Abstract
Recent studies support the existence of a common progenitor for the cardiac and endothelial cell lineages, but the underlying transcriptional networks responsible for specification of these cell fates remain unclear. Here we demonstrated that Ets-related protein 71 (Etsrp71), a newly discovered ETS family transcription factor, was a novel downstream target of the homeodomain protein, Nkx2–5. Using genetic mouse models and molecular biological techniques, we demonstrated that Nkx2–5 binds to an evolutionarily conserved Nkx2–5 response element in the Etsrp71 promoter and induces the Etsrp71 gene expression in vitro and in vivo. Etsrp71 was transiently expressed in the endocardium/endothelium of the developing embryo (E7.75-E9.5) and was extinguished during the latter stages of development. Using a gene disruption strategy, we found that Etsrp71 mutant embryos lacked endocardial/endothelial lineages and were nonviable. Moreover, using transgenic technologies and transcriptional and chromatin immunoprecipitation (ChIP) assays, we further established that Tie2 is a direct downstream target of Etsrp71. Collectively, our results uncover a novel functional role for Nkx2–5 and define a transcriptional network that specifies an endocardial/endothelial fate in the developing heart and embryo.
Keywords: cardiac progenitor cells, endocardium, Etsrp71, Tie2, cardiac development
Formation of the cardiovascular system involves a precisely orchestrated series of molecular and morphogenetic events whereby even subtle perturbations of this process can have catastrophic consequences in the form of congenital heart disease. Heart formation begins at approximately embryonic day (E)7.5 in the mouse when progenitor cells migrate through the primitive streak to the anterior-proximal side of the embryo to form the cardiac crescent (1, 2). In response to permissive and repressive cues, the cardiac progenitors migrate in a dorsomedial direction, fuse, and form the linear heart tube and ultimately the 4-chambered heart. While heart formation has been extensively described morphologically (1–3), the transcriptional networks that govern fate determination in the heart are relatively unknown. Of particular interest is the recent finding that multipotent cardiac progenitors are capable of daughtering alternative lineages (i.e., myocardial, smooth muscle, and endocardial lineages) during cardiogenesis although the transcriptional networks that govern these fate decisions remain incompletely defined (4–6).
The homeobox transcription factor, Nkx2–5 is the vertebrate homolog of Drosophilla homeodomain protein Tinman, which is necessary for heart formation in the fly (7). Nkx2–5 is one of the earliest transcription factors expressed in the cardiac lineage of developing vertebrate embryos. Targeted disruption of Nkx2–5 results in perturbed heart morphogenesis, severe growth retardation, and embryonic lethality at approximately E9.5 (8, 9). One of the distinguishing features of the mutant heart is the absence of an endocardial cushion (8, 9). Studies using Nkx2–5-Cre lineage tracing strategies showed reporter expression and endogenous Nkx2–5 expression in the endocardial lineage of the developing heart (10). Moreover, recent studies revealed that Nkx2–5 was expressed in multipotent progenitors during cardiac morphogenesis (4–6). Collectively, these studies support the notion that Nkx2–5 regulates endocardial/endothelial development. Although several Nkx2–5 downstream targets have been identified that are expressed in the myocardium (9, 11–13), no target genes have been identified to date that are restricted to the endocardium.
Ets-related protein 71 (Etsrp71) is a newly discovered ETS domain-containing transcription factor that has extensive conservation with pointed, a Drosophila ETS-domain factor (14, 15). The conserved ETS domain is responsible for DNA binding activity of the ETS protein family (14). Etsrp71 has homology with other family members only within the ETS domain but no homology to sequences outside this domain. Furthermore, limited mechanistic information exists regarding the functional role(s) for the Ets family members during embryogenesis.
In this study, we demonstrate that Etsrp71 is a novel downstream target gene of Nkx2–5. Etsrp71 is transiently expressed in the endocardium/endothelium of the developing embryo and is extinguished during the latter stages of development and in the adult heart. We also demonstrate that Etsrp71 null embryos lack endocardial and endothelial lineages and are lethal early during embryogenesis. Furthermore, we demonstrate that Tie2, which is expressed in endothelial progenitors, is a direct downstream target of Etsrp71. Collectively, these data complement and extend our understanding of the mechanisms whereby Nkx2–5-expressing multipotent cardiac progenitors promote an endocardial/endothelial fate in the developing heart and embryo.
Results
Downregulation of Etsrp71 Expression in the Nkx2–5 Null Heart.
