Abstract
Elucidation of the early events in amyloidogenesis is key to understanding the pathology of, and developing therapies for, amyloid diseases. Critical informants about these early events are amyloid assembly proteins that facilitate the transition from monomer to amyloid fiber. Curli are a functional amyloid whose in vivo polymerization requires a dedicated nucleator protein, CsgB, and an assembly protein, CsgF. Here we demonstrate that without CsgF, curli subunits are released from the cell into the media and are inefficiently polymerized, resulting in fewer and mislocalized curli fibers. CsgF is secreted to the cell surface, where it mediates the cell-association and protease-resistance of the CsgB nucleator, suggesting that CsgF is required for specific localization and/or chaperoning of CsgB for full nucleator activity. CsgF is thus critical to achieve localized and efficient nucleation of fiber subunits into functional, cell-associated amyloid.
Keywords: Escherichia coli, fiber biogenesis, extracellular matrix
Amyloids were originally identified as aberrantly folded, disease-causing proteins. However, the list of functional amyloids has grown in recent years to include bacterial (1, 2), fungal (3), arachnid (4), fish (5), and mammalian proteins (6). Among these are curli, a unique class of extracellular fimbriae produced by Escherichia coli and Salmonella spp (1, 7). Curli are an important component of the extracellular matrix in biofilms, mediating initial attachment to surfaces, cell-cell contacts, and formation of three-dimensional, mature biofilms (8–10). Curli also provide significant protection against environmental stresses, such as desiccation and antimicrobial agents (11, 12).
In addition to their roles in bacterial physiology, curli are also a powerful molecular model to study mechanisms of directed amyloid formation. Curli fibers are composed of 2 subunits, CsgA and CsgB, each of which contain a 5-fold imperfect glutamine/asparagine-rich repeat motif (R1–R5) and display the biochemical properties of amyloid (1, 13, 14). Polymerization of purified CsgA, the major curli subunit, exhibits nucleation-dependent kinetics and includes a transient structural intermediate common during the polymerization of disease-related amyloids (15). Thus, CsgA polymerization in vitro occurs unassisted, perhaps via a folding pathway that is common among many amyloids. In vivo however, CsgA requires additional molecular machinery for its efficient and directed assembly at the bacterial cell surface (1). CsgG is an oligomeric lipoprotein located in the outer membrane that is responsible for stabilizing and exporting both the major subunit, CsgA, and the minor subunit, CsgB, to the surface of the cell (16, 17). Once secreted outside the cell, CsgB initiates CsgA polymerization via an extracellular nucleation-polymerization pathway, and both subunits are incorporated into the resulting amyloid fiber (18). In the absence of CsgB, CsgA is secreted to the extracellular milieu as a soluble protein and does not polymerize into fibers (18, 19). CsgE and CsgF are chaperone-like proteins with no significant homology to other proteins in the National Center for Biotechnology Information nonredundant database. Both CsgE and CsgF are required for efficient curli assembly, and have been shown to interact with CsgG at the outer membrane, yet little is known about the specific roles of these proteins in curli biogenesis (1, 16).
Previous work suggesting a role for CsgF in the nucleation of curli fibers was based on the phenotype of the csgF mutant. In the absence of CsgF, CsgA is largely secreted away from the cell, where much of it remains in an SDS-soluble state, and very little CsgA polymerizes into cell-associated curli fibers (1). In an interbacterial complementation assay, in which a donor strain (e.g., a csgB mutant) secretes soluble CsgA that is polymerized on the surface of an acceptor strain (e.g., a csgA mutant) when they are grown in close proximity, a csgF mutant is able to donate CsgA to an acceptor strain, but it is unable to assemble CsgA provided by a donor strain (1). This indicates that the defect in the csgF strain is in the nucleation step, and that CsgF is potentially interacting with or facilitating the nucleator function of CsgB.
As the process of amyloid nucleation remains poorly understood, the dedicated nucleator and assembly proteins of the curli system represent a unique opportunity to determine the molecular mechanisms required to initiate amyloid polymerization. In this study, we describe the role of CsgF in bacterial amyloid nucleation. We show that CsgF, like both curli subunits, depends on CsgG and CsgE for stability and is secreted across the outer membrane. Furthermore, CsgF plays a critical role in proper localization of the nucleator protein, CsgB, and is required for efficient and properly localized fiber formation. We also show that CsgF mediates CsgB protease-resistance, and that this correlates with nucleator function. Our work indicates that CsgF and CsgB act cooperatively on the surface of the outer membrane to initiate curli subunit polymerization near the cell surface, and provides new insights into mechanisms of functional amyloid biogenesis.
Results
Whole-Cell CsgF Levels Are Dependent on CsgG and CsgE.
