Abstract
Two forms of a plant-specific RNA polymerase (Pol), PolIV(PolIVa) and PolV(PolIVb), currently defined by their respective largest subunits [NRPD1(NRPD1a) and NRPE1(NRPD1b)], have been implicated in the production and activity of 24-nt small RNAs (sRNAs) in RNA-directed DNA methylation (RdDM). Prevailing models support the view that PolIV(PolIVa) plays an upstream role in RdDM by producing the 24-nt sRNAs, whereas PolV(PolIVb) would act downstream at a structural rather than an enzymatic level to reinforce sRNA production by PolIV(PolIVa) and mediate DNA methylation. However, the composition and mechanism of action of PolIV(PolIVa)/PolV(PolIVb) remain unclear. In this work, we have identified a plant-specific PolV(PolIVb) subunit, NRPE5a, homologous to NRPB5a, a common subunit shared by PolI-III and shown here to be present in PolIV(PolIVa). Our results confirm the combinatorial diversity of PolIV(PolIVa)/PolV(PolIVb) subunit composition and indicate that these plant-specific Pols are eukaryotic-type polymerases. Moreover, we show that nrpe5a-1 mutation differentially impacts sRNAs accumulation at various PolIV(PolIVa)/PolV(PolIVb)-dependent loci, indicating a target-specific requirement for NRPE5a in the process of PolV(PolIVb)-dependent gene silencing. We then describe that the triad aspartate motif present in the catalytic center of PolV(PolIVb) is required for recapitulation of all activities associated with this Pol complex in RdDM, suggesting that RNA polymerization is important for PolV(PolIVb) to perform its regulatory functions.
Keywords: Arabidopsis, silencing, RNA polymerase
Multimeric DNA-dependent RNA polymerases (Pol) are widely distributed in living organisms, in which they ensure the transcription of all eukaryotic, archaeal, and bacterial genomes. They share a common origin, as reflected in their conserved subunit composition (1–3). Indeed, all multimeric Pols are made of 2 evolutionarily conserved large subunits that are specific of each enzyme class and a set of more or less conserved smaller polypeptides. The bacterial Pol, made of the β′, β, ω, and α2 subunits, is regarded as the founding member of this family, because orthologues of these subunits are present in both archaeal/eukaryotic Pols (A; A190; RPB1; C160 for the largest subunit β′), (B; A135; RPB2; C128 for the second-largest subunit β), (K; RPB6 for the ω subunit), and (D/L; AC40/AC19; RPB3/RPB11 for the pair of bacterial α subunits). In addition to these core subunits, the archaeal and nuclear Pols (PolI–III) have a more sophisticated organization, with at least 8–12 additional subunits (1–3). Reminiscent of their common origin, PolI–III shared 5 universal subunits (RPB5, -6, -8, -10, and -12) that, with the exception of RPB8, have direct orthologues in the archaeal Pol. Recent structural data indicate that the 5 core subunits underlie a conserved architecture surrounding the enzyme active site, whereas the archaeal/eukaryotic-specific subunits are located at the periphery of the enzyme core (1–3).
In eukaryotes, the discovery of small RNAs (sRNAs) with specific regulatory roles has added an extra layer of complexity to gene regulation. In Arabidopsis thaliana, sRNAs function both in transcriptional gene silencing (TGS) by guiding DNA and histone methylation and in posttranscriptional gene silencing by targeting endogenous mRNAs for cleavage or translational repression (4). The most abundant sRNA class in Arabidopsis corresponds to 24-nt sRNAs that guide sequence-specific cytosine methylation and TGS at repeated DNA loci in a process known as RNA-directed DNA methylation (RdDM) (5). Their production is known to involve a set of conserved RNA silencing genes that include RNA-dependent RNA polymerase 2 (RDR2) and Dicer-like 3 (DCL3) (6), and these sRNAs accomplish their regulatory functions after their recruitment into a putative effector complex containing either ARGONAUTE4 or -6 (7–9). More recently, both forward and reverse genetic approaches have characterized plant-specific components of the RdDM pathway that include 2 SNF2-type chromatin remodeling proteins (DRD1 and Classy) (10, 11), as well as 2 forms of a fourth type of multimeric Pol, so-called PolIV(PolIVa) and PolV(PolIVb) (12–16). Phylogenetic studies indicate that PolIV(PolIVa)/PolV(PolIVb) are widely distributed in the plant kingdom, being present from algae to higher plants, and that they have emerged in a multistep manner from PolII (17).
