Abstract
Hydrogen atoms constitute about half of all atoms in proteins and play a critical role in enzyme mechanisms and macromolecular and solvent structure. Hydrogen atom positions can readily be determined by neutron diffraction, and as such, neutron diffraction is an invaluable tool for elucidating molecular mechanisms. Joint refinement of neutron and X-ray diffraction data can lead to improved models compared with the use of neutron data alone and has now been incorporated into modern, maximum-likelihood based crystallographic refinement programs like CNS. Joint refinement has been applied to neutron and X-ray diffraction data collected on crystals of diisopropyl fluorophosphatase (DFPase), a calcium-dependent phosphotriesterase capable of detoxifying organophosphorus nerve agents. Neutron omit maps reveal a number of important features pertaining to the mechanism of DFPase. Solvent molecule W33, coordinating the catalytic calcium, is a water molecule in a strained coordination environment, and not a hydroxide. The smallest Ca–O–H angle is 53°, well beyond the smallest angles previously observed. Residue Asp-229, is deprotonated, supporting a mechanism involving nucleophilic attack by Asp-229, and excluding water activation by the catalytic calcium. The extended network of hydrogen bonding interactions in the central water filled tunnel of DFPase is revealed, showing that internal solvent molecules form an important, integrated part of the overall structure.
Keywords: enzyme mechanism, H/D exchange
Hydrogen atoms are among the most important elements in biological systems, yet they are very difficult to directly observe in 3D structures obtained by X-ray crystallography due to their single electron and their relative mobility. Only a limited number of hydrogen atoms can be resolved in the highest-resolution X-ray structures (≈1.0 Å and better resolution). In this context, neutron crystallography has been invaluable in the study of protein structure and enzyme mechanisms, because it is one of the only techniques able to directly visualize hydrogen atoms in structures at moderate resolutions (<2.5 Å) (1–8). In contrast to X-rays, neutrons are scattered by atomic nuclei, and the primary atoms in proteins (H, C, N, O, S) have similar neutron-scattering lengths. Of particular importance, the 2 isotopes of hydrogen, H and D, have opposite coherent scattering lengths; H is negative, and D is positive. The scattering length of D is comparable with that of oxygen, nitrogen, and carbon atoms with respect to sign and magnitude. These scattering properties allow one to identify the positions of hydrogen atoms, and to identify labile H atoms that can be exchanged for D and the extent of this replacement. Thus, the protonation states of catalytically important residues can be determined (9). The H/D exchange characteristics derived from neutron scattering provide a tool for the study of protein dynamics, complementary to NMR methods (4, 10, 11). An additional advantage of this scattering property is that water molecules, in the form of D2O, can be precisely oriented in nuclear density maps, whereas hydrogen atoms in water molecules are usually invisible in electron density maps. This allows the unambiguous assignment of hydrogen bonding interactions.
Nevertheless, the practical application of neutrons to protein crystallography has been limited in the past by the relatively weak neutron flux of beam lines compared with synchrotrons for X-ray experiments and the requirement for large crystals (>1 mm3). The low neutron flux leads to extended data acquisition times that in addition to the very small number of available sources limits the availability of beam time for individual experiments. A further challenge is the refinement of the structural model against neutron data, due to its limited resolution and the low data-to-parameters ratio. The data-to-parameters ratio is particularly critical, because the addition of hydrogen atoms in a macromolecule roughly doubles the number of atoms whose position and temperature factor are being refined. Inclusion of both X-ray and neutron data in refinement may aid in increasing the effective number of observations. A series of studies ≈25 years ago demonstrated the potential of the joint X-ray and neutron refinement technique. The implementation of joint X-ray and neutron refinement into programs such as CNS (12) [in the form of a patch version called nCNS (13)] and PHENIX (14, 15) has now made it possible to combine this approach with recent developments in macromolecular structure refinement, such as cross-validated maximum likelihood and simulated annealing.
As a test of the utility of this development, we used nCNS to refine the structure of diisopropyl fluorophosphatase (DFPase) from the squid Loligo vulgaris (EC 3.1.8.2) using both X-ray and neutron crystallographic data. DFPase, a 35 kDa, calcium-dependent phosphotriesterase, efficiently hydrolyzes a wide variety of highly toxic organophosphorus compounds, although its physiological substrate remains unknown (16, 17). In addition to hydrolyzing diisopropyl fluorophosphate (DFP), the enzyme detoxifies a range of organophosphorus nerve agents, including Tabun, Sarin, Soman, and Cyclohexylsarin, through hydrolysis of the bond between the phosphorus and the leaving group (17). DFPase can be highly expressed, is thermally stable, and as such, is a prime candidate for enzymatic decontamination of these highly toxic agents.
