Abstract
F-box proteins are substrate-recognition components of the Skp1-Rbx1-Cul1-F-box protein (SCF) ubiquitin ligases. In plants, F-box genes form one of the largest multigene superfamilies and control many important biological functions. However, it is unclear how and why plants have acquired a large number of F-box genes. Here we identified 692, 337, and 779 F-box genes in Arabidopsis, poplar and rice, respectively, and studied their phylogenetic relationships and evolutionary patterns. We found that the plant F-box superfamily can be divided into 42 families, each of which has a distinct domain organization. We also estimated the number of ancestral genes for each family and identified highly conservative versus divergent families. In conservative families, there has been little or no change in the number of genes since the divergence between eudicots and monocots ≈145 million years ago. In divergent families, however, the numbers have increased dramatically during the same period. In two cases, the numbers of genes in extant species are >100 times greater than that in the most recent common ancestor (MRCA) of the three species. Proteins encoded by highly conservative genes always have the same domain organization, suggesting that they interact with the same or similar substrates. In contrast, proteins of rapidly duplicating genes sometimes have quite different domain structures, mainly caused by unusually frequent shifts of exon-intron boundaries and/or frameshift mutations. Our results indicate that different F-box families, or different clusters of the same family, have experienced dramatically different modes of sequence divergence, apparently having resulted in adaptive changes in function.
Keywords: birth-and-death evolution, F-box protein, multigene family, plant evolution
Recent studies of developmental biology and comparative genomics have shown that physiological and morphological characters are generally controlled by genes belonging to multigene families (1). Investigations of the evolution of multigene families are thus an important step for understanding the evolution of phenotypic characters. Much has been known about the relationship between the evolutionary and functional changes within multigene families. In particular, it has been shown that most multigene families are subject to birth-and-death evolution, although the rates of gain and loss of genes vary considerably among families (2). In the families that have experienced rapid birth-and-death evolution, the number of genes may be quite different between closely related species, or even between individuals of the same species (3–5). The reason for the great variation in gene number is not always clear. However, several recent studies suggest that the number of genes in a family was initially determined by their functional requirement, but after the number reached a sufficient level, the number can increase or decrease by chance (2, 4, 6, 7).
The development and functioning of an organism require cellular response to a variety of internal and external signals. One mechanism for such responses is to change the abundance of key regulators via protein degradation by the proteosome (8). Protein degradation by the proteosome is a relatively conserved process, and requires the attachment of multiple ubiquitin molecules to target proteins. The attachment of ubiquitin to target proteins is accomplished by the sequential action of three enzymes, E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligases). E1 and E2 are relatively nonspecific, whereas different E3 enzymes recognize different substrates for ubiquitination (9). One type of best-characterized E3 ubiquitin ligases are the SCF protein complexes formed by Skp1, Cullin, Rbx1, and F-box proteins (10). Within each SCF complex, Cullin and Rbx1 form a core scaffold and Skp1 connects the scaffold to an F-box protein, which in turn confers substrate specificity (11, 12).
Since the discovery of the first F-box protein (Cyclin F) from human (13), numerous F-box proteins have been identified by the presence of a well-conserved N-terminally located ≈60-aa F-box domain (14). Interestingly, the number of F-box genes varies greatly from species to species (13–15). In budding yeast, nematode, fruit-fly, and human, 14, 337, 24, and 38 F-box genes have been reported, respectively (14). In plants, at least 694 and 687 F-box genes have been identified in the Arabidopsis thaliana (hereafter called Arabidopsis) and Oryza sativa (hereafter called rice) genomes (15, 16), respectively, making the F-box superfamily one of the largest in plants. Most likely because of their roles in protein ubiquitination and degradation, plant F-box genes have been found genetically to control many crucial processes such as embryogenesis, hormonal responses, seedling development, floral organogenesis, senescence, and pathogen resistance (17). Therefore, it is important to investigate how F-box genes have evolved in plants and how and why plants have acquired so many F-box genes.
