal and X-linked genes in a sexual population with an unequal sex ratio (e.g., Nordborg and Krone, 2002; Hartl and Clark, 2007). The expected times to coalescence given in equations (8–11) also reflect that two lineages sampled from different demes must enter the same deme to coalesce, and then coalesce at a rate expected for an X-linked or autosomal locus in a population with an unequal sex ratio.
X)13 are familiar: they are half the reciprocals of the variance effective population size of autosomal and X-linked genes in a sexual population with an unequal sex ratio (e.g., Nordborg and Krone, 2002; Hartl and Clark, 2007). The expected times to coalescence given in equations (8–11) also reflect that two lineages sampled from different demes must enter the same deme to coalesce, and then coalesce at a rate expected for an X-linked or autosomal locus in a population with an unequal sex ratio.
Using the expected times to coalescence for loci sampled in the same deme or in different demes, we can calculate FST at autosomal and X-linked loci in our model, as in equations (12) and (13). The forms of (12) and (13) are 1/(1 + 4Neme), where Ne = 4NmNf/(Nm + Nf) = 4Nr(1 − r) and me = (mm + mf)/2 for autosomal loci, and Ne = 9NmNf/(4Nm + 2Nf) = 9Nr(1 − r)/[2(2 − r)] and me = (2mf + mm)/3 for X-linked loci.
When Mm = Mf = M, then FST,A = 1/(1 + 8M) and FST,X = 1/(1 + 6M), with FST,X being greater than FST,A. When the number of female migrants per generation is greater than the number of male migrants, and these values are less than 1, then FST,X can become less than FST,A for some values of r on (0,1), as shown in Figure 2A, where FST,X crosses FST,A at r = 0.5076 and r = 0.9949. If the number of male migrants exceeds the number of female migrants per generation, then FST,X > FST,A; in Figure 2B these values become closer for larger values of r.
Figure 2.
FST at X-linked and autosomal loci (equations (12) and (13)) as r, the female fraction of the population, varies on [0,1]. The dashed line is FST,X and the solid line is FST,A. A: Mf = Nf mf = 1 migrant per generation, while the number of male migrants per generation is 0.01. B: Mm, the number of male migrants per generation, is equal to 1, while Mf = 0.01.
Using observed values of FST from the Human Genome Diversity Panel at X-linked and autosomal loci, we then use equations (14) and (15) to estimate the ratio of female to male migrants Mf/Mm. We assume equal numbers of males and females when calculating the estimates in Tables 2 and 3. In this model, there are no differences between the rates of reproductive success in males and females. However, the consequence of differences between reproductive success in males and females is an important question for further investigation (see, for example, Helgason et al., 2003, and Wilder et al., 2004b).
When r = 0.5, global FST values across HGDP populations can be explained by requiring a ratio of female to male migrants only slightly larger than 1. Regional values vary a great deal, both when r = 0.5 and when r varies over [0.4, 0.6] (Figure 3). In the Middle East, Central/South Asia, East Asia, and the Americas, assuming r = 0.5, observed FST values require a greater number of female migrants than male migrants to be explained in our model, while in Africa, Europe, and Oceania, the analysis finds support for more male migrants than female migrants. This could be due to differences in reproductive success for males and females in these regions, or to some other assumption made in our model.
Figure 3.
Global and regional estimates of the ratio of female to male migrants (the ratio of equation (14) to equation (15)) as r varies over [0.4, 0.6], based on FST values calculated using 952 individuals from the Human Genome Diversity Panel.
Although we are not able to draw strong empirical conclusions from these data, we have rigorously derived FST and expected times to coalescence under this new model, making explicit how population differentiation, as measured by FST, depends on the number of males and females in a population and on the migration rates of the sexes. A scenario with males migrating more than females (Figure 2B) creates a bigger discrepancy between FST,A and FST,X than the reverse situation, producing differences that are much larger than those observed between FST values for the autosomes and for the X chromosome in the HGDP dataset. In combination with other tools, our results may assist in further investigations of the contributions of males and females to the history of human migration.
