Abstract
Certain features such as small vascular lesions seen in human MRI are detected reliably only in postmortem histological samples by microscopic imaging. Co-registration of these microscopically detected features to their corresponding locations in the in-vivo images would be of great benefit to understanding the MRI signatures of specific diseases. Using non-linear Polynomial transformation, we report a method to co-register in-vivo MRIs to microscopic images of histological samples drawn off the postmortem brain. The approach utilizes digital photographs of postmortem slices as an intermediate reference to co-register the MRIs to microscopy. The overall procedure is challenging due to gross structural deformations in the postmortem brain during extraction and subsequent distortions in the histological preparations. Hemispheres of the brain were co-registered separately to mitigate these effects. Approaches relying on matching single-slices, multiple-slices and entire volumes in conjunction with different similarity measures suggested that using four slices at a time in combination with two sequential measures, Pearson correlation coefficient followed by mutual information, produced the best MRI-postmortem co-registration according to a voxel mismatch count. The accuracy of the overall registration was evaluated by measuring the 3D Euclidean distance between the locations of microscopically identified lesions on postmortem slices and their MRI-postmortem co-registered locations. The results show a mean 3D displacement of 5.1 ± 2.0 mm between the in-vivo MRI and microscopically determined locations for 21 vascular lesions in 11 subjects.
I. Introduction
A microscopic examination of histological samples is often the gold standard to detect pathology. For example, in diseases such as Alzheimer Disease or Vascular Dementia, small lesions may be visualized during in-vivo MRIs, but are detected reliably only at postmortem. With the objective of improving the detection and characterization of such small lesions seen in human MRIs but ascertained only at postmortem, we have developed a method to co-register lesions detected in the in-vivo MRIs of subjects when they were alive, to those detected by a microscopic imaging of histological samples drawn off the postmortem brain. Thus the technique achieves co-registration between in-vivo MRIs, which have a nominal resolution of 1mm, to microscopic images, which have a nominal resolution of 1μm.
Previous work to co-register in-vivo MRIs to microscopically examined samples has been reported mostly for small animal studies [Jacobs et al. 1999; Jiang et al. 2004; Meyer et al. 2006; Anderson et al. 2006], a limited number of primates [Malandain et al. 2004; Kaufman et al. 2005; Dauguet et al. 2007] and relatively few human studies [Schormann et al. 1995; Mega et al. 1997; Toga et al. 1999; Kenwright et al. 2003]. Most studies involve brain imaging. One of the earlier animal studies was based on a two-step approach where a surface-matching initial co-registration was followed by a nonlinear thin plate splines based warping [Jacobs et al., 1999]. Subsequent studies have relied mostly on maximizing mutual information [Jiang et al., 2004; Meyer et al., 2006] or an image intensity based non-rigid registration [Anderson et al., 2006]. Animal studies have an advantage over human studies in that animals can be sacrificed immediately after an in-vivo scan, and the postmortem brain can be extracted rapidly and scanned in dedicated animal scanners without any significant scheduling delays. Then the postmortem brain can be frozen and sliced for a microscopic histological examination. Thus very little time elapses between the in-vivo, postmortem, and microscopic examinations, thereby minimizing distortions in the shape and structure of the brain viewed by the different modalities. Also, in animal studies it is logistically straightforward to obtain an in-vivo scan of the premortem brain and the full postmortem brain immediately after extraction. Consequently MR images of postmortem brain slices are available to co-register with the microscopic examination, providing additional information that may not be available in human studies.
A reported early human study [Schormann et al. 1995] relied on a landmark based approach where histological sections were deformed to match local regions within the MRIs. Not only is it difficult to define landmarks in the histological sections, but warping the high-resolution histological images to match relatively low-resolution specific MRI regions introduces significant interpolation errors. In another set of studies, the postmortem brain was cryogenically frozen, micro-sectioned, stained and then elastically warped either to in-vivo PET images [Mega et al. 1997] or to MRIs [Toga et al. 1999]. The cryosectioning approach, however, is not readily available and introduces non-linear distortions during the freezing process itself, which requires additional warping procedures and significantly increases the computational expenses. Another relatively recent human study relied on mostly rigid-body registration [Kenwright et al. 2003], which is also not optimal as there are bound to be significant non-linear distortions between the postmortem and in-vivo images. The approach reported by us utilizes non-linear warping based on a second-order polynomial transformation and does not require cryosectioning. Thus the approach presented here would be applicable to co-register postmortem to in-vivo MRI where either cryosectioning equipment is not available or in those instances where there are existing databases of postmortem image photographs that were acquired without cryosectioning.
An important consideration in human studies is that frequently there is a large time-gap, on the order of 1–3 years, between the in-vivo MRIs and the time when the person comes to postmortem, which is likely to introduce significant changes between the in-vivo MRIs and the brain at death. Also logistically it is difficult to arrange for a MRI of a human postmortem brain on short notice. Thus postmortem brain MRIs, which are readily available in animals immediately after in-vivo studies and greatly simplify the co-registration procedure, are usually unavailable for humans. In the work reported here, no MRIs of the postmortem brain were available. Instead, digital photographs of slices of the postmortem brain, which can be obtained quite easily, formed the basis of this work. Give these practical constraints, the objective of this work was to co-register in-vivo MRIs (taken when the person was alive and may be 1–3 years before death) to digital photographs of postmortem slices and subsequent microscopic images of samples drawn off the postmortem slices. Postmortem images in this paper refer specifically to digital photographs of the sliced postmortem brain and not to any postmortem MRIs.
The overall co-registration strategy relied on using the postmortem photographs as an intermediate reference, onto which the in-vivo MRIs and the microscopic images were co-registered separately and superimposed to obtain, in effect, a co-registration between the in-vivo MRIs and the microscopic images.
Co-registering photographs of the postmortem slices to corresponding in-vivo MR slices is a challenging task for a number of reasons: 1) The time lapse between the in-vivo MRI and the postmortem brain creates structural discrepancy, 2) the procedure to extract the brain from the cranial vault leads to multiple deformations such as collapse of the ventricles and other regions, distortions while slicing, and deformations due to dehydration, and 3) the variability in the thickness and orientation of the postmortem brain slices makes dense 3D-volume reconstruction of the postmortem brain difficult.
Current image registration approaches such as SPM (http://www.fil.ion.ucl.ac.uk/spm/) perform efficiently if evenly sliced and orientation-preserved slices are available such that an accurate 3D reconstruction of the image volume is possible. The application of these existing registration methods to the problem at hand is severely limited by the difficulty in forming an accurate 3D-volume with the postmortem photographs due to significant interslice gaps and severe geometrical distortions. The registration task becomes more challenging due to pixel-voxel size mismatch, scaling, global and local geometrical distortions and regions present in one but missing from the other modality.