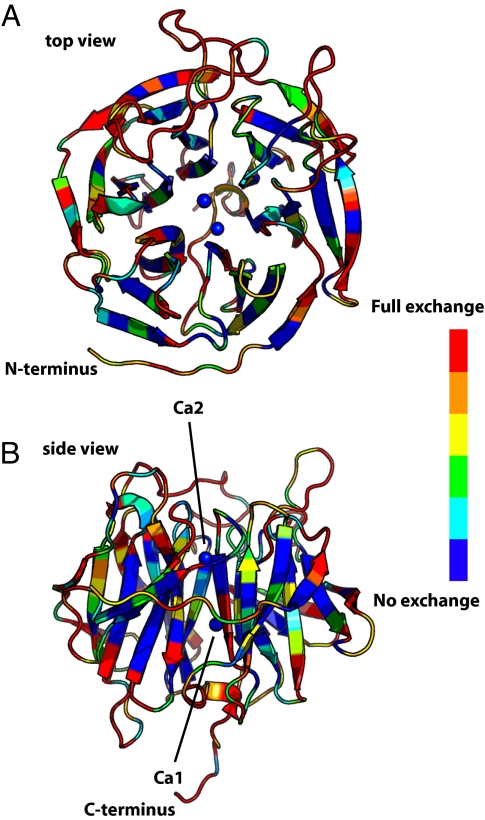
The overall structure of DFPase is a 6-bladed pseudosymmetrical ß-propeller with a central water filled tunnel (Fig. 1). A high-affinity calcium ion, Ca2, is located in the center of the tunnel and is essential for structural integrity. A second calcium ion, Ca1, located at the base of the active site pocket, is crucial for catalysis; its removal results in a folded, yet inactive enzyme (18). The active-site pocket is elongated and primarily hydrophobic in character, with an apparently rigid structure, as only minor conformational rearrangements are observed upon binding of a substrate analogue (19).
Fig. 1.
Overall structure of DFPase and backbone H/D exchange characteristics. (A) Top view, with the 2 calcium ions represented as blue spheres. (B) Side view, rotated 90° from a). The occupancy of backbone amide hydrogens is scaled from blue (H, unexchanged) via green and yellow (partially exchanged) to red (D, exchanged).
A number of enzymes with a similar overall fold and active site environment have been identified, including drug resistance protein 35 (Drp35) from Staphylococcus aureus (20), and human serum paraoxonase (PON) (21), a component of the high-density lipoprotein (HDL) complex. Notably, the active sites of DFPase and PON possess a conserved arrangement of amino acids around the catalytic calcium and, like DFPase, PON also shows activity against selected organophosphate triesters (22, 23).
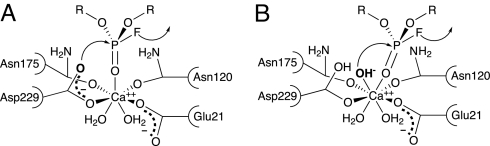
Recently, we proposed a possible mechanism, on the basis of the 1.73-Å X-ray crystal structure of DFPase in complex with a substrate analogue, kinetic and mutagenesis data, and H218O isotope labeling studies. We identified calcium-coordinating residue Asp-229 as the nucleophile, which in concert with direct substrate activation by the calcium ion, generates a phosphoenzyme intermediate (Fig. 2A). This intermediate is subsequently hydrolyzed by attack at the Asp-229 carboxyl carbon by water (19).
Fig. 2.
Schematic representation of possible mechanisms for DFPase. (A) Direct nucleophilic attack of Asp-229 on the substrate, with a phosphoenzyme intermediate and a fluoride leaving group. (B) Mechanism involving a calcium-bound hydroxide ion as the active nucleophile.
A second possible mechanism involves activation of calcium-bound water by the metal, resulting in a hydroxide nucleophile that can then attack the substrate (Fig. 2B). However, because the active-site metal in DFPase can be exchanged for other divalent metal ions such as magnesium and barium, with little change in catalytic activity, this mechanism has been deemed to be highly unlikely (19). Nevertheless, a similar mechanism was proposed for the lactonase activity of the structurally homologous enzyme Drp35 (20). Here, the calcium-bound water is activated by one of the metal-coordinating aspartate residues resulting in a hydroxide nucleophile.
To discriminate between these proposals, it is necessary to accurately determine the protonation states of the active-site residues, and the orientations and identities of critical solvent molecules in the active site vicinity. In addition to assigning the protonation states of residues Asp-229 and Glu-21 in the active site, a particularly important question with a direct bearing on mechanism is whether the solvent molecule W33, bound to the catalytic calcium, is a water molecule or a hydroxide ion. For our proposed mechanism, we expect Asp-229 to be deprotonated and W33 to be a water molecule, whereas in the case of metal-assisted water activation, we expect W33 to be a hydroxide ion. Although the X-ray structure of the holoenzyme has been determined at 0.85-Å resolution (24, 25), it was not possible to unambiguously discern the protonation states of residues in the active site region or to assign the hydrogen bond network in the active site or the water tunnel.
We present here the results of a neutron protein crystallography study of DFPase. In this work, we have successfully collected time-of-flight Laue neutron data to 2.2 Å from a small (0.43 mm3) crystal equilibrated against D2O-containing mother liquor, using the PCS beam line (26) at the Los Alamos Neutron Science Center (LANSCE) spallation source. Because H atoms account for nearly half of the atoms in a protein, adding H atoms in a neutron structure refinement increases the number of parameters and reduces the data-to-parameter ratio, increasing the danger of overfitting and decreasing the accuracy of the optimized model. We have therefore used the approach, originally developed by Wlodawer and Hendrickson (27), of combining the neutron data with 1.8-Å X-ray data in a joint (XN) structure refinement. We demonstrate the effectiveness of the joint refinement in nCNS at determining the protonation states of residues and solvent molecules in the active site of DFPase. We show the orientation of water molecules and the associated hydrogen bonding network in the water tunnel of DFPase.