Unlike the 3 nuclear Pols, PolIV(PolIVa) and PolV(PolIVb) share the same second-largest subunit, NRPD2, but differ in their largest subunit, named NRPD1(NRPD1a) and NRPE1(NRPD1b), respectively (12–15). Biochemical analyses have confirmed that PolIV(PolIVa) and PolV(PolIVb) form independent multimeric complexes that are mutually stable in the single nrpd1 and nrpe1 mutants (15). Further analysis of the nrpd1 and nrpe1 mutants has led to the proposal that PolIV(PolIVa), in concert with RDR2 and to some extend DCL3, is involved in the production of nearly all 24-nt sRNAs, whereas PolV(PolIVb), which is not mandatory for sRNAs accumulation, probably acts downstream in the RdDM pathway by targeting DNA methylation (7, 14–16, 18, 19). Consistently, recent studies have revealed that a specific interaction between the effector protein AGO4 and a WG/GW-rich domain present in the extended C-terminal region of PolV(PolIVb) is critical for RdDM (20). However, the exact function of PolV(PolIVb) in RdDM remains unclear, and several models favor a structural rather than an enzymatic role for PolV(PolIVb) in this pathway (14, 19). Moreover, to date, both PolIV(PolIVa) and PolV(PolIVb) are defined only by their largest and second-largest subunits, and their precise subunit composition is not yet known.
Here, we have devised a bioinformatics-based approach aimed at characterizing subunits of PolIV(PolIVa)/PolV(PolIVb) complexes in Arabidopsis. In particular, we show that the RPB5-type coding genes have expanded in plants, owing to the emergence of a group of related subunits, hereafter referred to as NRPE5, whose prominent member NRPE5a is physically and functionally linked to PolV(PolIVb). We also show that NRPE5a is replaced by the common subunit NRPB5a in PolIV(PolIVa), therefore confirming the combinatorial diversity of PolIV(PolIVa)/PolV(PolIVb) subunit composition. Finally, we show that nrpe1-3/drd3-3, a substitution mutant in the aspartate triad motif conserved in the catalytic site of multimeric Pols, is a stable, loss-of-function allele of NRPE1(NRPD1b) that is functionally indistinguishable from nrpe1-11, a null T-DNA insertion mutant allele lacking NRPE1(NRPD1b)/PolV(PolIVb). These results strongly support the notion that PolV(PolIVb) is a functional eukaryotic-type enzyme whose activity is important for its function in RdDM.
Results and Discussion
High Complexity of the RPB5-Type Subunit Family in Plants.
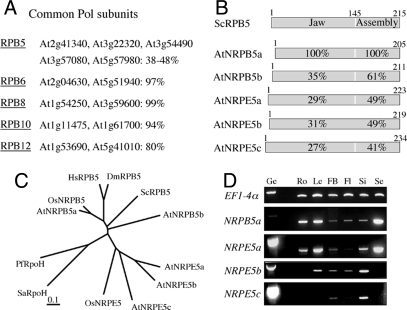
All conserved nuclear Pols share a similar subunit organization, with 2 related specific large subunits and a various number of smaller polypeptides, including 5 common subunits (RPB5, -6, -8, -10, and -12) that are often encoded by single genes (2). As a starting point of this study, we reasoned that it is likely that plant-specific variant forms of the common subunits might be present in the PoIIV(PolIVa)/PolV(PolIVb) enzymes. We therefore performed an exhaustive search of the Arabidopsis genome to identify all genes encoding for putative common-type Pol subunits. Beside close homologues of the RPB6, -8, -10, and -12-type Pol subunits encoded by 2 genes respectively, we identified 5 different genes for RPB5-type proteins that exhibit 38–48% identity over their entire sequences (Fig. 1A). This result is in agreement with a previous report that noticed the existence of an extended RPB5-like Pol subunit family in Arabidopsis (21). All RPB5-type proteins characterized in our study harbor the bipartite organization typical of Saccharomyces cerevisiae (Sc) RPB5, with a eukaryote-specific N-terminal Jaw domain and a C-terminal Assembly domain resembling the archaeal Pol subunit H (22, 23) ([Fig. 1B and Fig. S1). To assess evolutionary relationships among these genes, phylogenetic trees for RPB5, -6, -8, -10, and -12 subunits were constructed with plant sequences and related proteins from yeast, animal, and archaea. Their comparison indicates that eukaryotic sequences corresponding to all subunits but RPB5-type ones form a monophyletic group that is sister to the corresponding archaea group, suggesting that plants do not have variant of RPB6, -8, -10, and -12 Pol subunits (Fig. S2). In contrast, the phylogenetic tree constructed with RPB5-type sequences consists of 3 independent groups, each represented by typical eukaryotic RPB5 (including 2 members of the Arabidopsis RPB5-type protein family, hereafter referred to as AtNRPB5a and AtNRPB5b), archaeal Pol subunit H, and plant-specific RPB5-type proteins (including 3 RPB5-type proteins, hereafter referred to as AtNRPE5a, AtNRPE5b, and AtNRPE5c according to our current data) (Fig. 1C). RT-PCR analysis indicated that all 3 plant-specific NRPE5 genes are expressed in Arabidopsis organs, with NRPE5a presenting the largest pattern of expression (Fig. 1D). Taken together with previous results, our data indicate that plants express 2 classes of RPB5-type Pol subunits.