The general approach reported here is based on registering multiple postmortem slices to a warped 3D space within the MRI volume. Warping relies on a modified nth-order non-linear polynomial approach initially developed by one of us for a different application [Singh et al. 1979]. The warping algorithm was adapted to this application by evaluating seven different voxel similarity measures to discover a similarity measure optimal for this problem. The overall registration methodology was evaluated by identifying specific vascular lesions called lacunes (irregular cavities containing strands of fibrillar connective tissue) in brain MRIs of patients diagnosed with vascular dementia and correlating these to microscopic images of histological samples after death. Details of the registration methodology and results showing a relatively good match between the locations of lesions obtained from the MRI-postmortem co-registration and their locations identified by microscopic imaging are presented in this paper.
II. Method
The approach relies upon co-registration of digital photographs of the postmortem brain to in-vivo MRIs of the person when he or she was alive and mapping suspected lesions from the MRI to the corresponding postmortem slice. Samples drawn from a small region surrounding the location of mapped lesions in the postmortem slice were stained and examined microscopically to identify lesions. The locations of the microscopically detected lesions were marked on the histological samples. These marked samples were then co-registered back onto the postmortem slice. Thus the postmortem brain slices became the common frame-of-reference onto which the in-vivo MRI and the histological exam were mapped to achieve co-registration between MRI and microscopy.
A. Preparation of Brain Slices
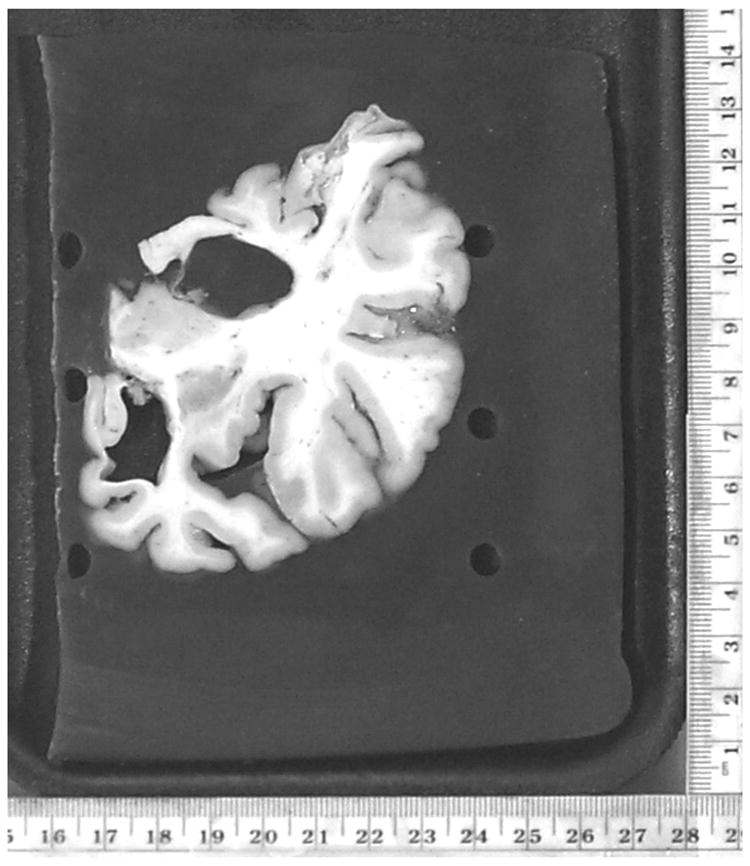
Institutional Review Board (IRB) approved specimens of postmortem brains were fixed in 10% neutral formalin for at least two weeks, and sectioned into 25–30 5mm thick coronal slices using a motor-driven rotary slicer. Coronal orientation was selected because it matched the orientation of the high-resolution in-vivo MR images that were available for these subjects (see Section B below). Each slice was photographed digitally and stored in Kodak PhotoCD RGB color format at a resolution of 3072×2048, keeping an in-plane spatial resolution of approximately 0.6mm. The brain was extracted from the background of the RGB postmortem photographs by selectively thresholding RGB intensity values and converted to gray-scale by eliminating the hue and saturation while retaining the luminance information.
Maintaining a specific distance between the two hemispheres of the postmortem brain and maintaining the geometric relationships between the two hemispheres is difficult in coronal sections as only the corpus callosum (which is relatively thin and flexible) connects the hemispheres. To mitigate this problem, we separated the brain into hemispheres by slicing through the corpus callosum and registered each hemisphere separately. Confining the coregistration to hemispheres only instead of the whole brain was not a particular concern in this work as the objective was to co-register specific lesions that are found only in the subcortical gray matter. Also, in conventional neuropathological readings, the pons is sliced perpendicular to its long axis and the cerebellum is sectioned sagittally. As the lesions of interest are not found in the pons or the cerebellum, these two regions were removed for additional pathological examination before coronal slicing. Moreover, removing the pons at the level of the third nerve allows a more accurate separation of the hemispheres through the corpus callosum.
To minimize distortions of mechanical slicing and dehydration, and to reduce the variability in the planar thickness and orientation of the postmortem slices, each hemisphere was embedded in 5% agar gel. An example of a coronal brain slice is shown in Fig. 1. A 3D volume was reconstructed from these coronal slices after aligning them with respect to each other with the help of six fiducial markers arranged in two columns (see the black fiducial markers on the left and right sides of the brain in Fig 1). More details of this standardized procedures for gel-embedding and handling the postmortem brain have been reported elsewhere [Zarow et al. 2004].
Figure 1.
B. MRI Data Acquisition and Processing
Three different pulse sequences were used to acquire MRI data on a GE 1.5T Signa System: coronal T1-weighted, transaxial Proton Density (PD), and transaxial T2-weighted scans respectively. The T1-weighted 3D coronal images were acquired using a spoiled gradient recall (SPGR) sequence with TR=24ms, TE=5ms, flip angle=45°, field of view=24×24cm2, 124 slices, slice thickness=1.5mm, and 0.86×0.86 mm2 in-plane resolution. The PD and T2-weighted images were acquired using a dual-echo turbo spin echo (TSE) sequence with TR=45ms, TE1=14ms, TE2=85ms, 51 slices, slice thickness=3mm, and 1.0×1.0mm2 in plane resolution. The transaxial PD and T2 images were used to detect vascular lesions that are best seen in these images due to their peculiar MR characteristics. The coronal T1-weighted MR images were used to form the 3D reference volume for the co-registration as these images had the best overall spatial resolution among all of the acquired images.
