Abstract
Non-LTR retrotransposons (NLRs) are a unique class of mobile genetic elements that have significant impact on the evolution of eukaryotic genomes. However, the molecular details and functions of their encoded proteins, in particular of the accessory ORF1p proteins, are poorly understood. Here, we identify noncanonical RNA-recognition-motifs (RRMs) in several phylogenetically unrelated NLR ORF1p proteins. This provides an explanation for their RNA-binding properties and clearly shows that they are not related to the retroviral nucleocapsid protein Gag, despite the frequent presence of CCHC zinc knuckles. In particular, we characterize the ORF1p protein of the human long interspersed nuclear element 1 (LINE-1 or L1). We show that L1ORF1p is a multidomain protein, consisting of a coiled coil (cc), RRM, and C-terminal domain (CTD). Most importantly, we solved the crystal structure of the RRM domain, which is characterized by extended loops stabilized by unique salt bridges. Furthermore, we demonstrate that L1ORF1p trimerizes via its N-terminal cc domain, and we suggest that this property is functionally important for all homologues. The formation of distinct complexes with single-stranded nucleic acids requires the presence of the RRM and CTD domains on the same polypeptide chain as well as their close cooperation. Finally, the phylogenetic analysis of mammalian L1ORF1p shows an ancient origin of the RRM domain and supports a modular evolution of NLRs.
Keywords: genome evolution, LINE-1, nucleic acid chaperone, RNP, crystal structure
The mammalian LINE-1 (L1) element is an active retrotransposon and probably one of the most significant players in the evolution of the mammalian genome (1–3). More than 17% of the human genome consists of interspersed L1 sequences (4). In nonmammalian vertebrates, L1 is present as well, but to a much lesser extent (3, 5, 6). The effects of L1 on genome composition and gene expression are numerous. Apart from an implication in genetic disease and tumorigenesis, L1 retrotransposition also generates allelic heterogeneity and new possibilities for genetic recombination. Furthermore it is responsible for the mobilization of nonautonomous retrotransposons (e.g., primate-specific Alu elements) and for pseudogene formation (1, 2).
L1 is a non-LTR retrotransposon (NLR). As such, it lacks the long terminal repeats (LTRs) that are present in retroviruses and LTR retrotransposons. L1 integrates via target-primed reverse transcription (TPRT), where the RNA intermediate is reverse transcribed in the nucleus at the spot of genomic integration (7, 8). This mechanism is fundamentally different from the retroviral integration mechanism and much less understood in its molecular details (9).
L1 RNA contains two open reading frames (ORF1 and ORF2). The reverse transcriptase (RT) necessary for TPRT is part of the 150-kDa multidomain protein encoded by ORF2 [L1ORF2p (3)]. L1ORF2p also contains two other domains: an N-terminal APE1-like endonuclease (EN), which nicks the chromosomal target DNA (10, 11), and a C-terminal CCHC zinc knuckle (3).
The exact function of L1ORF1p remains unclear. It binds single-stranded RNA and DNA with high affinity (12, 13) and is essential for retrotransposition of L1 (14). Like ORF2p, it shows a remarkable cis preference, that is, it associates preferentially with its encoding transcript (15, 16). L1ORF1p can be localized in the cytoplasm (in putative stress granules) as well as in the nucleus (17, 18) and can also be identified in large L1 ribonucleoprotein particles (RNPs) fractionated from cytoplasmic extracts (16, 19, 20).
L1ORF1p is a 40-kDa protein in humans. Its size varies across species as the N-terminal region is not conserved regarding both sequence and length (5, 21). Sedimentation studies and atomic force microscopy indicate that purified murine L1ORF1p forms unusual, dumbbell-shaped trimers that are held together by a coiled coil (cc) formed between sequences in the N-terminal halves of the monomers (22, 23). The other, well-conserved half of murine L1ORF1p is highly basic and binds nucleic acids, but does not display any classical sequence motifs that would be indicative of RNA binding (21). Also, the recent NMR structure of the murine L1ORF1p C-terminal domain (CTD) does not relate L1ORF1p to any other protein of known function and shows a rare αβββαα fold (24). Because, additionally, the isolated CTD binds RNA only weakly (24), it remained unclear how L1ORF1p achieves its high RNA affinity.
The mammalian L1 element belongs to the L1 clade of NLRs, which is just one of at least 14 defined phylogenetic clades (9). Many of these clades contain ORF1p proteins that are unrelated to mammalian L1ORF1p but have similar functional properties (25–29). These proteins frequently contain CCHC zinc knuckles that are also found in the retroviral nucleocapsid protein Gag (30). This has fueled speculations on a more general functional (25, 26, 31) and structural (27, 28) similarity between NLR ORF1p and retroviral nucleocapsid proteins. Clearly, structural information is required to gain general insight into common ORF1p functions, into the phylogenetic relations among NLRs and into their differences to LTR retrotransposons and retroviruses.
