Abstract
The CpG island methylator phenotype (CIMP) with widespread promoter CpG island methylation is a phenotype in colorectal cancer, associated with microsatellite instability (MSI) and BRAF mutation. Genome-wide hypomethylation may also play an important role in genomic instability. However, the relation between global DNA methylation level and methylation in individual CpG islands remains uncertain. Utilizing 869 population-based colorectal cancers, we measured LINE-1 (long interspersed nucleotide element-1) methylation level by Pyrosequencing, which correlates with global DNA methylation level. We quantified DNA methylation in 8 CIMP-specific promoters (CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1) by real-time PCR (MethyLight technology). LINE-1 methylation levels in tumors were approximately normally distributed (mean 61.4%, median 62.3%, standard deviation 9.6%). Among the 869 tumors, 128 (15%) were classified as CIMP-high (≥6/8 methylated promoters). The mean LINE-1 methylation level was higher in CIMP-high tumors (65.1%, p<0.0001) than non-CIMP-high tumors (60.7%), and higher in MSI-high tumors (64.7%, p<0.0001) than non-MSI-high tumors (60.7%). When tumors were stratified by MSI/CIMP status, compared to non-MSI-high non-CIMP-high tumors (mean LINE-1 methylation level 60.4%), the mean LINE-1 methylation level was higher in MSI-high CIMP-high (64.8%, p<0.0001), MSI-high non-CIMP-high (64.6%, p=0.03) and non-MSI-high CIMP-high tumors (66.1%, p=0.0003). In addition, 18q loss of heterozygosity in non-MSI-high tumors was correlated with LINE-1 hypomethylation (p=0.004). In conclusion, both CIMP-high and MSI-high are inversely associated with LINE-1 hypomethylation, suggesting that CIMP/MSI and genomic hypomethylation may represent different pathways to colorectal cancer. Our data also support a possible link between global hypomethylation and chromosomal instability.
Keywords: colon cancer, methylation, epigenomics, CIMP, LINE-1
INTRODUCTION
Epigenomic aberrations are important mechanisms in human carcinogenesis.1-3 A number of tumor suppressor genes are silenced by promoter CpG island methylation in colorectal cancer.1-4 A subset of colorectal cancers exhibit widespread promoter CpG island methylation, which is referred to as the CpG island methylator phenotype (CIMP).1, 4, 5 CIMP-high colorectal tumors have distinct features, such as associations with proximal tumor location, female gender, poor differentiation, BRAF mutation, wild-type TP53, stable chromosomes and JC virus T antigen expression.6-12 Many of these associations are independent of microsatellite instability (MSI) status.13-15 Molecular classification of colorectal cancer based on MSI and CIMP status is increasingly important,16 because MSI and CIMP status reflect global genomic and epigenomic aberrations in tumor cells.
Genome-wide DNA hypomethylation is considered to play an important role in genomic instability and a carcinogenic process.17-23 An unresolved question is whether methylation in individual promoter CpG islands is related to global methylation level in neoplastic cells. Attempts to answer this question have resulted in contradictory findings, with some studies20, 21 supporting and others24-27 refuting this relationship, perhaps in part due to different techniques used to measure global DNA methylation level. It has been shown that LINE-1 (long interspersed nucleotide element-1) methylation measured by Pyrosequencing is reproducible and well correlated with global DNA methylation level.28, 29 In addition, Pyrosequencing technology can detect subtle differences in average LINE-1 methylation levels among different subtypes of colorectal cancer (e.g., MSI vs. non-MSI).30 These data emphasize the importance of the use of an accurate and precise method to measure methylation in LINE-1 (which normally shows high-level methylation), in order to demonstrate potentially subtle differences in average LINE-1 methylation levels.
In this study using quantitative DNA methylation analyses (Pyrosequencing for LINE-1 and MethyLight for CIMP analysis) and a large number of population-based colorectal cancers, we have demonstrated that LINE-1 methylation level is correlated positively with both MSI and CIMP. Since LINE-1 methylation level provides a surrogate marker for global DNA methylation level,28, 29, 31 our data support an inverse relation between CIMP/MSI and genome-wide hypomethylation.
