Abstract
d-Amino acids exist in living organisms as specialized components of many different machineries. Biosynthesis of d-amino acids from racemization of predominant l-enantiomers is catalyzed by a single enzyme. Here, we report the finding of a novel 2-component amino acid racemase for d-to-l inversion in d-arginine metabolism of Pseudomonas aeruginosa. From DNA microarray analysis, the putative dauBAR operon (for d-arginine utilization) of unknown functions was found to be highly induced by d-arginine. The importance of the dau operon in d-arginine metabolism was demonstrated by the findings that strains with a lesion at dauA or dauB failed to use d-arginine as sole carbon source. Two lines of evidence suggest that DauA and DauB are required for d-to-l racemization of arginine. First, growth complementation of an l-arginine auxotroph by d-arginine was abolished by a lesion at dauA or dauB. Second, d-arginine induced l-arginine-specific genes in the parental strain PAO1 but not in its dauA or dauB mutants. This hypothesis was further supported by activity measurements of the purified enzymes: DauA catalyzes oxidative deamination of d-arginine into 2-ketoarginine and ammonia, and DauB is able to use 2-ketoarginine and ammonia as substrates and convert them into l-arginine in the presence of NADPH or NADH. Thus, we propose that DauA and DauB are coupled catabolic and anabolic dehydrogenases to perform d-to-l racemization of arginine, which serves as prerequisite of d-arginine utilization through l-arginine catabolic pathways.
Keywords: amino acid, arginine dehydrogenase, racemase
Although l-amino acids are the predominant amino acids in protein synthesis, d-amino acids serve as specialized components of many types of machineries in living organisms. In mammals, d-serine and d-aspartate are associated with cell aging and neural signaling (1, 2). In bacteria, some d-amino acids are essential ingredients of cell wall synthesis (3). Endogenous d-amino acids are produced by racemization from the prevalent l-amino acids through the action of racemases. Amino acid racemases are classified into 2 groups: pyridoxal 5′ phosphate-dependent and phosphate-independent enzymes (4). Completely different reaction mechanisms have been proposed for these 2 groups of enzymes for the spatial rearrangement of α-hydrogen in the corresponding amino acids. Nevertheless, racemization of amino acids reported so far is catalyzed by a single enzyme.
When provided in excess, some d-amino acids can be used as nutrients to support growth by bacteria. In most cases, d-amino acid oxidase or dehydrogenase catalyzes the oxidative deamination as the first step in catabolism. Pseudomonas aeruginosa, an opportunistic human pathogen with an enormous catabolic capacity, is capable of growing on d-arginine as the sole source of carbon and nitrogen (5). The presence of an inducible d-arginine dehydrogenase activity in this organism was initially reported by Haas and coworkers (6), and 2-ketoarginine derived from this reaction could be converged into the arginine transaminase (ATA) pathway (7, 8), 1 of the 4 pathways for l-arginine catabolism in pseudomonads (Fig. 1). In fact, it has been proposed that l-arginine might be converted into d-arginine via racemization (6), reminiscent of l-alanine utilization through a catabolic alanine racemase and d-alanine dehydrogenase in Escherichia coli and many bacteria (9, 10). Existence of an arginine racemase in P. aeruginosa was supported by growth complementation of arginine auxotrophs with d-arginine (6). However, the activity of P. aeruginosa arginine racemase has never been demonstrated in vitro, presumably because of the instant decomposition of both l- and d-arginine in extracts.
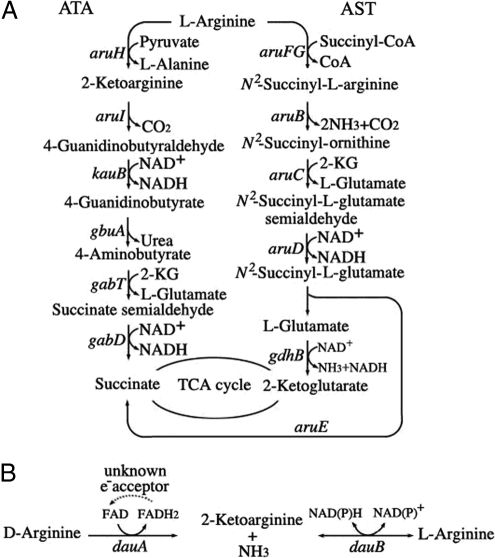
Fig. 1.
Arginine catabolic pathways in P. aeruginosa PAO1. (A) Relevant intermediates and genes of the ATA and AST pathways were shown. (B) Proposed functions of dauA and dauB in conversion of d-arginine to l-arginine.
Under aerobic conditions, l-arginine is preferentially catabolized by the arginine succinyltransferase (AST) pathway, followed by the ATA pathway (7, 11). Enzymes of the AST pathway are encoded by the aruCFGDBE operon (12), which is induced by exogenous l-arginine in the presence of a functional arginine regulator, ArgR (13). The ArgR protein belongs to the AraC family of transcriptional regulators. Depending on the location of its binding sites, ArgR serves as a repressor or activator of ArgR regulon in arginine and glutamate metabolism. Thus, when the AST pathway is absent or remains uninduced (e.g., in the argR mutant), the ATA pathway then takes charge as the auxiliary route of l-arginine utilization. Genes of the ATA pathway are composed of several modules that are subjected to sequential induction by different intermediate compounds in the pathway (6). Expression of the aruHI genes encoding the first 2 enzymes of the ATA pathway is controlled by the AruRS 2-component systems (7) in response to l-arginine, and the gbuA gene encoding 4-guanidinobutyrase is controlled by GbuR and 4-guanidinobutyrate (14).