Here, we identify noncanonical RRM domains, the most common eukaryotic RNA-binding domain (32, 33), in both mammalian-type L1ORF1p proteins and Gag-like NLR ORF1p proteins. We focus our analysis on human L1ORF1p, which emerges as a true multidomain protein and we determine the crystal structure of its middle domain, which adopts a distinct RRM fold. We also show trimerization of the full-length protein, generalizing this function for all homologues, and we demonstrate that the specific binding to single-stranded nucleic acid requires the close cooperation of the RRM and CTD domains. Stably base-paired nucleic acids are a poor substrate and base pairing is not prevented by the presence of the L1ORF1p RNA-binding fragment. This is consistent with a postulated nucleic acid chaperone activity.
Results
ORF1p Proteins from Many NLR Clades Contain RRM Domains.
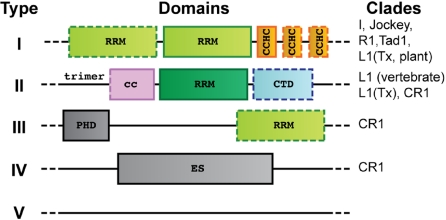
To identify structured domains within NLR ORF1p proteins, we subjected individual sequences to sensitive searches for remote protein domain homologues [HHpred (34)]. This revealed potential RRM domains in nearly all of the major NLR clades that contain an ORF1p (Fig. 1 and Tables S1 and S2). Because no classic RNA-binding domains could be identified in these proteins in the past, the discovery of RRM domains was unexpected. It provides an explanation for the RNA-binding properties of many NLR ORF1p proteins and clearly establishes that they are not related to the Gag proteins encoded by retroviruses and LTR-retrotransposons (9). According to the arrangement of the predicted structural domains, we can roughly distinguish five types of NLR ORF1p proteins.
Fig. 1.
Identification and organization of RRM domains in phylogenetically unrelated NLR-ORF1p proteins. Type I ORF1p is widespread and contains Gag-like CCHC zinc-knuckles. Type II ORF1p is found in the human L1 element and trimerizes via a coiled coil (cc). Other types are described in the main text (see also Tables S1 and S2). CTD, C-terminal domain; PHD, plant homeodomain; ES, esterase domain. Some domains may not always be present (dotted outlines).
Type I ORF1p (Fig. 1) is the most widespread type and contains at least one RRM domain immediately upstream of a Gag-like CCHC zinc knuckle. A second RRM domain and additional zinc knuckles are frequent. The close association of the zinc-knuckle and RRM modules suggests a functional cooperation as observed frequently in other RRM proteins (32, 33). Type I ORF1p is found from vertebrates to plants across at least five different clades (9, 35), which indicates its ancient origin.
Type II ORF1p (Figs. 1 and 2) is found in the human L1 element. It contains a single RRM domain that is preceded by additional conserved amino acids leading to a trimerization of the molecule via a coiled coil (Fig. S1, see below). The CTD domain (24) is conserved in vertebrate type II ORF1p proteins and characterizes the lineage of modern L1 elements (also referred to as mammalian-type L1 elements). This lineage is distinct from ancient members of the L1 clade that are found in amphibians (36), fish (37), insects (38), and plants (39) and that contain an ORF1p of type I.
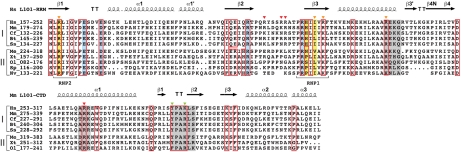
Fig. 2.
Phylogenetic conservation of mammalian-type L1 ORF1p. Structure-based sequence alignments of the RRM (L1O1-RRM, top) and CTD (L1O1-CTD, bottom) domains show highly conserved residues boxed in red. Surface residues only conserved in placental mammals (group I) or only outside of placental mammals (group II) are boxed separately. Residues forming the conserved salt bridges are shaded in blue. Residues providing aromatic, RNA-binding side-chains in canonical RRMs are shaded in yellow. Triangles mark residues mutated in this study with a strong (red), moderate (orange) or negligible (green) effect on RNA-binding. Additional motifs mutated in a previous study (14) are shaded in gray. The C-terminal sequences of Sp and Nv cannot be confidently aligned to the mammalian-type CTD domain. Gene identifiers: Hs, Homo sapiens (gi:307098); Mm, Mus musculus (gi:198644); Cf, Canis familiaris (gi:116175029); Bt, Bos taurus (gi:66734172); Ss, Sus scrofa (gi:148645275); Me, Macropus eugenii (gi:151302550); Xt, Xenopus tropicalis (gi:85740540); Ol, Oryzias latipes (gi:3746501), Sp, Strongylocentrotus purpuratus (gi:111740418); Nv, Nematostella vectensis (gi:149338150).