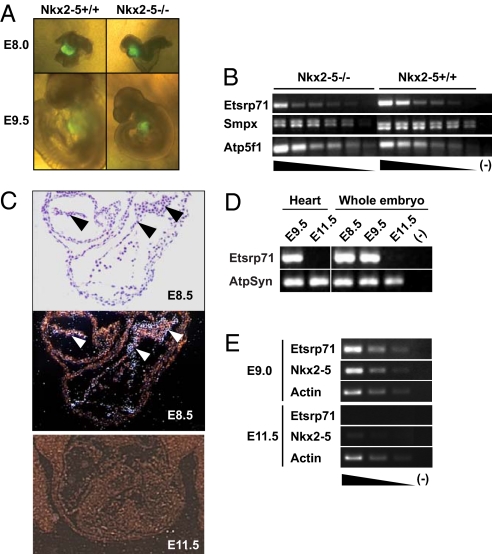
We previously engineered a transgenic mouse model using a 6-kb upstream enhancer region of the Nkx2–5 gene to direct an enhanced yellow fluorescent protein (EYFP) reporter to the cardiac progenitor cell population early during development (E7.75), which recapitulates endogenous Nkx2–5 expression (16). To identify Nkx2–5 direct downstream target genes, we examined differential gene expression in the cardiac progenitors at discrete developmental stages in the presence and absence of Nkx2–5. We mated the Nkx2–5 heterozygous (8) and the Nkx2–5-EYFP transgenic:Nkx2–5 heterozygous mice to produce wild-type (WT), heterozygous, and null littermate embryos with EYFP expression in the cardiac progenitors of the developing heart (Fig. 1A). Although the Nkx2–5 null embryos were indistinguishable from the WT littermates at E8.0, a severe growth retardation and perturbed cardiac development were observed in the E9.5 null embryos as previously described (Fig. 1A) (8).
Fig. 1.
Transient expression of Etsrp71 in the endocardial/endothelial lineage is downregulated in Nkx2–5 null cardiac progenitors. (A) Six-kilobase Nkx2–5-EYFP Tg:Nkx2–5± and Nkx2–5± mice were mated to generate EYFP+ Nkx2–5+/+ and EYFP+ Nkx2–5−/− littermates at E8.0 and E9.5. Note that the WT and Nkx2–5 mutant embryos are indistinguishable at E8.0 while at E9.5 the Nkx2–5−/− embryo has severe growth retardation compared to the WT littermate. (B) Semiquantitative RT-PCR in 6-kb Nxk2–5-EYFP Tg:Nkx2–5 null vs. 6-kb Nxk2–5-EYFP Tg:WT cardiac progenitors. Note decreased Etsrp71 transcript expression in the Nkx2.5−/− cardiac progenitors vs. the WT controls. (C) In situ hybridization techniques for Etsrp 71 expression at E8.5. The bright (Top) and dark (Bottom) fields of the transverse section of an E8.5 heart are shown and arrowheads indicate endocardium. Note an absence of Etsrp71 expression in the E11.5 embryo. (D) RT-PCR analysis of Etsrp71 expression in the developing heart and embryo. RNA was extracted at the indicated stages from the developing heart and embryo to analyze the expression of Etsrp71. ATP synthase (ATPSyn) was used as a loading control. (E) Coexpression of Etsrp71 and Nkx2–5 in endothelial progenitor cells of developing embryos. Tie2-GFP+ cells were isolated from developing embryos using FACS for semiquantitative RT-PCR (25 cycles). Actin was used as a loading control and was analyzed after 15 cycles.
Individual WT and null littermate embryos from each stage were dissociated and EYFP+ cells were FACS sorted as previously described (16). RNA was extracted from EYFP+ cells, amplified, and processed for transcriptome analysis (16). A total of 86 and 113 transcripts were significantly downregulated at E8.0 and E8.5 stages of development in the Nkx2–5 null cardiac progenitors, respectively [see supporting information (SI) Fig. S1A]. Among them, 18 transcripts were downregulated at both developmental stages (Fig. S1). Two time periods were analyzed to identify transcripts commonly downregulated to enrich for authentic Nkx2–5 downstream targets and minimize the selection of a candidate transcript that was not a Nkx2–5 target but was downregulated in the null heart as a consequence of an anatomical (secondary) defect. For example, we observed that Nppa, Nppb, Smpx, and Carp, which have been previously defined as direct downstream target genes of Nkx2–5 (9, 11, 17), were downregulated in the Nkx2–5 null cardiac progenitors at both developmental stages, which further supports the authenticity of the screen (Fig. 1B and Fig. S1B). Furthermore, we observed that in addition to Nppa and Nppb, Etsrp71 was also significantly downregulated in the Nkx2–5 null progenitor cell population at both developmental stages (Fig. S1B). We confirmed these transcriptome results using semiquantitative RT-PCR analysis of RNA isolated from an independent WT and Nkx2–5 null progenitor cell population (Fig. 1B). These transcriptome and RT-PCR results support the hypothesis that Etsrp71 is a novel downstream target gene of Nkx2–5.
Etsrp71 Is Expressed in the Endothelium/Endocardium of the Developing Heart.
Nkx2–5 has been previously shown to be expressed in the cardiac crescent (E7.75), the endocardium, and the myocardium of the embryonic and fetal heart (8–10). In contrast, the temporal and spatial expression pattern of Etsrp71 is unknown. Using in situ hybridization, we observed that Etsrp71 expression was restricted to the endothelium/endocardium of the E8.5 heart (Fig. 1C and Fig. S2) and that expression was completely extinguished at E11.5 (Fig. 1C) and no myocardial expression was observed. We further isolated RNA from embryonic hearts and whole embryos (including the heart) and used RT-PCR analysis to demonstrate that Etsrp71 was expressed only in the early developing heart and embryos (E8.5 and E9.5) and not in the midgestational E11.5 heart or whole embryo (Fig. 1D). These results further confirm the temporal expression pattern of Etsrp71, which was observed using in situ hybridization techniques.