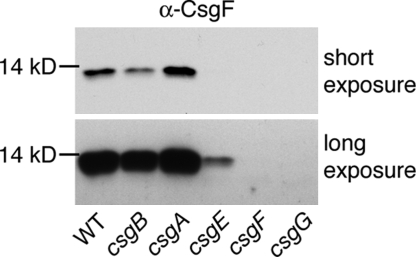
Whole-cell CsgF protein levels were examined by immunoblot analysis in the wild-type E. coli strain MC4100 and each of the following curli mutant strains: csgB, csgA, csgE, csgF, csgG [Table S1]. All strains were grown under curli-inducing conditions (i.e., 48 h on YESCA agar at 26 °C). The csgB and csgA mutants showed minor, but reproducible, differences in whole-cell CsgF levels compared to wild type. The csgB strain had less total CsgF protein than wild type, while the csgA strain had more CsgF protein than wild type (Fig. 1, short exposure). More dramatic differences were observed in the csgE and csgG mutants. In the csgE strain, very little CsgF protein was detected and none was observed in the csgG strain (see Fig. 1, long exposure). No CsgF protein was detected in the negative control csgF strain.
Fig. 1.
CsgF steady-state protein levels. Whole cells of wild type (WT, MC4100), csgB (MHR261), csgA (LSR10), csgE (MHR480), csgF (MHR592), and csgG (LSR1) were analyzed by α-CsgF immunoblot. A long exposure is included to visualize faint bands.
CsgF Localization.
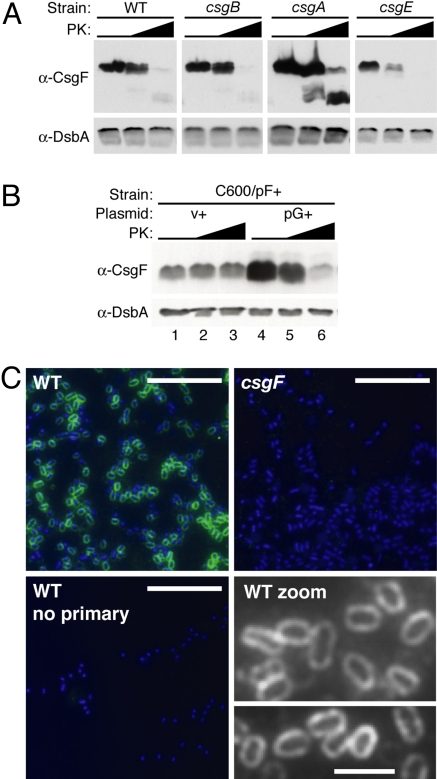
CsgF localizes to the outer membrane where it directly interacts with CsgG, the outer membrane secretion conduit for curli subunit proteins (16, 17, 20). To determine if CsgF is also secreted to the cell surface, whole cells grown under curli-inducing conditions were incubated with increasing concentrations of proteinase K and analyzed by immunoblot. CsgF was degraded with increasing proteinase K treatment in wild-type cells, suggesting that CsgF was exposed on the cell surface in this strain (Fig. 2A, α-CsgF). The periplasmic chaperone protein, DsbA, was not degraded, demonstrating that intracellular proteins were not accessible to proteinase K during the whole-cell protease experiments (see Fig. 2A, α-DsbA). Similar to wild-type cells, CsgF was also protease-sensitive in the csgB, csgA, and csgE mutants, showing that surface localization of CsgF was not dependent on the presence of these curli proteins or on the secretion or assembly of curli subunits.
Fig. 2.
CsgF localization. (A) Whole cells of wild type (WT, MC4100), csgB (MHR261), csgA (LSR10), and csgE (MHR480) were collected after 48 h of growth on YESCA agar. Whole cells were treated with 0, 5, or 50 μg/ml PK, and then analyzed by α-CsgF immunoblot. A long exposure of the csgE blot is shown to visualize faint bands. (B) C600/pF+/v+ (C600/pLR75/ptrc99a) and C600/pF+/pG+ (C600/pLR75/pMC1) whole cells were treated with 0, 10, or 200 μg/ml PK and analyzed by α-CsgF immunoblot. α-DsbA immunoblots were used in (A) and (B) as a positive control for cell integrity. (C) Whole cells of wild type (WT, MC4100) and csgF (MHR592) probed with rabbit polyclonal α-CsgF antibody or PBS (no primary), then FITC conjugated goat α-rabbit antibody. Each image is an overlay of the FITC channel (green, CsgF) and the DAPI channel (blue, DNA, Hoechst dye), except for “WT zoom,” where images are of the FITC (CsgF) channel only, viewed in greyscale. PK, proteinase K; v, empty matched vector; “+” indicates an over-expression vector. (Scale bars: 10 μm in all panels, except in “WT zoom,” where scale bar: 2 μm.)