MATERIALS AND METHODS
Study group
We utilized the databases of two large prospective cohort studies; the Nurses' Health Study (NHS, N = 121,700 women followed since 1976),32, 33 and the Health Professionals Follow-up Study (HPFS, N = 51,500 men followed since 1986).33 Informed consent was obtained from all participants prior to inclusion in the cohorts. A subset of the cohort participants developed colorectal cancers during prospective follow-up. Thus, these colorectal cancers represented a population-based sample. Previous studies on the cohorts have described baseline characteristics of cohort participants and incident colorectal cancer cases, and confirmed that our colorectal cancers were representative as a population-based sample.32, 33 Clinical information was obtained through chart review by physicians. We collected paraffin-embedded tissue blocks from hospitals where cohort participants had undergone resections of primary tumors. We obtained specimens (hematoxylin and eosin-stained slides, unstained slides and/or paraffin blocks) from 648 cases (retrieval rate 76%) in the HPFS and 662 cases (retrieval rate 58%) in the NHS. We excluded cases if patients were preoperatively treated with radiation and/or chemotherapy or adequate tumor materials were unavailable. As a result, a total of 869 colorectal cancer cases (394 from the men's cohort and 475 from the women's cohort) were included. Clinical characteristics of the cases are described in Table 1 (on the left). Among our cohort studies, there was no significant difference in demographic features between cases with tissue available and those without available tissue.33 Most tumors have previously been characterized for CIMP, MSI, 18q LOH, KRAS and BRAF gene status and pathologic features.13, 15, 34, 35 However, we have not examined LINE-1 methylation level. Tissue collection and analyses were approved by the Dana-Farber/Harvard Cancer Center and Brigham and Women's Hospital Institutional Review Boards.
Table 1.
LINE-1 methylation level and clinical features of colorectal cancer
| N (%)* | LINE-1 methylation level mean (%) | 95% confidence interval (CI) | ||
|---|---|---|---|---|
| All cases | 869 | 61.4 | 60.8-62.0 | |
| Sex | Male | 394 (45%) | 61.8 | 60.8-62.8 |
| Female | 475 (55%) | 61.0 | 60.2-61.8 | |
| Age | ≤59 | 193 (23%) | 60.5 | 59.0-62.0 |
| 60-69 | 359 (43%) | 61.7 | 60.8-62.6 | |
| ≥70 | 292 (35%) | 61.6 | 60.5-62.7 | |
| Tumor location | Proximal | 370 (44%) | 61.9 | 61.0-62.8 |
| Distal | 465 (56%) | 61.1 | 60.2-61.9 | |
| Stage | I | 196 (25%) | 61.5 | 60.2-62.8 |
| II | 253 (33%) | 62.5 | 61.4-63.7 | |
| III | 225 (29%) | 60.1 | 58.9-61.4 | |
| IV | 103 (13%) | 59.8 | 57.6-61.9 |
There is no significant relation between LINE-1 and any of the clinical features examined.
(%) indicates the proportion of cases with a specific clinical feature.
Pathologic evaluation
Hematoxylin and eosin (H&E) stained tissue sections were examined by a pathologist (S.O.) blinded from clinical and other laboratory data as previously described.35 The following features were evaluated. 1) Tumor differentiation was categorized as well/moderate (≥50% gland formation) vs. poor (<50% gland formation). 2) The degree of tumor infiltrating lymphocytes (TILs) was graded as absent/1+ vs. 2+. 3) The presence and extent of extracellular mucin were categorized as 0% (no mucin), 1-49% or ≥ 50% of the tumor volume. 4) The presence and extent of signet ring cells were categorized as 0% (no signet ring cells), 1-49% or ≥ 50% of the tumor volume.