The Brain Surface Extraction (BSE) Algorithm [Sandor and Leahy 1997], which extracts the brain regions using a morphological algorithm, was used to strip off the skull and scalp in MR images. In addition, the regions of pons and cerebellum were removed manually from each MR slice, since these regions were absent in the postmortem slices. The extracted MR brain volume was also cut into two hemispheres through the inter-hemispheric fissure to match the postmortem slices.
C. Initial Alignment of 3D MRI and Postmortem Volumes
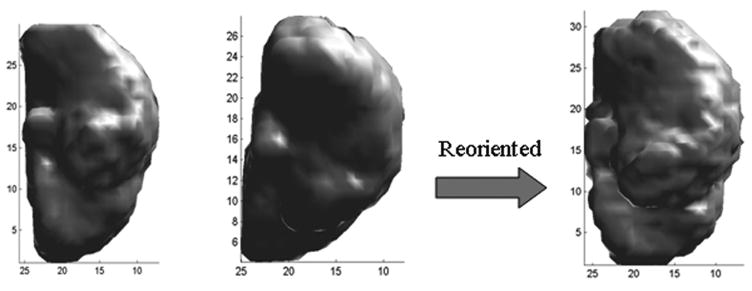
The 3D volumes reconstructed from the coronal MRI slices of each hemisphere were initially aligned to the corresponding hemispheric postmortem volumes using rigid-body rotation, translation and scalar multiplication. This coarse alignment was achieved by a first order Polynomial transformation (Singh et al. 1979), where the coefficients were estimated iteratively by minimizing the mean squared error differences between the intensities of corresponding pixels. This procedure was performed to minimize effects of differences in spatial resolution and orientation between the two modalities. An example of this step is shown in Fig. 2. Fig. 2(left) shows the postmortem volume of one hemisphere, Fig. 2(middle) the initial volume formed by the coronal MR images of the same hemisphere and Fig. 2(right) the volume obtained after rigid-body alignment. It can be seen that the two volumes are in better alignment after the registration.
Figure 2.
D. MRI and Postmortem Volume Co-registration
D.1 General Approach
Most existing volume-to-volume co-registration procedures, including for example SPM (http://www.fil.ion.ucl.ac.uk/spm/), rely on co-registering the complete or almost complete 3D volumes acquired by individual modalities. However in this work, postmortem images were available only of the separated hemispheres, with no information available on the left-right hemispheric alignment to form a 3D volume of the whole brain. In addition severe non-linear distortions were apparent between the two modalities and our attempts to achieve direct volume-to-volume co-registration between the MR and postmortem hemispheric volumes using standard procedures such as SPM were unsuccessful. Instead we investigated a procedure where one or multiple slices of one modality are co-registered to a limited volume of the other modality. This procedure allows for greater flexibility in correcting local distortions that may prevail in some regions. The MRI volume formed by coronal slices was warped locally and resliced in three dimensions incorporating a second order polynomial as described below. This procedure matched one or more postmortem slices (that were retained in their original frame of reference) to a warped sub-volume within the MR volume.
The polynomial based warping [Singh et al. 1979, Kim et al. 2000 ] may be described by the following equations.
| (1) |
where (u,v,w) are the new coordinates of a voxel in the 3D MRI volume, (i,j) are a pixel’s coordinates within a postmortem image, and ai, bi, and ci are the coordinate transformation coefficients. This transformation translates the coordinates of pixels in the 2D postmortem slices to voxels in the 3D MRI volume. The polynomials represent a surface whose location and shape can be varied by changing the transformation coefficients.
The initial location of a specific postmortem slice within the 3D MR volume can be calculated in a straightforward manner from the known thickness and number of postmortem slices in conjunction with the rigid-body transformations shown in Fig. 2. Beginning with initial locations within the MRI volume for a given number of postmortem slices, the general co-registration procedure was to iteratively warp 2D sections lying within the 3D MRI volume until best-matching MR sections were found by optimizing a registration cost.
A standard approach to estimate the transformation coefficients for the warping is to minimize the sum of differences between the given postmortem images and the iteratively extracted MR sub-volume, i.e.,
| (2) |
where e is a function defining a registration cost, P a set of postmortem images, and M the set of extracted MRI sub-volume. The parameter ‘d’ is the inter-slice distance when multiple postmortem slices are used.
D.2. Similarity Measures
The registration cost function was evaluated in terms of seven commonly used similarity measures to determine the most appropriate cost function for this application. These similarity measures are based on image intensities, information measures, and object patterns, and have been described in detail by Hajnal [Hajnal et al. 1995] and Holden [Holden et al. 2000]. Specifically, the measures investigated here were: (1) Mean-Squared Difference (MSD) [Viola 1995, Kim et al. 2000]; (2) Pearson Cross Correlation (PCC) [Viola 1995, Lemieux et al. 1998]; (3) Mutual Information (MI) [Maes et al. 1997, Kim et al. 1997, Kim et al. 1999, Kjems et al. 1999]; (4) Normalized Mutual Information (NMI) [Studholme et al. 1997, Studholme et al. 2000a, Studholme et al. 2000b]; (5) Entropy of Difference Image (EDI) [Buzug et al. 1997]; (6) Ratio Image Uniformity (RIU) and Modified RIU (MRIU) [Woods et al. 1998, Singh et al. 1998, Holden et al. 2000] and (7) Pattern Intensity (PI) [Weese et al. 1997].
The voxel mismatch count or VMC [Freeborough et al. 1996] in the difference images was used as the metric to evaluate the accuracy of co-registration. The intensities of pixels were scaled to achieve the same mean intensity in both images before computing their difference, and the number of mismatched pixels was determined by counting pixels whose intensity in the difference image was >20% of the mean intensity as defined in [Freeborough et al. 1996]. In addition to VMC, visual identification of common anatomical landmarks (e.g., anterior commissure, pillars of fornix, perivascular spaces, and optic chiasm) by experienced anatomists was used to verify co-registration. Data from the first five subjects entered in this study were used for this evaluation in conjunction with a second order polynomial warping. Evaluations were done for n = 2–4 slices near the central portion of the volume by co-registering one postmortem slice at a time.
The VMC, expressed as percent changes for five cases used to develop the methodology are summarized in Table 1. The VMCs in all cases were normalized by the total number of voxels of the brain and expressed in percent changes to make statistical comparisons across slices and subjects. Although the overall trend in Table 1 suggests that PCC seems to be the best cost, there was an exception for case C1. It was found that when there was relatively less distortion in the postmortem brain slices, as was the case in C1, similarity measures based on probability density such as MI, NMI, and EDI performed better than any other measure. On the other hand, when severe distortions were present in postmortem slices, similarity measures depending on voxel intensity such as MSD and PCC were found to be more sensitive in matching the location, overall shape, and size of the postmortem brain slices, suggesting that these measures are more sensitive for correcting geometrical distortions.