CsgF protein is completely undetectable in the csgG mutant of the MC4100 strain when grown under typical, curli-inducing conditions (48 h on YESCA agar at 26 °C) (see Fig. 1). However, we found that CsgF was detectable when it was over-expressed from an arabinose-inducible promoter (pF+) during growth in LB broth in the E. coli strain C600 (Fig. 2B). Thus, the dependence of CsgF localization on CsgG was tested in C600/pF+ cells in the presence and absence of CsgG expression from an isopropyl-fl-D-thiogalactopyranoside (IPTG)-inducible plasmid (pG+). After growth and induction, whole cells were subjected to proteinase K treatment and analyzed by α-CsgF immunoblot. Under these conditions, CsgF was not accessible to proteinase K digestion in the absence of CsgG (see Fig. 2B, Lanes 1–3). Only in the presence of CsgG was CsgF proteinase K sensitive, demonstrating that CsgG was required for cell-surface localization of CsgF (see Fig. 2B, lanes 4–6). Interestingly, CsgF protein was produced at low steady-state levels without CsgG expression, and at much higher steady-state levels with CsgG expression (see Fig. 2B, compare lanes 1 and 4). The expression of CsgF was driven by an inducible promoter in both cases, indicating that the decrease in CsgF protein levels in the absence of CsgG was likely the result of a decrease in CsgF protein stability and not because of a change in CsgF expression. This argues that CsgG contributes to CsgF protein stability.
In addition, CsgF was localized on whole cells with an α-CsgF antibody and visualized by immunofluorescent microscopy (IFM). In wild-type cells, CsgF staining was unevenly distributed around the circumference of the cell, as opposed to polar or discrete punctate staining (Fig. 2C WT and WT zoom). Similar staining patterns were observed on cells deficient in fiber formation (Fig. S1). No staining was observed in the csgF mutant, or in a no primary antibody control (see Fig. 2C, csgF, WT no primary).
Curli Fibers and Subunits Are Mislocalized in the Absence of CsgF.
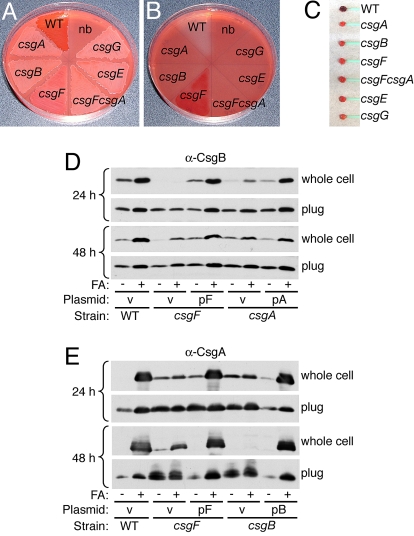
Congo red indicator plates can provide a measure of curli fiber formation, where wild-type curli-producing strains stain bright red and noncurli-producing strains remain white (19). A csgF mutant exhibits an intermediate light pink phenotype on a Congo red indicator plate (1). The pink coloring of a csgF mutant can be attributed to polymerization of CsgA, as a csgFcsgA double mutant appeared white on a Congo red plate (Fig. 3A). Removal of curli-producing wild-type bacteria from a Congo red indicator plate revealed a clearing effect in the agar because of uptake of the Congo red dye by curli fibers produced within the bacterial lawn (Fig. 3B), as described previously (13). In contrast to wild type, removing csgF bacteria from a Congo red indicator plate revealed distinct Congo red staining in the agar (see Fig. 3B), a phenotype that could be complemented by expression of CsgF from a plasmid (Fig. S2). No change in agar color was observed beneath any of the other curli mutant strains (see Fig. 3B, compare csgA, csgB, csgFcsgA, csgE, csgG to nb). Additionally, whole csgF cells removed from the Congo red agar plates were as deficient in red coloring as all of the other curli mutant strains (Fig. 3C). Thus, the pink coloring observed when the csgF mutant strain was grown on Congo red agar was because of CsgA-dependent Congo red staining within the agar. Congo red staining of the agar suggests that most of the curli fibers produced in the csgF strain were mislocalized to the agar and were not cell-associated.
Fig. 3.
