Abstract
Transcription initiation is a dynamic process in which RNA polymerase (RNAP) and promoter DNA act as partners, changing in response to one another, to produce a polymerase/promoter open complex (RPo) competent for transcription. In Escherichia coli RNAP, region 1.1, the N-terminal 100 residues of σ70, is thought to occupy the channel that will hold the DNA downstream of the transcription start site; thus, region 1.1 must move from this channel as RPo is formed. Previous work has also shown that region 1.1 can modulate RPo formation depending on the promoter. For some promoters region 1.1 stimulates the formation of open complexes; at the Pminor promoter, region 1.1 inhibits this formation. We demonstrate here that the AT-rich Pminor spacer sequence, rather than promoter recognition elements or downstream DNA, determines the effect of region 1.1 on promoter activity. Using a Pminor derivative that contains good σ70-dependent DNA elements, we find that the presence of a more GC-rich spacer or a spacer with the complement of the Pminor sequence results in a promoter that is no longer inhibited by region 1.1. Furthermore, the presence of the Pminor spacer, the GC-rich spacer, or the complement spacer results in different mobilities of promoter DNA during gel electrophoresis, suggesting that the spacer regions impart differing conformations or curvatures to the DNA. We speculate that the spacer can influence the trajectory or flexibility of DNA as it enters the RNAP channel and that region 1.1 acts as a “gatekeeper” to monitor channel entry.
Transcription initiation is a multistep process that requires both recognition of promoter DNA and structural isomerization of the RNA polymerase (RNAP)/promoter complex to form a machine competent for transcription (reviewed in refs. 1–4). This process must be flexible enough to initiate transcription at a variety of promoter sequences yet rigid enough to provide specificity. In bacteria, the σ subunit of RNAP holoenzyme is the primary factor that sets this specificity. Although bacteria can have multiple σ factors, the primary σ, such as Escherichia coli σ70, is responsible for the expression of housekeeping genes during exponential growth (5, 6). All σ factors share related regions 2, 3, and 4, but only primary σ proteins have a related, negatively-charged N-terminal portion, region 1.1 (6).
Transcription initiation begins with the initial binding of RNAP to dsDNA elements to form the polymerase/promoter closed complex (RPc) (7–9) (reviewed in ref. 1) (Fig. 1A). In RPc, polymerase interacts with a fully ds promoter (P). Promoter recognition can arise from interactions between the C-terminal domains (CTDs) of the α-subunits (α-CTDs) and ds promoter sequences between −40 and −60 (UP elements), between σ70 region 4 and a −35 element, between σ70 region 3 and sequences at −15, −14 (the extended −10 motif), and between σ70 region 2.4 (a portion of region 2) and sequences at −12/−11 (the 5′ end of the −10 element) (reviewed in ref. 1). The RPc, which is usually unstable and competitor sensitive, gives an abbreviated protection footprint that does not include DNA downstream of the transcription start site (7, 9, 10). Creation of the stable polymerase/promoter open complex (Rpo) requires bending and unwinding of the DNA (11) and major conformational changes (isomerization) of the polymerase (Fig. 1A) (12–14). The result of these changes generates a complex in which the promoter is unwound from −11 to around +3, and the protection footprint extends to around +25 (9, 11, 15–21). In addition, RPo is normally competitor resistant, although RPo at the very strong ribosomal promoters does not follow this rule (22, 23).
Fig. 1.
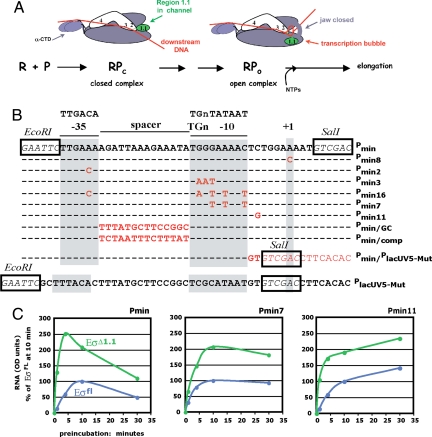
Process of transcription initiation, promoter sequences, and transcription with Pmin, Pmin7, and Pmin11 with Eσfl and EσΔ1.1 (A) Diagram depicting polymerase promoter contacts in RPc and RPo with core polymerase (β, β′, α2, and ω) in purple, σ70 regions 2–4 in white, σ70 region 1.1 in green, and DNA in red. R is RNA polymerase; P is the promoter DNA. The transcriptional start site is designated +1. Interactions between the α-CTDs and the UP element(s), σ70 region 4 and the −35 element, σ70 region 3 and the −15TGn−13 element, and σ70 region 2 and the −10 element are indicated. In RPc, the dsDNA has not yet entered the primary channel; full entry of DNA into the channel is blocked by σ70 region 1.1. In RPo, σ70 region 1.1 has moved, the DNA is bent and is unwound from −11 to +3, the template strand has descended into the active site of polymerase, and a portion of β,β′, called jaws, has secured the downstream DNA. (B) Sequences of Pmin, Pmin derivatives, and PlacUV5-Mut. Consensus sequences for the σ70-dependent −35, TGn, and −10 promoter elements are shown at the top. The EcoRI and SalI restriction sites used for plasmid constructions are boxed. σ70 elements are shaded in gray, and base-pair substitutions in the Pmin derivatives are in red. (C) Effect of promoter mutations on activity with Eσfl or EσΔ1.1. Single-round transcription reactions were performed as described in Materials and Methods using Eσfl (blue) or EσΔ1.1 (green). The amount of RNA from the indicated promoter (relative to the amount of RNA obtained at the 10-min time point with Eσfl × 100) is plotted versus the length of the incubation of polymerase with the DNA (in min) before the addition of rNTPs and heparin.