Genomic DNA extraction, whole genome amplification, and sequencing of KRAS and BRAF
Genomic DNA was then extracted using QIAmp DNA Mini Kit (Qiagen, Valencia, CA USA) 13. Whole genome amplification (WGA) of genomic DNA was performed by PCR using random 15-mer primers for subsequent genetic analyses.13 Previous studies by us and others showed that WGA did not significantly affect downstream genetic analyses.13, 36 PCR and Pyrosequencing targeted for KRAS codons 12 and 13, and BRAF codon 600 were performed as previously described.13
Real-time PCR (MethyLight) to determine CIMP status
Sodium bisulfite treatment on genomic DNA was performed as previously described.37 Utilizing ABI 7300 instrument (Applied Biosystems, Foster City, CA, USA) and MethyLight technology (real-time PCR),38 we quantified DNA methylation in 8 CIMP-specific promoters (CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1).9, 13, 34 We have shown that these 8 markers are sensitive and specific markers for CIMP diagnosis.34 COL2A1 was used to normalize for the amount of bisulfite-converted DNA.37 Primers and probes were previously described.9 The PCR condition was initial denaturation at 95C for 10 min followed by 45 cycles of 95C for 15 sec and 60C for 1 min. A standard curve was made in each PCR plate by duplicating PCR for COL2A1 on bisulfite-converted human genomic DNA at 4 different concentrations (in a 5-fold dilution series). The percentage of methylated reference (PMR, i.e., degree of methylation) at a given locus was calculated by dividing the GENE:COL2A1 ratio of template amounts in a sample by the GENE:COL2A1 ratio of template amounts in SssI-treated human genomic DNA (presumably fully methylated) and multiplying this value by 100.38 A PMR cutoff value of 4 (except for 6 in CRABP1 and IGF2) was based on previously validated data.37 Precision and performance characteristics of bisulfite conversion and subsequent MethyLight assays were evaluated and the assays have been validated.37
CIMP-high was defined as the presence of ≥6/8 methylated promoters, CIMP-low as 1/8-5/8 methylated promoters and CIMP-0 as the absence (0/8) of methylated promoters based on the fact that CIMP-high and CIMP-low are associated with BRAF and KRAS mutations, respectively, while CIMP-0 is associated with wild-type BRAF/KRAS.34, 39
Pyrosequencing to measure LINE-1 methylation
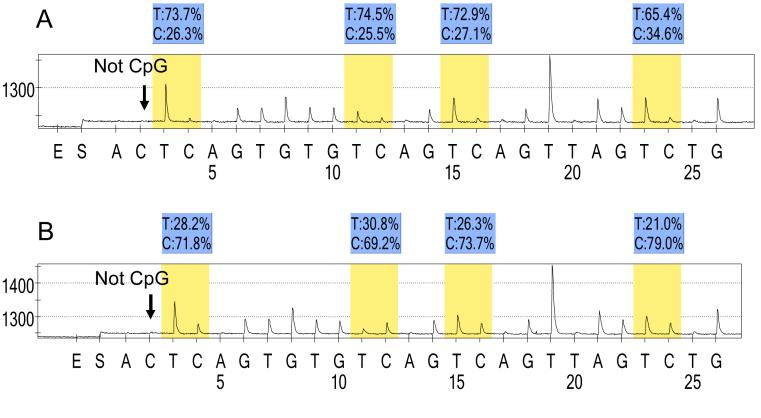
A difference in average LINE-1 methylation levels among different subtypes of colorectal cancers can be subtle.30 In order to accurately quantify relatively high methylation levels, we utilized Pyrosequencing technology (Figure 1). PCR and subsequent Pyrosequencing for LINE-1 were performed using the PyroMark kit (Biotage, Uppsala, Sweden). The PCR condition was 45 cycles of 95C for 20 sec, 50C for 20 sec and 72C for 20 sec, followed by 72C for 5 min. The biotinylated PCR product was purified and made single-stranded to act as a template in a pyrosequencing reaction, using the Pyrosequencing Vacuum Prep Tool (Biotage). Pyrosequencing reactions were performed in the PSQ HS 96 System (Biotage). The nucleotide dispensation order was: ACT CAG TGT GTC AGT CAG TTA GTC TG. Complete conversion of cytosine at a non-CpG site ensured successful bisulfite conversion (Figure 1, arrows). The amount of C relative to the sum of the amounts of C and T at each CpG site was calculated as percentage. The average of the relative amounts of C in the 4 CpG sites (Figure 1, blue shades) was used as overall LINE-1 methylation level in a given sample. Pyrosequencing to measure LINE-1 methylation has been previously validated.29, 30
Figure 1. Pyrosequencing for LINE-1 methylation.