Table 1.
| Mean (Standard Deviation) of (VMC/Total No. Brain Voxels)*100 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Case | MSD | PCC | MI | NMI | EDI | MPI | RIU | MRIU |
| C 1 (n=4) | 21.99(1.87) | 20.57(0.73) | 20.15(1.70) | 19.96(1.70) | 19.96(1.71) | 39.09(40.61) | 78.81(42.37) | 60.81(45.27) |
| C 2 (n=2) | 17.81(4.43) | 17.82(4.73) | 22.73(2.87) | 23.35(6.11) | 27.65(2.78) | 56.97(60.85) | 56.89(60.86) | 56.85(60.97) |
| C 3 (n=2) | 30.17(2.40) | 30.73(1.74) | 31.07(1.18) | 31.78(1.79) | 35.01(1.57) | N/A* | N/A* | N/A* |
| C 4 (n=2) | 17.67(0.57) | 15.79(1.01) | 18.85(4.88) | 18.93(2.90) | 22.65(0.39) | 16.49(4.14) | 59.75(56.92) | 56.68(61.26) |
| C 5 (n=3) | 8.646(3.05) | 3.53(0.52) | 19.23(3.19) | 18.35(7.26) | 22.86(1.53) | 44.63(48.33) | 71.89(48.70) | N/A* |
| Total (n=13) | 18.86(7.46) | 17.04(9.15) | 21.82(4.80) | 21.79(6.00) | 24.54(5.49) | 49.01(42.07) | 74.17(40.34) | 74.64(33.67) |
No covergence was obtained.
D.3. Two-Step Augmented Co-registration
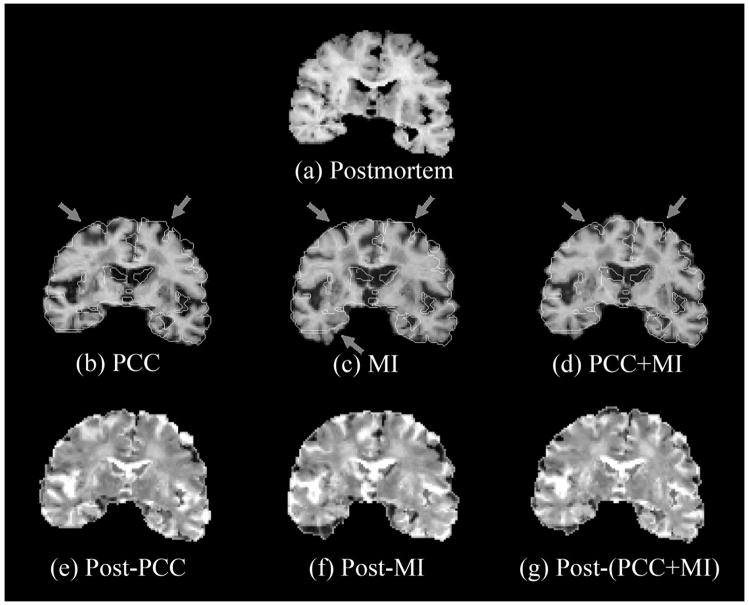
Based on above observations, we devised a two-step augmented registration approach where geometrical distortions were corrected in a first step by co-registering images with MSD or PCC (thus matching the size and shape of the brain), and then, using the preestimated transformation parameters, registration was improved further by matching internal structures and intensity distribution using MI or NMI. Fig. 3 shows typical results of co-registration using the augmented similarity measures MSD/PCC followed by MI/NMI where registration parameters from the first step were used as initial values for the second step. Either MSD or PCC was selected for the first step depending on which method produced the lower VMC. The postmortem image is shown in the top row. Edges within the postmortem image superimposed on co-registered images obtained with PCC alone, MI alone, and the PCC followed by MI augmented procedure are shown in the middle row, and the respective difference images are shown in the bottom row. Edges were detected using the “Canny” method [Canny 1986], which locates edges by looking for local maxima of the gradient of the image. Thresholds, low and high, were set to 0.0 and 0.5, respectively, so as to ignore weak edges. It can be seen that there is an improvement in matching edges with the augmented approach (the arrows point to mismatches with PCC or MI alone that are better matched with the two-step approach) and also the augmented measure qualitatively shows more uniform gray patches in the difference image of the two-step compared to the single-step approaches. A visual inspection of anatomical features in the registered images with PCC+MI in Fig. 3 shows that the following features match closely in the postmortem and registered images: the outside contours of insula (particularly the left), and the details of the hippocampus (right), the amygdala (left), and the nuclei of the subcortical grey (both hemispheres).
Figure 3.
Consistent with visual observations, the voxel mismatch counts for the two-step augmented approach, expressed in percents in Table 2, also show a significant reduction over the single-step approaches. Although marginal reduction in the counts was obtained with the augmented cost compared to PCC or MSD alone, significant reduction was obtained compared to those of MI or NMI alone. To obtain a measure of the statistical significance of reduction in the counts for the augmented costs (i.e., AMI and ANMI), we performed the paired t-test with one-tail at the 95% confidence level against other costs [Kanji et al. 1993]. The registration results using AMI were found to be better than those of MSD, MI, and NMI significantly (P=0.0086, 0.0027, and 0.0035) and so were the results using ANMI (P=0.0068, 0.0021, and 0.0027). When the results using AMI and ANMI were compared with the results using PCC, the improvement however was not so significant (P=0.0825 and 0.0697). The two-step approach increased the total computation time by only about 10–15% over that for MSD or PCC alone.
Table 2.