We discuss the implications of our results for the phosphotriesterase mechanism of DFPase, including a comparison of the competing potential reaction mechanisms for this class of enzymes. As we proposed earlier, both DFPase and PON may share the same phosphotriesterase mechanism in light of the highly conserved coordination environment around the catalytic calcium. Therefore, mechanistic insights derived from the XN structure of DFPase may aid in the understanding of the phosphotriesterase activity of PON.
Results and Discussion
Joint Neutron and X-Ray Refinement (XN).
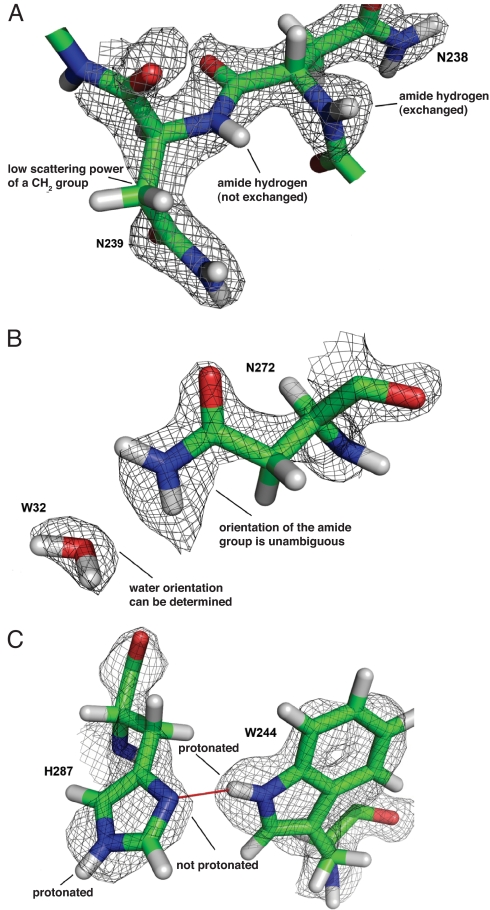
X-ray and neutron crystallographic data provide complementary structural information. Labile hydrogen atoms, including those on solvent molecules, hydroxyl groups, acidic hydrogens that might be absent due to deprotonation, and other exchangeable hydrogens that might be substituted by deuterium, are accurately located through neutron diffraction data. These hydrogens are often invisible in electron density maps. Conversely, X-ray data can also help define heavy atom positions and a limited number of hydrogens at high resolution. For example, methylene groups are rarely visible in neutron maps, because the opposite signs of the scattering lengths of C and H cancel each other out (Fig. 3A). In this case X-ray data not only defines the position of the carbon atom, but from inclusion of prior knowledge about bond lengths and angles in the refinement it also helps to define the hydrogen positions. Joint refinement can thus lead to improved phases and better electron and nuclear density maps. Wlodawer and Hendrickson observed a quarter-century ago during their refinement of ribonuclease A, that refinement of neutron data on its own was often unstable, and that including the X-ray data in a joint refinement greatly reduced errors in the model (27).
Fig. 3.
Signature features of nuclear density in proteins. (A) Unexchanged backbone amide hydrogens can be identified by the lack of positive nuclear density for these atoms as shown for Asn-238 (exchanged) and Asn-239 (not exchanged). The sidechain of Asn-239 also shows the weak neutron scattering power of a CH2 group. Additional information from electron density in joint refinement helps to correctly position the carbon atom. (B) Density for the side-chain amide of Asn-272 allows for unambiguous assignment of the oxygen and nitrogen due to the greater neutron scattering of the NH2 group and the bean shaped density of W32 allows determining the orientation of that water molecule. Also in this case additional information from electron density in joint refinement helps to correctly position the oxygen atom. (C) Nuclear density for active site residues His-287 and Trp-244 clearly indicating the protonation states of the imidazole and indole ring nitrogen atoms. Hydrogen bonding is indicated by a red line.
Despite the advantages of joint X-ray and neutron refinement, to date most neutron structures have been determined by using programs, such as SHELX (28) and CNS (12), which allow the use of either X-ray or neutron data, but not both at the same time. In addition, these programs were originally developed for X-ray crystallography and are not completely suitable for macromolecular neutron crystallography. Joint X-ray and neutron refinement for biological macromolecules has now been implemented in a practical setting, through the development of nCNS (13), which allows for a robust, user-friendly crystallographic refinement environment.
XN Structure of DFPase.