Curli fibers and subunits are mislocalized in the absence of CsgF. (A–C) Congo red supplemented YESCA agar plate containing wild type (WT, MC4100), csgA (LSR10), csgB (MHR261), csgF (MHR592), csgFcsgA (NDH58), csgE (MHR480), and csgG (LSR1) after 48 h of growth (A) and the same plate after bacterial lawns were removed (B). One section of the plate contained no bacterial lawn (nb) as a color control. Bacterial cells (C) removed from the agar plate in (A). (D and E) Whole cells and plugs of wild type/v (MC4100/pLR1), csgF/v (MHR592/pLR1), csgF/pF (MHR592/pLR73), csgA/v (LSR10/pLR1), csgA/pA (LSR10/pLR5), csgB/v (MHR261/pLR1) and csgB/pB (MHR261/pLR8) strains were collected after 24 and 48 h of growth on YESCA agar and either directly resuspended in SDS sample buffer or pretreated with formic acid (FA). Samples were then analyzed by α-CsgB (D) or α-CsgA (E) immunoblot. v, empty matched vector. [See Fig. S2 for Congo red YESCA plate of the strains used in (D) and (E)]
In wild-type cells, curli remain cell-associated when cells are collected from YESCA agar plates after 48 h of growth. After treatment with formic acid to depolymerize curli, both curli subunits (CsgB and CsgA) can be detected in the whole-cell fraction by immunoblot (17–19). csgA mutant bacteria do not form curli fibers; however, this strain still produces cell-surface localized, nucleation-competent CsgB protein after 48 h of growth (17, 19). We investigated whether CsgB was cell-associated in the csgF strain, at both 48 h and at an earlier stage of growth, 24 h. Both the wild-type and csgA mutant strains contain CsgB protein in the whole-cell fraction at 24 and 48 h of growth (Fig. 3 D and E). In contrast, little-to-no CsgB was detected in the whole-cell fraction of the csgF mutant after 24 h (see Fig. 3D). CsgB protein was detectable in plug samples (whole cells and underlying agar) of the csgF mutant at 24 h, demonstrating that CsgB was stable, but secreted away from csgF cells into the agar media at earlier stages of growth (see Fig. 3D). By 48 h, CsgB was detected in the whole-cell fraction of csgF bacteria after formic acid treatment (see Fig. 3D). To determine if the cell-associated CsgB protein found in csgF bacteria at 48 h was localized on the cell surface where is it proposed to act (17–19), we performed a whole-cell proteinase K assay. In all strains tested, CsgB was found to be completely sensitive to proteinase K (see Fig. S2). Thus, we conclude that small amounts of CsgB become surface-exposed in the csgF mutant at 48 h. However, the surface-localized CsgB in the csgF mutant is unable to serve as an acceptor in the interbacterial complementation assay (1), and thus is deficient in nucleation activity. In contrast, in the csgA mutant, CsgB was cell-associated and surface-localized at 24 and 48 h, and can serve as an acceptor of secreted CsgA in an interbacterial complementation assay (17, 19), and is therefore nucleation-competent.
We went on to determine the localization of the major curli subunit, CsgA, in the csgF mutant. For the wild-type and complemented strains, CsgA was almost completely SDS-insoluble in the whole-cell fractions after 24 h of growth, indicating most of the CsgA in these strains was already polymerized into a curli fiber at this time (see Fig. 3E). In contrast, low levels of SDS-soluble CsgA were detectable in the whole-cell fraction of the csgF strain at 24 h. This was similar to the low levels of SDS-soluble CsgA present in the nucleator-deficient csgB strain, indicating that some unpolymerized CsgA remains cell-associated at early stages of curli production, even in the absence of the curli nucleator, CsgB (see Fig. 3E). By 48 h, CsgA was no longer detected in the whole-cell fraction of the csgB strain, as shown previously (19), and only a small amount of CsgA was found in the whole-cell fraction of the csgF strain (see Fig. 3E), consistent with previous findings that only a small amount of curli fibers are detected in a whole-cell fraction of this strain by electron microscopy (1). The majority of CsgA produced by the csgF strain was located in the agar plug sample at 48 h, indicating that CsgA produced in the absence of CsgF predominantly localizes to the underlying agar, where Congo red staining was detected (see Fig. 3 B and E). Furthermore, in contrast to wild-type or complemented strains, some of the cell-associated CsgA produced by the csgF mutant remained SDS-soluble, indicating that this protein had not polymerized into a curli fiber (see Fig. 3E). Thus, a greater proportion of SDS soluble CsgA is found in both the whole-cell and plug samples in a csgF mutant compared to wild type (see Fig. 3E) (1). Taken together, these data show that in the absence of CsgF, there is a defect in CsgB cell-association at an early stage of growth, resulting in CsgA polymerization that is both incomplete and mislocalized by later stages of biogenesis.
Complex Role of CsgF.
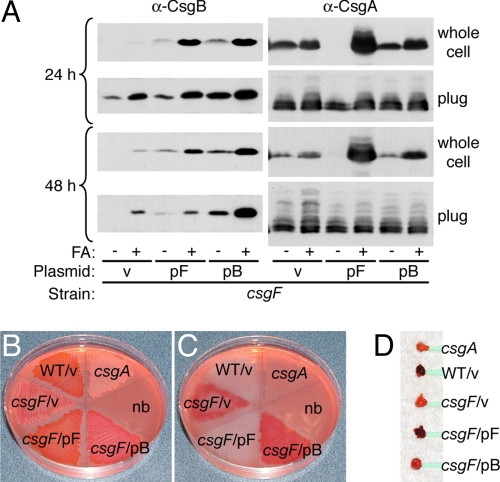
To address the possibility that the function of CsgF in promoting CsgB cell-association was coupled to an additional activity that facilitates formation of a functional CsgB nucleator, we sought to rescue the CsgB cell-association defect in the csgF mutant and look for complementation of fiber formation. We found that over-expression of CsgB in the csgF mutant strain restored whole-cell CsgB protein levels back to that of wild type after both 24 and 48 h of growth (Fig. 4A). As before, this CsgB protein was determined to be cell-surface localized by whole-cell proteinase K assay where it is thought to nucleate CsgA (data not shown). However, rescuing the CsgB cell-association defect did not restore wild-type curli formation in the csgF strain. Similar to the vector control, very little CsgA was observed in the whole-cell fraction of the csgF strain expressing CsgB from a plasmid, indicating that CsgA was still largely mislocalized to the agar media in this strain (Fig. 4B). Furthermore, this strain only exhibited Congo red staining of the agar media and not of bacterial cells, indicating curli fibers were mislocalized to the agar media as they are in the csgF mutant (Fig. 4 B–D). CsgB expressed from the same pB plasmid in the wild-type MC4100 strain still exhibited a wild-type Congo red binding phenotype (i.e., bright red cells and clearing of underlying agar), indicating that increased CsgB expression was not somehow inhibiting CsgA cell-surface polymerization (data not shown). Therefore, the overall defect in curli fiber formation was not rescued despite restoration of CsgB cell-association to the csgF strain at both time points. Thus, while CsgF plays a role in CsgB cell-association (see Fig. 3D), it is not absolutely required for CsgB cell-association (see Fig. 4A) and must play an additional or more specific role in curli biogenesis.