| Mean (Standard deviation) | ||
|---|---|---|
| Case | AMI | ANMI |
| C 1 (n=4) | 20.36 (0.94) | 19.97 (1.37) |
| C 2 (n=2) | 18.25 (4.18) | 18.08 (4.56) |
| C 3 (n=2) | 13.96 (0.04) | 13.69 (0.18) |
| C 4 (n=2) | 16.05 (0.69) | 15.86 (0.55) |
| C 5 (n=3) | 3.28 (0.49) | 3.34 (0.41) |
| Total (n=13) | 14.45 (6.89) | 14.24 (6.77) |
D.4. Single Slice verses Simultaneous Multiple-Slices Co-registration
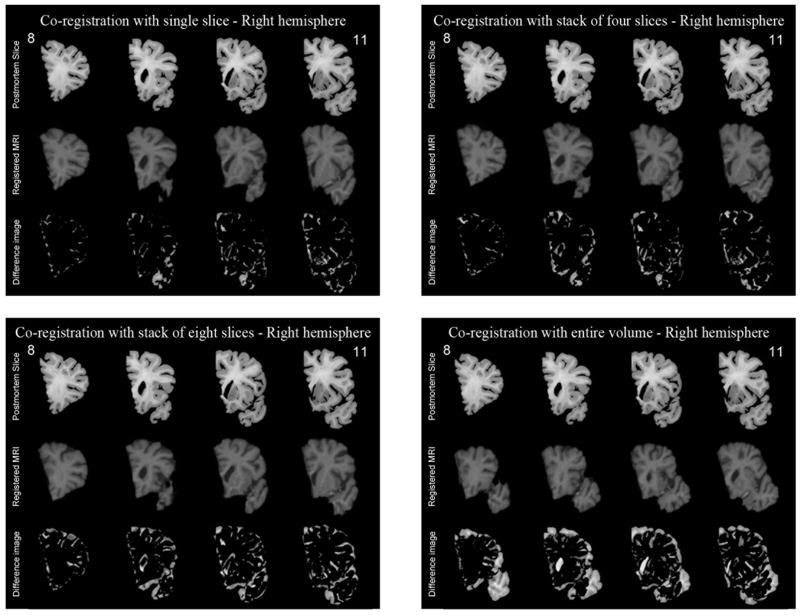
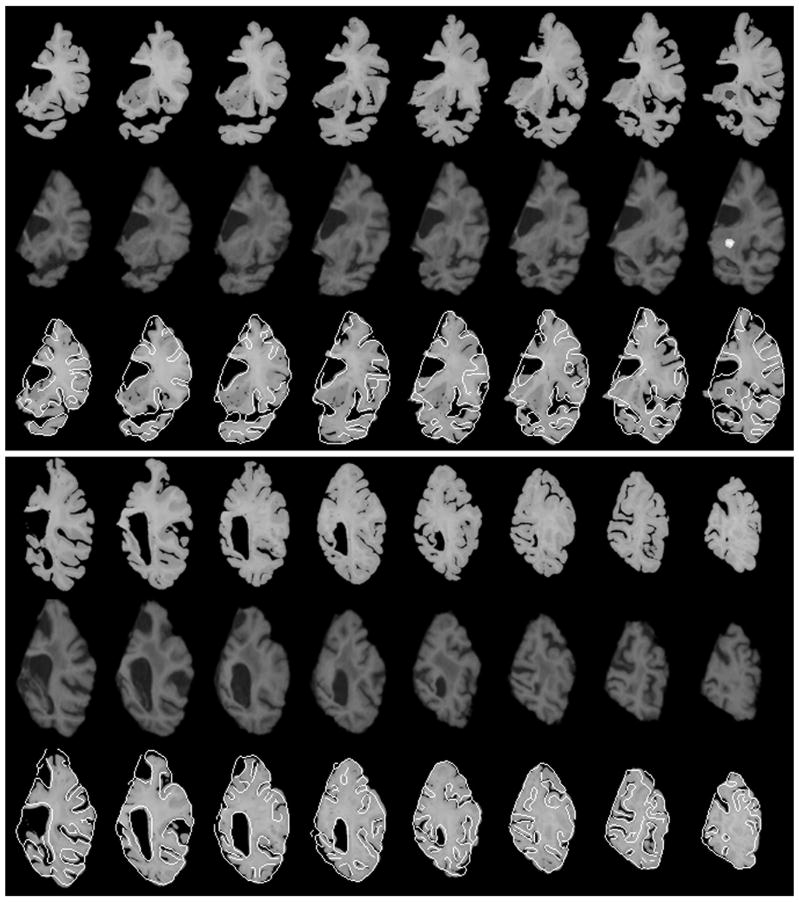
Though maximum flexibility to correct non-linear local distortions would be attainable if individual postmortem slices were co-registered one-at-a-time to the 3D MR volume, in practice this approach is computationally very slow and also carries the risk that more than one slice could map onto the same MR section. To reduce the possibility of multiple slices matching the same MR volume, we compared the performance of co-registering a stack of postmortem slices, i.e., multiple postmortem slices, to a corresponding MR volume that would be partitioned in accordance with the spacing of individual slices within the stack. The results of single-slice and stacks of four, eight and the whole volume simultaneously are presented in Figure 4. Visually it is apparent from Fig. 4 that co-registration using eight slices or the entire volume produces relatively large difference images, suggesting inferior co-registration compared to four or single slice matching. This is expected because when a large number of slices are used, it is difficult to correct local deformations efficiently.
Figure 4.
The co-registration accuracy was also evaluated quantitatively by computing the mutual information between the postmortem and the corresponding registered MR slices. An example using a total of 20 postmortem slices is shown in Table 3, which shows that the mutual information with four slices is very close to that of single slices, whereas the mutual information with 8 slices or the entire volume is significantly lower, indicating worse matching when a relatively large number of slices are used simultaneously for the co-registration. Given that computation is more efficient when four slices are used instead of one, and that the risk of more than one slice mapping to the same MR volume is minimized, we selected the approach of co-registering four slices simultaneously to a corresponding MR volume to conduct this work. An example of the four-slice co-registration applied to the entire right hemisphere of another subject is shown in Figure 5. It is clear that there is strong match in the size, shape, and internal structures including outer and inner contours of the ventricles, subcortical grey nuclei, hippocampus, gyri, and sulci.
Table 3.
| Mutual Information | ||||
|---|---|---|---|---|
| Slice Number | Stack of 1 slice | Stack of 4 slice | Stack of 8 slice | Stack of whole volume |
| 1 | 0.7177 | 0.7225 | 0.7019 | 0.5698 |
| 2 | 0.781 | 0.7777 | 0.7572 | 0.6312 |
| 3 | 0.8132 | 0.8138 | 0.7902 | 0.6701 |
| 4 | 0.8378 | 0.8337 | 0.812 | 0.7046 |
| 5 | 0.8986 | 0.9002 | 0.8735 | 0.7462 |
| 6 | 0.9384 | 0.9313 | 0.8993 | 0.7693 |
| 7 | 0.9835 | 0.9741 | 0.9288 | 0.7998 |
| 8 | 1.0214 | 1.0165 | 0.9698 | 0.8301 |
| 9 | 1.1025 | 1.0964 | 1.0375 | 0.8822 |
| 10 | 1.1394 | 1.1218 | 1.0676 | 0.9215 |
| 11 | 1.1352 | 1.1206 | 1.0709 | 0.9408 |
| 12 | 1.1013 | 1.0867 | 1.0354 | 0.9143 |
| 13 | 1.0843 | 1.0598 | 1.0094 | 0.9109 |
| 14 | 1.0663 | 1.0664 | 1.0014 | 0.9167 |
| 15 | 1.0233 | 1.0338 | 0.9706 | 0.8936 |
| 16 | 1.0363 | 1.0229 | 0.9806 | 0.9009 |
| 17 | 0.9926 | 0.9681 | 0.9796 | 0.9044 |
| 18 | 1.0126 | 0.9843 | 0.9917 | 0.9146 |
| 19 | 0.9406 | 0.9509 | 0.95 | 0.8766 |
| 20 | 0.9107 | 0.9224 | 0.9204 | 0.8447 |
| Average | 0.976835 | 0.9702 | 0.93739 | 0.827115 |
Figure 5.