Neutron diffraction on a crystal of DFPase, exchanged against deuterated mother liquor by vapor diffusion, allowed for the determination of the complete structure of the enzyme, including H atoms and the ordered solvent network. Compared with the X-ray structure, the model has approximately double the number of atoms, due to the inclusion of H atoms. To account for the larger number of parameters, the refinement of the structure used a patched version of CNS (nCNS) that allows for refinement of the model against both X-ray and neutron diffraction data. As such, the inclusion of X-ray and neutron data increased the data-to-parameters ratio and improved the accuracy of the model, while reducing the possibility of overfitting of data. Details of the joint refinement are described in SI Text, and data collection and refinement statistics are summarized in Table S1.
Inspection of the neutron maps, at a resolution of 2.2 Å, showed clear features for nuclear density. Because of the cancellation of the scattering of H and N atoms, unexchanged backbone amides show a lack of positive nuclear density (Fig. 3A). Water molecules, in the form of D2O, appear as bean-shaped density (Fig. 3B). The protonation state of histidine residues can be seen (Fig. 3C), whereas ND2 and ND3+ groups at the termini of side chains appear as strong nuclear density (Fig. 3B).
The degree of backbone amide exchange was determined by individual occupancy refinement of main-chain amide hydrogens (Fig. 1). Labile hydrogens in the crystal were exchanged for deuteriums for a period of less than 1 month before the start of data collection. The observed pattern of exchange mirrors the results obtained from NMR studies, which showed that ≈30% of the backbone amides did not show any detectable exchange (29). Although internal portions of the protein are generally most resistant to exchange (30), surprisingly, a number of surface residues do not undergo exchange. These residues correspond to β-strand amide hydrogens involved in hydrogen bonding with adjacent β-strands, whereas amide protons pointing toward the solvent are exchangeable.
The final model is essentially identical to the room temperature X-ray structure (PDB: 2GVW), with a root mean squared deviation (rmsd) of 0.086 Å over Cα atoms. The differences between the joint refined structure and 2GVW lie primarily in residues with larger side chains; for example, the presence of strong nuclear density for the terminal ND3+ groups on lysines allows for unambiguous placement of such side chains. There are greater differences between the joint refined structure and the ultrahigh resolution structure (Protein Data Bank ID code: 1PJX), with a rmsd of 0.258 Å over the Cα atoms, attributable to the different temperature of data collection (100 K vs. room temperature) and slightly different cell dimensions.
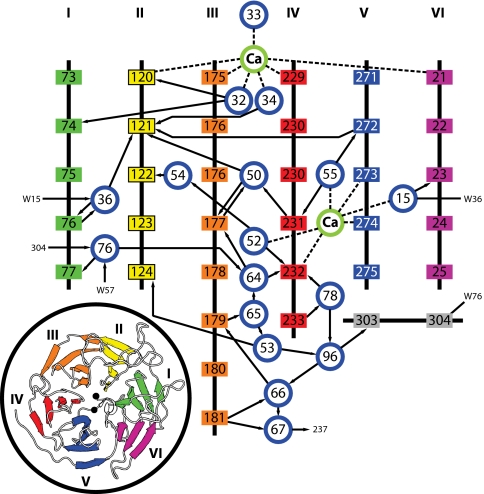
DFPase is the first protein for which neutron diffraction data are reported that contains an extended network of internal water molecules, connected by hydrogen bonds in form of a water tunnel in the center of the propeller. Despite the highly ordered water orientations within the tunnel and the series of “gates” in the tunnel such as the 2 calcium ions and the Asp-121/Asn-272 hydrogen bond, all waters in the tunnel have exchanged for D2O within the time scale of the experiment. The tunnel segment between structural calcium Ca2 and catalytic calcium Ca1 shows a complex network of interactions (Fig. 4), involving a variety of side chain–water and main chain–water hydrogen bonds (Fig. S1). The active site end of the water tunnel is occupied by catalytic calcium Ca1. Water molecules W34 and W32, coordinating with Ca1 on the inside of the tunnel, are unusual because they interact only with Ca1 and the protein atoms on the sides of the tunnel wall and can therefore be considered as trapped or at least isolated. W34 donates a hydrogen bond to Oδ2 of Asp-121. The carboxylate group of Asp-121 is the starting point of an extended and intricate network of hydrogen bonded water molecules that passes through the protein and incorporates structural calcium Ca2, before forking into 2 exit channels. Asp-121 is connected to Ca2 through double water bridges that form hexagonal and pentagonal faces that share common edges, reminiscent of a clathrate structure. Perhaps significantly, all of the water molecules in this motif have hydrogen bond interactions only with main chain amide and carbonyl groups, and therefore this motif may be resistant to some mutations. This in agreement with the limited degree of sequence identity among β-propeller proteins (31).
Fig. 4.
Schematic representation of hydrogen bonding interactions in the central water tunnel of DFPase. Arrows point from H-bond donor to acceptor. Dashed lines indicate direct metal coordination. Blue circles represent water molecules, amino acids are boxed colored according to the propeller blade they belong to.
Active Site Environment and Mechanistic Implications.
The DFPase active site is an elongated cleft with the catalytic calcium Ca1 in the center. The active site is oriented such that 2 acidic amino acids, Asp-229 and Glu-21, coordinate the calcium on one side of the cleft, whereas Asn-120 and Asn-175 are oriented so that their side chain Oδ1 atoms coordinate to Ca1 (Fig. 5A). Above the catalytic calcium is W33. Hydrogen bonds from the amino groups of Asn-120 and Asn-175 to W33 are possible in terms of distance and at least for Asn-120 in terms of geometry, although such interactions between a metal coordinated amide and a metal coordinated water molecule have rarely been reported (32).