A. MSS CIMP-low tumor. B. MSI-high CIMP-high tumor. The % numbers (in blue shade) are proportions of C and T at each CpG site after bisulfite conversion. Thus, the methylation level of each CpG site is estimated by the proportion of C (%). An overall LINE-1 methylation level is calculated as the average of the proportions of C (%) at the 4 CpG sites. The first, third and fourth CpG sites follow stretches of Ts, resulting in higher T peaks (in yellow shade) than the second CpG site, and the proportion of C (%) has been adjusted accordingly. The arrows indicate no residual C at the non-CpG site, ensuring complete bisulfite conversion.
CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable.
Microsatellite instability (MSI) and 18q loss of heterozygosity (LOH) analyses
Methods to determine MSI status have been previously described.13 In addition to the recommended MSI panel consisting of D2S123, D5S346, D17S250, BAT25 and BAT26,40 we also used BAT40, D18S55, D18S56, D18S67 and D18S487 (i.e., 10-marker panel). A “high degree of MSI” (MSI-H) was defined as the presence of instability in ≥ 30% of the markers. A low degree of MSI (MSI-L) was defined as the presence of instability in < 30% of the markers, and “microsatellite stable (MSS)” tumors were defined as tumors without an unstable marker.
For 18q LOH analysis using microsatellite markers D18S55, D18S56, D18S67 and D18S487,15 we duplicated PCR reaction in each sample to exclude allele dropouts of one of two alleles. LOH at each locus was defined as ≥40% reduction of one of two allele peaks in tumor DNA relative to normal DNA. Overall 18q LOH positivity was strictly defined as the presence of ≥2 markers with LOH, and overall 18q LOH negativity as the presence of ≥2 informative markers and the absence of LOH in all markers.
Tissue microarrays (TMAs) and immunohistochemistry for p53
Tissue microarrays (TMAs) were constructed as previously described,33 using the Automated Arrayer (Beecher Instruments, Sun Prairie, WI, USA). We examined at least two tissue cores (each 0.6mm) in immunohistochemical analysis. We examined whole tissue sections in cases for which no tissue block was available for TMAs or results were equivocal in TMAs. Immunohistochemistry for p53 was performed as previously described.34 p53 positivity was defined as 50% or more of tumor cells with unequivocal strong nuclear staining. Appropriate positive and negative controls were included in each run of immunohistochemistry. All immunohistochemically-stained slides were interpreted by one of the investigators (S.O.) blinded from clinical and other laboratory data.
Statistical analysis
We used SAS program (version 9.1, SAS Institute, Cary, NC, USA) to compute means with 95% confidence intervals (CIs) of LINE-1 methylation levels, to perform linear regression analysis, to compare means by two-sample t-test assuming unequal variances, to compare variances by F test, and to compare proportions by the chi-square test (or Fisher's exact test for categories with an N value of less than 10). All p values were two-sided, and statistical significance was set as p ≤ 0.05.
RESULTS
LINE-1 methylation levels in colorectal cancer and normal mucosa
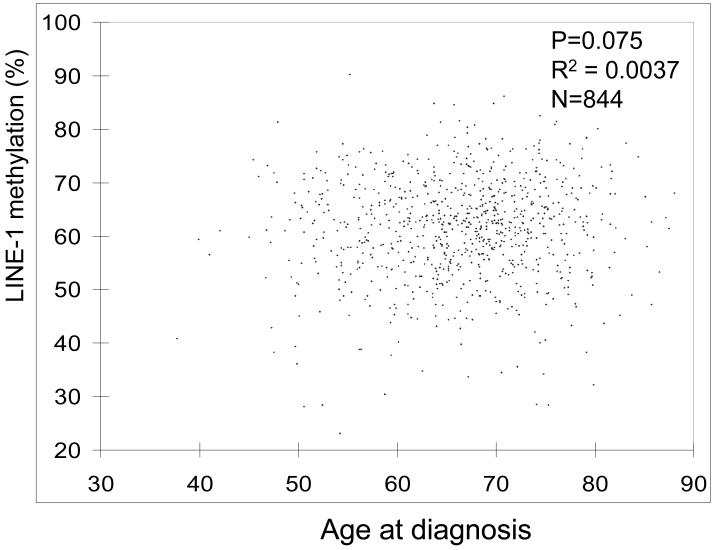
We quantified LINE-1 methylation in 869 population-based colorectal cancers by Pyrosequencing technology (Figure 1), which could provide precise data for LINE-1 methylation levels.28-30 LINE-1 methylation levels in the 869 tumors were distributed approximately normally [mean, 61.4(%); median, 62.3; standard deviation (SD), 9.6; range, 23.1-90.3; 10th percentile, 49.3; 90th percentile, 73.0]. Figure 2 shows a distribution of tumors according to age at diagnosis and LINE-1 methylation level. LINE-1 methylation level was not significantly correlated with age (p=0.075, R2=0.0037). There was no significant correlation between LINE-1 methylation and any of the clinical features examined (Table 1).