Fig. 4.
Restoring CsgB cell-association in the csgF strain does not restore wild-type fiber formation. (A) Whole cells and plugs of csgF/v (MHR592/pLR1), csgF/pF (MHR592/pLR73) and csgF/pB (MHR592/pLR8) strains were collected after 24 and 48 h of growth on YESCA agar, treated with or without FA, and analyzed by α-CsgB and α-CsgA immunoblot. v, empty matched vector. (B–D) Congo red supplemented YESCA agar plate containing the same strains in (A) after 48 h of growth (B), and the same plate after bacterial lawns were removed (C). Bacterial cells (D) removed from the agar plate in (B). csgA (LSR10) and no bacteria (nb) are included as negative controls.
CsgF Influences the Protease Resistance of CsgB.
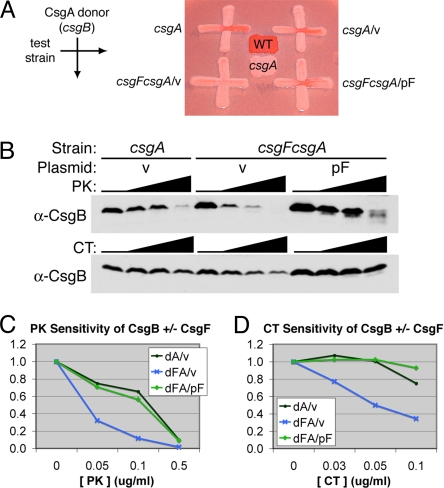
Our data indicate that CsgF mediates a modification of CsgB that is required for nucleation activity. Thus, we investigated the degree of protease resistance of CsgB in the presence and absence of CsgF expression. The following strains were examined: the csgA mutant harboring an empty vector (csgA/v, F+), the csgFcsgA double mutant harboring an empty vector (csgFcsgA/v, F–), and the csgFcsgA double mutant expressing CsgF from a plasmid (csgFcsgA/pF, F+). The csgA mutant background was chosen to eliminate protease resistance of CsgB because of incorporation into curli fibers. All 3 strains were able to produce cell-surface-localized CsgB, as determined by whole-cell protease assays (Fig. S3). The nucleation activity of CsgB produced by each of these strains was assessed by interbacterial complementation. Both F+ strains were able to polymerize exogenously provided CsgA from the donor strain (csgB mutant), indicating these strains were producing nucleation-competent CsgB (Fig. 5A, Right). In contrast, the F– strain did not polymerize CsgA from the donor strain, indicating a lack of nucleation-competent CsgB (see Fig. 5A, Lower Left). We then analyzed these strains for the level of CsgB protease resistance, using 2 proteases: proteinase K and chymotrypsin. Whole cells were collected from YESCA agar after 48 h of growth, then exposed to low levels of extracellular proteases and analyzed by α-CsgB immunoblot. A representative experiment is shown (Fig. 5B). To normalize the initial CsgB levels between the 3 strains, the National Institutes of Health (NIH) ImageJ program was used to calculate the intensity of the CsgB band at each treatment. The calculated intensities were then plotted as a percentage of the CsgB band intensity at 0-μg/ml protease treatment for each strain. Only the full-length CsgB band and not proteolytic fragment bands were included in this measurement. The graphs in Fig. 5 C and D represent the ImageJ calculations for the blots shown in Fig. 5B. After treatment with 0.05 μg/ml proteinase K, the F+ strains retained >70% of total CsgB, while the F– strain retained only one-third of total CsgB; a similar disparity was observed at the 0.1-μg/ml proteinase K treatment (see Fig. 5C). Chymotrypsin-treated cells exhibited the same trend, with minimal-to-no CsgB degradation observed at 0.05 and 0.1 μg/ml chymotrypsin in the F+ strains, while ≤50% of total CsgB remained in the F– strain at these protease concentrations (see Fig. 5D). Thus, both strains expressing CsgF produced nucleation-competent CsgB that was more resistant to proteolytic degradation than nucleation-inactive CsgB produced in the absence of CsgF. These data suggest that CsgF is mediating the increased protease resistance of CsgB, perhaps via a direct protein-protein interaction or by facilitating the conversion of CsgB into a more protease-resistant conformation.