For type III ORF1p, we predict an occasional C-terminal RRM module in addition to a previously described N-terminal plant homeodomain (PHD) (40). Type III ORF1p is found in the heterogeneous CR1 clade, which also harbors ORF1p proteins of type IV. These contain a functional esterase domain that enhances retrotransposition (40, 41). Finally, there are numerous NLR ORF1p proteins (type V) that cannot be classified so far (Fig. 1).
Human L1ORF1p Consists of Three Distinct Domains and Forms Nonspherical Trimers.
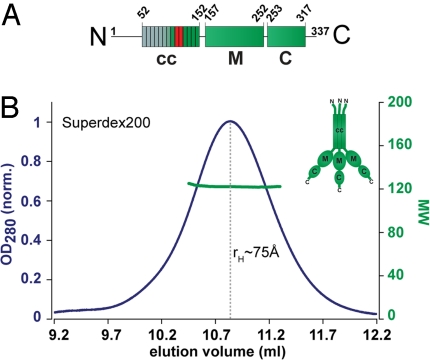
Including the predicted RRM domain, human L1ORF1p shows three distinct domains (Fig. 3). In addition to the CTD (C) domain (24) and the predicted RRM (M) domain there is a less well-conserved N-terminal cc domain (Fig. 3A). For murine L1ORF1p, the cc domain is required for the trimerization of the protein (22, 23). However, because of the variability of the cc domain, it was not clear whether this function could be generalized for all type II ORF1p proteins.
Fig. 3.
Domain structure and trimerization of human L1 ORF1p. (A) Schematic organization of the monomer into the coiled-coil (cc), RRM (M) and CTD (C) domains. Heptad repeats in the cc-domain are indicated. The unconserved part of the protein is in gray, the positions of the trimerization motifs are indicated red. (B) Trimeric state of the full-length protein analyzed by size-exclusion chromatography (rH ≈ 75 Å) and MALLS (Mr = 120 kDa). (Inset) Schematic representation of the trimer adapted from (22).
We therefore aligned the seven C-terminal heptad repeats of the type II ORF1p cc domain (Fig. S1). This revealed conserved RhxxxhE motifs (h, hydrophobic; x, any amino acid) that were demonstrated in other proteins to induce a parallel trimeric coiled coil (42). Consequently, trimerization of type II ORF1p seems conserved for functional reasons. Furthermore, we purified recombinant human L1ORF1p (hL1ORF1p) and did size exclusion chromatography followed online by multiangle static laser-light scattering (MALLS). The protein elutes as a single peak forming particles with a molecular weight (Mr) of 120 kDa. This is consistent with a trimeric state of the protein. The hydrodynamic radius (rH) of approximately 75 Å indicates a nonspherical shape, because a globular trimer would have an rH of only approximately 45 Å (Fig. 3B).
The largest monomeric fragment (hL1ORF1p-MC) that we could identify by deletion analysis comprises both RRM- and CTD-domains and is rather globular as indicated by an rH of approximately 20 Å (Fig. S2). This represents a significantly larger portion of the protein as compared with previous studies with the murine protein (23, 24, 43). Furthermore, we can show that the predicted RRM- and CTD domains (hL1ORF1p-M and hL1ORF1p-C, respectively) are soluble independently from each other and remain monomeric at concentrations up to 100 μM. When mixed at these concentrations, they also do not detectably interact with each other (see Fig. 4A and data not shown).
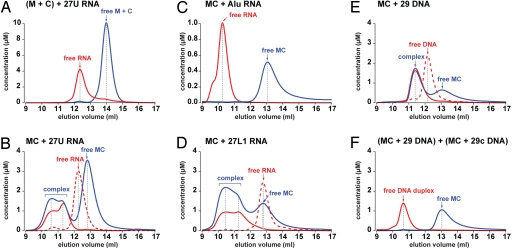
Fig. 4.
Nucleic acid-binding properties of human L1 ORF1p. Size exclusion chromatography was done with various nucleic acid substrates (red lines) in the absence (dashed lines) or in the presence (solid lines) of protein (blue solid lines). Elution volumes of the complexes and of the free components are indicated by arrows and dashed gray lines, while apparent concentrations are calculated from the relative absorption properties of the components. M, hL1ORF1p-M; C, hL1ORF1p-C; MC, hL1ORF1p-MC; AluRNA, SA86 (46).
The Crystal Structure of the RRM Domain in Human L1ORF1p Shows Extended Loops and Noncanonical RNP Motifs.
To reveal the molecular details of the RRM domain we determined the crystal structure of hL1ORF1p-M (Fig. 5 and Fig. S3). The structure was solved by single-wavelength anomalous dispersion (SAD) from seleno-methionine substituted protein and was refined at 1.4-Å resolution to an Rfree of 18.5% (Table S3).
Fig. 5.