To further examine the expression pattern of Nkx2–5 and Etsrp71 in endocardial and endothelial progenitor cells, we used FACS to isolate GFP+ cells from Tie2-GFP transgenic (Tg) embryos. Tie2, the vascular endothelial-specific receptor tyrosine kinase, is expressed in endocardial and endothelial progenitors during embryogenesis (18). Tie2-GFP Tg mice (19) were mated with WT mice and GFP+ cells were isolated by FACS from E9.0 and E11.5 embryos. RNA was extracted, amplified, and analyzed for gene expression using semiquantitative RT-PCR techniques (16). As shown in Fig. 1E, both Etsrp71 and Nkx2–5 were expressed in GFP+ cells isolated from E9.0 embryos and essentially absent from E11.5 embryos, suggesting that Tie2-expressing endothelial/endocardial cells temporally coexpress Etsrp71 and Nkx2–5. These data further support the notion that Nkx2–5 is a transcriptional activator of Etsrp71 in the endothelium/endocardium of the developing heart.
Endocardial Expression of Etsrp71 Is Severely Attenuated in the Absence of Nkx2–5.
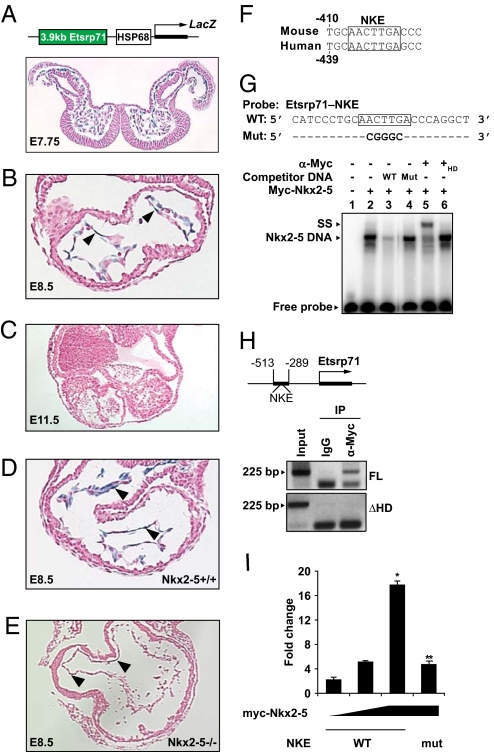
To further explore the regulatory interaction between Nkx2–5 and Etsrp71, we used a 3.9-kb Etsrp71 promoter fragment that contained evolutionary conserved regions between mouse and human. We generated Tg mice using this 3.9-kb Etsrp71 promoter fragment to drive the lacZ reporter gene (Fig. 2A) (20). Expression of the lacZ reporter in transgenic embryos recapitulates the endogenous expression of Etsrp71. We observed lacZ reporter expression in the cardiac crescent (E7.75) (Fig. 2A) and expression was restricted to the endocardial/endothelial lineages at E8.5 (Fig. 2B) and E9.5 (data not shown) and then extinguished at E11.5 (Fig. 2C). Therefore, our results indicate that the 3.9-kb upstream promoter segment of Etsrp71 harbors regulatory elements that direct Etsrp71 expression to the endocardial/endothelial lineages early during embryogenesis. To address whether Nkx2–5 is an essential upstream transcriptional activator of Etsrp71, we mated Nkx2–5± and Nkx2–5±:3.9-kb Etsrp71 Tg adult mice. Age-matched WT:3.9-kb Etsrp71 Tg and Nkx2–5 null:3.9-kb Etsrp71 Tg embryos were harvested and the expression of β-galactosidase was evaluated. We observed that cardiac expression of β-galactosidase was severely reduced in the Nkx2–5 null embryo (Fig. 2 D–E). These results further support the hypothesis that Nkx2–5 lies genetically upstream of Etsrp71 and functions as a transcriptional activator of Etsrp71. An alternative explanation is that the loss of Etsrp71 promoter activity could be due to a secondary effect. Therefore, further studies were undertaken to examine this hypothesis.
Fig. 2.