Fortunately, the genomes of several plant species have been (nearly) completely sequenced and carefully annotated. This makes it possible to perform genome-wide comparisons to investigate the patterns of gene number variation and gene family expansion within the plant F-box superfamily. In this study, we have conducted detailed evolutionary studies of F-box genes from Arabidopsis, Populus trichocarpa (hereafter called poplar) and rice, the three plant species whose genomes have been best annotated. In angiosperms, Arabidopsis and poplar are representatives of the eudicot lineage, whereas rice is a member of the monocot lineage. Molecular data have suggested that the divergence between monocots and eudicots and that between Arabidopsis and poplar occurred ≈145 and ≈100 million years ago (Mya), respectively (18). Therefore, the comparison between the three species has allowed us to understand the general pattern of the evolution of plant F-box genes over much of the angiosperm history.
Results
Number and Domain Organization of F-Box Proteins.
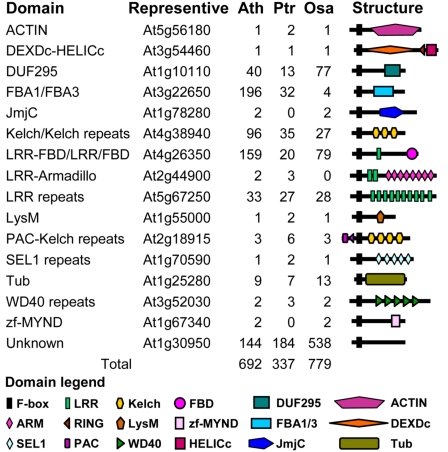
We performed BLAST and HMMer searches and identified 698, 337, and 858 putative F-box proteins from Arabidopsis, poplar, and rice, respectively, with E-values ≤10.0. SMART and Pfam analyses further reduced the numbers to 692, 337, and 779 (Fig. 1; and see Dataset S1). The number of F-box genes in Arabidopsis was very close to that reported previously (694) (15), whereas that in rice was 13.4% greater than the previously reported number (687) (16). Rice has a slightly larger (12.6%) collection of F-box genes than Arabidopsis, whereas poplar has dramatically fewer (51.3%) genes than Arabidopsis. The predicted proteomes of poplar and rice contain 58,036 and 66,710 proteins, 70.3% and 95.3% larger than that of Arabidopsis (34,151), respectively. This suggested that the numbers of F-box proteins were not proportional to the sizes of the predicted proteomes.
Fig. 1.
Number and domain structure of F-box proteins from Arabidopsis (Ath), poplar (Ptr), and rice (Osa).
In addition to the F-box domain, many plant F-box proteins have other domains in the C-terminal regions (14). These additional domains have been shown, or were predicted, to interact with various substrates (14). Using the SMART and Pfam databases, we confirmed the existence of all previously identified C-terminal domains in plant F-box proteins (Fig. 1). We also found that proteins with some domains (e.g., Actin, JmjC, and LysM) were small in number in all three species, whereas those with other domains (e.g., FBA, Kelch, and LRR) were consistently large in number. F-box proteins with not-yet-defined (or unknown) C-terminal domains were also large in number.
Highly Conservative and Divergent F-Box Genes.
To understand the evolution of the plant F-box superfamily, we conducted phylogenetic analyses with all 1,808 F-box genes from Arabidopsis, poplar and rice. Although the bootstrap values for many interior branches were low because of the large number of sequences and the small size of the F-box domain, the topology and groupings were generally reasonable because proteins with the same or similar domain organization usually clustered together (Fig. S1). Based on phylogenetic relationships and domain organizations, we divided the F-box gene superfamily into 42 groups, or families, each of which contained genes encoding proteins with the same or similar domain organizations. Interestingly, proteins with unknown domains either formed their own families, or were scattered in the families formed by proteins with other domains. This implied that the evolutionary histories of the proteins with unknown C-terminal domains might be complex.