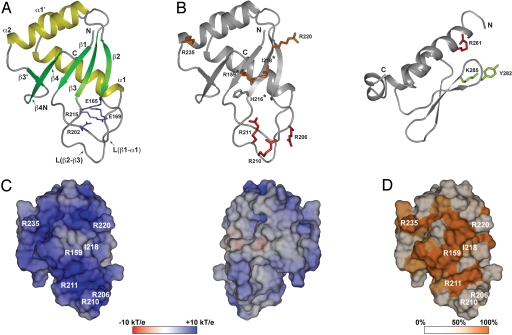
Crystal structure of the RRM domain of human L1 ORF1p. (A) Ribbons representation with α-helices in yellow and β-strands in green. The side chains forming the conserved salt-bridges are shown as sticks (blue). (B) Localization of mutated side-chains. The RRM (Left) and CTD (Right) domains are shown as ribbons with selected side chains as sticks (for colors see triangles in Fig. 2; asterisks: aromatic side chain in canonical RRMs; murine CTD (PDB-ID 2jrb (24)) shown with human amino acid numbers). (C) Electrostatic potential mapped on the molecular surface of the RRM domain (pI = 10.6). Potentials are contoured from −10 kT/e (red) to + 10 kT/e (blue). (Left) View as in A, onto the surface of the β-sheet and the adjacent loop L(β2-β3). (Right) Backside view, 180° from (A). (D) Surface colored by sequence conservation. Sequence similarity among placental mammals (Fig. 2, group I) is color-ramped : white (50% or less) to orange (100%). All three-dimensional representations are done with PyMOL (http://www.pymol.org).
The protein shows a classical RRM fold with the typical βαββαβ topology, where the two α-helices are packed against one surface of the four-stranded, anti-parallel β-sheet (2.8 Å r.m.s.d. over 76 Cα positions compared with the classical U1A-RRM; PDB-ID: 1oia). In the present case, there is an additional small β-hairpin (β3′/β4N) that is located between helix α2 and strand β4, and an extra α-helix α1′ within the loop L(α1-β2). Whereas the β-hairpin is occasionally observed in other RRM domains, the helix α1′ has not been seen before. The two salt bridges (E165-R215 and E169-R202) that are formed between loop L(β1-α1) and the extended loop L(β2-β3) are another unique feature of the hL1ORF1p RRM domain that is well-conserved among type II NLR ORF1ps. These salt bridges stabilize the structures of the loops and fix their relative orientations (Figs. 2, 5A, and S3). They likely are of functional importance, because a single point mutation (E165G) results in a strong nucleolar localization of the protein (17). Interestingly, the unique parts of the RRM-domain interact with each other in the crystal [helix α1′ fits nicely into the cleft between the loops L(β1-α1) and L(β2-β3)], but there is no evidence so far that this might be physiologically relevant.
Canonical RRM domains are characterized by two conserved sequence signatures, RNP1 ([RK]-[G]-[FY]-[GA]-[FY]-[ILV]-[X]-[FY]) located on β-strand β3, and RNP2 ([ILV]-[FY]-[ILV]-X-N-L) located on β-strand β1. These strands provide aromatic side chains on the surface of the β-sheet (positions 3, 5 in RNP1, and position 2 in RNP2) that are frequently involved in base-stacking or in hydrophobic interactions with nucleic acid substrates (32). In the human L1ORF1p RRM domain the RNP1 (P-R-H-I-I-V-R-F) and RNP2 (L-R-L-I-G-V) sequences deviate significantly from the consensus signature (Figs. 2 and 5B). This may explain why this RRM domain was not identified earlier and raises the question if and how the β-sheet surface of this domain is involved in nucleic acid binding.
Sequence Conservation and the Distribution of Surface Charge Indicate the Interface Involved in Nucleic Acid Binding.
The C-terminal half of L1ORF1 is highly positively charged but the isolated CTD domain is not sufficient to mediate strong nucleic acid binding (24). As a classical single-strand specific nucleic acid binding domain the presently identified RRM domain may therefore play a major role. The structure shows a highly asymmetric distribution of charges with a strongly basic surface that includes the canonical β-sheet but also the adjacent surface of the extended loop L(β2-β3) that is unique to the present RRM domain (Fig. 5C).
Furthermore, we analyzed the sequence conservation of the RRM domain among five placental mammals and found that the most highly conserved surface side chains cluster on and around the basic β-sheet surface (Figs. 2 and 5D). These side-chains include N157 and D252 that link the N-and C-termini of the RRM domain (Fig. S3), R159 on strand β1, H216, I218, and R220 on strand β3 and K227, E228, and R235 on helix α2. Many of those residues do not fulfill any obvious structural roles and are likely conserved for functional reasons. To test if they are important for nucleic acid binding we constructed a series of point mutants (see below).
Efficient Nucleic Acid Binding Requires the Cooperation of the RRM and CTD Domains.