Nkx2–5 is a transcriptional activator of Etsrp71. (A–C) Schematic of the construct used to generate an Etsrp71 transgenic mouse line. Within the developing transgenic heart, β-galactosidase expression was observed in endocardial precursors at E7.75–E8.5 and was extinguished at E11.5. (D and E) Etsrp71-LacZ Tg mice were mated with Nkx2–5± mice and their progeny analyzed. Transverse sections of the looping E8.5 heart from Nkx2–5+/+ and Nkx2–5−/− embryos are shown. Arrowheads denote the β-galactosidase-positive endothelium/endocardium. Note that β-galactosidase expression was severely attenuated in the Nkx2–5 null heart. (F) Alignment of the mouse and human Etsrp71 promoter sequence revealing conservation of the Nkx2–5 response element. (G) Single-strand sequence of the Etsrp71 promoter harboring the WT (shown in box) and mutant (Mut) NKE (shown in boldface type) were used as radiolabeled probes. Electrophoretic mobility shift assay (EMSA) revealed the formation of a stable Nkx2–5-DNA complex (lane 2) that could be competed with WT but not with Mut probe (lanes 2–4) and supershifted (lane 5). Note that heat denaturation (+HD) (control for the supershift assay) of the myc antibody is unable to supershift the complex (lane 6). (H) Schematic of the upstream promoter region of Etsrp71 containing the evolutionary conserved NKE (black bar). ChIP assay for in vivo binding of Nkx2–5 to the Etsrp71 promoter used the myc-tagged full-length (FL) and mutant (lack of DNA-binding domain, ΔHD) Nkx2–5. DNA purified before immunoprecipitation (IP) was used as input. Note IP of Etsrp71 promoter harboring the NKE only from cells expressing FL Nkx2–5. (I) Nkx2–5 transcriptionally activates Etsrp71 gene expression in C2C12 myoblasts. Overexpression of the CMV-myc-Nkx2–5 plasmid results in a dose-dependent activation of the 2-kb-Etsrp71-luc promoter construct. Mutation of NKE in the 2-kb Etsrp71 promoter significantly attenuated trancriptional activation by Nkx2–5 (n = 6 for each sample; data presented are mean ± SEM; *, P < 0.001). Fold change represents luciferase activity normalized to lacZ expression.
Nkx2–5 Binds to the Etsrp71 Promoter and Transactivates Its Expression.
Analysis of the 3.9-kb upstream sequence of the Etsrp71 gene revealed an evolutionary conserved Nkx2–5 responsive element (NKE) (Fig. 2F). We examined the binding of myc-tagged Nkx2–5 fusion protein to a radiolabeled probe containing the NKE, using a gel-shift assay, and competed that binding with cold probe containing WT but not the mutant (Mut) NKE (Fig. 2G). The protein–DNA complex could be supershifted while the addition of heat-inactivated antibody was unable to supershift the complex (lanes 2, 5, and 6), suggesting that the protein–DNA complex observed in lane 2 was specific for the myc-tagged Nkx2–5 fusion protein. Promoter occupancy of Nkx2–5 to this evolutionary conserved site in vivo was confirmed using chromatin immunoprecipitation (ChIP) assays (20, 21). As shown in Fig. 2H, we were able to PCR amplify the Etsrp71 promoter fragment harboring the NKE by specific primers when immunoprecipitation of chromatin fragments was performed with the anti-myc serum but not with control IgG serum. Moreover, we were able to PCR amplify the Etsrp71 promoter only from cells expressing full-length Nkx2–5 (Fig. 2H). Using gel-shift and ChIP assays, we further demonstrated that the endogenous Nkx2–5 in C2C12 myoblast cells also binds to this NKE (Fig. S3). These data support our in vivo observation (see Fig. 2 D and E) that endogenous Nkx2–5 binds to this NKE in the Etsrp71 promoter and transactivates gene expression.
Having established that Nkx2–5 binds to the Etsrp71 promoter in vivo, we then assessed the physiological relevance of the binding of Nkx2–5 to the Etsrp71 promoter and Etsrp71 gene expression using quantitative (q)RT-PCR. Transfection of C2C12 cells with Nkx2–5 or induction of Nkx2–5 expression in an inducible ES/EB system exhibited an activation of endogenous Etsrp71 gene expression (Fig. S4). We further undertook transcriptional assays to define the specificity of Nkx2–5 as a transcriptional regulator of Etsrp71 gene expression. We fused the upstream fragment, which harbors the evolutionary conserved NKE, of the Etsrp71 promoter to the luciferase reporter. Using transcriptional assays, we observed that Nkx2–5 expression resulted in a significant dose-dependent activation of Etsrp71 gene expression while mutation of the NKE significantly attenuated the activation of gene expression (Fig. 2I). These results further support our hypothesis that Nkx2–5 is an important transcriptional activator of the Etsrp71 gene.
Etsrp71 Specifies the Endocardial/Endothelial Lineage in the Developing Embryo.