From DNA microarray analysis, a 4-gene locus (PA3862 to PA3865 of the annotated genome project) of unknown functions was found highly inducible by d-arginine in P. aeruginosa PAO1. We provide evidence that these genes participate in d-arginine catabolism. The promoters of PA3862–PA3864 and the convergent PA3865 are inducible by exogenous d-arginine but not by l-arginine. PA3863 encodes a d-arginine dehydrogenase, and in concert with PA3862 encoding an anabolic l-arginine dehydrogenase, these 2 genes play essential roles in d- to l-arginine conversion. As a result, this unique racemization reaction is the prerequisite for d-arginine utilization through l-arginine catabolic pathways. From here on, PA3862–PA3865 are designated as dauBART, for d-arginine utilization.
Results
DNA Microarray Analysis.
P. aeruginosa grows on d-arginine as the sole source of carbon and nitrogen. To understand the metabolic flow of d-arginine, we conducted DNA microarray experiments and compared the transcriptional profiles of strain PAO1 grown in the minimal medium in the absence or presence of d-arginine. As shown in Table 1, the dauBART genes were significantly induced by d-arginine. Surprisingly, d-arginine exerts the same effects as l-arginine (15) on all genes in the ArgR regulon (as represented by genes in group II for the activation effect and genes in group IV for the repression effect of Table 1).
Table 1.
Microarray analysis of representative genes for arginine metabolism in P. aeruginosa PAO1
| Gene no.* | Gene name | Absolute signal value |
|
|---|---|---|---|
| Glu† | Glu + d-arginine‡ | ||
| Group I: d-Arginine-specific | |||
| PA3862 | dauB | 199 | 9209 |
| PA3863 | dauA | 80 | 3247 |
| PA3864 | dauR | 185 | 2194 |
| PA3865 | dauT | 173 | 4840 |
| Group II: The AST pathway | |||
| PA0895 | aruC | 925 | 5442 |
| PA0896 | aruF | 442 | 4071 |
| PA0897 | aruG | 419 | 3680 |
| PA0898 | aruD | 350 | 2805 |
| PA0899 | aruB | 637 | 3965 |
| PA0901 | aruE | 786 | 2835 |
| PA3068 | gdhB | 1681 | 7026 |
| Group III: The ATA pathway | |||
| PA0265 | gabD | 796 | 1500 |
| PA0266 | gabT | 943 | 1315 |
| PA1421 | gbuA | 78 | 943 |
| PA4976 | aruH | 169 | 152 |
| PA4977 | aruI | 93 | 105 |
| PA5312 | kauB | 1099 | 1357 |
| Group IV: Arginine and glutamate biosynthesis | |||
| PA3537 | argF | 1715 | 267 |
| PA4588 | gdhA | 373 | 56 |
*Gene numbers are from the Pseudomonas Genome Project (www.pseudomonas.com).
†Cells were grown in minimal medium P with l-glutamate.
‡Cells were grown in minimal medium P with l-glutamate and d-arginine.
Deamination by a variety of enantiomer-specific enzymes is the most common first step of d-amino acid catabolism. In the case of d-arginine, 2-ketoarginine has been identified as a product of an inducible d-arginine dehydrogenase in PAO1 (6). Therefore, one would expect that genes in the ATA pathway (group III of Table 1) of 2-ketoarginine utilization must be induced by d-arginine; however, that was not the case. These results provide the first line of evidence to support the hypothesis of d-to-l racemization, instead of deamination, as the prerequisite of d-arginine catabolism.
Bioinformatic Analysis.
Based on genome annotations (www.pseudomonas.com), DauBARs were all hypothetical proteins of unknown functions. By Pfam database predictions, DauB belongs to the ornithine cyclodeaminase/mu-crystallin family (PF02423; E value 9e-26) and exhibits a limited sequence similarity to an alanine dehydrogenase from an archeabacterium (TIGRFAM accession no. 02371). DauA was predicted as an FAD-dependent oxidoreductase (PF01266; E value 6e-24), and DauR possesses a PAS domain (PF08348; E value 2e-32) of cell signaling at the N terminus. By using a BLAST search against the Protein Data Bank, we found that the C terminus of DauR (R162-E204) exhibits limited similarity to a helix-turn-helix DNA-binding domain (R144-E181) of γδ resolvase (16). Downstream of the putative dauBAR operon, the convergent dauT gene encodes a polypeptide that shows 80% sequence similarity to the AotJ protein (17), a periplasmic arginine/ornithine-binding protein of an ABC transport system in P. aeruginosa. The dauBART locus is conserved among 4 strains of P. aeruginosa and 2 strains of Pseudomonas fluorescens with a completed genomic sequence. Without dauT, a putative dauBAR operon can also be found in Pseudomonas mendocina, but not in Pseudomonas putida, Pseudomonas stutzeri, and Pseudomonas syringae.
Promoter Induction by d-Arginine.
To examine the possible effects of exogenous arginine on dauBART expression, lacZ fusion plasmids carrying the putative promoter regions of dauB and dauT were constructed and introduced into strain PAO1, and the cells were grown in the minimal medium in the absence and presence of exogenous l- and d-arginine. As shown in Table 2, both dauB and dauT promoters were significantly induced by d-arginine. In comparison, exogenous l-arginine exerted a repression effect on the dauT promoter but showed no effect on the dauB promoter. These results support the hypothesis that DauBART could be related to d-arginine catabolism and uptake.
Table 2.