To test for stable nucleic acid binding under constant buffer conditions we used analytical size exclusion chromatography, monitored by triple wavelength UV absorption spectroscopy. We estimated the concentrations of the individual protein and RNA components as they eluted from the column, providing insight into the stoichiometry of the complexes (Fig. 4).
No interaction was detected between the isolated RRM domain (hL1ORF1p-M) and a 27-mer poly(U) RNA substrate (27U RNA), at concentrations up to 75 μM. Similarly, we did not detect any interaction with the isolated CTD domain (hL1ORF1p-C) or with a protein sample where the individual RRM and CTD domains were premixed at equimolar concentrations (Fig. 4A and data not shown). With both RRM and CTD domains on a single polypeptide chain (hL1ORF1p-MCH6), however, the RNA substrate was bound quantitatively. The majority of this RNA (27U RNA) was found in an equimolar complex with the human L1ORF1p-MCH6 fragment. Even with an excess of protein only a small fraction of the RNA bound additional protein molecules (probably up to three, see below) (Fig. 4B). The enhanced RNA affinity of the RRM-CTD fragment over the mixture of the individual domains can be explained by their cooperation and by the extremely short linker sequence that probably constrains the relative positions of the two domains (44).
Point mutations of selected surface side-chains confirm that both domains participate in RNA-binding (27U RNA). As a result, in size exclusion chromatography, the RNA no longer co-elutes with the mutated protein (strong effect) or elutes significantly later than in the complex with the wild-type RRM-CTD fragment (intermediate effect). The most severe effects are shown by the R206A/R210A/R211A triple mutant on the extended loop L(β2-β3) of the RRM domain and by the R261A mutant on helix α1 of the CTD domain. The single R220A, R159A, I218Y, and R235A mutants on the RRM domain have an intermediate effect, while the Y282A/K285A mutant on the loop L(β1-β2) of the CTD domain behaves quasi identically to the wild-type protein (Figs. 2, 5B, and S4).
Although none of the mutants abolished RNA binding completely, the results confirm the importance of the basic protein surface of the RRM domain for RNA binding and show that cooperation with the CTD domain is essential. Many of the exchanged arginine residues may solely make contacts to the phosphate-ribose backbone of the RNA, thereby fixing its conformation. R159 and R261, however, appear particularly important, as they are invariant in sequence alignments and are functionally required at several steps in retrotransposition (14, 16, 17, 45). They may be involved in multiple contacts, possibly stacking on bases or locking the relative orientations of the RRM and CTD domains on the RNA. Furthermore, the shallow surface cavity centered over the hydrophobic I218 seems essential, because the tyrosine substitution frequently found in canonical RRM domains (position 3 in the RNP1 motif) reduces RNA binding. The negligible effect of the Y282A/K285A mutation on RNA binding indicates that the Y282PAKLS motif in the CTD domain probably does not interact directly with RNA. It rather plays a structural role and the original alanine substitution of the entire motif is likely to affect the structural integrity of the CTD (14, 24). A similar effect can be expected for the original alanine substitution of the R235EKG motif (14) on the RRM domain, although we see an RNA-binding defect for the single R235A substitution alone.
The Monomeric RRM-CTD Fragment Binds Single-Stranded Nucleic Acid and Competes with the Formation of Base-Paired Structures.
To exclude that complex formation simply results from electrostatic attraction of the negatively charged RNA backbone by the positively charged protein surface, we tested highly structured AluRNA (SA86) (46) as a substrate in size exclusion chromatography. Most of the phosphate-ribose backbone of this 86-mer RNA is conformationally fixed and most of its nucleotides are involved in base-pair interactions. In the gel filtration assay it did not bind to the RRM-CTD fragment (Fig. 4C).
To test whether weak secondary structures or the nucleotide composition of the RNA substrate affect the interaction with the RRM-CTD fragment, we selected an alternative 27-mer (5′ UAACAAUAUUAACUUUAAAUAUAAAUG 3′) derived from the human L1 RNA (27L1 RNA). It corresponds to the 3′-terminal nucleotides of a longer 41-mer that specifically copurifies with endogenous human L1ORF1p (12). In size exclusion chromatography 27L1 RNA is delayed with respect to 27U RNA, indicating that it folds into a more compact stem-loop structure. Nevertheless, 27L1 RNA also binds quantitatively to hL1ORF1p-MCH6. In contrast to 27U RNA, each 27L1 RNA molecule recruits at least two or even three protein monomers (Fig. 4D). This shows that the RRM-CTD fragment can distinguish between RNA sequences and will consequently have preferential binding sites on longer RNA substrates.