Fig. 1.
Higher plants contain 2 classes of RPB5-type Pol subunits. (A) Genes identified for each common Pol subunit in the Arabidopsis genome. Identical amino acid percentages between the Arabidopsis sequences are given for each universal subunit. (B) Diagrams show ScRPB5 and its homologues in Arabidopsis. The overall amino acid identity between NRPB5a and its homologues is indicated for the Jaw and the Assembly domains. AtNRPB5a and -b correspond to At3g22320 and At5g57980, respectively, whereas AtNRPE5a, -b, and -c correspond to At3g57080, At2g41340, and At3g55490. (C) Evolutionary relationships between NRPB5-type proteins and related factors. The unrooted phylogenetic tree was inferred from the full-length protein alignment from Fig. S1. Sc, Saccharomyces cerevisiae; At, Arabidopsis thaliana; Os, Oryza sativa; Hs, Homo sapiens; Dm, Drosophila melanogaster; Pf, Pyrococcus furiosus; Sa, Sulfolobus acidocaldarius. (D) RT-PCR analysis of NRPB5a and NRPE5s expression in various Arabidopsis tissues, including roots (Ro), leaves (Le), flower buds (FB), flowers (Fl), siliques (Si), and seeds (Se). EF1-4α was used as a control. Ge represents the genomic DNA control.
NRPE5a Is a PolV(PolIVb) Subunit.
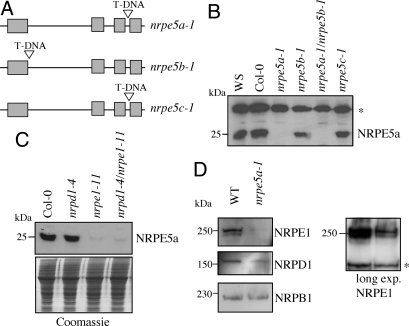
As a first step toward the understanding of NRPE5 family function, 2 different peptide antibodies were raised against NRPE5a and NRPE5c proteins, and we characterized Arabidopsis lines containing the T-DNA-disrupted mutant alleles nrpe5a-1, nrpe5b-1, and nrpe5c-1 (Fig. 2A). Plants homozygous for these mutant alleles were identified by PCR genotyping and were used as controls in Western blot experiments. For the 3 homozygous mutants, RT-PCR analysis indicated the absence of the corresponding full-length NRPE5 transcript (Fig. S3). In wild-type flower extracts, a protein with an apparent mass similar to the NRPE5a predicted size (≈25 kDa) was specifically detected with anti-NRPE5a antibody as judged by the absence of signal in corresponding KO line (Fig. 2B). Although the anti-NRPE5a antibody was prone to detect a recombinant NRPE5b (data not shown), no specific protein corresponding to NRPE5b was detected by Western blot in the nrpe5a-1 mutant (Fig. 2B). Likewise, Western blot analysis performed on wild-type vs. nrpe5c-1 null mutant using a functional anti-NRPE5c antibody failed to detect any specific signal corresponding to this protein, suggesting that NRPE5c is expressed at a very low level in flowers (data not shown). Taken together, our results suggest that NRPE5a is the prominent NRPE5-type subunit expressed in Arabidopsis.
Fig. 2.
Mutual stability of NRPE5a and NRPE1(NRPD1b) in various null mutants. (A) Diagram of the AtNRPE5-type genes in Arabidopsis. Translated exons are indicated with open gray boxes. Vertical open arrowheads indicate the location of the T-DNA insertions. (B) Western blot analysis of total extracts from wild-type (Ws/Col-0) and independent nrpe5 single/double mutants using the anti-NRPE5a antibodies. A nonspecific cross-reacting band (indicated by an asterisk) was used as a loading control. (C) Stability of the NRPE5a protein in single nrpd1-4 and nrpde1-11 and corresponding double mutants. Coomassie blue staining was shown as a loading control. (D) Stability of the NRPE1(NRPD1b) subunits in nrpe5a-1 mutant. NRPB1, the largest subunit of PolII, and a nonspecific anti-NRPE1(NRPD1b) cross-reacting band (indicated by an asterisk) were used as loading controls. Long exp, longer exposure.