Effect of d- or l-arginine on dauB and dauT promoter activities in PAO1
| Plasmid (promoter) | C & N source | Specific activities, nmol/min per mg* |
|---|---|---|
| pCR1 (dauB) | Glu | 34 |
| Glu + d-arg | 680 | |
| Glu + l-arg | 63 | |
| pZY4 (dauT) (7) | Glu | 619 |
| Glu + d-arg | 6006 | |
| Glu + l-arg | 233 |
C & N, carbon and nitrogen; Glu, cells were grown in MMP-glutamate; Glu + d-arg, cells were grown in MMP-glutamate supplemented with 5 mM d-arginine; and Glu + l-arg, cells were grown in MMP-glutamate supplemented with 20 mM l-arginine.
*Specific activities of β-galactosidase expressed from plasmids were averages of 2 measurements for each growth condition, with standard errors of less than 5%.
Growth Phenotypes.
The knockout mutants of dauBART were constructed to investigate the potential functions of these genes in d-arginine catabolism. As shown in Table 3, the parental strain PAO1 grew on d-arginine as the sole carbon source. Growth on d-arginine was completely abolished in either dauB or dauA mutants but was sustained in dauR and dauT mutants. When supplied as the sole nitrogen source, only the dauA mutant showed no growth on d-arginine. In comparison, none of these mutants exhibited any growth defects on l-arginine or l-glutamate.
Table 3.
Growth phenotype of PAO1 and its mutants
| Strains (genotype) |
d-Arg |
l-Arg |
||
|---|---|---|---|---|
| C source* | N source† | C & N source‡ | ||
| PAO1 (wild type) | + | + | + | |
| PAO5801 (dauB) | −§ | + | + | |
| PAO5802 (dauA) | − | − | + | |
| PAO5803 (dauR) | + | + | + | |
| PAO5804 (dauT) | + | + | + | |
| PAO4558 (aruF) | + | + | + | |
| PAO5602 (aruF aruH) | − | + | − | |
| PAO5603 (aruF aruI) | − | + | − | |
*d-Arginine as the sole carbon (C) source. Cells were grown in minimal medium P with 5 mM NH4Cl and 10 mM d-arginine.
†d-Arginine as the sole nitrogen (N) source. Cells were grown in minimal medium P with 20 mM glucose and 5 mM d-arginine.
‡l-Arginine as the sole carbon and nitrogen (C&N) source, cells were grown in minimal medium P with 20 mM l-arginine.
§No growth for at least 48 h.
Effects of dau Mutations on d-Arginine Dehydrogenase Activity.
The results from growth phenotype analysis suggested that the dau genes are involved in d-arginine utilization. Haas and coworkers reported the presence of d-arginine-inducible d-arginine dehydrogenase (DADH) activity in P. aeruginosa (6) to generate 2-ketoarginine and ammonia (Fig. 1). We therefore conducted experiments to measure DADH activities in PAO1 and congenic dau mutants.
Consistent with a previously published report (6), the DADH of PAO1 was specifically induced by the addition of d-arginine (Table 4); no induction by l-arginine could be observed (data not shown). The dauT mutant retained the induction effect of d-arginine on DADH. However, in the dauB and dauA mutants that lost the capability to grow on d-arginine as the sole carbon source, no d-arginine-inducible DADH could be detected. In contrast, the dauR mutant exhibited a high basal level of DADH in the absence of d-arginine. In conjunction with genome annotations as described above, we hypothesized that DADH might be composed of DauA and/or DauB, and that DauR serves as a transcriptional repressor of the dauBAR operon.
Table 4.
Effect of d-arginine on DADH activity of PAO1 and its mutants
| Strain (genotype) | Specific activities, nmol/min per mg* |
|
|---|---|---|
| Glu | Glu + d-arg | |
| PAO1 (wild type) | 20 | 261 |
| PAO5801 (dauB) | 12 | 11 |
| PAO5802 (dauA) | ND | ND |
| PAO5803 (dauR) | 140 | 174 |
| PAO5804 (dauT) | 22 | 251 |
Glu, cells were grown in glutamate minimal medium; Glu + d-arg, cells were grown in glutamate minimal medium supplemented with 5 mM d-arginine; ND, not detectable.
*Specific activities were averages of 2 measurements for each growth condition, with standard errors of less than 5%.
DauA Exhibits DADH Activity.
To test the proposed biochemical function of DauA and DauB, we constructed recombinant strains of E. coli for overexpression of DauA and DauB separately, with a hexa-histidine tag fused to the N terminus. Although DauA and DauB tended to form inclusion bodies when overexpressed, at least 50% of recombinant proteins remained soluble. Measurements of the DADH activity from the crude extracts of these recombinant strains clearly indicated that DauA but not DauB possesses DADH activity (data not shown, but see below).
The recombinant DauA protein was purified to homogeneity, and the results of SDS/PAGE and size exclusion column chromatography revealed that the histidine-tagged DauA protein is monomeric in its native form. The purified DauA had a distinct yellowish color, suggesting the presence of a bound chromophore. The specific activity of the recombinant DauA was estimated to be 18.8 μmol/min per mg when d-arginine (20 mM) serves as substrate, and it was negligible with l-arginine as substrate.