Much work has been done to understand the conformational changes that occur as RPc transitions to RPo (reviewed in ref. 1). In addition, structures of σ, core polymerase, and holoenzyme from thermophilic bacteria (4, 24–29) or portions of E. coli σ70 (30, 31) have provided 3D scaffolds on which to model these steps. Kinetic analyses using the λ promoter PR have revealed transcriptional intermediates in the pathway from RPc to RPo (refs. 12–14, 20, and 32 and references therein). Initially, the ds promoter DNA is thought to lie across the polymerase, making sequence-specific contacts with σ70 and the α-CTDs. The interaction of the DNA with the downstream DNA channel (portions of β and β′) generates an early intermediate (I1), which, like RPc, is unstable and competitor sensitive. The DNA then moves deeper into the DNA channel through extensive interactions with portions of β and β′, forming a competitor-resistant intermediate, I2. Finally, the DNA around the +1 site begins to melt, and σ70 region 2.3 contacts single-stranded (ss)DNA bases at positions −10 through −7 on the nontemplate strand. For some promoters, contract(s) between residues in σ70 region 1.2 and ss bases at −5 and −6 occurs also (22, 33). The protein/ssDNA interactions stabilize the polymerase/promoter complex, allowing the template strand to descend into the active site of core and the dsDNA downstream to fully enter the downstream channel. RPo is achieved when portions of β and β′, designated the polymerase “jaws,” close onto the downstream DNA, securing the DNA within polymerase.
σ70 region 1.1 does not contact DNA, but is thought to play a crucial role in the transition from RPc to RPo (34, 35). At some promoters (λPR, Ptac, PRNAI) region 1.1 is needed for efficient formation of the open complex (34, 35). However, at the Pminor promoter the rate of RPo formation is actually inhibited by region 1.1 (34). Although the structure of region 1.1 of Thermophilus maritima has been reported (59), the structure of σ70 region 1.1 has yet to be determined, presumably because its flexibility has made crystallization difficult. However, FRET data modeled with structural analyses indicate that in holoenzyme, region 1.1 lies within the channel that will be occupied by the downstream DNA when RPo is formed (Fig. 1A) (36). Consequently, region 1.1 must move for the DNA to occupy the channel as it does in RPo. Kinetic data with λPR suggests that a conformational rearrangement, occurring at the I1 ↔ I2 transition, is consistent with the movement of 1.1 out of the channel (12), and FRET analyses have suggested that region 1.1 moves to a portion of core called the β pincer tip (36). It has been proposed that region 1.1 could facilitate DNA entry into the channel by holding the jaws open for free incoming DNA and/or by supplying energy needed for closure of the jaws by 1.1 movement (12).
In this article, we have investigated which features of Pminor are responsible for inhibition by region 1.1. We find that changing the promoter recognition elements of Pminor do not change this effect. However, substituting the Pminor spacer with a more GC-rich spacer or the complement sequence of this spacer generates promoters that are equally active with polymerase lacking region 1.1, EσΔ1.1 or polymerase with full-length σ70 (Eσfl). The Pminor spacer is an AT-rich sequence with features that can affect DNA conformation, and we demonstrate that the presence of this spacer affects the mobility of Pminor DNA during gel electrophoresis. We speculate that it is the DNA conformation, perhaps the trajectory or bend, as set by the spacer region, that influences the effect of region 1.1 on the initiation process. Furthermore, our DNase I footprinting indicates that the stable, open complex at Pminor is similarly and fully protected to +27 whether region 1.1 is present or absent. Our data suggest that movement of region 1.1 is not an obligatory step for the closing of the β/β′ jaws onto the downstream promoter DNA.
Results
Effect of σ70 Region 1.1 on Promoter Activity Is Not Determined by Promoter Class or Promoter Recognition Elements.
Although early work led to the idea that σ70 region 1.1 is required for efficient open complex formation (35), this work was performed using only 2 promoters, λPR and Ptac. Subsequent work showed that the effect of σ70 region 1.1 varies depending on the specific promoter tested (34). For PuvsX-sigma, region 1.1 has little effect on the rate of open complex formation. For another promoter, Pminor, region 1.1 significantly inhibits formation of RPo.
Deletion analyses indicate that a minimal Pminor promoter, Pmin, which contains Pminor sequences from only −35 to +4 (Fig. 1B), is also inhibited by region 1.1 (Fig. 1C). Our previous work demonstrated that Pminor and Pmin belong to a newly identified promoter class, − 35/TGn, that is characterized by a requirement for both an excellent −35 element and an extended −10 TGn sequence to compensate for a poor −10 element (1, 37, 38). To investigate whether the effect of region 1.1 with Pminor reflects a general characteristic of −35/TGn promoters, we compared the formation of active transcription complexes at Pmin with that at Pmin16, a derivative with perfect −35 and −10 elements, and at Pmin7, a derivative with a good −35 and a perfect TGn and −10 element (Fig. 1B). In addition, we tested Pmin derivatives with more subtle modifications: Pmin2, which has a perfect −35 element, and Pmin3, which has a poor extended −10, but an improved −10 element (Fig. 1B). We incubated EσΔ1.1 or Eσfl with the DNA for various times. Ribonucleoside triphosphates (rNTPs) and the polyanion heparin were then added together, allowing a single round of transcription and providing a read-out of the amount of transcriptionally-competent complexes present at any given time.