Previous works have shown that NRPB5a, as expected for a common Pol subunit, is associated with the purified forms of PolI–III (21, 24) (Fig. S4). In contrast, NRPE5a was shown to be absent in PolII and PolIII (21) (Fig. S4), therefore raising questions about the nature of its Pol partner. To test whether NRPE5a is associated with PolIV(PolIVa)/PolV(PolIVb), we first analyzed the costability of this protein in mutant backgrounds lacking PolIV(PolIVa), PolV(PolIVb), or both forms (15). These experiments indicated that NRPE1(NRPD1b) stabilizes NRPE5a, because most of the signal disappeared in single nrpe1-11 and double nrpd1-4/nrpe1-11 mutants but remained mostly unaffected in the single nrpd1-4 mutant (Fig. 2C). Conversely, we then tested the impact of NRPE5a depletion on the stability of NRPD1(NRPD1a)/NRPE1(NRPD1b) proteins. As shown in Fig. 2D, the level of NRPE1(NRPD1b) protein was significantly reduced (see longer exposure), whereas the accumulation of NRPD1(NRPD1a) seemed at most slightly affected, considering the loading controls. These results suggest that a complex formation between NRPE1(NRPD1b) and NRPE5a is necessary to maintain their mutual stability in vivo. This conclusion is also consistent with the structural data indicating that RPB5-type proteins bind the large subunit of multimeric Pol (3, 22).
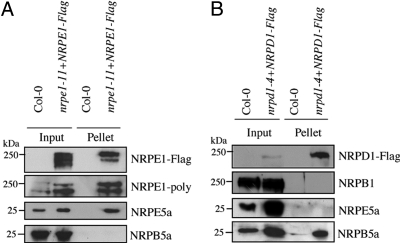
Our results suggest that the majority of the pool of NRPE5a protein is specifically associated with PolV(PolIVb) in vivo. However, the association of a low amount of NRPE5a with PolIV(PolIVa) cannot be excluded. We then specifically immunoprecipitated either PolIV(PolIVa) or PolV(PolIVb) from flower extracts of complemented nrpd1-4 (Fig. S5) or nrpe1-11 (20) transgenic lines expressing functional Flag-tagged version of NRPD1(NRPD1a) or NRPE1(NRPD1b), respectively. Both Flag-tagged versions of NRPD1(NRPD1a) and NRPE1(NRPD1b) and their associated NRPD2 subunit (data not shown) were specifically pulled down from transformed plant extracts (Fig. 3), indicating that the tagged proteins retain their ability to form both PolIV(PolIVa) and PolV(PolIVb) in vivo. Analysis of the corresponding M2 affinity eluates using anti-NRPE5a antibody demonstrated the exclusive association of NRPE5a with PolV(PolIVb) (Fig. 3). Contamination of the NRPE1(NRPD1b)-Flag eluate by other Pols could be ruled out because NRPB5a, a subunit shared by the 3 conserved Pols (21, 24), was not recovered in the purified fraction (Fig. 3A). In contrast, NRPB5a but not NRPE5a was recovered in the NRPD1(NRPD1a)-Flag eluate (Fig. 3B), suggesting that NRPB5a is also associated with PolIV(PolIVa). Although PolII is not present in the NRPD1(NRPD1a)-Flag eluate as judged by the absence of NRPB1, we cannot formally rule out a contamination of the purified material by PolI or PolIII. In any case, these experiments demonstrate that the plant-specific NRPB5-like proteins define a bona fide class of Pol subunit associated to PolV(PolIVb), referred to as NRPE5, in keeping with the system of nomenclature commonly used for multimeric Pols (25). Taken together, these data extend our understanding of the repertory of PolIV(PolIVa)/PolV(PolIVb) subunits and reveal an unexpected diversity in these Pol subunit compositions.
Fig. 3.