Fig. 5.
CsgF enhances the protease resistance of CsgB. (A) Interbacterial complementation. The CsgA donor strain (csgB, MHR261) was streaked from top to bottom. The positive control for an acceptor strain, csgA (LSR10), is shown (Top Left). The following test strains were streaked from left to right: csgA/v (LSR10/pLR1), csgFcsgA/v (NDH58/pLR1), and csgFcsgA/pcsgF (NDH58/pLR73). Wild type (MC4100) and csgA (LSR10) are included as a positive and negative control for Congo red binding, respectively. (B–D) Whole cells of the test strains in (A) were collected after 48 h of growth on YESCA agar and treated with 0, 0.05, 0.1, or 0.5 μg/ml of proteinase K (PK) or 0, 0.03, 0.05 or 0.1 μg/ml chymotrypsin (CT), then analyzed by α-CsgB immunoblot (B). For the immunoblots shown in (B), ImageJ (NIH) was used to calculate CsgB band intensity at each treatment: proteinase K (C); chymotrypsin (D). For each strain, the band intensity at 0 μg/ml protease was set to 100%, or total CsgB, and the band intensity in protease-treated samples is plotted as a percentage of the total. Immunoblots shown are a representative trend of 3 experiments. v, empty matched vector.
Discussion
The molecular mechanisms that occur during the initial stages of amyloid fiber formation are still not completely understood. Amyloid assembly proteins that facilitate the transition from amyloid precursor to amyloid fiber represent molecular tools that can be used to understand and manipulate amyloid-folding pathways. The functional amyloid curli requires at least 2 assembly proteins for efficient and localized nucleation of an amyloidogenic monomer. We have investigated the role of one of these proteins, CsgF. Previous models of curli biogenesis assumed that CsgF was a periplasmic protein and did not provide a function for CsgF in CsgA polymerization (16, 17, 20, 21). We have discovered that CsgF is localized on the outer surface of the outer membrane, where it influences the localization, protease resistance, and activity of the CsgB nucleator. Our data are consistent with a model in which CsgF and CsgB act cooperatively to achieve CsgA polymerization near the cell surface. In the absence of CsgF, CsgB is secreted away from cells to the agar medium at early time points. Because of mislocalization of CsgB, CsgA escapes directed nucleation at the cell surface and is also secreted to the agar medium. Some polymerization of CsgA occurs within the agar over time, likely as a result of incidental CsgB-CsgA interactions as the subunits randomly diffuse through the agar. These CsgB-CsgA interactions occur at a lower frequency and in the wrong location, resulting in fewer and mislocalized curli fibers. Mislocalized curli fibers are less likely to function properly, (i.e., mediate cell-cell contacts within a biofilm) or provide protection for bacteria in harsh environments. Thus, CsgF determines where and how well curli are made, and is an essential component of a dedicated machinery to produce a functional amyloid.
A Chaperone-Like Role for CsgF.
A recent study showed that a C-terminally truncated version of CsgB (CsgBtrunc) exhibited a phenotype similar to that of the csgF strain, including CsgA and CsgBtrunc secretion to the agar media (13). The investigators proposed that the CsgB C terminus may act as an outer-membrane anchor, aiding the nucleation process by optimally localizing CsgB on the cell surface, near the highest concentrations of CsgA. Similarly, our data show that at least part of the role of CsgF in curli nucleation is enhancement of CsgB cell-association (see Fig. 3). However, results from additional genetic manipulations of the curli machinery suggest that the mechanism of CsgF function may be more involved than simple membrane anchoring of CsgB. Restoration of CsgB localization to the cell surface in the csgF strain was not sufficient to restore wild-type curli assembly (see Fig. 4). One hypothesis is that while over-expression of CsgB in the csgF mutant achieved an increase in general CsgB cell-association, CsgB may require a specific CsgF-directed localization for full functionality. An alternative hypothesis is that CsgF has chaperone-like activity, required for generation of fully active CsgB. CsgB has been shown to have an amyloid-like domain and is proposed to provide a folding template to drive the conversion of soluble CsgA into an amyloid fiber by interacting with sequence specific “CsgB-responsive domains” present in CsgA (13, 22, 23). The active CsgB nucleus is likely in an amyloid-like form, with amyloid properties such as protease resistance. We have shown that CsgF mediates CsgB protease resistance, and that this correlates with the ability to nucleate CsgA (see Fig. 5). Similar to the templating of CsgA by CsgB, CsgF may guide the folding or oligomerization of CsgB into an active nucleus with an amyloid-like conformation at the cell surface, thereby promoting CsgA nucleation immediately after CsgA has been secreted. Specific localization and structural guidance of CsgB are not mutually exclusive roles for CsgF, and both would entail a direct protein-protein interaction. It is interesting to note that the primary amino acid sequence of CsgF contains a high percentage of glutamine (Q) and asparagine (N) residues, similar to that of the two curli subunit proteins (9.2% Q and 13.4% N of the mature CsgF protein; 11.5% Q and 9.2% N of mature CsgB; 8.4% Q and 12.2% N of mature CsgA). Glutamine and asparagine residues have been shown to contribute to the amyloidogenicity of a protein, which is supportive of the idea that CsgF might be promoting the formation of amyloid structure in CsgB (24–26). Further experiments investigating the potential CsgB-CsgF interaction, as well as experiments to assess the effect of CsgF on polymerization of curli subunits in an in vitro assay, will help determine a more detailed mechanism of CsgF function and its potential chaperone activity.