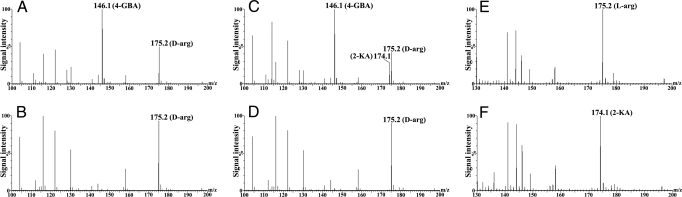
Generation of 2-ketoarginine from the proposed reaction was demonstrated by MS analysis, with phenazine methosulfate (PMS) as the artificial electron acceptor. PMS can be recycled by transferring the electron to oxygen and forming hydrogen peroxide. In the mass spectrum of the reaction mixture with active DauA alone (Fig. 2A), a molecular ion peak of m/z 146.1 was observed, which corresponds to that of 4-guanidinobutyric acid (4-GBA), the decarboxylation product of 2-ketoarginine by hydrogen peroxide (18). With the addition of catalase (Fig. 2C) to reduce the amount of hydrogen peroxide, another peak of m/z 174.1 appeared, which is identical to that of the authentic 2-ketoarginine. As expected, neither 4-GBA nor 2-ketoarginine was detected in the negative controls with heat-inactivated DauA (Fig. 2 B and D). It is essential to include PMS in the reaction to ensure DauA remains active with FAD; no product signal can be detected without PMS (data not shown).
Fig. 2.
ESI-MS analysis of the reaction products of DauA and DauB. (A and C) Analysis of reaction mixtures with active DauA without (A) or with (C) catalase. (E) Analysis of reaction mixture with active DauB. (B, D, and F) Corresponding controls with heat-denatured DauA or DauB. The signals with m/z values of 146.1, 175.2, and 174.1 were in accordance with the positive ions of 4-guanidinobutyric acid (4-GBA; A and C), d-arginine (D-Arg; A–D), or l-arginine (L-arg; E), and 2-ketoarginine (2-KA; C and F).
Participation of the AST and ATA Pathways in d-Arginine Catabolism.
Under aerobic conditions, the AST and ATA pathways are 2 major routes for l-arginine utilization as the sole source of carbon and nitrogen. Arginine succinyltranferase (encoded by aruF) and arginine transaminase (encoded by aruH) catalyze the first reaction of the AST and ATA pathways, respectively, and both enzymes take l-arginine, but not d-arginine, as substrate (8, 19). Because d-arginine is converted into 2-ketoarginine by DauA, one might expect that 2-ketoarginine can be further catabolized by 2-ketoarginine decarboxylase (encoded by aruI), and subsequent enzymes of the ATA pathway into succinate (Fig. 1). Accordingly, d-arginine utilization was tested in a series of mutants with lesions in the affected pathways, and the results are shown in Table 3.
Interestingly, growth on d-arginine as the sole carbon source was partially retarded in strain PAO4558 (aruF) (20) devoid of the AST pathway. No growth on d-arginine could be observed in strains PAO5602 (aruF, aruH) or PAO5603 (aruF, aruI) (7) in which both AST and ATA pathways were blocked. The growth defects of PAO4558 and PAO5602 on d-arginine do not fit into the proposed model of d-arginine utilization through 2-ketoarginine and the ATA pathway. Instead, these results suggest that d-arginine is mainly converted into l-arginine before being channeled into the AST and/or ATA pathways.
Role of dauA and dauB on the Induction of the AST Pathway by d-Arginine.
To test the hypothesis of d-to-l racemization of arginine, we conducted experiments to measure the promoter activity of aruC, the first gene of the aruCFGDBE operon for the AST pathway. As shown in Table 5, exogenous d-arginine exerted an induction effect slightly less than that by l-arginine in the parental strain PAO1; it was not clear whether this observed minor difference is of physiological significance. In strains PAO5802 (dauA) and PAO5801 (dauB), the induction effect of d-arginine was greatly abolished, whereas the aruC promoter remained inducible by l-arginine. The residual 2-fold induction by d-arginine in dauA and dauB mutants might imply possible cross-activation by d-arginine on ArgR, the transcriptional regulator of the aruC promoter. These findings strongly suggest that DauA and DauB are essential for the proposed d-to-l racemization of arginine.
Table 5.
Effect of d- or l-arginine on the aruC promoter activity in PAO1 and its mutants
| Strain (genotype) | C & N source | Specific activities, nmol/min per mg* | Fold change |
|---|---|---|---|
| PAO1 (wild type) | Glu | 501 | 1.0 |
| Glu + d-arg | 7007 | 14.0 | |
| Glu + l-arg | 11104 | 22.2 | |
| PAO5801 (dauB) | Glu | 349 | 1.0 |
| Glu + d-arg | 750 | 2.2 | |
| Glu + l-arg | 6950 | 19.9 | |
| PAO5802 (dauA) | Glu | 380 | 1.0 |
| Glu + d-arg | 610 | 1.6 | |
| Glu + l-arg | 7345 | 19.3 |
Glu, cells were grown in glutamate minimal medium; Glu + d-arg, cells were grown in glutamate minimal medium supplemented with 5 mM d-arginine; Glu + l-arg, cells were grown in glutamate minimal medium supplemented with 20 mM l-arginine.
*Specific activities of β-galactosidase expressed from the aruC::lacZ fusion plasmid pSC500 [Park SM, Lu CD, Abdelal AT (1997) J Bacteriol 79:5309–5317] were averages of 2 measurements for each growth condition, with standard errors of less than 5%.
Growth Complementation of an Arginine Auxotroph by d-Arginine Requires DauA and DauB.