E. Co-registration of MRI Detected Lesions to Locations on Postmortem Images
As the objective of this work was to co-register MRI detected lesions to their microscopic images at postmortem, the next step in this procedure was to map lesions from MRIs to locations within the postmortem images. Though this work was focused on specific types of small vascular lesions, it could be applied to any type of lesions or small regions of the brain. The vascular lesions of interest were best identified in the axial scans (PD and T2 weighted images described in Section B). After an expert marked the locations of these lesions, their locations were first mapped from axial to coronal sections by volume reslicing and then mapped onto the postmortem slices by using the Polynomial-transformation parameters found by the four-slices-at-a-time warping technique described above.
F. Co-registration of Microscopy Detected Lesions to Locations on Postmortem Images
Relatively large samples of the postmortem slices around locations indicated by the above MRI to postmortem co-registration were extracted and sent to a pathologist for stained microscopy. The pathologist sub-sliced the samples into 10–20 micron sections and through a microscopic examination, marked the regions where these lesions were identified histologically. Microscopy was performed at several magnifications. Frequently portions of the sample disintegrated during the staining and sub-slicing procedures. Thus another warping procedure was required to co-register the microscopy pictures back to the postmortem slices.
Co-registration of the microscopy pictures to corresponding postmortem slices was also accomplished by warping the microscopy pictures using a second order polynomial coordinate transformation as represented by eq (1). An edge-matching cost-function was used which minimized the pixel mismatch count between the intact edges of the microscopy image and edges in the postmortem image at corresponding boundaries. Since the regions from where the samples were extracted for microscopic examination were known, it was not difficult to initially start with a rough alignment of the intact boundaries in the microscopy images to corresponding boundaries in the postmortem slices. The missing boundaries were then masked and alignment was refined iteratively by the warping procedure.
G. Validation of the MRI to Microscopy Co-registration
The accuracy of the entire procedure to co-register in-vivo MRI to microscopic findings was evaluated and validated by quantifying the displacement between a) the locations of in-vivo MRI detected lesions mapped onto the postmortem slices and b) the location of presumably the same lesions identified through a microscopic examination and then co-registered back to their corresponding postmortem slices. Since the MRI marked lesions extended over several slices, the 3D coordinates of the centroid were used to compute the displacement. However, since the microscopic examinations were performed on successive 10–20 micron thick sections of a 5mm thick postmortem slice, the 3D coordinates corresponding to the middle layer of the postmortem slice were used in the displacement computation.
III. Results
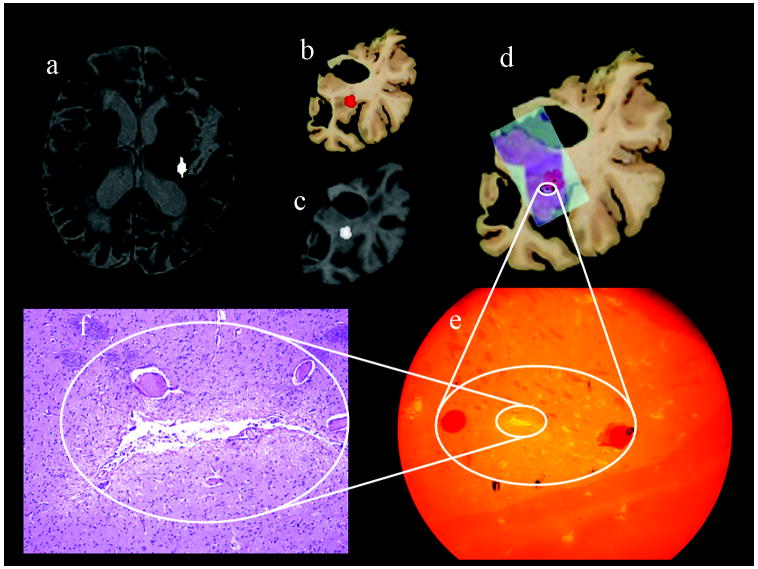
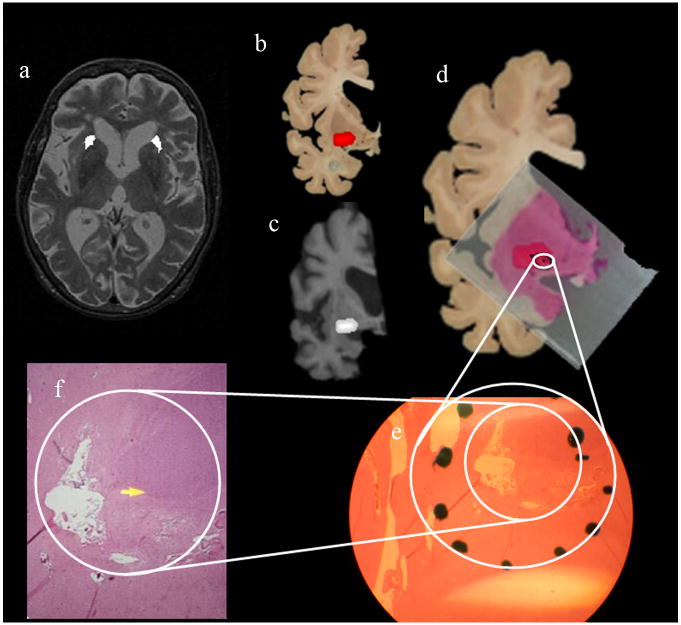
The complete MRI to microscopy co-registration procedure was carried out for 21 lesions detected in eleven human subjects. Two examples of the entire MR to microscopy procedure are presented in Figs 6 and 7. The locations of the lesions marked on axial T2 images by an expert are shown in Figs. 6–7(a). These lesions were transferred to the postmortem slices through co-registration. As an example, the locations of the lesions in one hemisphere are shown in Figs. 6–7(b). Their locations in corresponding warped MRI sections are shown in Figs. 6–7(c). Figs. 6–7(d) show the locations of the histology slides after warping boundaries in the stained sections to match their original boundaries in the postmortem brain.
Figure 6.
Figure 7.
The white circles correspond to the locations of the lesions identified through the microscopic examination of the histology slides. The red areas are the locations of the lesions through MRI-postmortem co-registration. Areas identified to contain a lesion during histology at a coarse microscopic resolution are shown in Figs 6–7(e), and high magnification microscopy images of the lesions within this region are shown in (f). The black dots within the white circles 6–7(d) correspond to the microscopic locations of the respective lesions. The 3D displacements between the microscopic locations and the centroids of the same lesions identified through MRI-postmortem co-registration are shown in Table 4.