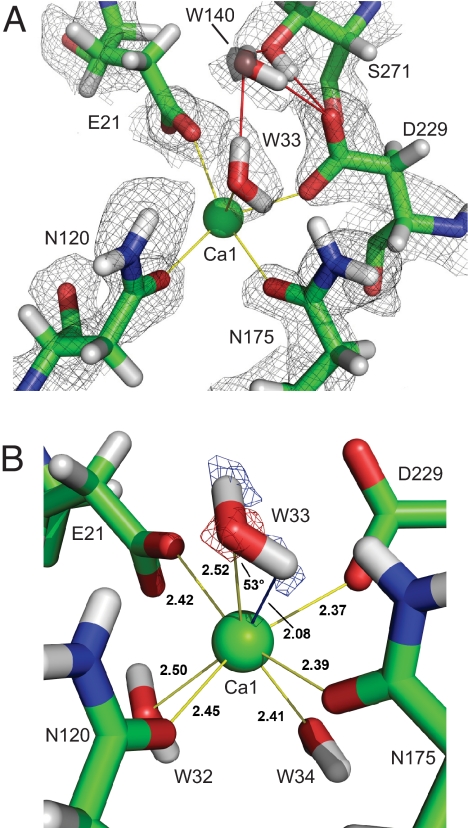
Fig. 5.
Active site environment. (A) Catalytic Ca1 with surrounding residues and positive nuclear density. Yellow lines indicate direct calcium coordination. Hydrogen bonding interactions are indicated in red. For the poorly resolved water W140 electron density is displayed (transparent gray sphere) showing the position of the oxygen atom. Nucleophilic Asp-229 is deprotonated. (B) A Fo-Fc nuclear density omit map (blue), with the 2 deuterium atoms of W33 omitted, contoured at 4.0 σ, showing identity of W33 as a water, and not a hydroxide ion. The electron density 2Fo-Fc map at 2.5 σ is superimposed (red), showing the location of the oxygen atom. Distances are shown in Å. The W33 hydrogen–Ca1 distance is indicated as a blue line. The Ca–O–H angle is 53°.
Oδ1 of Asp-229 and Oε2 of Glu-21 coordinate with Ca1. Oε1 of Glu-21 accepts H bonds from the amino group of Asn-272 and the amide D of Gly-22, and Oδ2 of Asp-229 accepts a hydrogen bond from the hydroxyl D of Ser-271 and from W140. The nuclear density clearly shows that Asp-229 is deprotonated, which is further confirmed by the hydrogen bonding network in the active site region. Again, the joint refinement clearly reveals the orientation of the side chains, the fact that Asp-229 and Glu-21 are charged, and the distribution of protons in the hydrogen bonding arrangement. At the bottom of the active site pocket W32 and W34 coordinate with Ca1 and donate hydrogen bonds to amino acid side chains.
In enzymes, metal ions can serve as water activators in general base-catalyzed mechanisms. Solvent molecule W33 coordinates the catalytic calcium, raising the question as to whether it is water or hydroxide. When interpreting 2Fo−Fc nuclear density during model building, W33 was clearly better modeled as a water molecule and not a hydroxyl ion (Fig. 5A). To unambiguously identify W33, a simulated annealing omit map was calculated, omitting the 2 deuteriums of W33. This omit map shows 2 difference Fo−Fc peaks of equal intensity appearing in the geometry of a water molecule, further evidence that W33 is a water, and not a hydroxide (Fig. 5B). The coordination of W33 is unexpected, with the hydrogen atom in close proximity to Ca1. The observed distance between Ca1 and the O of W33 is 2.52 Å, whereas the closest hydrogen atom of W33 is only 2.08 Å from Ca1 (Fig. 5B). This unfavorable orientation for the hydrogen atom appears to be a consequence of the environment around W33, such that even more unfavorable orientations are minimized. Einspahr and Bugg (33) concluded that the minimal limit for Ca–O–H angles in crystalline hydrates is 75°, based on neutron and X-ray diffraction of calcium salts. In our case we observe an angle of 53° (Fig. 5B). We attribute this to the special environment in the protein active site, which is not comparable to highly ordered small inorganic molecule complexes. Although what we observe is unusual, and to our knowledge is the first such example reported for a calcium-bound water molecule both in proteins and small molecules, the omit map Fo−Fc density is clear and unambiguous. It is important to note that in the present orientation of W33 there is no hydrogen bonding interaction with the Oδ1 of Asp-229, which could enhance water activation. Asp-229 is deprotonated in the structure as required for a nucleophilic attack on a substrate molecule, consistent with our earlier isotope labeling studies. Finally, this observed orientation would not have been seen with even the highest resolution X-ray structures available, but is unambiguous with neutron diffraction at moderate resolution.