To further investigate the proposed racemization, the growth phenotypes of an arginine auxotroph PAO303 (argB) (21) and its dauA and dauB mutants were determined. As expected, the growth defect of these 3 strains in the minimal medium due to a mutation on argB of arginine biosynthesis was complemented by exogenous l-arginine. When d-arginine was added as supplement, only PAO303 resumed growth, but its dauA and dauB mutants failed to grow. This is consistent with the proposed function of DauA and DauB in arginine racemization.
Anabolic l-Arginine Dehydrogenase Activity of DauB.
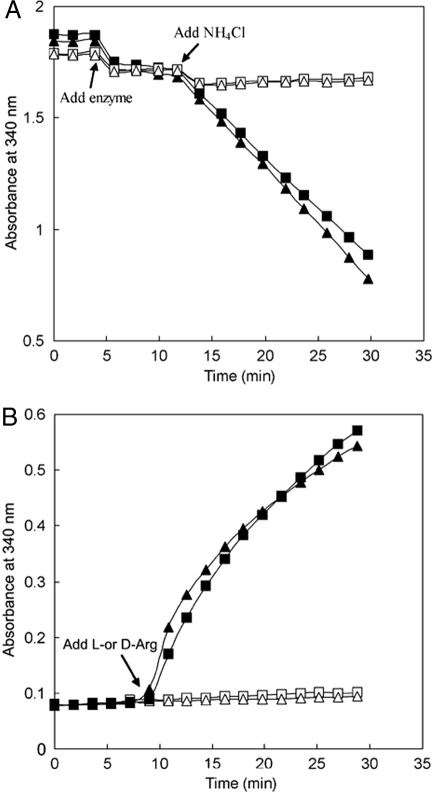
In the proposed racemization reaction, 2-ketoarginine and ammonia derived from d-arginine in the DauA-catalyzed reaction would be taken by DauB to regenerate l-arginine in a second anabolic dehydrogenase reaction. To test this hypothesis, we overexpressed and purified a histidine-tagged DauB. With the purified enzyme we were able to demonstrate the proposed reaction with NADH or NADPH as the reducing power. The reaction was 2-ketoarginine-dependent and ammonia-dependent; consumption of NADH or NADPH could only be observed when both substrates were present (Fig. 3A). We also tested the proposed reaction of DauB in the reverse direction and found that significant activity could only be detected when l-arginine instead of d-arginine was used as substrate (Fig. 3B).
Fig. 3.
Reversible l-arginine dehydrogenase activity of DauB. (A) The forward reaction for arginine synthesis with 2-ketoarginine and NH4Cl as substrates in the presence of NAD(P)H. The initial assay mixture contained either NADH (squares) or NADPH (triangles) with 2-ketoarginine (black symbols) or ddH2O (white symbols). (B) The reverse reactions with either l-arginine (black symbols) or d-arginine (white symbols) as substrates in the presence of NAD+ (squares) or NADP+ (triangles). The time points for addition of the missing components are marked by arrows. Consumption or generation of NAD(P)H in the reactions was monitored by absorbance at 340 nm.
Generation of l-arginine from 2-ketoarginine and ammonia was also demonstrated by MS analysis. Although the ion peak of 2-ketoarginine (m/z value of 174.1) was the predominant one in the control sample of heat-inactivated DauB (Fig. 2F), it was almost diminished in the mass spectrum of the reaction mixture with active DauB (Fig. 2E). Instead, one molecular ion peak was observed with an m/z value of 175.2, which is identical to that of the authentic l-arginine.
Discussion
Several lines of evidence were presented to indicate that P. aeruginosa PAO1 performs d-to-l racemization of arginine through 2 collaborative enzymes: a catabolic d-arginine dehydrogenase (encoded by dauA) and an anabolic l-arginine dehydrogenase (encoded by dauB). The proposed biochemical reactions of these 2 enzymes were demonstrated with purified DauA and DauB, and the importance of dauAB in arginine racemization was supported by the finding that growth complementation of an l-arginine auxotroph by exogenous d-arginine was completely abolished in strains with a dauA or dauB lesion.
We also established that d-to-l racemization is in fact a prerequisite for d-arginine utilization as the carbon source through l-arginine catabolic pathways. Without converting into l-arginine, d-arginine by itself is not able to induce genes of the AST pathway, and most likely genes of the ATA pathway as well. Therefore, exogenous d-arginine either cannot be used at all, as in strain PAO5802 (dauA), or it can only be used as nitrogen source, as in strain PAO5801 (dauB). When d-to-l inversion is blocked in the dauB mutant, ammonia released from the DauA-catalyzed reaction is sufficient to support growth on d-arginine as a nitrogen source, and accumulation of 2-ketoarginine is anticipated. Although 2-ketoarginine may be channeled into the ATA pathway, contribution of this route in the dauB mutant is minimized as the ATA pathway is induced by l-arginine. It has been reported that strain PAO1 grows well on exogenous 2-ketoarginine as a sole source of carbon and nitrogen, and this compound is able to induce DADH as well as 4-guanidinobutyraldehyde dehydrogenase and guanidinobutyrase of the ATA pathway (Fig. 1). Perhaps this discrepancy is due to racemization, and it suggests that 2-ketoarginine may be the signal of dauBART induction.