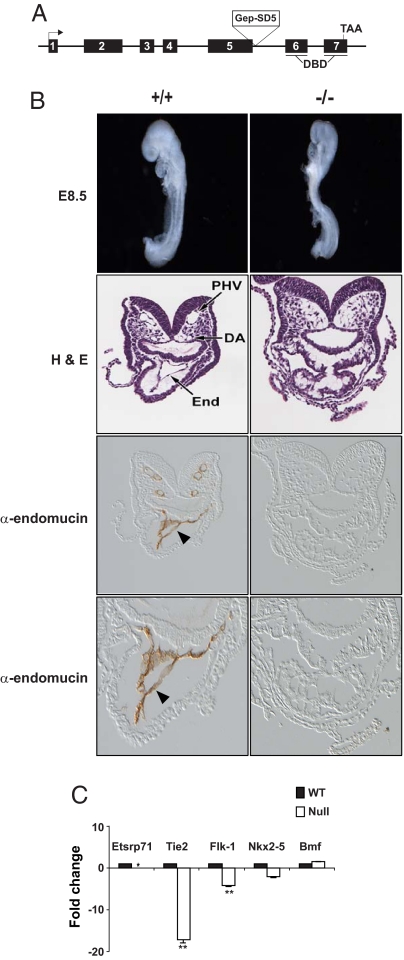
To examine the functional role for Etsrp71 during embryogenesis, we obtained ES cells that contained a trapping construct (pGep-SD5) to disrupt Etsrp71 (Fig. 3A) (22). We generated chimera mice using this targeted ES cell line, which was confirmed by PCR analysis. Analysis of the progeny from heterozygous matings revealed that Etsrp71 homozygous null mice were nonviable between E9.0 and E10.5 (Fig. S5 A and B). The Etsrp71−/− embryos were indistinguishable from the WT (+/+) littermates at E8.5 (Fig. 3B and Fig. S5B). However, histological and immunohistochemical analyses revealed the absence of an endocardial/endothelial lineage in the Etsrp71−/− heart and embryo (Fig. 3B). The absence of endocardial/endothelial lineage in the Etsrp71 null embryo was not due to increased cell death (Fig. S5C) or lack of cellular proliferation (Fig. S5D). These data clearly define an essential role for Etsrp71 in the genesis of the endocardial/endothelial lineage during development. To further examine our hypothesis, we used qRT-PCR analyses, which revealed a significant downregulation of endothelial/endocardial-specific transcripts (Flk-1), and Tie2 transcripts were essentially absent in the null compared to the WT heart (Fig. 3C). A modest decrease in Nkx2–5 expression in the null heart is consistent with our (Fig. 3C) and other (10) observations demonstrating Nkx2–5 expression in the endothelial/endocardial lineages. No significant change in Bcl2 modified factor (Bmf) expression suggests that downregulation of gene expression in the null heart was Etsrp71 specific. Collectively, these in vivo and in vitro results establish that Etsrp71 is important for the specification of the endocardial/endothelial lineage.
Fig. 3.
Etsrp71 is essential for cardiac morphogenesis and specification of the endocardial/endothelial lineage. (A) Schematic of the Etsrp71 gene and the targeted disruption. (B) (Top) Morphological appearance of E8.5 +/+ and Etsrp71 −/− embryo littermates. Note that the whole-mount WT and Etsrp71 mutant embryos are indistinguishable at E8.5. (Middle) Histological analyses of the WT and null embryo demonstrating an absence of the endocardial/endothelial lineage (End) and vasculature (PHV, primary heart vein; DA, dorsal aorta) in the null embryo compared to WT littermate. (Bottom) Immunohistochemical analysis of α-endomucin in the WT and Etsrp71 null embryo further demonstrating an absence of the endocardial/endothelial lineage (arrowhead shown in WT) and an absence of the vasculature in the null embryo. A higher magnification field reveals an absence of endocardium in the null heart compared to the WT littermate (α-endomucin expressing endocardium indicated by an arrowhead). (C) qRT-PCR analyses using purified RNA from individual Etsrp71 WT (black) and null (white) E8.5 hearts (performed in triplicate). Transcript level in the WT heart was considered to be 1 and Etsrp71 transcript was not detectable in the mutant heart (*). Note the significant downregulation of Flk1 transcripts and the essential absence of Tie2 transcripts in the Etsrp71 mutant heart (**, P < 0.001). Each assay was analyzed in triplicate and repeated twice.
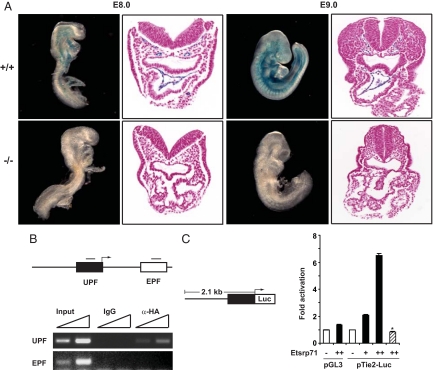
Having observed that the Etsrp71−/− embryo lacks an endocardial/endothelial lineage and is further characterized by the essential absence of the Tie2 transcript, we hypothesized that Tie2 is a direct downstream target of Etsrp71. We mated the Tie2-lacZ transgenic mouse (18) into the Etsrp71 null background. In contrast to the WT littermate controls that demonstrate β-galactosidase expression in the endocardial and endothelial lineages at E8.0 and E9.0, no reporter expression was observed in the Etsrp71 null embryos (Fig. 4A; n = 3 at each stage). These results further support our analysis of the Etsrp71 null embryo (see Fig. 3). Interestingly, embryonic and/or extraembryonic expression of β-galactosidase was not observed in the Etsrp71 null embryo at E7.75 (Fig. S6A). These results demonstrating the loss of Tie2 promoter activity in the absence of Etsrp71 could be a secondary consequence of the loss of endocardial/endothelial specification. Therefore, further studies were undertaken to examine whether Tie2 is a downstream target of Etsrp71. Previous studies have identified critical upstream and intronic modules of the Tie2 gene that direct Tie2 expression to the endothelial/endocardial lineage (18, 23) and both the upstream promoter fragment (UPF) and the intronic enhancer promoter fragment (EPF) harbor several evolutionally conserved Ets-binding elements (EBEs) (Fig. S6B). We used ChIP assays to demonstrate that Etsrp71 has the capacity to bind to the evolutionary conserved EBEs within the UPF but not the EPF of the Tie2 gene in vivo (Fig. 4B). We further undertook transcriptional assays using the upstream promoter fragment to define the specificity of Etsrp71 as a transcriptional regulator of Tie2 gene expression. We observed that Etsrp71 expression resulted in a significant dose-dependent activation of Tie2 gene expression (Fig. 4C) while mutation of the EBEs (Fig. S6B) completely abolished the activation of reporter gene expression (Fig. 4C). These results further support our hypothesis that Etsrp71 is an important transcriptional activator of the Tie2 gene.