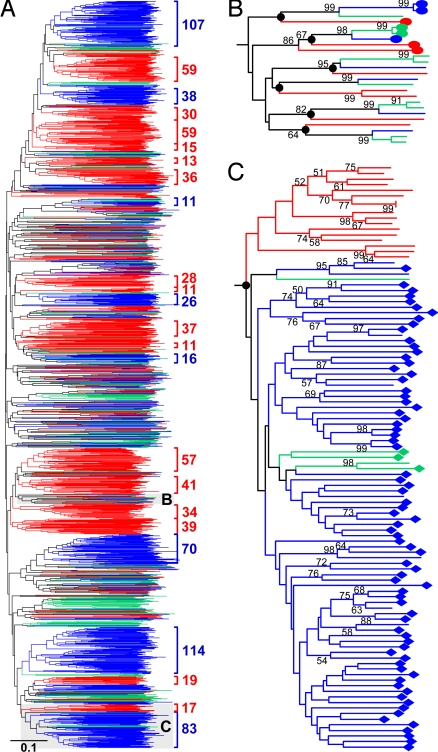
Phylogenetic analyses allowed us to identify evolutionarily conservative and divergent F-box genes. From Fig. 2A, it is clear that there were numerous gene families or clades that were composed of sequences from Arabidopsis, poplar, and rice. Many of these clades also contained similar numbers of genes from each species (Fig. 2B), suggesting that major expansion/contraction in gene number have not occurred since the divergence between eudicots (Arabidopsis and poplar) and monocots (rice). In 20 cases, each clade was composed of only three genes, one from each of the three species, and the gene tree was congruent with the species phylogeny (Table S1). In 63 other cases, there were fewer than three genes from each species, suggestive of limited, lineage-specific gene gains after the eudicot/monocot and/or Arabidopsis/poplar splits. Taken together, our phylogenetic analyses revealed 83 strongly supported conservative F-box gene clades. The fact that members of each clade usually have identical domain organization suggested that they function to interact with the same or similar substrates.
Fig. 2.
Phylogenetic relationships of F-box proteins from Arabidopsis, poplar, and rice. (A) A simplified version of the neighbor joining (NJ) tree, with sequences from Arabidopsis, poplar, and rice being color-coded blue, green, and red, respectively. Species-specific clusters with more than ten genes are highlighted with bracket. The full phylogeny is shown in Fig. S1. (B) Part of the tree in (A) showing examples of evolutionarily conserved gene clusters. The 24 genes belong to six gene clusters, each of which contains one or two genes from each species. The black dots stand for the ancestral genes in the most recent common ancestor (MRCA) of the three species. Genes encoding Kelch domain-containing proteins are labeled with ovals. (C) Part of the tree in (A) showing examples of extensively duplicating gene clusters. The 105 genes (83 from Arabidopsis, 5 from poplar, and 17 from rice) were derived from a single ancestral gene from the MRCA of the three species. Genes encoding FBA domain-containing proteins are labeled with diamonds.
Fig. 2A also shows that a great number of genes formed lineage-specific clusters. The largest of such clusters had 114 Arabidopsis genes and contained S-locus-like F-box genes that are involved in self recognition during pollination (17). The second largest lineage-specific cluster was composed of 107 Arabidopsis genes, including one that participates in pathogen response (17). When we only counted the lineage-specific clusters with more than five genes, Arabidopsis, poplar and rice contained 14, 13 and 23 such clusters, respectively. In total, the numbers of genes belonging to the lineage-specific clusters were 505 (73.0% of 692), 136 (40.4% of 337), and 564 (72.4% of 779) in Arabidopsis, poplar and rice, respectively. The large numbers of lineage-specific clusters in each species suggested that many subsets of the F-box gene superfamily have experienced extensive gene duplications.
Contrasting Changes in the Numbers of F-Box Genes.