Genome annotations suggest DauA as an FAD-dependent d-amino acid oxidase. We have observed that addition of FAD to the purified DauA indeed can increase the specific activity about 2-fold. In the proposed reaction of DauA, d-arginine was catabolized into 2-ketoarginine and ammonia, and the electrons released from the reaction were transferred to FAD on the enzyme. The reduced FADH2 needs to further transfer its electrons to an electron acceptor to reset the reaction cycle. It appears that oxygen is not the immediate electron acceptor of this reaction, because no hydrogen peroxide can be detected by a coupled reaction with horseradish peroxidase and a chromogenic substrate (data not shown). Therefore, DauA is a true dehydrogenase, not oxidase. With the native electron acceptor remaining unknown, electron transfers were carried out by PMS followed by p-iodonitrotetrazolium violet (INT) in the colorimetric assay.
d-alanine dehydrogenase of E. coli was characterized as a membrane-associated d-amino acid dehydrogenase containing FAD and nonheme iron-sulfur chromophore (22). In collaboration with an alanine racemase, this 2-enzyme couple is encoded in a single operon and is responsible for l-alanine catabolism in E. coli (10) and many other bacteria, including P. aeruginosa (7). In comparison, DauA is not a membrane-associated enzyme, because it remained in the soluble fraction after ultracentrifugation, and it is not required for dl-alanine utilization.
In the proposed racemization reaction, DauB is able to generate l-arginine from 2-ketoarginine and ammonia driven by NADH or NADPH. It produces l-arginine but not d-arginine, as supported by the finding that l-arginine is the preferred substrate in the reverse reaction. It is possible that DauA and DauB form a complex to streamline the racemization reaction. This process could provide at least 2 advantages in P. aeruginosa—to alleviate any potential adverse effect of d-arginine in translation and to use d-arginine through the sophisticated l-arginine catabolic pathways. Although more work is required to further characterize interactions between DauA and DauB, data presented in this report strongly support a unique approach of amino acid racemization that is completely different from those catalyzed by the conventional racemases. Whether this racemization reaction can be applied to other d-amino acids remains to be determined. Extended from this finding, one might predict racemization by a variety of coupled enzymes as long as products from the first reaction on one enantiomer can be used as substrates in the second reaction to make another enantiomer.
Materials and Methods
Strains and Growth Conditions.
Strains with designated lesions were derived from P. aeruginosa PAO1 (21). For strain and plasmid construction, Luria-Bertani (LB) medium was used with the following antibiotics as required: ampicillin, 100 μg/mL; tetracycline, 10 μg/mL for E. coli and 100 μg/mL for P. aeruginosa; carbenicillin, 100 μg/mL; and streptomycin, 500 μg/mL. The minimal medium P (21) was used for the growth of P. aeruginosa strains with specific C and N sources as indicated (20 mM, unless specified otherwise).
RNA Isolation, Generation of cDNA Probes, and Data Analysis.
Two independent sets of P. aeruginosa PAO1 cultures were grown aerobically in minimal medium P, with 350-rpm shaking at 37°C, in the presence of l-glutamate alone or with the addition of d-arginine at 20 mM. Cells were harvested at mid-log phase (OD600 = 0.5–0.6) by centrifugation at 4°C. Total RNA samples were isolated by using an RNeasy purification kit (Qiagen) following the instructions of the manufacturer. Reverse transcription for cDNA synthesis, fragmentation, labeling, and hybridization were performed according to the instructions of the manufacturer (Affymetrix). Data were processed by Microarray Suite 5.0 software (Affymetrix). Absolute expression signal values were normalized for each chip by globally scaling to a target intensity of 500. GeneSpring software (Silicon Genetics) was further used for the expression pattern analysis and comparison. Only genes showing consistent expression profiles in duplicates were selected for further analysis.
Construction of a dauB–lacZ Fusion.
A DNA fragment containing the regulatory region of dauB was amplified by PCR from the genomic DNA of strain PAO1 with pfu polymerase and the following synthetic oligonucleotides designed to generate HindIII restriction site on the forward primer: 5′-TAC AAG CTT CTG TAC AAC CTG GTG GCG CAG-3′ and 5′-GGC GCT CAT GCG ATC TCC GGA ATG GAT GTA-3′. The PCR product was purified from 1% agarose gel after electrophoresis, digested by HindIII, and ligated into the HindIII and SmaI sites of pQF50 (23), a broad host-range lacZ transcriptional fusion vector. The resulting plasmid was designated pCR1. The nucleotide sequence of the resulting construct was verified by nucleotide sequence determination.
Construction of Mutant Strains.
The EZ-Tn5 <TET-1> insertion system (Epicentre) was used for generation of knockout mutants. A 4.2-kb fragment covering the dauBART cluster was amplified from the genomic DNA of PAO1 with the following primers designed to generate BamHI and HindIII sites at the 5′ and 3′ ends of the PCR product, respectively: 5′-TTT GGA TCC GGC GAT CGT CGA GAT CGA GCC-3′ and 5′-GGG AAG CTT CGA CTA CGA CAT CGG CAA CGC-3′. The PCR product was purified and digested by BamHI and HindIII before being subcloned into the same sites of the conjugation vector pRTP2, a vector derived from pRTP1 (24) with deletion of EcoRI site. The resulting plasmid DNA was subjected to in vitro transposon mutagenesis under the conditions recommended by the manufacturer. After the reaction, the mixture was used to transform E. coli DH5α, and transformants were selected on LB plates with ampicillin and tetracycline. The transposon insertion sites of mutant clones were determined by nucleotide sequencings with transposon-specific flanking primers. For gene replacement, the resulting transposons insertion plasmids were first introduced into E. coli SM10 (25) and then mobilized into the desired recipient strains of P. aeruginosa by biparental plate mating (25). After incubation at 37°C for 6 h, transconjugants were selected on LB plates supplemented with tetracycline and streptomycin. The genotypes of all of the mutants constructed were verified by colony PCR.
Expression of dauB and dauA in E. coli.