Fig. 4.
Etsrp71 transcriptionally activates the Tie2 gene. (A) Tie2-lacZ Tg and Etsrp71± mice were mated to analyze β-galactosidase expression in WT (+/+) and Etsrp71 null (−/−) littermate embryos at the indicated developmental stages. Note the absence of β-galactosidase expression in the Etsrp71 null embryos. (B) Etsrp71 binds to the upstream promoter fragment of the Tie2 gene. Promoter occupancy of Etsrp71 in the upstream promoter fragment (UPF) and enhancer modules of the Tie2 gene was evaluated using a ChIP assay. Note the Etsrp71 binding to the UPF only (not the intronic enhancer of the Tie2 gene). (C) Etsrp71 transactivates Tie2 expression from UPF. Schematic of a 2.1-kb UPF fused to the luciferase (Luc) reporter is shown. Transcriptional assays in C2C12 myoblast cells reveal a dose-dependent induction of luc activity by Etsrp71 (black). Mutation of the Ets-binding site completely abolished the transcriptional activity (hatched) (*, P < 0.001). Each assay was analyzed in triplicate and repeated twice.
Discussion
The overall goal of our study was to decipher Nkx2–5-mediated transcriptional networks that govern cardiac morphogenesis. This study reports 3 important findings. First, using transgenic, transcriptome, and molecular biological technologies, we have identified Etsrp71 as a novel downstream target of Nkx2–5 in cardiac progenitor cells. Second, we have identified Etsrp71 as an essential regulator of the endocardial/endothelial cell fate of the Nkx2–5-expressing multipotent cardiac progenitor cells. Third, we have identified Etsrp71 as a direct upstream regulator of the Tie2 gene.
Using a gene disruption strategy, we and others have shown that embryos lacking Nkx2–5 have perturbed cardiac morphogenesis, which includes an absence of an endocardial cushion and perturbed endocardial development (8, 9). Recent reports revealing Nkx2–5 expression in the endocardium (10) and the endocardial fate of the Nkx2–5-expressing cardiac progenitors (4–6) support the hypothesis that Nkx2–5 is an important regulator of the endothelial/endocardial lineage during cardiogenesis although the molecular mechanisms are unknown. Recent studies in mice (24) and zebrafish (25, 26) have reported that Etsrp71/ER71 is essential for the genesis of endothelial/endocardial lineage in the developing embryos. However, the transcriptional regulation of Etsrp71 was unclear. In the present study, we defined a transcriptional network whereby Nkx2–5 regulates Etsrp71 to specify an endocardial/endothelial fate in the developing embryo. We also recognize that Nkx2–5 is not the only regulator of Etsrp71 gene expression as both the endocardial lineage and Etsrp71 expression were perturbed and decreased, respectively, in the Nkx2–5 null heart, but they were not absent, suggesting that other factors including Nkx family members may also regulate Etsrp71 gene expression in the presence and absence of Nkx2–5. Although transcriptional activation of several Nkx2–5 downstream target genes is essential during cardiogenesis (9, 11–13), the cooperative action of Nkx2–5 with other cellular factors is also essential for the maintenance of gene expression during cardiogenesis and hematopoiesis (27, 28). Therefore, the loss of Etsrp71 promoter activity in the Nkx2–5 null heart could be due to the loss of a cooperative function of Nkx2–5 in the multipotent cardiac progenitor cells. Importantly, our studies provide further support of a progenitor cell population that daughters alternative lineages during heart development.
Other studies support the existence of a bipotential precursor cell (29). Indeed, lineage-tracing studies in the mouse have demonstrated that the endocardium and a population of myocardium develop from common Flk-1+ precursor cells (30). Several other reports support the notion of common precursor cells for paraxial skeletal muscle cells and aortic smooth muscle cells (31), head skeletal muscle and myocardial cells of the outflow tract (32), myocardial and hematopoietic cells (16), and endothelial and vascular smooth muscle cells (33). These studies strongly suggest that a single multipotent stem/progenitor cell contributes to cells of diverse lineages within the heart and other organs. Additionally, lineage-tracing studies using the ES/EB system support the existence of Nkx2–5+/Flk-1+/isl1+ cardiovascular progenitor cells that can daughter all 3 cardiac lineages—myocardial, endothelial, and smooth muscle cells—and hematopoietic lineages (4–6). In addition, a common progenitor for the myocardial and endocardial lineages was also reported in zebrafish (34) and in the chick cell line, QCE-6 (29). While these progenitor cell populations have been identified, the molecular networks that direct the fate of their progeny remain unclear. In the present study, we identified Etsrp71 as a direct downstream target of Nkx2–5.