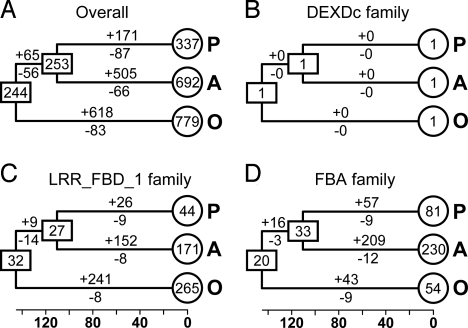
To better understand how F-box genes have evolved in plants, we estimated the number of F-box genes in the most recent common ancestor (MRCA) of eudicots and monocots and that of Arabidopsis and poplar (Table S2). Reconciliation of the gene trees with the species phylogeny suggested that there were ≈244 ancestral F-box genes in the MRCA of eudicots and monocots (Fig. 3A). When the numbers of ancestral genes were compared with those in Arabidopsis and rice, it appeared that the F-box superfamily has increased in size as much as tripling since the divergence of eudicots and monocots ≈145 Mya. However, the expansion was uneven among gene families and between plant species. In several families (e.g., Actin and JmjC), the number of genes remained unchanged, or nearly so, since the eudicot/monocot split (Fig. 3B; Table S2). In contrast, in other families, the number had increased dramatically. In the LRR_FBD_1 family, for example, there were 171, 44, and 265 genes in Arabidopsis, poplar, and rice, respectively, and the estimated number of genes in the MRCA of eudicots and monocots was 32 (Fig. 3C; Table S2). Arabidopsis, poplar, and rice have gained 161, 35 and 241 genes and lost 22, 23, and 8 genes, respectively, since their splits (Fig. 3C). Clearly, the numbers of genes gained in the Arabidopsis and rice lineages were much greater than that in the poplar lineage.
Fig. 3.
Evolutionary change of the number of F-box proteins in plants. The numbers in circles and rectangles represent the numbers of genes in extant and ancestral species, respectively. The numbers with plus and minus signs indicate the numbers of gene gains and losses, respectively, for each branch.
We also noted that, in several families, rapidly duplicating genes coexisted with conservative ones, suggestive of unequal rates in gene duplication. Therefore, the estimated birth rates for some lineage-specific clusters may be much higher than those for the whole family. In the aforementioned 114- and 107-gene clusters, for example, the numbers of genes in extant species (Arabidopsis) were 114 and 107 times the single gene in the MRCA of eudicots and monocots. These unusually high rates of gene birth were not only significantly greater than those estimated for the two families that they belong to, but also greater than those observed for many other well-known rapidly-duplicating genes, such as disease resistance (R) loci (19), Type I MADS-box genes (20), receptor-like kinase genes (21), and SKP1-like genes (22).
Great Contributions of Tandem Duplication.
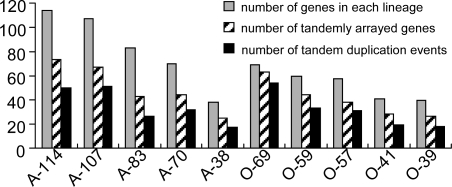
Previous studies have shown that F-box genes sometimes can form tandem arrays in the same chromosomal regions, suggestive of tandem duplication (15, 16). To investigate the contribution of tandem duplication to the expansion of the F-box superfamily, we combined the information of phylogenetic relationships and chromosomal locations. Tandem duplication was inferred if closely related genes were located within the same chromosomal region (i.e., fewer than 20 genes apart from each other). We started with the lineage-specific clusters, and found that 73 (64.0%) and 67 (62.6%) genes of the aforementioned 114- and 107-gene clusters belonged to tandem arrays (Fig. 4; Table S3). In total, the generation of ≈306 (44.3% of 692) Arabidopsis and 389 (50.0% of 779) rice F-box genes could be explained by tandem duplication.
Fig. 4.
Contribution of tandem duplication to the expansion of the five largest species-specific gene clusters in Arabidopsis and rice.