The expression vector pBAD-His-6, containing a 6-histidine tag followed by a SmaI site, was derived from pBAD-HisA (Invitrogen) by replacing the NcoI-XhoI region of pBAD-HisA with the following sequence: 5′-CC ATG GGT CAT CAT CAT CAT CAT CAT CCC GGG CTC GAG-3′. The dauB and dauA structural genes were amplified by PCR from the genomic DNA of PAO1 using the following primer pairs: 5′-AGC GCC GCC ACT CCC CTG ATC-3′ and 5′-CCC AAG CTT CAG CCT GCC TGG CGC AG-3′ for dauB; and 5′-ATC GAA GCG GAT TAC CTC-3′ and 5′-CCC AAG CTT CAG GGG GAC AGG CGG CG-3′ for dauA. The resulting PCR products were digested with HindIII, which is a unique restriction site at the 3′ end introduced by the primers, and cloned into the SmaI and HindIII sites of the expression vector so that the N terminus of DauA and DauB was fused in frame with the 6-histidine tag preceded by a ribosomal binding site and an arabinose-inducible promoter in the plasmid. The resulting plasmids, pCR2 and pCR3, were introduced into E. coli Top10. For overexpression of DauB and DauA, the recombinant strains of E. coli were grown in LB medium containing ampicillin (100 μg/mL) at 30°C until the optical density at 600 nm reached 0.5, at which point 0.2% arabinose (wt/vol; final concentration) was added to the cultures for induction. Culture growth was continued for another 3 h under the same conditions, and cells were harvested by centrifugation.
Purification of Hexa-Histidine-Tagged DauB and DauA.
The cell pellets were suspended in the phosphate buffer (20 mM sodium phosphate, 500 mM NaCl, and 20 mM imidazole, pH 7.4) plus 1 mM PMSF as a protease inhibitor, and the cells were ruptured by an Aminco French pressure cell at 17,000 psi. Cell debris was removed by centrifugation at 20,000 × g for 1 h, and the resulting cell-free crude extracts were further precipitated by streptomycin sulfate and centrifuged again. The supernatants from this step were applied to a HisTrap HP column (GE Healthcare) equilibrated with the sodium phosphate buffer described above. After washing off of the unbound proteins with equilibration buffer, His-tagged DauB or DauA was eluted with a linear gradient of 20–500 mM imidazole over 10 column volumes. For further purification, DauA protein was subjected to anion-exchange chromatography using a Mono Q HR 5/5 column (GE Healthcare) followed by gel filtration chromatography using a Superdex 200 HR 10/30 column (GE Healthcare).
Enzyme Assays.
For the measurements of enzyme activities with the crude extracts, cell cultures in the logarithmic phase were collected by centrifugation, and the cell pellets were washed once with 20 mM potassium phosphate buffer containing 1 mM EDTA (pH 7.6) and resuspended in the same buffer. Cells were broken with a French pressure cell at 17,000 psi, and soluble cell extracts were prepared after centrifugation at 16,000 × g for 30 min. Protein concentration was determined by the method of Bradford with BSA as standard. For the measurements of β-galactosidase activity, o-nitrophenyl-β-d-galactopyranoside was used as substrate. d-Arginine dehydrogenase activity was assayed with the addition of artificial electron acceptors, PMS and INT, as described previously (6). One unit of d-arginine dehydrogenase activity was defined as the amount of enzyme that led to the reduction of 1 nmol iodonitrotetrazolium chloride per minute under the standard assay conditions.
The forward activity of DauB in generation of l-arginine (Fig. 1) was determined by monitoring the consumption of NADH or NADPH at 340 nm using Cary 3E (Varian). The reaction mixture contained the following components in a volume of 2 mL: 50 mM Tris·Cl (pH 8.7), 0.35 mM NADH or NADPH, 40 mM NH4Cl, 10 mM 2-ketoarginine (KA) (for sample) or ddH2O (for control), and 20 μg of the purified DauB. The reaction mixtures without NH4Cl were preincubated at 37°C for about 6 min, and the reaction was initiated by adding NH4Cl. The reverse reaction of DauB in taking l- or d-arginine as the substrate was measured by monitoring the formation of NADH or NADPH at 340 nm using Cary 3E (Varian). The reaction mixture contained 10 mM l- or d-arginine, 0.5 mM NAD+ or NADP+, and 20 μg of the purified DauB in 50 mM Tris·Cl (pH 8.7) in a total volume of 2 mL. The mixtures without l- or d-arginine were preincubated at 37°C for about 9 min, and the reaction was initiated by adding the missing substrate.
2-KA was synthesized by following the published protocol (6) and lyophilized. Quality and purity of thusly prepared 2-KA has been analyzed by HPLC in a previous report (7) and was confirmed by electrospray ionization (ESI)-MS in this study (Fig. 2F). The 2-KA solution was prepared by dissolving an appropriate amount of the lyophilized powder in water, assuming more than 95% purity and a molecular weight of 174 g/mol.
ESI-MS Analysis of the Reaction Products.
For DauA, the reaction mixtures (500 μL) containing 5 μg of purified DauA, 5 mM d-arginine, and 0.8 mM PMS in 10 mM Tris·Cl buffer (pH 8.7) with or without catalase (50 μg) were incubated for 1.5 h at 37°C. For DauB, a reaction mixture (500 μL) containing 48 μg of purified DauB, 5 mM 2-ketoarginine, 40 mM NH4Cl, and 5 mM NADH in 10 mM Tris·Cl buffer (pH 8.7) was incubated overnight at 37°C. The samples were boiled for 10 min after incubation and centrifuged at 16,000 × g for 30 min. The resulting supernatants were submitted for ESI-MS analysis at the MS facility of Georgia State University. In the negative control experiments, heat-denatured recombinant DauA or DauB was used to prepare the reaction mixture. Samples were diluted in 50% methanol with the addition of 0.1% formic acid for positive ion analysis by TOF mass spectrometry.