Figure 2. Distribution of LINE-1 methylation levels in colorectal cancer and age at diagnosis.
LINE-1 methylation levels are approximately normally distributed, and not significantly correlated with age.
We also examined LINE-1 methylation levels in matched normal colonic mucosa in 31 colorectal cancer cases. Normal mucosa showed significantly higher levels of LINE-1 methylation (71.8, SD 7.5, p<0.0001 by paired t-test) than matched tumor tissue, which was in agreement with previous studies.27, 30
LINE-1, microsatellite instability (MSI) and CpG island methylator phenotype (CIMP)
We quantified DNA methylation in the 8 promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1) by MethyLight technology, which was analytically sensitive and suited for the determination of promoter methylation status.9, 37 The 8 loci were selected as good predictors of CIMP-high by screening of 195 CpG islands.9, 34 CIMP-high tumors (with ≥6/8 methylated promoters) showed a higher mean LINE-1 methylation level (65.1) than CIMP-low tumors (with 1/8-5/8 methylated promoters) (61.3, p<0.0001) and CIMP-0 tumors (60.2, p<0.0001) (Table 2). MSI-H tumors also showed a higher mean LINE-1 methylation level (64.7) than MSI-L (59.8, p=0.0004) and MSS (60.8, p<0.0001) tumors. Because MSI is correlated with CIMP, we stratified tumors according to MSI and CIMP status. Compared to MSI-L/MSS CIMP-low/0 tumors (mean LINE-1 methylation level, 60.4), the mean LINE-1 methylation level was significantly higher in MSI-H CIMP-high (64.8, p<0.0001), MSI-H CIMP-low/0 (64.6, p=0.03) and MSI-L/MSS CIMP-high tumors (66.1, p=0.0003) (Table 2), indicating that both MSI-H and CIMP-high are positively correlated with LINE-1 methylation.
Table 2.
LINE-1 methylation level and molecular features of colorectal cancer
| N | LINE-1 methylation mean (%) | 95% CI | P value | ||
|---|---|---|---|---|---|
| MSI | MSI-H | 124 | 64.7 | 63.1-66.3 | Referent |
| MSI-L | 70 | 59.8 | 57.5-62.0 | 0.0004 | |
| MSS | 661 | 60.8 | 60.1-61.5 | <0.0001 | |
| MSI-L/MSS | 731 | 60.7 | 60.0-61.4 | <0.0001 | |
| CIMP | CIMP-high | 128 | 65.1 | 63.7-66.6 | Referent |
| CIMP-low | 333 | 61.3 | 60.3-62.3 | <0.0001 | |
| CIMP-0 | 408 | 60.2 | 59.3-61.2 | <0.0001 | |
| CIMP-low/0 | 741 | 60.7 | 60.0-61.4 | <0.0001 | |
| CIMP (by the Weisenberger et al.'s panel and criteria* {Weisenberger, 2006 #705}) | CIMP(+) | 171 | 64.6 | 63.3-65.9 | Referent |