NRPE5a is a plant-specific subunit of PolV(PolIVb). Physical interaction between NRPE5a and PolV(PolIVb) (A) or PolIV(PolIVa) (B) detected by immunoprecipitation. Protein extracts prepared from either wild-type or complemented NRPE1(NRPD1b)/NRPD1(NRPD1a)-Flag transgenic lines were immunoprecipitated using anti-Flag M2 antibody, and the input/pellet fractions were analyzed by Western blot. Membranes were analyzed to detect the Flag epitope, NRPE1(NRPD1b), NRPE5a, and NRPB5a Pol subunits (A) or Flag epitope, NRPB1, NRPE5a, and NRPB5a Pol subunits (B).
Partial Loss of Methylation and Silencing of RdDM Targets in nrpe5a-1 Mutant.
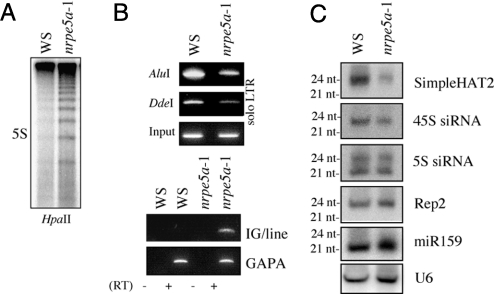
The association of NRPE5a with PolV(PolIVb) raises questions concerning the ability of the PolV(PolIVb) complex lacking NRPE5a to direct sRNA-dependent DNA methylation and silencing. Methylation-sensitive restriction enzymes were used for the examination of DNA methylation of the 5S rDNA and solo LTR loci, 2 PolV(PolIVb) targets, in wild-type vs. nrpe5a-1 lines. CpG methylation of the 5S rDNA loci was reduced in nrpe5a-1 mutant plants (Fig. 4A). Similarly, both AluI and DdeI enzymes cut the solo LTR element more efficiently in nrpe5a-1 than in wild type, indicating moderate loss of asymmetric methylation at these sites (Fig. 4B Top). However, although neither enzyme digested solo LTR to completion, as observed with the null nrpe1-11 mutant (Fig. 5C), the loss of DNA methylation in nrpe5a-1 was sufficient to activate the expression of the adjacent IG/LINE transcript (Fig. 4B Bottom) (26). To extend this analysis, we assessed the impact of NRPE5a KO mutation at PolIV(PolIVa)/PolV(PolIVb)-dependent loci that depend on PolV(PolIVb) for the production of 24-nt sRNAs (Fig. S6) (15, 19). We found that sRNAs corresponding to 2 of these loci, SimpleHAT and 45S rDNA, were reduced in nrpe5a-1 plants, whereas the levels of the 5S and Rep2 sRNAs were not substantially altered (Fig. 4C). Taken together, these results indicate that the silencing/methylation defects noted in nrpe5a-1 could be linked to a defect in sRNA production/accumulation at some loci (Rep2 and 45S loci) and also to an intrinsic defect of PolV(PolIVb) to act downstream of the sRNA to direct high levels of DNA methylation (5S loci). The differential outcomes on DNA methylation and sRNA production may result from target-specific requirements for NRPE5a function in RdDM, which is reminiscent of the gene-specific activation defects reported for the ScRPB5 mutant, rpb5-9 (27). Because the nrpe5a-1 mutation does not completely destabilize NRPE1(NRPD1b), it remains possible that the remaining NRPE1(NRPD1b) is sufficient to sustain PolV(PolIVb) activity at unaffected loci.
Fig. 4.
Partial loss of methylation and silencing of RdDM targets in nrpe5a-1 mutant. (A) Southern blot analysis of 5S rDNA loci on genomic DNA digested with methylation-sensitive enzyme HpaII in wild-type (WS) and nrpe5a-1. (B) Partial loss of CNN methylation at the solo LTR locus in wild-type vs. nrpe5a-1 plants (Top). AluI-, DdeI-, and nondigested DNA (Input) were used as a template for PCRs using solo LTR primers. Derepression of the solo LTR-driven IG/LINE expression in the nrpe5a-1 mutant was assessed by semiquantitative RT-PCR using primers located in the IG/LINE. The constitutively expressed GAPA gene is used as control (Bottom). (C) Differential effect of NRPE5a loss on sRNA accumulation. Total RNA (30 μg) from inflorescences was successively probed with various sRNAs, U6, and miR-159 as loading controls.
Fig. 5.