To investigate whether binding to hL1ORF1p-MCH6 is limited to RNA we also tested a 29-mer DNA (29 DNA) (31) as well as its reverse complement (29c DNA) (31). In the absence of protein, each sample elutes as a single peak at the same position as the other, indicating an extended conformation without secondary structure. In the presence of a slight molar excess of hL1ORF1p-MCH6 29 DNA is bound with equimolar stoichiometry (Fig. 4E). The same is true for 29c DNA (data not shown). When both complexes are mixed together, the DNA strands readily anneal to form a duplex, quantitatively liberating the bound protein (Fig. 4F).
In conclusion, hL1ORF1p-MCH6 preferably binds flexible, single-stranded nucleic acid, and the identical elution volumes of the 27U and 27L1 RNA complexes indicate that weakly base-paired structures like 27L1 RNA can be unwound by the protein. As a consequence, the RRM-CTD fragment could help resolve kinetically trapped nucleic acid structures, providing a path to the thermodynamically most favorable conformation.
The Ancient Origin of the RRM Domain in Type II ORF1p Supports a Modular Evolution of NLRs.
So far, type II ORF1p has only been described in vertebrate members of the L1 clade. We were therefore surprised to identify homologues of the RRM domain in NLRs of the starlet see anemone Nematostella vectensis (a non-bilaterian animal) and of the purple sea urchin Strongylocentrotus purpuratus (a deuterostomian animal) (Fig. 2 and Tables S1 and S2). This indicates a deeply rooted origin of this RRM-domain before the emergence of bilaterians approximately 750 million years ago and, possibly, a selective loss from the branch of protostomian animals. The respective NLRs do not seem to contain an equivalent for the CTD domain, and according to their reverse transcriptases they belong to the Tx group of the L1 clade and to the CR1 clade (9, 35). The existence of such chimerical elements strongly supports the idea of a modular evolution of NLRs. Furthermore, the modular nature of the ORF1p and ORF2p proteins and their respective combinations can be exploited to clarify ambiguous relations among NLRs and can ultimately help to regroup their phylogenetic tree with higher resolution.
Discussion
Identification of RRM Domains in NLRs and Their Significance for Retrotransposition.
For the last twenty years, NLR ORF1p proteins were studied in the absence of detailed structural information, and it was rather obscure how ORF1p would bind RNA. The present identification of RRM modules opens a new perspective and relates the observed cytoplasmic RNPs (16, 19, 20, 25–27) to cellular hnRNPs and mRNPs rather than to viral nucleocapsid-like RNPs. Apart from their structural role in RNP formation the RRM domains in NLR ORF1p proteins likely have specialized functions as well, assisted by accessory domains like the CTD or the CCHC zinc knuckles. This is indicated by point mutations that have rather moderate effects on RNP formation (16, 45) or on the localization to cytoplasmic foci (17), but nevertheless affect retrotransposition activity strongly.
One of these specialized functions may be the promotion of RNA folding, helping it to overcome kinetic barriers on the way to the thermodynamically most stable structure. Consistent with such nucleic acid chaperone activity (25, 31, 43, 45), we observe that the monomeric RRM-CTD fragment binds nucleic acids as single strands, but does not prevent them from forming base-paired structures that cannot be bound anymore. On the molecular level this could be achieved by an interaction mainly with the flexible phosphate-ribose backbone of a single-stranded nucleic acid, leaving the bases exposed for interactions. Furthermore, the local crowding of nucleic acids bound in the context of the L1ORF1p trimer could facilitate their mutual annealing. Nucleic acid chaperone activity could assist in target-primed reverse transcription (31), or in the folding of the RNA into structures that are competent for transport (29) and inaccessible to nucleases and small RNAs from the defense systems of the host (47).
Domain Structure and Nucleic Acid Binding of Human L1ORF1p.
Similar to the NLR ORF2p proteins, ORF1p proteins are multidomain proteins that may have evolved in a modular fashion. The present identification of the RRM domain in human L1ORF1p leads to a redefinition of L1ORF1p structure and is in conflict with previous reports, which assumed an unstructured linker in the center of the protein. The significance of results obtained with protein constructs that contain less than half of the RRM domain therefore needs to be revisited, as we do not expect the RRM domain to get folded in this context (23, 24, 43).
For RNA binding, the RRM domain is assisted by the CTD. Furthermore, the protein forms unusual trimers. As a consequence, RNA interactions of this RRM domain probably deviate from the canonical mode. Experimentally, a whole series of mutations on and around the β-sheet surface affect nucleic acid binding, but we could not identify a single point mutation that abolishes binding completely. Dependent on the RNA sequence, we find that 27-mer RNA substrates can recruit up to probably three copies of the RRM-CTD fragment. To fully understand the complex nucleic acid binding properties of L1ORF1p, we therefore seek to determine high quality structures of the trimeric state of the protein in complex with various nucleic acid substrates.
The presently identified domain architecture of NLR ORF1p proteins provides crucial insight on the way toward this goal and will undoubtedly fuel the further characterization of non-LTR retrotransposition and related processes.