Our second discovery using gene disruption technologies was that Etrsp71 was important for the specification of the endocardial/endothelial lineage during embryogenesis. Embryonic expression of several other Ets family transcription factors has been shown to be expressed in the endothelial cells but the temporal-spatial expression patterns were much broader (35), suggesting that Etsrp71 might have a more restrictive role than other family members during development. We and others (24) have demonstrated that Etsrp71 is expressed early during development in the endothelial/endocardial lineage and mutant embryos had perturbed heart development and lacked the endocardial/endothelial lineages. However, our study is the first to demonstrate the essential role of an Nkx2–5 downstream target gene in the specification of endocardial/endothelial fate during development.
Targeted disruption of the Tie2 gene resulted in embryonic lethality, severely decreased numbers of endothelial cells, and perturbed cardiac morphogenesis, which further emphasizes the critical interaction between the endocardial/endothelial lineage and cardiac myocytes during development (36). However, the transcriptional regulation of Tie2 within the endothelial lineage was unclear. Our third discovery supports the hypothesis that Tie2 is a direct downstream target of Etsrp71. Our data reveal that Etsrp71 binds to the EBE in the upstream promoter of the Tie2 gene and regulates its expression, which is further supported by studies demonstrating that the deletion (23) or mutation (37) of the EBEs in the upstream promoter sequence significantly abolishes Tie2 promoter activity in vivo. Future studies will define additional Etsrp71 targets and the role they play in establishing the endocardial/endothelial lineage during embryogenesis. Thus, our study has uncovered a previously undescribed transcriptional network of Nkx2–5, which is essential for the specification and genesis of an endocardial/endothelial fate in the developing embryo.
Materials and Methods
Generation of Transgenic Mice.
Etsrp71 transgenic mice were generated as described (16, 20), except that a 3.9-kb upstream region of Etsrp71 was PCR amplified and cloned into the hsp68-LacZ reporter plasmid (38). Methods for identification of transgenic mice were performed as described (16, 20). Generation of Nkx2–5-EYFP transgenic (16), Nkx2–5 heterozygous (8), Tie2-GFP (19), and Tie2-LacZ mice (18) was previously described. All mice were maintained in the animal facility at University of Texas Southwestern Medical Center or University of Minnesota according to the guidelines of Institutional Animal Care and Use Committee and the Animal Resource Center at each institute.
Generation and Analysis of Etsrp71 Chimera Mice.
Etsrp71 chimera mice were generated using the ES cell line (141.1H7), which contains the insertion of the gene-trapping construct (pGep-SD5; International Gene Trap Consortium) at the exon–intron boundary of exon5 of Etsrp71/Etv2. Germline transmission of one copy of the potentially trapped Etsrp71 allele was confirmed by PCR analysis (see SI Materials and Methods).
Morphological Analyses of the Etsrp71−/− Embryo.
All embryos were harvested in ice-cold PBS, fixed in 4% paraformadehyde overnight at 4 °C, and then processed as described (16). Immunostaining for endomucin, an endothelial-specific sialomucin, was performed using rat anti-endomucin antibody (clone V.7C7, gift from Dietmar Vestweber) and detected according to previously published methods (39). Specimens were subsequently dehydrated and cleared before coverslipping with permount.
Promoter Binding and Transcriptional Assays.
EMSAs were performed as described previously (refs. 20 and 21; see SI Materials and Methods). ChIP assays were performed as described (20, 21), with the exception that anti-Nkx2–5 and/or anti-myc and anti-HA sera were used to immunoprecipitate the Nkx2–5-DNA and HA-Etsrp71-DNA complexes to analyze the promoter occupancy, respectively. Transcriptional assays were performed as described with several modifications (refs. 20 and 21; see SI Materials and Methods). Each assay was performed in triplicate and repeated 3 times. Additional methods for plasmids, transcriptome analyses, in situ hybridization, generation of Nkx2–5-inducible ES cell lines, and RT-PCR analyses are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments.
The authors acknowledge Dr. R. Janknecht for providing the Etsrp71 cDNA and John Shelton (University of Texas Southwestern Medical Center) for assistance with the histological analyses. Funding for these studies was from the American Heart Association, GlaxoSmithKline, and the March of Dimes Association. D.J.G. is an Established Investigator of the American Heart Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807583106/DCSupplemental.