The promoter derivatives Pmin2, Pmin7, and Pmin16 have improved polymerase recognition sequences (Fig. 1B) and, as expected, yield greater promoter activity than Pmin (ref. 37 and data not shown). However, each of these promoter variants is still inhibited by region 1.1 (Fig. 1C and Fig. S1A). Our transcription analyses also indicate that Pmin3, which lacks the TGn element, is inhibited by region 1.1 (Fig. S1A). Thus, the effect of region 1.1 on forming a heparin-resistant complex at Pminor appears to be determined by promoter context, i.e., something other than the promoter recognition elements (−35 element, extended-10 sequence, and −10 element) themselves.
Effect of σ70 Region 1.1 on Promoter Activity Is Not Determined by a Short Polymerase/Promoter Half-Life, a Transcription “Stutter” Start, or Sequences Downstream of the −10 Element.
Besides its unusual promoter class, Pminor has other peculiar properties. First, the open complex at Pminor has a half-life of only a few minutes whether it is made with EσΔ1.1 or Eσfl (34). However, the Pmin derivatives, Pmin2, Pmin7, and Pmin16, which have improved binding elements, form heparin-resistant polymerase/promoter complexes that remain stable for at least 30 min in contrast to the polymerase/Pmin complexes that decrease after a few minutes (Fig. 1C and Fig. S1A). Thus, the increased activity of EσΔ1.1 with these promoters is not associated with a short complex half-life.
Pminor is also unusual in that it has a stutter start, in which ≈3 nontemplated nucleotides are incorporated at the 5′ end of its transcript (37). To examine whether this start was responsible for the inhibition of Pminor by region 1.1, we tested a Pminor derivative, Pmin8, that contains a +1 A to C mutation (Fig. 1B). Although this change eliminates the stutter start at Pminor when using Eσfl (37) or EσΔ1.1 (Fig. S1B), the Pmin8 promoter is still inhibited by region 1.1 (Fig. S1A). Therefore, the greater activity of Pminor with EσΔ1.1 is not determined by its unusual start.
Because region 1.1 is thought to lie within the DNA downstream channel of holoenzyme and must move out of this channel during the formation of open complex, we tested two other Pmin derivatives, besides Pmin8, with mutations downstream of the −10 element. First, we examined Pmin/PlacUV5-Mut (Fig. 1B). This is a promoter in which Pmin sequences downstream of position −7 have been replaced with the corresponding sequences of PlacUV5-Mut, a promoter that is more active in the presence of region 1.1 (Fig. S1A). Although the Pmin/PlacUV5-Mut mutation changes nearly every base pair downstream from position −6, it is still inhibited by region 1.1 (Fig. S1A). We also tested Pmin11, which contains a −5 C to G mutation (Fig. 1B). A comparable mutation in the ribosomal promoter rrnB P1 has been shown to increase RPo half-life via interaction with σ70 region 1.2 residue M102 and, in addition, the absence of region 1.1 increases the detection of this contact (22, 33). Pmin11 is also inhibited by region 1.1 (Fig. 1C). We conclude that the downstream region is not responsible for the increased activity of Pmin with EσΔ1.1.
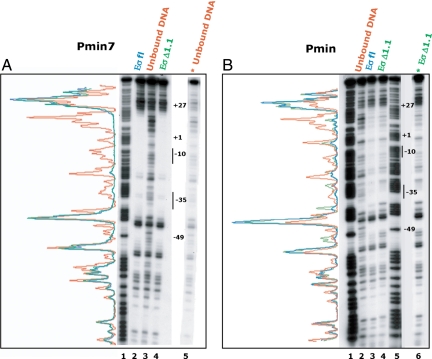
The RPo complex is characterized by a transcription bubble surrounding the start of transcription, stability upon heparin challenge, and a DNase I protection footprint extending to around +25 (11, 19, 32). Previous kinetic analyses using the strong lambda promoter PR have suggested that the movement of region 1.1 is coupled to the late folding of the β/β′ jaws of core onto the downstream DNA and that this movement could provide the energy needed for jaw closure and stable RPo formation (12). Thus, as another way to assess the effect of region 1.1 on the downstream DNA and on RPo formation and jaw closure, we determined the DNase I protection patterns of heparin-resistant complexes with Eσfl and EσΔ1.1 at Pmin and at the more consensus Pmin7 promoter. Previous KMnO4 footprinting analyses have shown that these complexes contain the expected transcription bubble (ref. 37 and data not shown). We find that DNase I footprints at Pmin7 with either polymerase are quite similar and in both cases extend to +27, as expected for RPo (Fig. 2A). Likewise, Pmin is also protected to +27 with either polymerase although the footprint at Pmin is not as strong as at Pmin7 (Fig. 2B). This finding is reasonable because Pmin is a weaker promoter than Pmin7 and open complexes at Pminor decay with time (ref. 34 and Fig. 1C). This analysis suggests that for these promoters, the polymerase jaws close sufficiently to protect the downstream DNA from DNase I cleavage, with or without the movement of region 1.1. Thus, we conclude that the movement of region 1.1 is not a prerequisite for jaw closure at all promoters.
Fig. 2.
Absence of σ70 region 1.1 does not affect DNase I protection of Pmin or Pmin7 downstream DNA. 5′-32P fragments, labeled on the nontemplate strand, containing Pmin7 (A) or Pmin (B) were incubated with polymerase and treated with DNase I, and the DNA bound with Eσfl or EσΔ1.1 (as indicated) or the unbound DNA was obtained and run on a 7 M urea, 6% polyacrylamide, denaturing gel. Lane 1 of A and lanes 1 and 5 of B are G+A marker ladders; lane 3 of A and lane 4 of B represent alternate exposures of lane 5 A and lane 6 B, respectively, and are shown for better comparison of the protection patterns. Positions −49, +1, and +27 within the promoters and the −10 and −35 elements are indicated on the gels. To the left of each gel is the trace for unbound DNA (red), and bound DNA with Eσfl (blue) or EσΔ1.1 (green).