In addition to tandem duplication, segmental duplications also played a role in the expansion of the F-box superfamily. By investigating duplicate gene pairs within conservative clusters, we found that at least 18 pairs of paralogous genes were likely results of segmental duplication because they were located in the chromosomal segments with clear synteny (Fig. S2; Table S1). However, this was probably an underestimate because segmental duplications in large, lineage-specific clusters may have been obscured by repeated tandem duplications.
Strong Correlation Between Evolutionary Pattern and Gene Function.
The distinct modes of evolutionary patterns in the F-box superfamily raised the question whether genes with a certain type of function tend to evolve through specific mechanisms (23). To address this question, we reviewed the literature for functionally characterized F-box genes, and investigated the relationship between gene function and evolutionary pattern. We classified the F-box genes with known function into two categories: (i) those that are involved in the relatively conserved processes (such as embryogenesis, seedling development, circadian rhythms, floral development, and senescence); and (ii) those that are involved in relatively specialized processes (such as pollen recognition and pathogen response). We found that genes belonging to the first category tended to have experienced little or no change in gene number (Table S4). In contrast, genes belonging to the second category usually formed lineage-specific clusters and experienced frequent gene duplications (Table S4). This clearly demonstrated that the evolutionary patterns of F-box genes were related with the type of their functions.
Conservation and Variation in Domain Composition and Organization.
We have shown that domain composition and organization of F-box proteins were usually similar among members of the same family. However, in some families, especially in those that contain large, lineage-specific gene clusters, proteins with the predominant domain organizations usually grouped together with those that had related, but distinct, domain structures (Fig. S1). For example, among the 470 members of the LRR_FBD_1 family, 134 (28.5%) had both LRR2 and FBD domains, 55 (11.7%) and 54 (11.5%) had LRR2 or FBD domain, respectively, and 227 (48.3%) had neither domain. Because the presence of both domains was likely to be the ancestral state, it seemed that in 336 (71.5% of 470) proteins, LRR2 and/or FBD domains had undergone some degree of degeneration and were thus no longer detectable (Fig. 5A; Fig. S1). Similar phenomena were found in many other families (Fig. 5 B and C; Fig. S1), suggesting that the loss of the characteristic C-terminal domains was a relatively easy process. This also implied that some of the proteins with unknown domains lacked any defined domains from inception, whereas others were derived from progenitors with known domains.
Fig. 5.
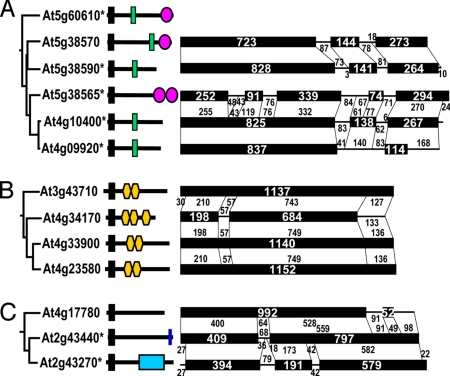
Phylogenetic relationships, domain organization, and exon-intron structure of representatives of recently duplicated F-box genes. (A) Six genes of the LRR_FBD_1 family, showing the loss of the LRR and FBD domains in two and three cases, respectively, as well as the gain of an additional FBD domain in one case. Note that the presence of both LRR and FBD domain is likely to be the ancestral state. (B) Four genes of the Kelch_2 family, showing the gain of an additional Kelch domain in one case. (C) Three genes of the FBA family, showing the gain of the transmembrane motif and the FBA domain, respectively. These genes are phylogenetically closely related, and are highly comparable at the genomic sequence level. However, because of shifts in exon-intron boundary and/or out-of-frame insertions/deletions, the protein products of them have evolved distinct domain organization. Annotated genes with expressed sequence tag (EST) supports are labeled with asterisks.