References
- 1.Olson EN, Srivastava D. Molecular pathways controlling heart development. Science. 1996;272:671–676. doi: 10.1126/science.272.5262.671. [DOI] [PubMed] [Google Scholar]
- 2.Fishman MC, Chien KR. Fashioning the vertebrate heart: earliest embryonic decisions. Development. 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- 3.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Wu SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 6.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 7.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 8.Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 10.Stanley EG, et al. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2–5. Intl J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 11.Durocher D, Chen CY, Ardati A, Schwartz RJ, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–4655. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou Y, et al. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2–5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]
- 13.Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol Cell Biol. 2003;23:9222–9232. doi: 10.1128/MCB.23.24.9222-9232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown TA, McKnight SL. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez AD, Shi W, Wilson BA, Skeath JB. Pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development. 2003;130:3015–3026. doi: 10.1242/dev.00488. [DOI] [PubMed] [Google Scholar]
- 16.Masino AM, et al. Transcriptional regulation of cardiac progenitor cell populations. Circ Res. 2004;95:389–397. doi: 10.1161/01.RES.0000138302.02691.be. [DOI] [PubMed] [Google Scholar]
- 17.Palmer S, et al. The small muscle-specific protein Csl modifies cell shape and promotes myocyte fusion in an insulin-like growth factor 1-dependent manner. J Cell Biol. 2001;153:985–997. doi: 10.1083/jcb.153.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlaeger TM, et al. Uniform vascular-endothelial-cell-specific gene expression in both embryonic and adult transgenic mice. Proc Natl Acad Sci USA. 1997;94:3058–3063. doi: 10.1073/pnas.94.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motoike T, et al. Universal GFP reporter for the study of vascular development. Genesis. 2000;28:75–81. doi: 10.1002/1526-968x(200010)28:2<75::aid-gene50>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 20.Meeson AP, et al. Sox15 and Fhl3 transcriptionally coactivate Foxk1 and regulate myogenic progenitor cells. EMBO J. 2007;26:1902–1912. doi: 10.1038/sj.emboj.7601635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin CM, et al. Hypoxia-inducible factor-2a transactivates Abcg2 and promotes cytoprotection in cardiac side population cells. Circ Res. 2008;102:1075–1081. doi: 10.1161/CIRCRESAHA.107.161729. [DOI] [PubMed] [Google Scholar]
- 22.Stanford WL, Epp T, Reid T, Rossant J. Gene trapping in embryonic stem cells. Methods Enzymol. 2006;420:136–162. doi: 10.1016/S0076-6879(06)20008-9. [DOI] [PubMed] [Google Scholar]
- 23.Schlaeger TM, Qin Y, Fuziwara Y, Magram J, Sato TN. Vascular endothelial cell lineage-specific promoter in transgenic mice. Development. 1995;121:1089–1098. doi: 10.1242/dev.121.4.1089. [DOI] [PubMed] [Google Scholar]
- 24.Lee D, et al. ER71 acts downstream of MBP, notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sumanas S, et al. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pham VN, et al. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiratori H, et al. Two-step regulation of left-right asymmetric expression of Pitx2:initiated by nodal signaling and maintenance by Nkx2. Mol Cell. 2001;7:137–149. doi: 10.1016/s1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- 28.Han Z, Olson EN. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development. 2005;132:3525–3536. doi: 10.1242/dev.01899. [DOI] [PubMed] [Google Scholar]
- 29.Eisenberg CA, Bader D. QCE-6: a clonal cell line with cardiac myogenic and endothelial cell potentials. Dev Biol. 1995;167:469–481. doi: 10.1006/dbio.1995.1042. [DOI] [PubMed] [Google Scholar]
- 30.Ema M, Takahashi S, Rossant J. Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood. 2006;107:111–117. doi: 10.1182/blood-2005-05-1970. [DOI] [PubMed] [Google Scholar]
- 31.Esner M, et al. Smooth muscle of the dorsal aorta shares a common clonal origin with skeletal muscle of the myotome. Development. 2006;133:737–749. doi: 10.1242/dev.02226. [DOI] [PubMed] [Google Scholar]
- 32.Tirosh-Finkel L, Elhanany H, Rinon A, Tzahor E. Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development. 2006;133:1943–1953. doi: 10.1242/dev.02365. [DOI] [PubMed] [Google Scholar]
- 33.Ema M, et al. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 2003;17:380–393. doi: 10.1101/gad.1049803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee RK, Stainier DY, Weinstein BM, Fishman MC. Cardiovascular development in the zebrafish. II. Endocardial progenitors are sequestered within the heart field. Development. 1994;120:3361–3366. doi: 10.1242/dev.120.12.3361. [DOI] [PubMed] [Google Scholar]
- 35.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 36.Dumont DJ, et al. Dominant-negative and targeted null mutations in the endothelial receptor tyrosine kinase, tek, reveal a critical role in vasculogenesis of the embryo. Genes Dev. 1994;8:1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- 37.Minami T, et al. Ets motifs are necessary for endothelial cell-specific expression of a 723-bp Tie-2 promoter/enhancer in Hprt targeted transgenic mice. Arterioscler Thromb Vasc Biol. 2003;23:2041–2047. doi: 10.1161/01.ATV.0000089326.63053.9A. [DOI] [PubMed] [Google Scholar]
- 38.Kothary R, et al. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 39.Brachtendorf G, et al. Early expression of endomucin on endothelium of the mouse embryo and on putative hematopoietic clusters in the dorsal aorta. Dev Dyn. 2001;222:410–419. doi: 10.1002/dvdy.1199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.