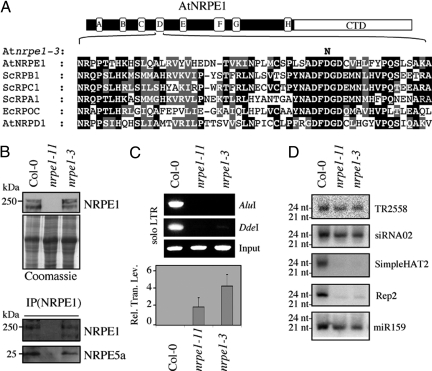
PolV(PolIVb) active site requirement for RdDM. (A) Sequence alignments in the conserved box D region of various Pol large subunits. The substitution in the catalytic triad of nrpe1-3/drd3-3 mutant protein is indicated. (B) Western blot analysis of total extracts from wild-type, nrpe1-11, and nrpe1-3/drd3-3 plants with the anti-NRPE1(NRPD1b) antibody (Top). The corresponding Coomassie staining is shown as a loading control. Immunoprecipitation of NRPE5a using anti-NRPE1(NRPD1b) antibody in wild-type, nrpe1-11, and nrpe1-3/drd3-3 extracts followed by the Western blot analysis of eluates with anti-NRPE1(NRPD1b) and anti-NRPE5a antibodies (Bottom). (C) Loss of CNN methylation at the solo-LTR locus in wild-type, nrpe1-11, and nrpe1-3/drd3-3 plants. Real-time PCR analysis of IG/LINE expression in wild-type, nrpe1-11, and nrpe1-3/drd3-3 plants. (D) Analysis of sRNA accumulation by Northern blot done with 30 μg of total RNA from inflorescences from wild-type, nrpe1-11, and nrpe1-3/drd3-3 plants.
PolV(PolIVb) Active Site Requirement for RdDM.
Previous studies have suggested that PolV(PolIVb) acts downstream of PolIV(PolIVa) at repeated DNA loci to amplify sRNA production and direct DNA methylation (7, 14, 15, 18, 19). To date, however, the catalytic competence of either PolIV(PolIVa)/PolV(PolIVb) form has not been directly assessed, and prevailing models favor a structural rather than an enzymatic role for PolV(PolIVb) in RdDM (14, 19). In an attempt to assess PolV(PolIVb) active site requirement for sRNA production and RdDM, we further characterized nrpe1-3/drd3-3, an EMS-induced nrpe1 allele formerly recovered in a genetic screen for loss-of-RdDM (14). This mutant allele carries a D→N substitution in the invariant metal-coordinating motif (DFDGD) named aspartate triad (Fig. 5A). This substitution affects metal-binding and transcriptional abilities of multimeric Pols while preserving the overall geometry of the active site (28). As shown in Fig. 5B, neither the accumulation of the triad mutant protein nor its ability to interact with NRPE5a was affected in nrpe1-3/drd3-3, as opposed to the strong NRPE1/PolV(PolIVb) complex destabilization observed in the null T-DNA insertion mutant nrpe1-11 (15). This result supports the assumption that nrpe1-3/drd3-3 mutation does not impact significantly the folding and the capacity of the triad mutant protein to interact with other PolV(PolIVb) subunits. However, both the nrpe1-3/drd3-3 and nrpe1-11 mutant plants exhibited similar loss of asymmetric cytosine (CNN) DNA methylation at 3 PolV(PolIVb)-dependent loci (solo LTR, 5S repeats, and AtSN1) (Fig. 5C and Fig. S7). Real-time PCR analysis indicates that both nrpe1-11 and nrpe1-3/drd3-3 mutant plants present a strong increase of solo LTR-dependent IG/LINE expression that is correlated with the decrease in cytosine methylation at solo LTR element (Fig. 5C). Additionally, the analysis of the levels of sRNAs corresponding to PolIV(PolIVa)/PolV(PolIVb)-dependent loci do not differ substantially between nrpe1-11 and nrpe1-3/drd3-3, indicating that both mutations affect PolV(PolIVb) activity to a similar extent (Fig. 5D). Taken together, these results show that nrpe1-3/drd3-3 is a stable, loss-of-function allele of NRPE1(NRPD1b) that probably compromises the catalytic activity of PolV(PolIVb).