Acknowledgments
We thank Jeremy Van Cleve and Daniel Garrigan for helpful discussions, and Jon Wilkins and an anonymous reviewer for comments on earlier versions of this manuscript. This work was supported by the William F. Milton Fund of Harvard University, NSF grant DEB-0609760, and NIH grants GM-28016 and GM-081441.
Appendix 1: The migration components of transition matrix entries
Recall is the probability that a sample of two males are currently found in i number of demes (i = I,II) and were found in j number of demes (j = I,II) in the previous generation; states i and j refer to whether the sampled lineages were in the same deme (denoted as I) or different demes (II). For any sample, gi,j will depend on the backwards migration rate and the population sizes of males (and/or females, depending on the individuals from which lineages were sampled), but will not depend on whether the sampled locus is X-linked or autosomal. Note that sampling of individuals is done without replacement. Thus, gi,j for a sample of two males is given below:
The corresponding probabilities for a sample of two females are obtained by substituting mf and Nf for mm and Nm, respectively, in the equations above.
For a sample with one male lineage and one female lineage:
Note that (the probability alleles sampled from two individuals with sexes k and l in state i in the present were in individuals in either the same or different demes in the previous generation) is 1 for all i, k, l.
To give an example of the decomposition of terms in matrices (1) and (2) according to equation (3), let us examine (PA)35 more closely. (PA)35 is the probability that two alleles sampled from two females in the same deme in the present were in one male and one female in the same deme one generation ago.
Substituting mf = Mf/Nf = Mf/(Nr),
Using the definition of BA from equation (3), we get
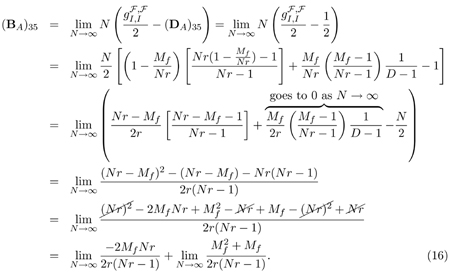 |
The second term of the right hand side of equation (16) above goes to 0 as N → ∞; using L’Hospital’s Rule on the first term, then
Let EN,A denote the error matrix EN from equation (3) of the autosomal transition matrix PA. Then
Appendix 2. The derivation of GX and GA using Möhle’s (1998) result
From equation (3), as N approaches infinity,
Using the above and RX given in matrix (5), the product matrix
where .
To obtain the terms ( X)ij given in matrix (6) in the main text,
X)ij given in matrix (6) in the main text,
The autosomal matrices DA, RA, BA, and GA are all 10 × 10 matrices, with states as in Table 1. Using PA given by matrix (1), from equation (3) DA = limN→ ∞ PA is
where .
The product matrix
where .
To obtain the terms ( A)ij given in matrix (7) in the main text
A)ij given in matrix (7) in the main text
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Pilkington MM, Wilder JA, Cox MP, Soodyall H, Strassmann B, Destro-Bisol G, de Knijff P, Novelletto A, Friedlaender J, Hammer MF. Inferring human population sizes, divergence times and rates of gene flow from mitochondrial, X and Y chromosome resequencing data. Genetics. 2007;177:2195–2207. doi: 10.1534/genetics.107.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. Sinauer; Sunderland, MA: 2007. pp. 124–125. [Google Scholar]
- Hedrick PW. Sex: differences in mutation, recombination, selection, gene flow, and genetic drift. Evolution. 2007;61:2750–2771. doi: 10.1111/j.1558-5646.2007.00250.x. [DOI] [PubMed] [Google Scholar]
- Helgason A, Hrafnkelsson B, Gulcher JR, Ward R, Stefánsson K. A populationwide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: evidence for a faster evolutionary rate of mtDNA lineages than Y chromosomes. Am J Hum Genet. 2003;72:1370–1388. doi: 10.1086/375453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Choi Y, van Oven M, Mona S, Brauer S, Trent RJ, Suarkia D, Schiefenhövel W, Stoneking M. The impact of the Austronesian expansion: evidence from mtDNA and Y-chromosome diversity in the Admiralty Islands of Melanesia. Mol Biol Evol. 2008;25:1362–1374. doi: 10.1093/molbev/msn078. [DOI] [PubMed] [Google Scholar]