Acknowledgments.
We thank Zhe Yang for assistance in microarray analysis; Siming Wang and Lupei Du for MS analysis; and Phang C. Tai, Ahmed Abdelal, Dieter Haas, and Sydney Kustu for encouragement of the manuscript. The project was supported by National Science Foundation grant 0415608.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Snyder SH, Kim PM. D-amino acids as putative neurotransmitters: Focus on D-serine. Neurochem Res. 2000;25:553–560. doi: 10.1023/a:1007586314648. [DOI] [PubMed] [Google Scholar]
- 2.Wolosker H. NMDA receptor regulation by D-serine: New findings and perspectives. Mol Neurobiol. 2007;36:152–164. doi: 10.1007/s12035-007-0038-6. [DOI] [PubMed] [Google Scholar]
- 3.Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: New insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimura T, Esak N. Amino acid racemases: Functions and mechanisms. J Biosci Bioeng. 2003;96:103–109. doi: 10.1016/s1389-1723(03)90111-3. [DOI] [PubMed] [Google Scholar]
- 5.Haas D, Matsumoto H, Moretti P, Stalon V, Mercenier A. Arginine degradation in Pseudomonas aeruginosa mutants blocked in two arginine catabolic pathways. Mol Gen Genet. 1984;193:437–444. doi: 10.1007/BF00382081. [DOI] [PubMed] [Google Scholar]
- 6.Jann A, Matsumoto H, Haas D. The fourth arginine catabolic pathway of Pseudomonas aeruginosa. J Gen Microbiol. 1988;134:1043–1053. doi: 10.1099/00221287-134-4-1043. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Lu CD. Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J Bacteriol. 2007;189:3945–3953. doi: 10.1128/JB.00261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Lu CD. Characterization of an arginine:pyruvate transaminase in arginine catabolism of Pseudomonas aeruginosa PAO1. J Bacteriol. 2007;189:3954–3959. doi: 10.1128/JB.00262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaczorowski G, Shaw L, F-entes M, Walsh C. Coupling of alanine racemase and D-alanine dehydrogenase to active transport of amino acids in Escherichia coli B membrane vesicles. J Biol Chem. 1975;250:2855–2865. [PubMed] [Google Scholar]
- 10.Lobocka M, Hennig J, Wild J, Klopotowski T. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of D-amino acid dehydrogenase and the catabolic alanine racemase. J Bacteriol. 1994;176:1500–1510. doi: 10.1128/jb.176.5.1500-1510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jann A, Stalon V, Wauven CV, Leisinger T, Haas D. N-Succinylated intermediates in an arginine catabolic pathway of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1986;83:4937–4941. doi: 10.1073/pnas.83.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh Y. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7280–7290. doi: 10.1128/jb.179.23.7280-7290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SM, Lu CD, Abdelal AT. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J Bacteriol. 1997;179:5309–5317. doi: 10.1128/jb.179.17.5309-5317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakada Y, Itoh Y. Characterization and regulation of the gbuA gene, encoding guanidinobutyrase in the arginine dehydrogenase pathway of Pseudomonas aeruginosa PAO1. J Bacteriol. 2002;184:3377–3384. doi: 10.1128/JB.184.12.3377-3384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu CD, Yang Z, Li W. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J Bacteriol. 2004;186:3855–3861. doi: 10.1128/JB.186.12.3855-3861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 17.Nishijyo T, Park SM, Lu CD, Itoh Y, Abdelal AT. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5559–5566. doi: 10.1128/jb.180.21.5559-5566.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlessis AA, Bartos D, Trunkey D. Importance of spontaneous alpha-ketoacid decarboxylation in experiments involving peroxide. Biochem Biophys Res Commun. 1990;170:1281–1287. doi: 10.1016/0006-291x(90)90532-r. [DOI] [PubMed] [Google Scholar]
- 19.Tricot C, Vander Wauven C, Wattiez R, Falmagne P, Stalon V. Purification and properties of a succinyltransferase from Pseudomonas aeruginosa specific for both arginine and ornithine. Eur J Biochem. 1994;224:853–861. doi: 10.1111/j.1432-1033.1994.00853.x. [DOI] [PubMed] [Google Scholar]
- 20.Itoh Y, Nakada Y. In: Pseudomonas: Biosynthesis of Macromolecules and Molecular Metabolism. Ramos JL, editor. New York: Kluwer Academic/Plenum Publishers; 2004. pp. 243–272. [Google Scholar]
- 21.Haas D, Holloway BW, Schambock A, Leisinger T. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol Gen Genet. 1977;154:7–22. doi: 10.1007/BF00265571. [DOI] [PubMed] [Google Scholar]
- 22.Olsiewski PJ, Kaczorowski GJ, Walsh C. Purification and properties of D-amino acid dehydrogenase, an inducible membrane-bound iron-sulfur flavoenzyme from Escherichia coli B. J Biol Chem. 1980;255:4487–4494. [PubMed] [Google Scholar]
- 23.Farinha MA, Kropinski AM. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stibitz S, Black W, Falkow S. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene. 1986;50:133–140. doi: 10.1016/0378-1119(86)90318-5. [DOI] [PubMed] [Google Scholar]
- 25.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]