Conclusion
The bioinformatic approach we followed has provided insights into the composition and function of PoIV(PolIVb) in Arabidopsis. First, we have found that both PolIV(PolIVa) and PolV(PolIVb) contain an RPB5-type subunit of a different nature; PolIV(PolIVa) harbors the universally shared NRPB5a subunit, whereas PolV(PolIVb) is associated with NRPE5a, a plant-specific variant of NRPB5a. These results corroborate and extend a previous phylogenetic analysis of the NRPD1(NRPD1a)/NRPE1(NRPD1b) subunits in plants (17), indicating that PolIV(PolIVa)/PolV(PolIVb) are eukaryotic-type multimeric Pols. Second, we found that the nrpe5a-1 mutation differentially impacts sRNAs production and accumulation by PolV(PolIVb) at various RdDM-dependent loci. One interpretation of this finding is that there is a target-specific requirement for NRPE5a in the process of PolV(PolIVb)-dependent gene silencing. The basis for this distinction could lie in the selective/facilitated recruitment of PolV(PolIVb) through its NRPE5a subunit via protein–protein or protein–DNA interactions. In this regard, previous studies have shown that the RPB5 subunit is endowed with the ability to contact DNA template (29, 30) and to interact with several transcriptional gene regulators (31, 32). Third, because a plant-specific variant has only been identified for NRPB5-type subunit, it is likely that the other common subunits (i.e., NRPDB6, -8, -10, and -12) are present in the 5 types of nuclear Pols. However, it remains also possible that PolIV(PolIVa)/PolV(PolIVb), like some virus-encoded multimeric RNA polymerases (33), had selectively lost a subset of these subunits in the course of its evolution. Future studies will be necessary to clarify this point and extend the repertory of PolIV(PolIVa)/PolV(PolIVb) subunits.
Magnesium ion plays a crucial role in nucleic acid polymerization driven by multimeric Pol, and its binding site has been mapped to a evolutionarily conserved triad of aspartate residues that is present in all known Pol large subunits, including NRPD1(NRPD1a) and NRPE1(NRPD1b). Despite the conservation of this motif, the catalytic competences of PolIV(PolIVa)/PolV(PolIVb) have not been directly assessed, and prevailing models favor a structural rather than an enzymatic role for PolV(PolIVb) in RdDM (14, 19). Results presented here on the drd3-3/nrpe1-3 mutant indicate that the conserved aspartate triad and consequently the catalytic center of NRPE1(NRPD1b) are essential for the function of PolV(PolIVb) in the RdDM pathway. Considering that catalytic triad mutants of Pol are capable of template recognition and DNA melting (34), our data do not favor a structural role of PolV(PolIVb) in which it would only create and stabilize an open DNA complex to facilitate the action of the DNA methyltransferase. In contrast, we found that the integrity of the triad motif is required to sustain all known PolV(PolIVb) activities associated with RdDM (i.e., de novo DNA methylation, sRNA accumulation at PolIV(PolIVa)/PolV(PolIVb)-dependent loci, and silencing). This suggests that a PolV(PolIVb)-nascent transcript is an integral component of the sRNA silencing pathway. Because AGO4–sRNA complex is bound to the PolV(PolIVb) C-terminal domain (18, 20), the nascent PolV(PolIVb) transcript could act as a template for AGO4-dependent recognition and cleavage, 2 steps that have been shown to be essential for sRNA production at specific loci (8). Interestingly, one can find a good correlation between the loci that require AGO4 catalysis (8) and PolV(PolIVb) activity (Fig. 5D), reinforcing the hypothesis that these 2 events are functionally linked. Future biochemical and functional studies will be necessary to test this model.
Materials and Methods
Plant Materials.
Characterization of the nrpe5 insertion mutant lines is described in the SI Materials and Methods. nrpd1-4 plants complemented with a NRPD1(NRPD1a)-tagged variant were obtained by using the strategy reported by El-Shami et al. (20). NRPD1 (NRPD1a) promoter and coding sequence were amplified using primers 352/465 and 462/463, respectively. The primers are listed in Table S1.
Bioinformatics.
Sequence alignment and phylogenetic analysis were derived using CLUSTAL W with default parameters as described elsewhere (35).
RNA Isolation and Analysis.
Total RNA were isolated from leaf tissues of nrpd1-4, nrpe1-11, and wild-type Arabidopsis (ecotype Columbia) using TRIzol reagent (Invitrogen). After DNase treatment, cDNA were obtained with an Affinity Multitemperature cDNA synthesis kit (Stratagene) using oligo-dT primer with 1 μg of RNA according to the manufacturer's instructions. Details on PCR analysis as well as real-time PCR amplification are presented in the SI Materials and Methods.
For sRNA analysis, total RNA extraction and Northern blots were performed as described by Pontier et al. (15), except that NX-Hybond membranes (Amersham) and EDC (Sigma)-mediated cross-linking were used. The probes are described in Table S1.
Plant Protein Extraction and Immunoblots.