- Laporte V, Charlesworth B. Effective population size and population subdivision in demographically structured populations. Genetics. 2002;162:501–519. doi: 10.1093/genetics/162.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Handley LJ, Perrin N. Advances in our understanding of mammalian sex-biased dispersal. Mol Ecol. 2007;16:1559–1578. doi: 10.1111/j.1365-294X.2006.03152.x. [DOI] [PubMed] [Google Scholar]
- Möhle M. A convergence theorem for Markov chains arising in population genetics and the coalescent with partial selfing. Adv Appl Prob. 1998;30:493–512. [Google Scholar]
- Mountain JL, Ramakrishnan U. Impact of human population history on distributions of individual-level genetic distance. Hum Genomics. 2005;2:4–19. doi: 10.1186/1479-7364-2-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Krone SM. Separation of time scales and convergence to the coalescent in structured populations. In: Slatkin M, Veuille M, editors. Modern Developments in Theoretical Population Genetics. Oxford University Press; Oxford: 2002. pp. 194–232. [Google Scholar]
- Oota H, Settheetham-Ishida W, Tiwawech D, Ishida T, Stoneking M. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal resident. Nat Genet. 2001;29:20–21. doi: 10.1038/ng711. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Rosenberg NA, Zhivotovsky LA, Feldman MW. Robustness of the inference of human population structure: a comparison of X-chromosomal and autosomal microsatellites. Hum Genomics. 2004;1:87–97. doi: 10.1186/1479-7364-1-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation in populations with different classes of individuals. Theor Popul Biol. 1999;55:297–308. doi: 10.1006/tpbi.1998.1406. [DOI] [PubMed] [Google Scholar]
- Schaffner SF. The X chromosome in population genetics. Nat Rev Genet. 2004;5:43–51. doi: 10.1038/nrg1247. [DOI] [PubMed] [Google Scholar]
- Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20:278–280. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Inbreeding coefficients and coalescence times. Genet Res. 1991;58:167–175. doi: 10.1017/s0016672300029827. [DOI] [PubMed] [Google Scholar]
- Vitalis R. Sex-specific genetic differentiation and coalescence times: estimating sex-biased dispersal rates. Mol Ecol. 2002;11:125–138. doi: 10.1046/j.0962-1083.2001.01414.x. [DOI] [PubMed] [Google Scholar]
- Wang J. Effective size and F-statistics of subdivided populations. II. dioecious species. Genetics. 1997;146:1465–1474. doi: 10.1093/genetics/146.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Effective size and F-statistics of subdivided populations for sex-linked loci. Theor Popul Biol. 1999;55:176–188. doi: 10.1006/tpbi.1998.1398. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nature Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic Data Analysis II. Sinauer; Sunderland, MA: 1996. [Google Scholar]
- Wilder JA, Kingan SB, Mobasher Z, Pilkington MM, Hammer MF. Global patterns of human mitochondrial DNA and Y-chromosome structure are not influenced by higher migration rates of females versus males. Nat Genet. 2004a;36:1122–1125. doi: 10.1038/ng1428. [DOI] [PubMed] [Google Scholar]
- Wilder JA, Mobasher Z, Hammer MF. Genetic evidence for unequal effective population sizes of human females and males. Mol Biol Evol. 2004b;21:2047–2057. doi: 10.1093/molbev/msh214. [DOI] [PubMed] [Google Scholar]
- Wilkins JF. Unraveling male and female histories from human genetic data. Curr Opin Genet Dev. 2006;16:611–617. doi: 10.1016/j.gde.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Marlowe FW. Sex-biased migration in humans: what should we expect from genetic data? BioEssays. 2006;28:290–300. doi: 10.1002/bies.20378. [DOI] [PubMed] [Google Scholar]