Table 4.
| Case # | 3D Euclidean dist (cm) |
|---|---|
| 1 | 0.8634 |
| 2 | 0.6001 |
| 0.5406 | |
| 3 | 0.8132 |
| 4 | 0.3117 |
| 5 | 0.5638 |
| 0.2753 | |
| 6 | 0.3623 |
| 7 | 0.3478 |
| 0.7324 | |
| 8 | 0.8329 |
| 0.5810 | |
| 0.4965 | |
| 9 | 0.6499 |
| 0.1672 | |
| 0.5417 | |
| 10 | 0.3194 |
| 0.3508 | |
| 0.5507 | |
| 11 | 0.3256 |
| 0.4893 | |
| Average= | 0.5103 |
| std dev = | 0.1957 |
IV. Discussion
Although pathological information at the cellular level can be derived from postmortem brain tissues, the natural progression of disease can be monitored more efficiently through longitudinal MR scans. The progression of a disease can then be compared to the pathology of brain tissue by co-registering microscopically observed specific features in postmortem slices to their corresponding MR slices. Using the postmortem slices as a common frame of reference, the purpose of this work is to provide a methodology for such co-registration between features detected in in-vivo MRIs (when the person was alive) and microscopic images at postmortem. As a knowledge of MR signal changes of pathological substrates may add significant information about the disease to the histological analysis of tissue itself, the MRI to microscopy co-registration procedure should be of high significance to many fields.
In a previous attempt to co-register postmortem slices to MRI, Thompson and Toga developed a method that relies on cryosectioning [Toga et al. 1999] the postmortem brain. Cryosectioning is more accurate than mechanical slicing, however, the equipment is not readily available at many sites and no such equipment was available to us. Also freezing introduces additional distortions requiring warping models [Toga et al. 1999] that are computationally demanding. The significance of our approach lies in its practicability, cost-effectiveness, and minimal expert human intervention. The postmortem brain preparation in gel can be done easily without sophisticated set-ups, there is no need for high-resolution postmortem image volume, no landmark localization or structural model generation is required, and once postmortem and MR images are pre-processed, no human intervention is needed to find the matching slices. In general targeted regions reside only in a few postmortem slices. Thus only the desired slices need to be registered using a single-slice or multiple-slice mode. The single-slice mode allows an option of co-registration when accurate realignment into a stack is not possible.
It is generally understood that mutual information is the optimal choice for registering multi-modal medical images, especially when images reflect different contrasts. However our results in this study suggest that the correlation measure could also be a good choice to register postmortem photographs to MRI. This observation could be explained by the intensity transformation between postmortem photographs and MRI, which has a linear relationship, thus producing good results as suggested by Viola [Viola, 1995]. Thus our two-step approach, which combines the advantages of the correlation and mutual information measures, would be expected to perform better that a single measure as suggested in our work.
In this study, we have tested the 2nd-order polynomial transformation allowing the following transformations in 3D: translation, rotation, scaling, skewness, and perspective [Singh et al. 1979, Schowengerdt et al., 1983]. It is not clear whether further expansion of polynomial order would produce better co-registration. In the case of MRI-MRI registration where structural deformation is not significant, Woods et al. showed marginal improvement of registration with higher number of coefficients [Woods et al., 1998]. In our opinion too, higher order polynomials provide diminishing returns as higher order polynomials complicate parameter estimation and demand more computation time. Future work should investigate the relation of model order versus registration performance.
Although we adopted the voxel mismatch count (VMC) as an indicator of image matching, we acknowledge that it might not be a very accurate reflection of the degree of co-registration and has limitations, since VMC is not a bias-less arbiter of each co-registration cost. Due to the nature of the problem, neither voxel or pixel intensity differences measures, nor visual inspection alone reflect the degree of correct co-registration.
Though we did not encounter this situation, it is possible that multiple lesions may be observed at histology that lie close to each other, making it difficult to pin-point the precise location of the lesion correlated to the MRI signature. However in such cases MRI would detect the combined effect of the cluster of lesions and then relating the MRI detected centroid to the microscopy detected centroid of multiple lesions would provide the needed correlation.
V. Conclusion
We have developed a technique to co-register human in-vivo MRIs to microscopic images of targeted regions at postmortem. The approach relies on using digital photographs of postmortem slices as a reference upon which the in-vivo MRIs and microscopic histological exams are co-registered. The accuracy of the procedure was evaluated by computing the 3D displacement between the in-vivo MRI identified lesions to their corresponding locations determined by a microscopic exam of histological sections in postmortem brain slices. Results reveal that the co-registration identifies the locations of 21 lesions in 11 subjects within an error of 5.1 ± 2.0 mm.
Acknowledgments
This work is supported in part by grants NIA-NIH 1PO1AG 12453 and NIA-NIH P50 AG05142.