Total plant protein extracts were obtained according to the method described by Pontier et al. (15). Immunoblot analyses were performed using the dilution 1:1,000 for anti-NRPB1 (8WG16 from AbCam) and anti-Flag M2 monoclonal antibody (Sigma–Aldrich) and 1:5,000 for anti-NRPB5a. The antibodies anti-NRPE1(NRPD1b) and anti-NRPD1(NRPD1a) were used as described by Pontier et al. (15). Rabbit antisera were raised against peptides designed in NRPE5a and NRPE5c (Table S1) (Eurogentec), affinity-purified, and diluted 1,000-fold for Western blot analysis.
Immunoprecipitation.
Total extracts from inflorescences were obtained as described by Li et al. (18) and incubated with Anti-Flag M2 affinity gel (Sigma) for 2 h at 4°C. After centrifugation at 1,800 × g for 1 min, the resin was washed with extraction buffer and resuspended in SDS-PAGE loading buffer. Immunoprecipitations using the anti-NRPE1(NRPD1b) antibody were performed by binding the antibodies onto ProteinA Sepharose CL-4B (Amersham) according to the manufacturer's instructions. After washes with PBS, total extracts prepared as described above were added (“input”). After a 2-h incubation at 4°C, the resin was centrifuged, washed, and the bound material (“pellet”) was recovered by direct resuspension in SDS/PAGE loading buffer.
DNA Methylation Analysis.
Genomic DNA was extracted from leaves using the Wizard Genomic kit (Promega). Southern blot analysis of 5S rDNA was digested with methylation-sensitive enzymes HaeIII or HpaII and hybridized to a 5S probe (15). Cytosine methylation of the solo LTR acting as a promoter for IG/LINE was studied using enzymes sensitive to CNN methylation (AluI and DdeI) in wild-type and nrpe5a-1 mutant. Disappearance or reduced levels of a fragment after digestion with a given enzyme indicate loss of methylation at that site. Primers used were LTR-F and LTR-R as described by Huettel et al. (26).
Note Added in Proof.
To facilitate the naming of the PolIVa/PolIVb subunits, a nomenclature has been adopted for these enzymes using PolIV and PolV in place of the PolIVa and PolIVb terminology used previously. Accordingly, and in agreement with the existing nomenclature for RNA polymerase subunits, the PolIV(PolIVa)/PolV(PolIVb)-specific subunits will be renamed using the prefixes “Nuclear RNA Polymerase D (NRPD)” and “Nuclear RNA Polymerase E (NRPE),” respectively. However, to facilitate the reading of this article, we have kept both nomenclatures: PolIVa appears as PolIV-(PolIVa), PolIVb as PolV(PolIVb), NRPD1a as NRPD1(NRPD1a), NRPD1b as NRPE1(NRPD1b).
Supplementary Material
Acknowledgments.
We thank J. Saez-Vasquez (Université de Perpignan) for the gift of the NRPB5a antibody and N. Picault, G. Mitta, and T. Pelissier for technical assistance. Research in the Lagrange laboratory is supported by Centre National de la Recherche Scientifique and Agence Nationale de la Recherche Grant NT05-3_45717.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810310106/DCSupplemental.
References
- 1.Ebright RH. RNA polymerase: Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 2.Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 3.Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008;451:851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pontes O, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 8.Qi Y, et al. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 9.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanno T, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 11.Smith LM, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 13.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Kanno T, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 15.Pontier D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA. 2007;104:4536–4551. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J, Hall B. A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol. 2007;64:101–112. doi: 10.1007/s00239-006-0093-z. [DOI] [PubMed] [Google Scholar]
- 18.Li CF, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 19.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larkin RM, Hagen G, Guilfoyle TJ. Arabidopsis thaliana RNA polymerase II subunits related to yeast and human RPB5. Gene. 1999;231:41–47. doi: 10.1016/s0378-1119(99)00090-6. [DOI] [PubMed] [Google Scholar]
- 22.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 23.Todone F, Weinzierl RO, Brick P, Onesti S. Crystal structure of RPB5, a universal eukaryotic RNA polymerase subunit and transcription factor interaction target. Proc Natl Acad Sci USA. 2000;97:6306–6310. doi: 10.1073/pnas.97.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saez-Vasquez J, Pikaard CS. RNA polymerase I holoenzyme-promoter interactions. J Biol Chem. 2000;275:37173–37180. doi: 10.1074/jbc.M006057200. [DOI] [PubMed] [Google Scholar]
- 25.Young RA. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 26.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and PolIV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyao T, Woychik NA. RNA polymerase subunit RPB5 plays a role in transcription activation. Proc Natl Acad Sci USA. 1998;95:15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sosunov V, et al. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T, et al. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 31.Cheong JH, Yi MK, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and plays a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soutourina J, et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Zaychikov E, et al. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 35.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.