The Spacer Sequence Affects Promoter Activity with EσΔ1.1.
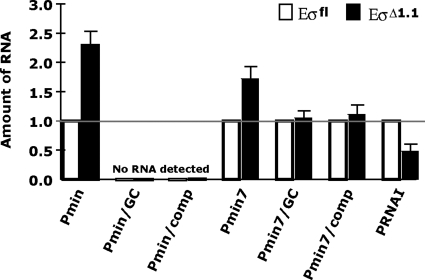
A comparison of various promoter sequences indicated that the AT richness of the spacer sequence correlates with the effect of region 1.1 on promoter activity (Fig. S2). Although the length of a promoter spacer dictates the distance between the promoter elements and is an important factor in promoter usage, the spacer is not known to make sequence-specific contact with RNAP (reviewed in refs. 1 and 4) and exactly how the spacer affects the interaction of RNAP with a promoter is not fully understood (39–41). The spacer in Pminor is the preferred length for a σ70-dependent promoter, eliminating the possibility that simply its length makes Pminor unusual. To test whether the Pminor spacer sequence (−29AGATTAAAGAAATA−16) affects inhibition by region 1.1, we replaced this sequence with either the corresponding GC-rich 14 bp of the PlacUV5-Mut spacer to generate Pmin/GC, or with the sequence complementary to the Pminor spacer to generate Pmin/comp. Transcription assays indicated that both the GC and the comp spacer substitution essentially eliminate transcription from Pmin with either Eσfl or EσΔ1.1 after incubations of polymerase and the DNA for 1 min (Fig. 3), 10 min (Fig. S3A), or even 30 min (data not shown).
Fig. 3.
The Pminor spacer region affects the modulation of stable complex formation by EσΔ1.1. Except for Pmin/GC and Pmin/comp, in which no RNA was detected with either Eσfl or EσΔ1.1, the amount of RNA seen with Eσfl and a particular promoter was set to 1. Values and standard deviations for EσΔ1.1 were determined from 3 or more independent single-round in vitro transcription reactions, performed as in Fig. 1C, except Pmin, Pmin/GC, Pmin/comp (or Pmin7 set) templates were prepared to produce transcripts of 3 different lengths, allowing the promoters to be assayed in the same reaction. The total template DNA (0.02 pmol) is the same as that used in Fig. 1C, with the polymerase/DNA ratio maintained at 10:1. PRNAI is present on each template DNA and serves as an internal control; this promoter is more transcriptionally active with Eσfl than with EσΔ1.1 (34). Polymerase was incubated with the template DNA for 1 min before the addition of rNTPs and heparin.
Previously, it has been shown that a spacer can influence promoter activity (38, 40–45). In particular, a GC-rich spacer can result in reduced promoter activity compared with that seen with an AT-rich spacer (39, 40). Spacers with runs of Ts (46) or As (47) have been shown to dramatically increase the overall activity of a promoter. Thus, the decrease in transcription observed with Pmin/GC is not surprising. However, Pmin/comp has the same base-pair composition as Pmin, yet yields no detectable transcription, indicating that something other than the overall AT richness of the Pmin spacer is determining Pmin activity.
Although the GC and comp spacers had a dramatic effect on Pmin activity, the transcriptional levels were too weak to assess the effect of region 1.1; therefore, we tested the spacer exchanges within Pmin7 because this promoter is recognized well by both Eσfl and EσΔ1.1. In this experiment, the DNA templates were prepared to produce transcripts of three different lengths, allowing the promoters to be assayed in the same reaction. As shown in Fig. 3 and Fig. S3, the relative amounts of Pmin7 RNA with Eσfl and EσΔ1.1 seen under these conditions are similar to those observed when Pmin7 is the only template (Fig. 1C), i.e., the presence of region 1.1 lowers the amount of RNA from Pmin7 ≈2-fold. However, the presence of region 1.1 has no significant effect on the amount of RNA from the Pmin/GC or Pmin/comp promoters (Fig. 3 and Fig. S3). As expected (34), the level of PRNAI RNA is greater when using Eσfl than when using EσΔ1.1 (Fig. 3). Taken together, these results indicate that the Pmin spacer region determines the inhibition by region 1.1 and that this property is not merely a result of the spacer's AT richness.