Furthermore, there are a number of supporting chemical arguments why W33 is water, and not hydroxide. It was reported in the literature that in the case of magnesium complexes water is indeed activated by coordination to the metal; however, the presence of even a single carboxylic ligand in the complex increases the free energy of the deprotonation step by ≈80 kcal/mol (34). In the case of DFPase there are 2 carboxylates in the coordination sphere of the catalytic calcium, further increasing the free energy of deprotonation. The interpretation of W33 as a water molecule is also consistent with the finding that little change in catalytic activity is observed when Ca2+ is replaced by ions such as Mg2+ or Ba2+ (19). If the mechanism proceeds through ion-mediated water activation, we would expect the catalytic activity to be influenced by the ionic radii of the metal species. A further argument against W33 being the active nucleophile is the requirement for the substrate to coordinate to the metal ion. This would imply 8-fold coordination of the Ca2+ ion, which although in principle is possible, is very unlikely, because the substrate occupies nearly the full volume of the active site. If Ca2+ is exchanged for Mg2+, neither 7- nor 8-fold coordination is possible, because Mg2+ coordinates a maximum of 6 ligands in an octahedral geometry with only very rare exceptions.
Next to W33 lies W140 (Fig. 5A), which is rather weakly defined in the neutron map. This water appears to be important for the orientation of W33, because it accepts a hydrogen bond from W33 with a distance of 2.11 Å, and W140 further donates a hydrogen bond to Asp-229, with a distance of 1.98 Å, and to the hydroxyl oxygen of Ser-271 (1.76 Å). For the substrate to bind, both W33 and W140 must be displaced; the X-ray crystal structure of the substrate analogue in complex with DFPase (19) shows no remaining waters in the active site.
By demonstrating that Asp-229 is deprotonated, and solvent molecule W33 is water, we have eliminated the possibility of an activated water nucleophile in the catalytic mechanism. Based on the body of evidence presented here, and together with our previous isotope labeling experiments, Asp-229 is identified as the nucleophile in the DFPase mechanism.
The active site of the structurally related enzyme PON is characterized by a very similar arrangement of residues coordinating a central calcium ion. We proposed earlier that Asp-269, the homologue of Asp-229 in DFPase, may act as the nucleophile in the phosphotriesterase activity of PON (19, 35). Even though differences in the active site topology of PON and DFPase exist, the XN structure of DFPase might help to rule out direct water activation by the metal ion in PON. At present neutron diffraction experiments on PON do not seem feasible given the small crystal size reported for the solved PON structure and taking into account unit cell parameters and space group (21). The XN structure of DFPase might therefore serve as a valuable model and inspire further studies to establish the phosphotriesterase mechanism for PON.
Implications for Neutron Protein Crystallography.
Neutron structures have been reported for ≈20 individual proteins and nucleic acids to date, compared with 52,000 X-ray structures deposited in the Protein Data Bank as of the end of July 2008. The XN structure of DFPase is one of the first examples of joint X-ray and neutron structure refinement for a protein in combination with cross-validated maximum likelihood simulated annealing. The use of this approach has several notable advantages, including an increased data-to-parameters ratio and more accurate information about protein structure, than either X-ray or neutron crystallography on its own. However, we found that one of the clearest practical benefits of joint refinement in this particular case was during water building. Superposition of the electron and nuclear density maps clearly indicated the orientation of water molecules, whereas inspection of either map on its own was often ambiguous. Because X-ray diffraction data normally extend to higher resolution than neutron data, the refinement can potentially be biased toward the positions of the heavy atoms. This illustrates the importance of proper weighting factors for X-ray and neutron data during refinement. Wlodawer and Hendrickson observed that refinement of the neutron data on its own was sometimes unstable (27); thus the addition of the X-ray data helped ‘stabilize’ the structure, and the neutron data allowed for the refinement of the positions of the hydrogen atoms.
Currently, the number of neutron sources is a limiting factor, and there has been a significant increase in the number of people interested in screening crystals for neutron diffraction. Sample sizes have become smaller, and these factors will inevitably lead to difficult and weak data. The use of nCNS provides a convenient platform for joint refinement, and employs the robust maximum likelihood target function, ideally suited for datasets with relatively weak data, which is typically the case for neutron data collected with the time-of-flight technique at spallation sources. The useful features of omit maps calculated in nCNS are shown, which allowed for the determining the orientation of W33. Furthermore, we note that most neutron structures solved since 2007 have applied the joint refinement technique, and thus will be the choice method in the future for refinement of neutron diffraction data. The implementation we discuss here is thus a major step forward, providing a starting point for the development of new strategies to further improve the refinement of neutron structures. The Protein Data Bank is in the process of determining a standard format for the deposition of future joint refined structures.
In addition to the insights into the DFPase mechanism reported here, this work also represents a major technical advancement in neutron crystallography. The crystal used here was 0.43 mm3 in size, among the smallest sample size used so far for neutron crystallography (36). The crystal was exchanged for less than 1 month before the start of data acquisition. In light of these improvements in sample size requirements, data acquisition and refinement methods, and the availability of new neutron sources, neutron crystallography can be seen as an increasingly practical and useful method for the study of macromolecules and enzyme mechanisms.
This study therefore has demonstrated the robustness of the nCNS protocols in revealing details about the mechanism of DFPase, and new insights about solvent structure within internal pores and tunnels in proteins.