The frequent losses of C-terminal domains raised the question about the evolutionary fates of duplicated F-box genes. To investigate the mechanism by which duplicated F-box genes have diverged, we compared the genomic sequences of the recently duplicated gene pairs. We found that many closely related duplicates (paralogs) showed dramatic differences in the number and boundaries of intron(s) and exon(s). The intronic sequences in one gene may become exonic sequences in the other (exonization), and vice versa (pseudoexonization) (Fig. 5). In addition, we found that 55 (98.2%) of the 56 investigated pairs of recent paralogs had detectable shifts in exon-intron boundary (Fig. S3). This was a surprising finding. To verify the results with expression data, we searched for cDNAs and found that 17 (or 30.9%) of the 56 investigated pairs were supported by EST (short for “expressed sequence tag”) data for both paralogs. Apparently, exonization and/or pseudoexonization have been a prevalent mechanism for the divergence between duplicate F-box genes. In many cases, shifts in exon-intron boundary, as well as insertions and/or deletions in exons, had caused changes in reading frames and, therefore, resulted in dramatic alterations in domain composition and organization (Fig. 5). This suggested that proteins encoded by rapidly duplicating F-box genes may potentially be able to interact with distinct substrates.
Discussion
Birth-and-Death Evolution of Plant F-Box Genes.
In this study, we have established the evolutionary backbone of the F-box superfamily in plants, and uncovered patterns of duplication and diversification in each family. We showed that many F-box families have maintained stable copy numbers, whereas many others have experienced rapid birth-and-death evolution, with birth rates much higher than death rates. In several families or clusters, the estimated birth rates are much greater than that estimated for all other well-known rapidly-duplicating genes. The extraordinarily high birth rates raised the question why plants have recruited so many new genes during evolution. One hypothesis is that members of the rapidly-duplicating gene families/clusters function to interact with variable and/or changing targets. During plant evolution, duplications have generated a greater number of genes whose products need to be regulated by proteolysis. Consequently, an increased number of F-box genes might have evolved to allow plant cells to selectively recognize the targets for degradation in a precisely controlled fashion.
Support for this hypothesis was found in the FBA and LRR_FBD_1 families. In the FBA family, some members of the 114-gene, Arabidopsis-specific cluster have been shown to be related to genes that are critical for pollen recognition (24). In plants, the so-called self-incompatibility system allows non-self pollen grains to germinate on the pistil and prevents inbreeding. Because a new allele may permit the pollen to pollinate a pistil carrying any of the previous alleles, the plants carrying the new allele usually have the potential to spread rapidly within the population. Therefore, a large number of F-box genes might facilitate the generation of new alleles, possibly via unequal crossing over. However, because Arabidopsis is self-fertile, it is not clear why it has many members in the FBA family. In the LRR_FBD_1 family, the dramatic increase in gene number might be due to the selection by variable pathogen-borne targets, because a few members of this family are involved in defense responses (25).
Although there are some correlations between the evolutionary pattern and function of F-box genes, it is still possible that, at least in some families, the change in gene number was due to random events. Several recent studies have shown that the variation in gene number does not always result in the difference in fitness (3, 6, 26). In particular, random genomic drift has been shown to play key roles in the evolution of rapidly-duplicating genes, such as chemosensory receptor genes (27). In the F-box gene superfamily, however, it has been difficult to evaluate the extent of genomic drift because the Arabidopsis, poplar and rice genomes have diverged more than 100 Mya. Nevertheless, we cannot rule out the possibility that the gain or loss of some F-box genes were due to random events for two reasons. First, the extraordinarily high birth rates in some families/clusters make it possible that closely related species have quite different numbers of genes. Second, some recently duplicated F-box genes have been found to be functionally redundant (28, 29), suggesting that small changes in gene number do not necessarily cause changes in the physiological requirement. However, to address this question, F-box genes from closely related species, or individuals of the same species, need to be investigated.