| CIMP(-) | 698 | 60.6 | 59.8-61.3 | <0.0001 | |
| MSI/CIMP | MSI-H CIMP-high | 87 | 64.8 | 63.1-66.5 | <0.0001 |
| MSI-H CIMP-low/0 | 37 | 64.6 | 60.9-68.2 | 0.03 | |
| MSI-L/MSS CIMP-high | 41 | 66.1 | 63.3-69.0 | 0.0003 | |
| MSI-L/MSS CIMP-low/0 | 690 | 60.4 | 59.7-61.1 | Referent | |
| 18qLOH (only MSI-L/MSS tumors) | (+) | 230 | 59.1 | 58.0-60.2 | 0.004 |
| (-) | 185 | 61.8 | 60.4-63.2 | Referent | |
| CIMP/18q LOH (only MSI-L/MSS tumors) | CIMP-high LOH(+) | 8 | 64.6 | 56.5-72.8 | |
| CIMP-high LOH(-) | 14 | 65.1 | 59.0-71.2 | ||
| CIMP-low LOH(+) | 87 | 58.8 | 57.0-60.6 | 0.008 | |
| CIMP-low LOH(-) | 90 | 62.3 | 60.4-64.2 | Referent | |
| CIMP-0 LOH(+) | 135 | 59.0 | 57.5-60.5 | ||
| CIMP-0 LOH(-) | 81 | 60.7 | 58.5-62.8 | ||
| KRAS mutation | (+) | 319 | 61.2 | 60.2-62.3 | |
| (-) | 538 | 61.3 | 60.5-62.1 | ||
| BRAF mutation | (+) | 108 | 64.2 | 62.3-66.2 | 0.001 |
| (-) | 733 | 60.8 | 60.1-61.5 | Referent | |
| CIMP/BRAF | CIMP-high BRAF(+) | 73 | 66.7 | 64.8-68.6 | 0.01 |
| CIMP-high BRAF(-) | 52 | 62.9 | 60.7-65.1 | Referent | |
| CIMP-low/0 BRAF(+) | 35 | 59.2 | 55.1-63.2 | ||
| CIMP-low/0 BRAF(-) | 681 | 60.7 | 60.0-61.4 | ||
| p53 expression^ | (+) | 371 | 60.7 | 59.8-61.7 | |
| (-) | 488 | 61.8 | 60.9-62.7 |
Only significant p values are shown.
CI, confidence interval; CIMP, CpG island methylator phenotype; LOH, loss of heterozygosity; MSI-H, microsatellite instability-high; MSI-L, microsatellite instability-low; MSS, microsatellite stable.
By the Weisenberger et al.'s criteria, {Weisenberger, 2006 #705} CIMP(+) is defined as ≥3 methylated markers among 5 markers (CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1).
p53 status was determined by immunohistochemistry.
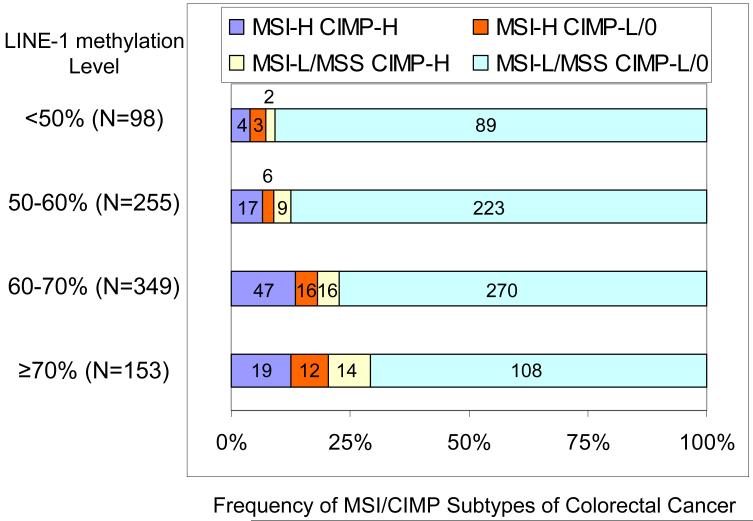
We also stratified tumors according to LINE-1 methylation levels, and examined the frequency of each MSI/CIMP subtype. The frequency of tumors with either MSI-H or CIMP-high or both (Figure 3, blue, red and yellow shades) increased as LINE-1 methylation level increased. The presence of either MSI-H or CIMP-high or both was significantly more common in tumors with ≥70% LINE-1 methylation (29%=45/153, p=0.0001) and 60-70% LINE-1 methylation (23%=79/349, p=0.002) than in tumors with <50% LINE-1 methylation (9.2%=9/98).