Materials and Methods
Data Collection, Processing, and Refinement.
Room-temperature neutron data were collected and processed as reported (36). Briefly, a single crystal measuring 2.4 × 0.5 × 0.36 mm in size was mounted in a glass capillary, and labile hydrogens were exchanged for deuteriums by vapor diffusion for 1 week before the start of data collection. Thirty-seven time-of-flight neutron diffraction images were collected at the PCS at LANSCE, with exposures of up to 24 h per image. Data were processed by using a version of d*TREK (37) modified for time-of-flight data (26). The structure was refined by using a version of CNS (12), called nCNS, modified for joint X-ray and neutron refinement (13). The refinement used neutron data between 20.0–2.2 Å resolution, accounting for a slight difference in the number of reflections than reported (36). A detailed treatment of the joint refinement method is described in the SI Text, and statistics of the joint refinement of DFPase are reported in Table S1.
Supplementary Material
Acknowledgments.
We thank Dr. Leighton Coates for expert advice on refinement and Mary Jo Waltman for technical assistance. We thank Drs. Jenny Glusker and Alexander Koglin for helpful discussions. This project was funded by the Hessisches Ministerium für Wissenschaft und Kultur and the German Ministry of Defense under Grants E/E590/6Z004/4F170 and E/UR3G/6G115/6A801. A travel grant was provided to M.-M.B. and J.C.-H.C. by the Glaxo Smith-Kline Foundation. The PCS is funded by the Office of Science and the Office of Biological and Environmental Research of the U.S. Department of Energy. M.M. and P.L. were partly supported by an National Institutes of Health–National Institute of General Medical Sciences-funded consortium (1R01GM071939–01) between Los Alamos National Laboratory and Lawrence Berkeley National Laboratory to develop computational tools for neutron protein crystallography.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3BYC).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807842106/DCSupplemental.
References
- 1.Kossiakoff AA, Spencer SA. Direct determination of the protonation states of aspartic acid-102 and histidine-57 in the tetrahedral intermediate of the serine proteases: Neutron structure of trypsin. Biochemistry. 1981;20:6462–6474. doi: 10.1021/bi00525a027. [DOI] [PubMed] [Google Scholar]
- 2.Wlodawer A, Sjölin L. Structure of ribonuclease A: Results of joint neutron and X-ray refinement at 2.0-A resolution. Biochemistry. 1983;22:2720–2728. doi: 10.1021/bi00280a021. [DOI] [PubMed] [Google Scholar]
- 3.Katz AK, et al. Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction. Proc Natl Acad Sci USA. 2006;103:8342–8347. doi: 10.1073/pnas.0602598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett B, et al. Neutron diffraction studies of Escherichia coli dihydrofolate reductase complexed with methotrexate. Proc Natl Acad Sci USA. 2006;103:18493–18498. doi: 10.1073/pnas.0604977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coates L, Erskine PT, Wood SP, Myles DAA, Cooper JB. A neutron Laue diffraction study of endothiapepsin: Implications for the aspartic proteinase mechanism. Biochemistry. 2001;40:13149–13157. doi: 10.1021/bi010626h. [DOI] [PubMed] [Google Scholar]
- 6.Blakeley MP, et al. Comparison of hydrogen determination with X-ray and neutron crystallography in a human aldose reductase-inhibitor complex. Eur Biophys J. 2006;35:577–583. doi: 10.1007/s00249-006-0064-8. [DOI] [PubMed] [Google Scholar]
- 7.Meilleur F, Snell EH, van der Woerd MJ, Judge RA, Myles DAA. A quasi-Laue neutron crystallographic study of D-xylose isomerase. Eur Biophys J. 2006;35:601–609. doi: 10.1007/s00249-006-0066-6. [DOI] [PubMed] [Google Scholar]
- 8.Kovalevsky AY, et al. Hydrogen location in stages of an enzyme-catalyzed reaction: Time-of-flight neutron structure of D-xylose isomerase with bound D-xylulose. Biochemistry. 2008;47:7595–7597. doi: 10.1021/bi8005434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niimura N, et al. Recent results on hydrogen and hydration in biology studied by neutron macromolecular crystallography. Cell Mol Life Sci. 2006;62:285–300. doi: 10.1007/s00018-005-5418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kossiakoff AA. Protein dynamics investigated by the neutron diffraction-hydrogen exchange technique. Nature. 1982;296:713–721. doi: 10.1038/296713a0. [DOI] [PubMed] [Google Scholar]
- 11.Bennett BC, Gardberg AS, Blair MD, Dealwis CG. On the determinants of amide backbone exchange in proteins: A neutron crystallographic comparative study. Acta Crystallogr D. 2008;64:764–783. doi: 10.1107/S0907444908012845. [DOI] [PubMed] [Google Scholar]