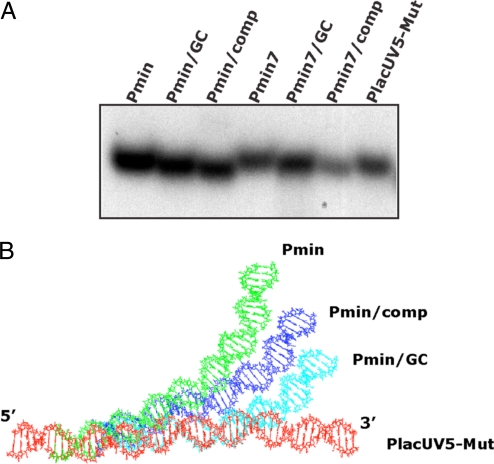
Previous work has indicated that AT-rich sequences, such as that present in the Pminor spacer, are apt to unwind more easily or be more flexible and easily distorted (48, 49). In particular, a T:A step within an A-tract has been shown to generate a flex point within the helix (49). KMnO4 footprinting using Pminor did not reveal unpaired Ts within the spacer region when using either Eσfl or EσΔ1.1 (ref. 37 and data not shown), suggesting that the spacer region itself is not grossly distorted. However, this analysis would not detect subtle or transient distortions in the spacer DNA. A-tracts can also result in an intrinsic bend in the DNA (50). However, a run of at least four contiguous A nucleotides is typically needed (50), and a T:A or G:C interruption, as here, has been shown to interrupt an A-tract bend (50, 51). To investigate the possibility that the Pminor sequence imparts a particular conformation or bend to the DNA, the mobilities of 51-bp fragments containing Pmin, Pmin/GC, Pmin/comp, the Pmin7 derivatives, or PlacUV5-Mut (sequences shown in Fig. 1B) were compared on native, polyacrylamide gels. This analysis revealed that these promoter DNAs migrate differently despite their identical length (Fig. 4A). In particular, Pmin and Pmin/comp have the same base-pair composition, as do Pmin7 and Pmin7/comp, yet have the most divergent migration rates, i.e., Pmin and Pmin7 migrate more slowly than the GC derivatives, whereas Pmin/comp and Pmin7/comp migrate faster. These results suggest that these spacer regions can impart differing conformations or curvatures to the DNA fragments. The sequences examined by electrophoresis were also analyzed in silico. The structures for Pmin, Pmin/GC, Pmin/comp, and PlacUV5-Mut DNAs were predicted by the “model it” (52) program and then aligned based on their identical sequence of GAATTC at the 5′ end (Fig. 4B). These models predict that the putative Pmin and Pmin/comp structures are significantly curved in the spacer region relative to PlacUV5-Mut and Pmin/GC, a result that is consistent with the native gel analysis. We conclude that the presence of the Pminor spacer results in a DNA promoter conformation that differs from that formed with either the GC or comp spacers.
Fig. 4.
Pminor spacer affects the conformation of Pminor DNA. (A) Mobilities of Pmin derivatives with different spacer sequences and PlacUV5-Mut (5′-32P end-labeled 51-bp fragments) upon electrophoresis in a native, 12% polyacrylamide gel. (B) Predicted structures of Pmin, Pmin/GC, Pmin/comp, and PlacUV5-Mut DNA. All structures were aligned using the common sequence GAATTC at the 5′ end. In addition, Pmin, Pmin/GC, and Pmin/comp have identical sequences except for their 14-bp spacer sequences (Fig. 1B).
Discussion
Region 1.1, the negatively-charged domain found at the N terminus of primary σ factors, such as σ70, is known to serve several important roles. In free σ70, the presence of this region prevents recognition of promoter DNA (53, 54). In holoenzyme, region 1.1 lies within a channel of core polymerase that will interact with dsDNA downstream of the transcription start site upon formation of the stable promoter/polymerase complex (36). Consequently, region 1.1 functions as a negatively-charged space keeper, which is replaced by the negatively-charged DNA. In addition, the interaction of region 1.1 with this channel increases the overall stability of the σ70/core interaction (55). Finally, in holoenzyme, the presence of σ70 region 1.1 modulates the rate of open complex formation, depending on the particular promoter (34, 35). At some promoters, σ70 region 1.1 stimulates the open complex formation, but at Pminor RPo formation is inhibited by region 1.1.
The important finding here is that the sequence of the Pminor spacer can decrease the activity of a promoter in the presence of region 1.1. How can a spacer influence the role of region 1.1? In the transition to RPo, the DNA around the start of transcription must melt, region 1.1 moves, the DNA is bent sharply into the channel, and the polymerase jaws close, stabilizing the complex (Fig. 1A) (reviewed in refs. 1–4). An AT-rich spacer, such as that in Pminor, may provide extra flexibility needed as the downstream region bends and unwinds (56). In particular, a T:A step within an A-tract, like position −17 of Pminor, can generate a flex point within the helix (49). In addition or alternatively, the spacer may provide a trajectory for the DNA that is conducive for channel entry. This possibility is consistent with the native gel and predicted promoter structure, which support the idea that the Pminor spacer imparts a particular curvature to the DNA. Therefore, the effect of region 1.1 may be caused by flexibility of the spacer in the context of RNA polymerase and/or promoter curvature via an intrinsic bend; in either case, region 1.1 would be inhibitory if the DNA is ready to enter the channel, but must wait for region 1.1 to exit. We speculate that Pminor exemplifies this type of promoter. Because the inhibition by region 1.1 is overcome when the Pminor spacer is replaced with either the GC-rich spacer or the AT-rich Pminor spacer complement (in the context of the Pmin7 promoter), we presume that the GC and complement spacers do not yield the same conformation as that of Pminor. It should be noted, though, that depending on the particular promoter, the presence of region 1.1 can change the rate of forming heparin-resistant complexes, the maximum amount of stable complexes that can be made, or both (Fig. 2, Fig. S1A, and Fig. S3B). Thus, further work is needed to determine what specific steps in initiation are affected by region 1.1.
It is interesting that the presence of the PlacUV5-Mut spacer within Pmin does not switch the sigma preference from σΔ1.1 to σfl even though the PlacUV5-Mut promoter is preferred by Eσfl. Thus, at PlacUV5-Mut, like λPR, Ptac, and PRNAI (34, 35), region 1.1 has a positive effect on the formation of stable polymerase/promoter complexes. This finding suggests that other properties of these promoters, not shared by Pmin, are needed for region 1.1 to stimulate promoter activity. We conclude that region 1.1 may have a pronounced inhibitory or “gatekeeper” function at promoters like Pminor, which have weak recognition elements coupled with a trajectory/conformation favorable for channel entry. In this way, region 1.1 may help polymerase discriminate against nonpromoter DNA and disfavor promoter sequences that are specific for alternate sigma factors, which lack region 1.1 (57). It is also worth noting that, unlike the promoter sequence elements, the spacer structure may be responsive to conditions (temperature) (41), DNA modifications (methylation), or transcription factors (CRP, MerR) (40, 58), and thereby provide an opportunity to regulate gene expression by modulating the formation of RPo.