Interestingly, SKP1-like genes have also been shown to evolve via a rapid birth-and-death process (22). In Arabidopsis and rice, there are 18 and 28 SKP1-like genes, respectively. Phylogenetic analysis suggested that these genes were derived from only one ancestral gene in the MRCA of the two species. Because Skp1-like proteins function to link the F-box proteins to Cullin and Rbx1, the rapid increase in the numbers of both Skp1 and F-box proteins would result in dramatic increases of the number of potential SCF complexes. The large number of possible SCF complexes would in turn allow the plants to respond to a wide array of intrinsic and extrinsic changes by modulating the abundance of the key regulatory proteins.
Domain Evolution in the F-Box Protein Superfamily.
The fact that some F-box gene lineages have experienced extensive duplications makes the F-box superfamily an excellent system for the study of the evolutionary fate of duplicated genes. In particular, sequence divergence via exonization of intronic sequences and pseudoexonization of exonic sequences occurred frequently, resulting in coding region differences of recently duplicated genes. However, this mechanism is not specific to the F-box genes. It has recently been reported in MADS-box gene family (30), and was believed to play key roles in the generation of the genes with distinct structures and novel functions. In MADS-box transcription factors, the N-terminal MADS-box domains are highly conserved and form DNA-binding dimmers. However, the C-terminal regions are variable but contain short, relatively conserved and lineage-specific motifs, and are involved in the formation of higher-order protein complexes. It was shown that the shift in exon-intron boundaries has resulted in the evolution of novel C-terminal motifs in MADS-box proteins (31, 32).
The C-terminal regions of some F-box proteins are critical for target recognition. Therefore, the differences caused by exon-intron boundary shifts and/or frameshift mutations may lead to the differences in function. TIR1 and AFB2, for example, are paralogs that were generated through a very recent duplication event, and are both involved in auxin signaling. However, small differences in the C-terminal regions are responsible for their isoform-specific roles in recognizing different members of the AUX/IAA family (28). To some extent, the selective interactions between F-box proteins and the variable targets are very similar to the arms races between hosts and pathogens. If so, then we would expect that some F-box proteins have evolved under positive selection. Indeed, in nematodes and plants, it has been suggested that some F-box proteins evolved under positive selection at sites in substrate-binding domains (33). However, we showed here that, because of the frequent frame-shift mutations caused by (pseudo)exonization and/or insertions/deletions, the C-terminal sequences may no longer be homologous even between closely related duplicate genes. Therefore, it is not always practical to accurately calculate the Ka/Ks values for parts of the C-terminal regions of F-box genes.
Methods
Sequence Retrieval and Domain Analysis.
F-box proteins were retrieved by BLASTP searches against the Arabidopsis, poplar and rice protein databases at the websites TAIR (http://www.arabidopsis.org/Blast/index.jsp), JGI (http://genome.jgi-psf.org/cgi-bin/runAlignment?db = Poptr1_1&advanced = 1), and TIGR (http://tigrblast.tigr.org/euk-blast/index.cgi?project = osa1), respectively, or by Hidden Markov Model (HMM) searches against the downloaded proteomes of the three species. Presence/absence of the F-box domain (PF00646) and other domains were checked in the SMART and Pfam websites. Note that the FBA_1 (PF07734) and FBA_3 (PF08268) domains were treated as the same domain, FBA, because: 1) they are very similar in sequence structure; 2) they are sometimes not distinguishable in SMART and Pfam analysis; and 3) proteins with the two domains are usually mixed together in phylogenetic trees.
Alignment and Phylogenetic Analysis.
Three strategies were tried to generate the alignment of the 1,808 F-box proteins from the three species. At the beginning, we used the CLUSTALX 1.81 (34) program and found that the alignment was not good because of too many obvious errors. Then, we adjusted the alignment manually and obtained a better alignment in which the F-box domain regions were aligned relatively accurately. However, because of the huge size of the data set and the uncertainty in sequence comparison, the new alignment was still not very accurate. Finally, we used the HMMalign program of the HMMer software package (35). We found that the alignment generated this way was of high quality because residues within the F-box domain regions were reasonably aligned. Phylogenetic analyses of the F-box proteins based on amino acid sequences were carried out using neighbor joining (NJ) methods in MEGA version 4 (36). NJ analyses were done on the F-box domain region only, using p-distance or Poisson Correction methods, pairwise deletion of gaps, and the default assumptions that the substitution patterns among lineages and substitution rates among sites were homogeneous. Support for each node was tested with 1,000 bootstrap replicates.