- 12.Brünger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 13.Mustyakimov M, Langan P. Los Alamos, NM: Los Alamos National Security; 2007. Copyright C-06, 104 Patch for CNS; nCNS an open source distribution patch for CNS for macromolecular structure refinement. [Google Scholar]
- 14.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 15.Adams PD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 16.Hartleib J, Rüterjans H. High-yield expression, purification, and characterization of the recombinant diisopropylfluorophosphatase from Loligo vulgaris. Protein Expr Purif. 2001;21:210–219. doi: 10.1006/prep.2000.1360. [DOI] [PubMed] [Google Scholar]
- 17.Scharff EI, Koepke J, Fritzsch G, Lücke C, Rüterjans H. Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris. Structure. 2001;9:493–502. doi: 10.1016/s0969-2126(01)00610-4. [DOI] [PubMed] [Google Scholar]
- 18.Hartleib J, Geschwindner S, Scharff EI, Rüterjans H. Role of calcium ions in the structure and function of the di-isopropylfluorophosphatase from Loligo vulgaris. Biochem J. 2001;353:579–589. doi: 10.1042/0264-6021:3530579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blum MM, Loehr F, Richardt A, Rüterjans H, Chen JCH. Binding of a designed substrate analogue to diisopropyl fluorophosphatase: Implications for the phosphotriesterase mechanism. J Am Chem Soc. 2006;128:12750–12757. doi: 10.1021/ja061887n. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka Y, et al. Structural and mutational analyses of Drp35 from Staphylococcus aureus: A possible mechanism for its lactonase activity. J Biol Chem. 2007;282:5770–5780. doi: 10.1074/jbc.M607340200. [DOI] [PubMed] [Google Scholar]
- 21.Harel M, et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- 22.Yeung DT, et al. Structure/function analyses of human serum paraoxonase (HuPON1) mutants designed from a DFPase-like homology model. Biochim Biophys Acta. 2004;1702:67–77. doi: 10.1016/j.bbapap.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Khersonsky O, Tawfik DS. Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry. 2005;44:6371–6382. doi: 10.1021/bi047440d. [DOI] [PubMed] [Google Scholar]
- 24.Koepke J, Scharff EI, Lücke C, Rüterjans H, Fritzsch G. Atomic resolution crystal structure of squid ganglion DFPase. Acta Crystallogr D. 2002;58:1757–1759. doi: 10.1107/s0907444902012714. [DOI] [PubMed] [Google Scholar]
- 25.Koepke J, Scharff EI, Lücke C, Rüterjans H, Fritzsch G. Statistical analysis of crystallographic data obtained from squid ganglion DFPase at 0.85 Å resolution. Acta Crystallogr D. 2003;59:1744–1754. doi: 10.1107/s0907444903016135. [DOI] [PubMed] [Google Scholar]
- 26.Langan P, Greene G. Protein crystallography with spallation neutrons: Collecting and processing wavelength-resolved Laue protein data. J Appl Crystallogr. 2004;37:253–257. [Google Scholar]
- 27.Wlodawer A, Hendrickson WA. A procedure for joint refinement of macromolecular structures with X-ray and neutron diffraction data from single crystals. Acta Crystallogr A. 1982;38:239–247. [Google Scholar]
- 28.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 29.Löhr F, Katsemi V, Hartleib J, Günther U, Rüterjans H. A strategy to obtain backbone resonance assignments of deuterated proteins in the presence of incomplete amide 2H/1H back-exchange. J Biomol NMR. 2003;25:291–311. doi: 10.1023/a:1023084605308. [DOI] [PubMed] [Google Scholar]
- 30.Kossiakoff AA, Ultsch M, White S, Eigenbrot C. Neutron structure of subtilisin BPN′: Effects of chemical environment on hydrogen-bonding geometries and the pattern of hydrogen-deuterium exchange in secondary structure elements. Biochemistry. 1991;30:1211–1221. doi: 10.1021/bi00219a008. [DOI] [PubMed] [Google Scholar]
- 31.Fülöp V, Jones DT. Beta propellers: Structural rigidity and functional diversity. Curr Opin Struct Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 32.McPhalen CA, Strynadka NCJ, James MNG. Calcium- binding sites in proteins: A structural perspective. Adv Prot Sci. 1991;42:77–144. doi: 10.1016/s0065-3233(08)60535-5. [DOI] [PubMed] [Google Scholar]
- 33.Einspahr H, Bugg CE. The geometry of calcium-water interactions in crystalline hydrates. Acta Crystallogr B. 1980;36:264–271. [Google Scholar]
- 34.Katz AK, Glusker JP, Markham GD, Bock CW. Deprotonation of water in the presence of carboxylate and magnesium ions. J Phys Chem B. 1998;102:6342–6350. [Google Scholar]
- 35.Blum MM, Timperley C, Williams G, Thiermann H, Worek F. Inhibitory potency against human acetylcholinesterase and enzymatic hydrolysis of fluorogenic nerve agent mimics by human paraoxonase 1 and squid diisopropyl fluorophosphatase. Biochemistry. 2008;47:5216–5224. doi: 10.1021/bi702222x. [DOI] [PubMed] [Google Scholar]
- 36.Blum MM, et al. Preliminary time-of-flight neutron diffraction study on diisopropyl fluorophosphatase (DFPase) from Loligo vulgaris. Acta Crystallogr F. 2007;63:42–45. doi: 10.1107/S1744309106052924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.