Materials and Methods
DNA.
According to convention, the transcriptional start site is designated +1; DNA downstream of +1 is positively numbered and corresponds to the RNA transcript; the DNA upstream of +1 is numbered in the negative, beginning with −1 (there is no 0 nucleotide). The PlacUV5-Mut plasmid, pFW11-P2, contains a PlacUV5 derivative promoter (called P2) inserted between the EcoRI–SalI site of pFW11-null as described (37). Construction of the Pmin plasmid, pXBJ402, which contains Pminor sequences from −35 to +4 inserted between the EcoRI–SalI site of pFW11-null and the plasmids containing the Pmin derivatives Pmin2 (pIH4022), Pmin3 (pIH4023), Pmin7 (pIH4027), and Pmin8 (pIH4028) have been described (37). The Pmin11, Pmin16, Pmin/GC, Pmin/comp, Pmin7/GC, Pmin7/comp, and Pmin/lacUV5-Mut plasmids were constructed similarly. Linear templates for in vitro transcriptions, which were digested using the indicated restriction enzymes, and the 5′-32P end-labeled, 156-bp fragments used for the DNase I footprinting were prepared as described (37). The 51-bp oligomers, containing Pmin or Pmin derivatives (positions −41 to +10) or PlacUV5-Mut (positions −44 to +7) (Fig. 1B) were synthesized by Operon Biotechnologies, labeled at the 5′ end of the nontemplate strand using [γ-32P]ATP and T4 polynucleotide kinase, and then annealed to obtain the ds fragments used for native gel analysis.
In Vitro Transcription and DNase I Footprinting.
For single-round in vitro transcriptions, RNA polymerase was first reconstituted by incubating 0.2 pmol of core (Epicentre) and 0.5 pmol of either σfl or σΔ1.1, purified as described (34), in a 1.95 μL solution containing 27 mM Tris·Cl (pH 7.9), 54 mM Tris·acetate (pH 7.9), 52 mM NaCl, 40% (vol/vol) glycerol, 0.9 mM EDTA, 0.007% Triton X-100, 0.24 mM DTT, 154 mM potassium glutamate, 4.1 mM magnesium acetate, and 103 μg of BSA/μL for 15 min at 37 °C. A 2.05 μL solution containing 0.02 pmol of linearized DNA template and 22 mM Tris·Cl (pH 7.9), 43 mM Tris·acetate (pH 7.9), 71 mM NaCl, 3.4% (vol/vol) glycerol, 0.5 mM EDTA, 0.15 mM DTT, 220 mM potassium glutamate, 5.8 mM magnesium acetate, 146 μg of BSA/μl, and 0.34 mM 2-mercaptoethanol was then added, and the solution was incubated for the indicated times at 37 °C. Transcription was initiated by adding 1 μL of a solution containing 1 mM each of GTP, CTP, ATP and 50 μM [α-32P]UTP (6 × 104 dpm/pmol) and 0.5 mg/mL heparin. After 8 min at 37 °C, reactions were stopped by the addition of 15 μL of gel loading solution (94% deionized formamide, 9.4 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol FF) and heating at 95 °C for 2 min before electrophoresis on 6% polyacrylamide, 7 M urea, denaturing gels run in 1 × Tris-borate- EDTA. For the reactions in Fig. 1C and Fig. S1A, templates were digested with BglI. For the reactions in Fig. 3, the Pmin and Pmin7 templates were digested with BglI, resulting in 220-nt runoff transcripts, whereas the Pmin/GC and Pmin7/GC DNAs were digested with Bsu36I, and Pmin/comp and Pmin7/comp were digested with BaeI, yielding transcripts of 290 and 282 nt, respectively. The 3 transcript lengths allowed promoters with variant spacers to be assayed in the same reaction.
DNase I footprinting reactions were assembled as described for in vitro transcription assays except that the reaction also contained 2.25 mM CaCl2. The labeled templates used in DNase I footprinting were 156-bp PCR products containing either the Pmin or Pmin7 promoter. PCR was performed with either pXBJ402 (Pmin) or pIH4027 (Pmin7) template, Pfu polymerase (Stratagene), and primers chosen to produce a fragment from position −99 to position +57, relative to the transcriptional +1 start. Primers were 5′ end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) before PCR. Each reaction contained labeled primer that annealed to one strand, and unlabeled primer that annealed to the other strand. The [γ-32P]-labeled PCR product was purified by gel electrophoresis. For the DNase I reactions, DNA and reconstituted RNAP were incubated at 37 °C for 10 min. Protein–DNA complexes were challenged with heparin for 30 s and then treated with DNase I (0.5 unit) for 30 s at 37 °C. Loading-stop solution [28 μL of 192 mM ammonium acetate, 32 mM EDTA, 0.14% (wt/vol) SDS, and 0.036 mg/mL calf thymus DNA] was added, and the mixture was immediately loaded onto 4% native-polyacrylamide gels. Retarded protein–DNA complexes, identified after autoradiography, were cut out of the gel, embedded in a 1% (wt/vol) agarose gel, electro-eluted onto NA45 membranes (Schleicher & Schuell), and eluted off the membranes by incubating in a solution of 10 mM Tris·HCl (pH 8), 1 mM EDTA, and 1 M NaCl for 30 min at 65 °C. The DNA, obtained after extraction with phenol and precipitation in ethanol, was then run on the denaturing gels.
After autoradiography, films were scanned by using a Powerlook 100XL densitometer and various species were quantified by using Quantity One software from Bio-Rad.
Native PAGE.