Determination of the Mechanisms for Gene Diversification.
Differences between duplicate genes were first observed from the alignments of protein sequences. Then, detailed comparisons of the genomic sequences between closely related paralogous genes were conducted to understand the mechanism by which a diverged C-terminal region was generated. During this process, the exon of one gene was aligned with its candidate counterpart (i.e., the exon of the other gene that are located at the same position) and the two adjacent introns (both upstream and downstream of the exon) to see whether the intronic sequence of one gene matched well to the exonic sequence of the other gene, or vice versa. Exonization was defined as the process in which intronic sequences became exonic sequences, and pseudoexonization was the opposite process.
Supplementary Material
Acknowledgments.
We thank Michael Axtell, Ke Bian, Brandon Gaut, Zhenguo Lin, Jongmin Nam, Masafumi Nozawa, Michael Purugganan, Hongyan Shan, Yujin Sun and Jianzhi Zhang for helpful suggestions and valuable comments. This work was supported by National Natural Science Foundation of China Grant 30530090 (to H.K.), Chinese Academy of Sciences Grant KSCX2-YW-R-135 (to H.K), and National Institutes of Health Grants GM020293–35 (to M.N.) and GM63871–01 (to H.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812043106/DCSupplemental.
References
- 1.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nei M. The new mutation theory of phenotypic evolution. Proc Natl Acad Sci USA. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nozawa M, Nei M. Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proc Natl Acad Sci USA. 2007;104:7122–7127. doi: 10.1073/pnas.0702133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci USA. 2007;104:20421–20426. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 8.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 9.Hellmann H, Estelle M. Plant development: Regulation by protein degradation. Science. 2002;297:793–797. doi: 10.1126/science.1072831. [DOI] [PubMed] [Google Scholar]
- 10.Cardozo T, Pagano M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 12.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 13.Bai C, Richman R, Elledge SJ. Human cyclin F. EMBO J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagne JM, et al. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain M, et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007;143:1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lechner E, et al. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Wikstrom N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proc R Soc Lond B Biol Sci. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 20.Nam J, et al. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci USA. 2004;101:1910–1915. doi: 10.1073/pnas.0308430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiu SH, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong H, et al. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50:873–885. doi: 10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- 23.Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comp Biol. 2006;2:e115. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, et al. Genome-wide analysis of S-Locus F-box-like genes in Arabidopsis thaliana. Plant Mol Biol. 2004;56:929–945. doi: 10.1007/s11103-004-6236-y. [DOI] [PubMed] [Google Scholar]
- 25.Kim HS, Delaney TP. Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell. 2002;14:1469–1482. doi: 10.1105/tpc.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young JM, et al. Extensive copy-number variation of the human olfactory receptor gene family. Am J Hum Genet. 2008;83:228–242. doi: 10.1016/j.ajhg.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in Mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 29.Schwager KM, et al. Characterization of the VIER F-BOX PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell. 2007;19:1163–1178. doi: 10.1105/tpc.105.040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu G, Kong H. Duplication and divergence of floral MADS-box genes in grasses: evidence for the generation and modification of novel regulators. J Integr Plant Biol. 2007;49:760–768. [Google Scholar]
- 31.Vandenbussche M, Theissen G, Van de Peer Y, Gerats T. Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 2003;31:4401–4409. doi: 10.1093/nar/gkg642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan H, et al. Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol Phylogen Evol. 2007;44:26–41. doi: 10.1016/j.ympev.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16:1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.