References
- Anderson AW, Choe AS, Stepniewska I, Colvin DC. Comparison of brain white matter fiber orientation measurements based on diffusion tensor imaging and light microscopy; Proc 28th IEEE EMBS Conf; 2006. pp. 2249–2251. [DOI] [PubMed] [Google Scholar]
- Buzug TM, Weese J, Fassnacht C, Lorenz C. Lecture Notes in Computer Science. Berlin, Germany: Speringer-Verlag; 1997. Image registration: Convex weighting functions for histogram-based similarity measures; pp. 203–212. [Google Scholar]
- Canny J. A Computational Approach to Edge Detection. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1986;PAMI-8(6):679–698. [PubMed] [Google Scholar]
- Dauguet J, Delzescaux T, Conde F, Manguin JF, Ayache N, Hantraye P, Frouin V. Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in-vivo MRI for whole baboon brain. J Neurosci Meth. 2007;164(1):191–204. doi: 10.1016/j.jneumeth.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Woods RP, Fox NC. Accurate registration of serial 3D MR brain images and its application to visualizing change in neurodegenerative disorders. J Comp Assisted Tomogr. 1996;20(6):1012–1022. doi: 10.1097/00004728-199611000-00030. [DOI] [PubMed] [Google Scholar]
- Hajnal JV, Saeed N, Soar EJ, Oatridge A, Young IR, Bydder GM. A registration and interpolation procedure for subpixel matching of serially acquired MR images. J Comp Assisted Tomogr. 1995;19:289–296. doi: 10.1097/00004728-199503000-00022. [DOI] [PubMed] [Google Scholar]
- Holden M, Hill DLG, Denton ERE, Jarosz JM, Cox TCS, Rohlfing T, Goodey J, Hawkes DJ. Pixel similarity measures for 3-D serial MR Brain Image Registration. IEEE Trans Med Imag. 2000;19(2):94–102. doi: 10.1109/42.836369. [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Windham JP, Soltanian-Zadeh H, Peck DJ, Knight RA. Registration and warping of magnetic resonance images to histological sections. Med Phys. 1999;26(8):1568–1578. doi: 10.1118/1.598671. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Pandya K, Smithies O, Hsu EW. Three-dimensional diffusion tensor microscopy of fixed mouse hearts. Magn Reson Med. 2004;52:453–460. doi: 10.1002/mrm.20191. [DOI] [PubMed] [Google Scholar]
- Kanji GK. 100 Statistical Tests. Sage Publications; Great Britain: 1993. [Google Scholar]
- Kaufman JA, Ahrens ET, Laidlaw DH, Zheng S, Allman JM. Anatomical analysis of an Aye-Aye brain combining histology, structural magnetic resonance imaging, and diffusion tensor imaging. Anat Rec A Discov Mol Cell Evol Biol. 2005;287(1):1026–1037. doi: 10.1002/ar.a.20264. [DOI] [PubMed] [Google Scholar]
- Kenwright C, Bardinet É, Hojjat SA, Malandain G, Ayache N, Colchester ACF. 2-D to 3-D refinement of postmortem optical and MRI co-registration. MICCAI, LNCS. 2003;2879:935–944. [Google Scholar]
- Kim B, Boes JL, Frey KA, Meyer CR. Mutual information for automated unwarping of rat brain autoradiography. Neuroimage. 1997;5(1):31–40. doi: 10.1006/nimg.1996.0251. [DOI] [PubMed] [Google Scholar]
- Kim B, Boes JL, Bland PH, Chenevert TL, Meyer CR. Motion correction in fMRI via registration of individual slices into an anatomical volume. Mag Res Med. 1999;41:96–972. doi: 10.1002/(sici)1522-2594(199905)41:5<964::aid-mrm16>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Kim TS, Singh M, Sungkarat W, Zarow C, Chui H. Automatic registration of postmortem brain slices to MRI reference volume. IEEE Trans Nucl Sci. 2000;47(4):1607–1613. [Google Scholar]
- Kjems U, Strother SC, Anderson J, Law I, Hansen LK. Enhancing the multivariate signal of [15O] water PET studies with a new nonlinear neuroanatomical registration algorithm. IEEE Trans Med Imag. 1999;8(4):306–319. doi: 10.1109/42.768840. [DOI] [PubMed] [Google Scholar]
- Lemieux L, Wieshmann UC, Moran NF, Fish DR, Shoovon SD. The detection and significance of subtle changes in mixed-signal brain lesions by serial MRI scan matching and spatial normalization. Med Image Anal. 1998;2(3):227–242. doi: 10.1016/s1361-8415(98)80021-2. [DOI] [PubMed] [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imag. 1997;16(2):187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- Malandain G, Bardinet E, Nelissen K, Vanduffel W. Fusion of autoradiographs with an MR volume using 2-D and 3-D linear transformations. NeuroImage. 2004;23(1):111–127. doi: 10.1016/j.neuroimage.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Mega MS, Chen SS, Thompson PM, Woods RP, Karaca TJ, Tiwari A, Vinters HV, Small GW, Toga AW. Mapping histology to metabolism: Coregistration of stained whole-brain sections to premortem PET in Alzheimer’s Disease. NeuroImage. 1997;5(2):147–153. doi: 10.1006/nimg.1996.0255. [DOI] [PubMed] [Google Scholar]
- Meyer CR, Moffat BA, Kuszpit KK, Bland PL, Mckeever PE, Johnson TD, Chenevert TL, Rehemtulla A, Ross BD. A methodology for registration of a histological slide and in-vivo MRI volume based on optimizing mutual information. Mol Imaging. 2006;5(1):16–25. [PMC free article] [PubMed] [Google Scholar]
- Sandor S, Leahy R. Surface-Based labeling of cortical anatomy using a deformable atlas. IEEE Trans Med Img. 1997;16(1):41–54. doi: 10.1109/42.552054. [DOI] [PubMed] [Google Scholar]
- Schormann T, Dabringhaus A, Zilles K. Statistics of deformation in histology and application to improved alignment with MRI. IEEE Trans Med Img. 1995;14(1):25–35. doi: 10.1109/42.370399. [DOI] [PubMed] [Google Scholar]
- Schowengerdt RA. Techniques for image processing and classification in remote sensing. Academic Press; New York: 1983. pp. 106–116. [Google Scholar]
- Singh M, Al-Dayeh L, Patel P, Kim TS, Guclu C, Nalcioglu O. Correction of head movements in multi-slice EPI and single-slice gradient-echo functional MRI. IEEE Trans Nucl Sci. 1998;45:2162–2167. [Google Scholar]
- Singh M, Frei W, Shibata T. A digital technique for accurate change detection in nuclear medicine images with applications to myocardial perfusion studies using thallium-201. IEEE Trans Nucl Sci. 1979;26:565–575. [Google Scholar]
- Studholme C. PhD thesis. University of London; London: 1997. Measures of 3D medical image alignment. [Google Scholar]
- Studholme C, Constable RT, Duncan JS. Accurate alignment of functional EPI data to anatomical MRI using a physics-based distortion model. IEEE Trans Med Imag. 2000;19(11):1115–1127. doi: 10.1109/42.896788. [DOI] [PubMed] [Google Scholar]
- Studholme C, Novotny E, Stokking R, Duncan JS, Zubal IG, Spencer D. Alignment of functional data acquired before and after intra-cranial electrode implantation using non-rigid anatomical MRI registration. Proc Intl Soc Mag Reson Med. 2000;8:585. [Google Scholar]
- Toga AW. An Introduction to brain warping. Brain Warping, Academic Press; San Diego: 1999. pp. 1–26. [Google Scholar]
- Viola PA. PhD thesis. Massachusetts Inst Technol; Cambridge: 1995. Alignment by maximization of mutual information. [Google Scholar]
- Weese J, Penney GP, Desmedt P, Buzug TM. Pixel-Based 2D/3D registration of fluoroscopy images and CT Scans for image-guided surgery. IEEE Trans Inform Technol Biomed. 1997;1(4):284–293. doi: 10.1109/4233.681173. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cheery SR, Mazziotta JC. Automated Image Registration: I. General methods and intrasubject, intramodality validation. J Comp Assisted Tomogr. 1998;22(1):139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Zarow C, Kim TS, Singh M, Chui H. A standarized method for brain cutting suitable for both stereology and MRI-brain co-registration. J Neurosci Meth. 2004;139(2):209–215. doi: 10.1016/j.jneumeth.2004.04.034. [DOI] [PubMed] [Google Scholar]