Annealed promoter fragments were diluted in 1 × T4 ligase buffer (NEBL) to 0.2 mM. 5 × loading dye (40% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol FF) was added, and the samples were loaded onto a 12% native gel (acrylamide/bis-acrylamide of 29:1). Electrophoresis was carried out in 1 × TAE, at 4 °C at 200 V for 26 h.
Modeling of DNA Fragments in Silico.
The 51-bp sequence of each DNA promoter fragment assessed by native-PAGE was copied into the “model.it” web server (http://hydra.icgeb.trieste.it/dna/model_it.html) (52). Parameters were set for “Electrophoresis (dinucleotide).” The resulting structure predictions were downloaded in pdb format and aligned in MacPyMOL. Each DNA sequence shares the common sequence GAATTC at the 5′ end, such that all structures are identical and perfectly align in this region. Pmin, Pmin/GC, and Pmin/comp have identical sequences except for their 14-bp spacer sequences.
Supplementary Material
Acknowledgments.
We thank R. Bonocora, T. James, K. Baxter, L. Knipling, L. Shern, S. Adhya, and R. Martin for helpful discussions. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808133106/DCSupplemental.
References
- 1.Hook-Barnard IG, Hinton DM. Transcription initiation by mix and match elements: Flexibility for polymerase binding to bacterial promoters. Gene Regul Syst Biol. 2007;2007:275–293. [PMC free article] [PubMed] [Google Scholar]
- 2.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu LM. Open season on RNA polymerase. Nat Struct Biol. 2002;9:502–504. doi: 10.1038/nsb0702-502. [DOI] [PubMed] [Google Scholar]
- 4.Murakami KS, Darst SA. Bacterial RNA polymerases: The wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 5.Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 7.Cook VM, Dehaseth PL. Strand opening-deficient E. coli RNA polymerase facilitates investigation of closed complexes with promoter DNA: Effects of DNA sequence and temperature. J Biol Chem. 2007;282:21319–21326. doi: 10.1074/jbc.M702232200. [DOI] [PubMed] [Google Scholar]
- 8.Buc H, McClure WR. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 9.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. Topography of intermediates in transcription initiation of E. coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacic RT. The 0 °C closed complexes between Escherichia coli RNA polymerase and two promoters, T7–A3 and lacUV5. J Biol Chem. 1987;262:13654–13661. [PubMed] [Google Scholar]
- 11.Sasse-Dwight S, Gralla JD. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- 12.Kontur WS, Saecker RM, Davis CA, Capp MW, Record MT., Jr Solute probes of conformational changes in open complex (RPo) formation by Escherichia coli RNA polymerase at the lambdaPR promoter: Evidence for unmasking of the active site in the isomerization step and for large-scale coupled folding in the subsequent conversion to RPo. Biochemistry. 2006;45:2161–2177. doi: 10.1021/bi051835v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saecker RM, et al. Kinetic studies and structural models of the association of E. coli sigma(70) RNA polymerase with the lambdaP(R) promoter: Large-scale conformational changes in forming the kinetically significant intermediates. J Mol Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 14.Kontur WS, Saecker RM, Capp MW, Record MT., Jr Late steps in the formation of E. coli RNA polymerase-lambda P R promoter open complexes: Characterization of conformational changes by rapid [perturbant] upshift experiments. J Mol Biol. 2008;376:1034–1047. doi: 10.1016/j.jmb.2007.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim HM, Lee HJ, Roy S, Adhya S. A “master” in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc Natl Acad Sci USA. 2001;98:14849–14852. doi: 10.1073/pnas.261517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kainz M, Roberts J. Structure of transcription elongation complexes in vivo. Science. 1992;255:838–841. doi: 10.1126/science.1536008. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen LH, Burgess RR. Comparative analysis of the interactions of Escherichia coli sigma S and sigma 70 RNA polymerase holoenzyme with the stationary-phase-specific bolAp1 promoter. Biochemistry. 1997;36:1748–1754. doi: 10.1021/bi961175h. [DOI] [PubMed] [Google Scholar]
- 18.Sasse-Dwight S, Gralla JD. Footprinting protein-DNA complexes in vivo. Methods Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- 19.Brodolin K, Zenkin N, Severinov K. Remodeling of the sigma70 subunit nontemplate DNA strand contacts during the final step of transcription initiation. J Mol Biol. 2005;350:930–937. doi: 10.1016/j.jmb.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 20.Davis CA, Bingman CA, Landick R, Record MT, Jr, Saecker RM. Real-time footprinting of DNA in the first kinetically significant intermediate in open complex formation by Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2007;104:7833–7838. doi: 10.1073/pnas.0609888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spassky A, Kirkegaard K, Buc H. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry. 1985;24:2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- 22.Haugen SP, et al. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: An additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 23.Gourse RL. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vassylyev DG, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6-Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 25.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme–DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 26.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4-Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 27.Campbell EA, et al. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 28.Young BA, Gruber TM, Gross CA. Views of transcription initiation. Cell. 2002;109:417–420. doi: 10.1016/s0092-8674(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang G, et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3-Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 30.Malhotra A, Severinova E, Darst SA. Crystal structure of a sigma 70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 31.Patikoglou GA, et al. Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J Mol Biol. 2007;372:649–659. doi: 10.1016/j.jmb.2007.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craig ML, et al. DNA footprints of the 2 kinetically significant intermediates in formation of an RNA polymerase-promoter open complex: Evidence that interactions with start site and downstream DNA induce sequential conformational changes in polymerase and DNA. J Mol Biol. 1998;283:741–756. doi: 10.1006/jmbi.1998.2129. [DOI] [PubMed] [Google Scholar]