Concerted Secretion of CsgA, CsgB, and CsgF.
In addition to facilitating fiber nucleation at the cell surface, CsgF may also modulate curli biogenesis at an earlier step. In the curli system, CsgA and CsgB secretion and stability are linked and depend on CsgG (16, 17). We demonstrated that CsgF secretion and stability are also CsgG-dependent (see Figs. 1 and 2B). Thus, CsgG is responsible for the stability and outer membrane secretion of 3 curli proteins, CsgB, CsgA, and CsgF. We also observed minor but reproducible variations in CsgF protein levels in the csgB and csgA strains (see Fig. 1). These differences are compatible with the notion that because all 3 curli proteins are secreted across the outer membrane in a CsgG-dependent manner, they potentially compete with or facilitate each other's secretion. Removal or modification of one secreted protein from the system could alter the secretion of the remaining proteins and manifest as changes in protein stability. Concerted secretion of CsgA, CsgB, and CsgF via CsgG may be a control mechanism to orchestrate optimal fiber biogenesis at the cell surface. Furthermore, the extracellular nucleation-precipitation model of curli biogenesis does not exclude the possibility that subunit interactions initiate fiber formation just before, or during, secretion across the outer membrane. It is possible that CsgF begins influencing CsgB structure or nucleator activity through interactions occurring before or during secretion.
Amyloid Chaperones.
There is precedent for proteins with amyloid chaperoning activity in other systems. ApoE is a lipid transport protein that has been implicated in the pathogenesis of Alzheimer's disease and cerebral amyloid angiopathy through interactions with amyloid-β (Aβ) pathogenic peptides (27–29). ApoE has been shown to bind both soluble forms of Aβ as well as amyloid deposits derived from many amyloid associated diseases; ApoE has also been shown to alter Aβ fibrillogenesis in in vitro polymerization experiments as well as in vivo mouse models of Alzheimer's disease and cerebral amyloid angiopathy (28, 30). It has been proposed that ApoE acts as a chaperone to Aβ peptides, influencing their transport and clearance, location of fibrillogenesis and level of deposition (27, 29, 31). It is interesting that CsgF seems to play an analogous role to ApoE by affecting the location and degree of curli subunit polymerization. Proteins that determine when and where amyloid is formed are key informants about amyloid-folding pathways and may provide us with ways in which to manipulate these pathways toward therapeutics for amyloid deposition diseases.
Amyloid chaperones have also been identified in functional amyloid systems. Sis-1, an Hsp40 chaperone, was found to protect yeast cells from toxicity because of Rnq1 over-expression by directly facilitating the conversion of unassembled Rnq1 into [RNQ+] amyloid, reducing the cytoplasmic pool of cytotoxic Rnq1 conformers (32). Numerous studies have implicated soluble protein oligomers and protofibrils, not amyloid fibers, as the primary cytotoxic agents responsible for the disease progression of amyloid-associated disorders (33–35). Yet, bacteria and yeast harbor the machinery to produce amyloid fibers in a timely and efficient manner, without cytotoxic consequences. Similar to the folding requirements of globular proteins, functional amyloid formation requires chaperone-like assembly proteins to direct proper localization and efficient folding into the native fiber product, eliminating intermediate complexes that may be harmful to the cell. Further study of the molecular mechanisms of these functional amyloid assembly proteins will offer a more detailed understanding of amyloid-folding pathways.
Materials and Methods
Strains, Plasmids, and Growth Conditions.
Strains and plasmids used in the present study are described in Table S1; primer sequences are provided in Table S2 (See also SI Methods). To induce curli expression, strains were grown on YESCA agar (19) for 24 or 48 h at 26 °C. For plug samples, bacteria were grown on 1- to 2-mm thick YESCA agar for 24 or 48 h at 26 °C. C600 harboring pLR75 and ptrc99a or pLR75 and pMC1 were grown in LB shaking cultures with 100-μg/ml ampicillin and 20-μg/ml chloramphenicol to OD600 0.8, induced with 0.1% arabinose and 0.2 mM IPTG for 1 h.
Immunoblot Analysis.
Whole cell and plug immunoblot analysis was performed essentially as described (1) (See also SI Methods).
Whole Cell Protease Assay.