Materials and Methods
Computational Identification of RRM Domains.
Individual NLRs and their ORF1p sequences (Tables S1 and S2) were identified by tBLASTn searches using queries from the literature or from RepBase (35). ORF1p sequences were analyzed for similarity to known domains using profile hidden Markov models as implemented in HHpred (34).
Sample Preparation.
DNA sequences corresponding to the respective human L1ORF1p constructs were PCR amplified from a plasmid (pJM130) encoding a functional human L1 element (48). They were inserted into the respective expression vectors, pETM11 [derived from pET24d (Novagen)] for hL1ORF1p (GAM1-K337), pGEX6p1 (GE Healthcare) for hL1ORF1p-MC (GPLGSN157-Q330), pET15b (Novagen) for hL1ORF1p-MCH6 (MGN157-Q330HHHHHH), pETM60 [derived from pET24d (Novagen)] for hL1ORF1p-M (GAMGN157-D252) and for hL1ORF1p-C (GAMVS254-R328). Proteins were expressed in the E. coli strain Rosetta 2(DE3) (Novagen) at 20°C overnight. They were purified from cleared cell lysates by Ni2+- or glutathione affinity steps. After proteolytic removal of the affinity tags (hL1ORF1p-MC, hL1ORF1p-M, hL1ORF1p-C), proteins were further purified by heparin-affinity chromatography (hL1ORF1p) and gel filtration.
Analytical Size-Exclusion Chromatography and Mass Determination.
Analytical size-exclusion chromatography was done on an ÄKTA™ Purifier-10 equipped with a Superdex75 10/300 GL column (GE Healthcare), monitoring optical density (OD) simultaneously at 230 nm, 260 nm, and 280 nm. Protein concentrations were estimated from the theoretical molar extinction coefficients ε280 at 280 nm. Nucleic acid concentrations were estimated from ε260 as provided by the manufacturers. The relative contributions of nucleic acid and protein to the total absorption at each wavelength were calculated assuming constant ratios of ε230/ε280 for each substance (49). Components were mixed in chromatography buffer (20 mM Tris/HCl, pH 8.0, 200–300 mM NaCl, and 0–10 mM MgCl2) using starting concentrations between 20 μΜ and 100 μΜ. After 5 min at 18°C, 100 μl were injected on the column (18°C) at a flow rate of 0.5 ml/min.
Multiangle static laser light-scattering experiments (MALLS) were done online with analytical size-exclusion chromatography using miniDAWN TREOS and Optilab rEX instruments (Wyatt Technologies) and the associated software (AstraV) for molecular weight determination. Hyrodynamic radii rH were determined from a calibration kit (GE Healthcare). hL1ORF1p was analyzed over a Superdex200 column (20 mM Tris/HCl, pH8.0, 20 mM MgCl2, 100 mM (NH4)2SO4, 300 mM NaCl).
Crystallization, Data Collection, and Refinement.
Crystalline clusters of hL1ORF1p-M (9.7 mg/ml in 5 mM Tris/Cl, pH 8.0, and 300 mM NaCl) were obtained by vapor diffusion (18°C) mixing 0.8 μl of protein solution with 0.8 μl of reservoir (2.2 M Na- malonate, pH 7.0) over a 500-μl reservoir. Crystals were optimized by hair-seeding (1.7–1.9 M Na-malonate) and flash frozen in liquid nitrogen without additional cryoprotection.
Crystals containing seleno-methionine were single and diffracted better than the initial crystals from the native protein. Diffraction data for the seleno-methionine derivative were collected at a single wavelength (0.97154 Å) on beamline PXII of the Swiss Light Source. Images were processed by XDS (50). The structure was solved by single anomalous dispersion (SAD). We used autoSHARP (51) to search for three selenium sites per molecule. Assignment of the correct hand and solvent flattening (optimum contrast at 51.6%) was done automatically. In the resulting map, ARP/wARP (52) was able to trace 92% of the final model and built 43% of the side chains. The model was completed manually in COOT (53), including alternative conformations. Refinement was done in REFMAC (54) and COOT iteratively, using anisotropic B-factors (see Table S3 for data collection, phasing, and refinement statistics).
Supplementary Material
Acknowledgments.
We thank beamline scientists at the Swiss Light Source for assistance with data collection, Heiko Keller for the preparation of AluRNA, Regina Büttner for excellent technical assistance, and Elisa Izaurralde and the members of the biochemistry department for helpful comments and discussions. E.K. and O.W. are supported by a personal VIDI grant (NWO-CW 700.54.427) to O.W. by the Dutch National Science Organization (NWO).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2w7a).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809964106/DCSupplemental.