- 33.Haugen SP, Ross W, Manrique M, Gourse RL. Fine structure of the promoter-sigma region 1.2 interaction. Proc Natl Acad Sci USA. 2008;105:3292–3297. doi: 10.1073/pnas.0709513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vuthoori S, Bowers CW, McCracken A, Dombroski AJ, Hinton DM. Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J Mol Biol. 2001;309:561–572. doi: 10.1006/jmbi.2001.4690. [DOI] [PubMed] [Google Scholar]
- 35.Wilson C, Dombroski AJ. Region 1 of sigma70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1997;267:60–74. doi: 10.1006/jmbi.1997.0875. [DOI] [PubMed] [Google Scholar]
- 36.Mekler V, et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase–promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 37.Hook-Barnard I, Johnson XB, Hinton DM. Escherichia coli RNA polymerase recognition of a sigma70-dependent promoter requiring a −35 DNA element and an extended −10 TGn motif. J Bacteriol. 2006;188:8352–8359. doi: 10.1128/JB.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thouvenot B, Charpentier B, Branlant C. The strong efficiency of the Escherichia coli gapA P1 promoter depends on a complex combination of functional determinants. Biochem J. 2004;383:371–382. doi: 10.1042/BJ20040792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auble DT, Allen TL, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase. Effects of substitutions in the spacer DNA separating the −10 and −35 regions. J Biol Chem. 1986;261:11202–11206. [PubMed] [Google Scholar]
- 40.Liu M, Tolstorukov M, Zhurkin V, Garges S, Adhya S. A mutant spacer sequence between −35 and −10 elements makes the Plac promoter hyperactive and cAMP receptor protein-independent. Proc Natl Acad Sci USA. 2004;101:6911–6916. doi: 10.1073/pnas.0401929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Repoila F, Gottesman S. Temperature sensing by the dsrA promoter. J Bacteriol. 2003;185:6609–6614. doi: 10.1128/JB.185.22.6609-6614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warne SE, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase. Effects of single base pair deletions and insertions in the spacer DNA separating the −10 and −35 regions are dependent on spacer DNA sequence. Biochemistry. 1993;32:6134–6140. doi: 10.1021/bi00075a003. [DOI] [PubMed] [Google Scholar]
- 43.Chan B, Spassky A, Busby S. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus −35 region sequences. Biochem J. 1990;270:141–148. doi: 10.1042/bj2700141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan B, Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989;84:227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- 45.Mellies J, Brems R, Villarejo M. The Escherichia coli proU promoter element and its contribution to osmotically signaled transcription activation. J Bacteriol. 1994;176:3638–3645. doi: 10.1128/jb.176.12.3638-3645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lozinski T, Adrych-Rozek K, Markiewicz WT, Wierzchowski K. Effect of DNA bending in various regions of a consensus-like Escherichia coli promoter on its strength in vivo and structure of the open complex in vitro. Nucleic Acids Res. 1991;19:2947–2953. doi: 10.1093/nar/19.11.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collis CM, Molloy PL, Both GW, Drew HR. Influence of the sequence-dependent flexure of DNA on transcription in E. coli. Nucleic Acids Res. 1989;17:9447–9468. doi: 10.1093/nar/17.22.9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mollegaard NE, Lindemose S, Nielsen PE. Uranyl photoprobing of nonbent A/T- and bent A-tracts. A difference of flexibility? Biochemistry. 2005;44:7855–7863. doi: 10.1021/bi0502083. [DOI] [PubMed] [Google Scholar]
- 49.Mack DR, Chiu TK, Dickerson RE. Intrinsic bending and deformability at the T-A step of CCTTTAAAGG: A comparative analysis of T-A and A-T steps within A-tracts. J Mol Biol. 2001;312:1037–1049. doi: 10.1006/jmbi.2001.4994. [DOI] [PubMed] [Google Scholar]
- 50.Crothers DM, Haran TE, Nadeau JG. Intrinsically bent DNA. J Biol Chem. 1990;265:7093–7096. [PubMed] [Google Scholar]
- 51.Koo HS, Wu HM, Crothers DM. DNA bending at adenine thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 52.Vlahovicek K, Kajan L, Pongor S. DNA analysis servers: Plot. it, bend.it, model.it, and IS. Nucleic Acids Res. 2003;31:3686–3687. doi: 10.1093/nar/gkg559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dombroski AJ, Walter WA, Gross CA. Amino-terminal amino acids modulate sigma-factor DNA-binding activity. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 54.Dombroski AJ, Walter WA, Record MT, Jr, Siegele DA, Gross CA. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 55.Hinton DM, Vuthoori S, Mulamba R. The bacteriophage T4 inhibitor and coactivator AsiA inhibits Escherichia coli RNA polymerase more rapidly in the absence of sigma70 region 1.1: Evidence that region 1.1 stabilizes the interaction between sigma70 and core. J Bacteriol. 2006;188:1279–1285. doi: 10.1128/JB.188.4.1279-1285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolasa IK, Lozinski T, Wierzchowski KL. Effects of distortions by A-tracts of promoter B-DNA spacer region on the kinetics of open complex formation by Escherichia coli RNA polymerase. Acta Biochim Pol. 2003;50:909–920. [PubMed] [Google Scholar]
- 57.Typas A, Hengge R. Role of the spacer between the −35 and −10 regions in sigmas promoter selectivity in Escherichia coli. Mol Microbiol. 2006;59:1037–1051. doi: 10.1111/j.1365-2958.2005.04998.x. [DOI] [PubMed] [Google Scholar]
- 58.Ansari AZ, Chael ML, O'Halloran TV. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature. 1992;355:87–89. doi: 10.1038/355087a0. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz EC, et al. A full-length bacterial sigma factor adopts a compact structure incompatible with DNA binding. Chem Biol. 2008;15:1091–1103. doi: 10.1016/j.chembiol.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.