Intact cells were scraped from YESCA agar or collected from broth culture and normalized by OD600 as described in SI Methods. Two optical density units of each strain was brought to 270 μl with 50-mM Tris-HCl pH 7.5, 5-mM CaCl2 (see Fig. 2 and Fig. S2) or PBS (see Fig. 5 and Fig. S3); 30 μl of water or a 10× protease stock in water was then added and vortexed to begin the reaction. After incubation for 2 h at 37 °C (see Fig. 2 and Fig. S2) or 20 min at RT (see Fig. 5 and Fig. S3), the reaction was quenched with 2-mM PMSF. Cells were pelleted, resuspended in SDS loading buffer, and analyzed by immunoblot. For α-CsgB immunoblots, cell pellets were pretreated with FA.
Immunofluorescence Microscopy.
IFM was performed essentially as described (36) (See also SI Methods).
Supplementary Material
Acknowledgments.
We thank Neal Hammer (University of Michigan) for supplying strains and reagents; Jennifer Elam and Jerry Pinkner of the Hultgren Laboratory (Washington University) for assistance in producing the CsgF antibody; Partho Ghosh (University of California at San Diego) for kindly supplying the pET28 expression plasmid; and Wandy Beatty and Darcy Gill of the Molecular Microbiology Imaging Facility (Washington University) for IFM technical advice. We would also like to thank Karen Dodson, Neal Hammer, Swaine Chen, and Molly Ingersoll for helpful discussions and review of this manuscript. This work was supported by National Institutes of Health Grant A148689, National Institutes of Health T32 GM07067 Training Program in Cell and Molecular Biology (to A.A.N.), and by the Lucille P. Markey Fellowship (to A.A.N.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences in this paper have been deposited in the GenBank database (accession nos. EU199782 and EU199783).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812143106/DCSupplemental.
References
- 1.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elliot MA, et al. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 2003;17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 4.Kenney JM, Knight D, Wise MJ, Vollrath F. Amyloidogenic nature of spider silk. Eur J Biochem. 2002;269:4159–4163. doi: 10.1046/j.1432-1033.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 5.Podrabsky JE, Carpenter JF, Hand SC. Survival of water stress in annual fish embryos: dehydration avoidance and egg envelope amyloid fibers. Am J Physiol Regul Integr Comp Physiol. 2001;280:R123–R131. doi: 10.1152/ajpregu.2001.280.1.R123. [DOI] [PubMed] [Google Scholar]
- 6.Fowler DM, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4(e6) doi: 10.1371/journal.pbio.0040006. 0100–0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Austin JW, Sanders G, Kay WW, Collinson SK. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol Lett. 1998;162:295–301. doi: 10.1111/j.1574-6968.1998.tb13012.x. [DOI] [PubMed] [Google Scholar]
- 9.Vidal O, et al. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 11.Ryu JH, Beuchat LR. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl Environ Microbiol. 2005;71:247–254. doi: 10.1128/AEM.71.1.247-254.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White AP, Gibson DL, Kim W, Kay WW, Surette MG. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J Bacteriol. 2006;188:3219–3227. doi: 10.1128/JB.188.9.3219-3227.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci USA. 2007;104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collinson SK, Parker JM, Hodges RS, Kay WW. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J Mol Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26(1):11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 18.Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marani P, et al. New Escherichia coli outer membrane proteins identified through prediction and experimental verification. Protein Sci. 2006;15:884–889. doi: 10.1110/ps.051889506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J Mol Biol. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283:21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulson HL, et al. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 25.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 26.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 27.Bales KR, Dodart JC, DeMattos RB, Holtzman DM, Paul SM. Apolipoprotein E, amyloid, and Alzheimer disease. Mol Interv. 2002;2:363–375. doi: 10.1124/mi.2.6.363. [DOI] [PubMed] [Google Scholar]
- 28.DeMattos RB. Apolipoprotein E dose-dependent modulation of beta-amyloid deposition in a transgenic mouse model of Alzheimer's disease. J Mol Neurosci. 2004;23:255–262. doi: 10.1385/JMN:23:3:255. [DOI] [PubMed] [Google Scholar]
- 29.Holtzman DM. Role of apoe/Abeta interactions in the pathogenesis of Alzheimer's disease and cerebral amyloid angiopathy. J Mol Neurosci. 2001;17:147–155. doi: 10.1385/JMN:17:2:147. [DOI] [PubMed] [Google Scholar]
- 30.Fryer JD, et al. Apolipoprotein E markedly facilitates age-dependent cerebral amyloid angiopathy and spontaneous hemorrhage in amyloid precursor protein transgenic mice. J Neurosci. 2003;23:7889–7896. doi: 10.1523/JNEUROSCI.23-21-07889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeMattos RB, et al. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- 32.Douglas PM, et al. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 34.Malisauskas M, et al. Does the cytotoxic effect of transient amyloid oligomers from common equine lysozyme in vitro imply innate amyloid toxicity? J Biol Chem. 2005;280:6269–6275. doi: 10.1074/jbc.M407273200. [DOI] [PubMed] [Google Scholar]
- 35.Moechars D, et al. Early phenotypic changes in transgenic mice that overexpress different mutants of amyloid precursor protein in brain. J Biol Chem. 1999;274:6483–6492. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- 36.Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 2007;4(e329):1949–1958. doi: 10.1371/journal.pmed.0040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.