References
- 1.Han JS, Boeke JD. LINE-1 retrotransposons: Modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 2.Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 3.Moran JV, Gilbert N. Mammalian LINE-1 Retrotransposons and Related Elements. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 836–869. [Google Scholar]
- 4.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Furano AV, Duvernell DD, Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Zingler N, Weichenrieder O, Schumann GG. APE-type non-LTR retrotransposons: Determinants involved in target site recognition. Cytogenet Genome Res. 2005;110:250–268. doi: 10.1159/000084959. [DOI] [PubMed] [Google Scholar]
- 7.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 8.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik HS, Eickbush TH. Origins and Evolution of Retrotransposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, D.C.: ASM Press; 2002. pp. 1111–1144. [Google Scholar]
- 10.Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 11.Weichenrieder O, Repanas K, Perrakis A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure. 2004;12:975–986. doi: 10.1016/j.str.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci USA. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 15.Wei W, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 17.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirilyuk A, et al. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 2008;36:648–665. doi: 10.1093/nar/gkm1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 21.Martin SL. The ORF1 protein encoded by LINE-1: Structure and function during L1 retrotransposition. J Biomed Biotechnol. 2006;2006:45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci USA. 2003;100:13815–13820. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basame S, et al. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J Mol Biol. 2006;357:351–357. doi: 10.1016/j.jmb.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 24.Januszyk K, et al. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J Biol Chem. 2007;282:24893–24904. doi: 10.1074/jbc.M702023200. [DOI] [PubMed] [Google Scholar]
- 25.Dawson A, Hartswood E, Paterson T, Finnegan DJ. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J. 1997;16:4448–4455. doi: 10.1093/emboj/16.14.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pont-Kingdon G, Chi E, Christensen S, Carroll D. Ribonucleoprotein formation by the ORF1 protein of the non-LTR retrotransposon Tx1L in Xenopus oocytes. Nucleic Acids Res. 1997;25:3088–3094. doi: 10.1093/nar/25.15.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rashkova S, Karam SE, Pardue ML. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc Natl Acad Sci USA. 2002;99:3621–3626. doi: 10.1073/pnas.032071999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rashkova S, Athanasiadis A, Pardue ML. Intracellular targeting of Gag proteins of the Drosophila telomeric retrotransposons. J Virol. 2003;77:6376–6384. doi: 10.1128/JVI.77.11.6376-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seleme MC, et al. In vivo RNA localization of I factor, a non-LTR retrotransposon, requires a cis-acting signal in ORF2 and ORF1 protein. Nucleic Acids Res. 2005;33:776–785. doi: 10.1093/nar/gki221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fawcett DH, Lister CK, Kellett E, Finnegan DJ. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986;47:1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- 31.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 33.Lunde BM, Moore C, Varani G. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 36.Garrett JE, Knutzon DS, Carroll D. Composite transposable elements in the Xenopus laevis genome. Mol Cell Biol. 1989;9:3018–3027. doi: 10.1128/mcb.9.7.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojima KK, Fujiwara H. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: Various multicopy RNA genes and microsatellites are selected as targets. Mol Biol Evol. 2004;21:207–217. doi: 10.1093/molbev/msg235. [DOI] [PubMed] [Google Scholar]
- 38.Biedler J, Tu Z. Non-LTR retrotransposons in the African malaria mosquito, Anopheles gambiae: Unprecedented diversity and evidence of recent activity. Mol Biol Evol. 2003;20:1811–1825. doi: 10.1093/molbev/msg189. [DOI] [PubMed] [Google Scholar]
- 39.Wright DA, et al. Multiple non-LTR retrotransposons in the genome of Arabidopsis thaliana. Genetics. 1996;142:569–578. doi: 10.1093/genetics/142.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapitonov VV, Jurka J. The esterase and PHD domains in CR1-like non-LTR retrotransposons. Mol Biol Evol. 2003;20:38–46. doi: 10.1093/molbev/msg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugano T, Kajikawa M, Okada N. Isolation and characterization of retrotransposition-competent LINEs from zebrafish. Gene. 2006;365:74–82. doi: 10.1016/j.gene.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 42.Kammerer RA, et al. A conserved trimerization motif controls the topology of short coiled coils. Proc Natl Acad Sci USA. 2005;102:13891–13896. doi: 10.1073/pnas.0502390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin SL, Li J, Weisz JA. Deletion analysis defines distinct functional domains for protein–protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J Mol Biol. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- 44.Shamoo Y, Abdul-Manan N, Williams KR. Multiple RNA binding domains (RBDs) just don't add up. Nucleic Acids Res. 1995;23:725–728. doi: 10.1093/nar/23.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin SL, et al. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Weichenrieder O, Wild K, Strub K, Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- 47.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 48.Moran JV, DeBerardinis RJ, Kazazian HH. Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 49.Müller M, Weigand JE, Weichenrieder O, Suess B. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J App Crystallgr. 1993;26:795–800. [Google Scholar]
- 51.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution With autoSHARP. Methods Mol Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 52.Cohen SX, et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D Biol Crystallogr. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.