Figure 3. Frequencies of MSI/CIMP subtypes of colorectal cancer according to LINE-1 methylation level.
The presence of MSI-H or CIMP-high or both (blue, red and yellow shades) is more frequent in tumors with ≥70% LINE-1 methylation (29%=45/153, p=0.0001) and 60-70% LINE-1 methylation (23%=79/349, p=0.002) than in <50% LINE-1 methylated tumors (9.2%=9/98).
CIMP, CpG island methylator phenotype; LOH, loss of heterozygosity; MSI-H, microsatellite instability-high; MSI-L, microsatellite instability-low; MSS, microsatellite stable.
LINE-1 methylation and 18q loss of heterozygosity (LOH)
We examined the relationship between LINE-1 methylation and 18q LOH because a link between global hypomethylation and chromosomal instability has been suggested.21 In MSI-L/MSS tumors, 18q LOH(+) tumors showed a lower mean LINE-1 methylation level (59.1) than 18q LOH(-) tumors (61.8, p=0.004) (Table 2). Since 18q LOH has been shown to be correlated with CIMP-0,15 we stratified tumors according to CIMP and 18q LOH status. 18q LOH was significantly correlated with a lower mean LINE-1 methylation level in CIMP-low tumors (p=0.008), but not in CIMP-high or CIMP-0 tumors.
LINE-1 methylation, BRAF, KRAS and p53
The mean LINE-1 methylation level was higher in BRAF-mutated tumors (64.2, p=0.001) than BRAF-wild-type tumors (60.8). Because BRAF mutation is associated with CIMP-high, we stratified tumors by CIMP and BRAF status (Table 2). BRAF mutation was significantly correlated with LINE-1 methylation in CIMP-high tumors (p=0.01), but not in CIMP-low/0 tumors. LINE-1 methylation was not correlated with KRAS mutation or p53 expression.
LINE-1 methylation and pathologic features of colorectal cancer
We examined the relationship between LINE-1 methylation level and pathologic features of colorectal cancer. LINE-1 methylation appeared to be correlated with the presence of tumor infiltrating lymphocytes (TILs) (p=0.05), ≥50% mucinous component (p<0.0001), and signet ring cells (p≤0.02) (Table 3). However, no significant correlation between TILs and LINE-1 methylation persisted when tumors were stratified by MSI status [since TILs were correlated well with MSI rather than CIMP35]. In contrast, the presence of ≥50% mucinous component was correlated with high LINE-1 methylation level after tumors were stratified by MSI status [since mucinous features were correlated well with MSI rather than CIMP35]. Because signet ring cells were correlated with CIMP rather than MSI,35 we stratified tumors by CIMP status and signet ring cells. The association between signet ring cells and high LINE-1 methylation level appeared to persist, although statistical significance was not reached within CIMP-high tumors (Table 3).
Table 3.
LINE-1 methylation level and pathologic features of colorectal cancer
| N | LINE-1 mean (%) | 95% CI | P value | ||
|---|---|---|---|---|---|
| Tumor differentiation | Well-moderate | 778 | 61.3 | 60.6-61.9 | |
| Poor | 79 | 62.3 | 59.8-64.8 | ||
| Tumor infiltrating lymphocytes (TILs) | Absent/1+ | 752 | 61.0 | 60.3-61.7 | Referent |
| 2+ | 97 | 63.1 | 61.1-65.2 | 0.05 | |
| TILs (in MSI-H tumors) | Absent/1+ | 70 | 64.8 | 62.6-67.0 | |
| 2+ | 53 | 64.7 | 62.3-67.1 | ||
| TILs (in MSI-L/MSS tumors) | Absent/1+ | 671 | 60.5 | 59.8-61.2 | |
| 2+ | 44 | 61.2 | 57.7-64.8 | ||
| Mucinous component | 0% | 452 | 59.7 | 58.9-60.6 | Referent |
| 1-49% | 189 | 61.3 | 59.8-62.7 | ||
| >50% | 119 | 64.1 | 62.4-65.8 | <0.0001 | |
| Mucinous component (in MSI-H tumors) | 0% | 40 | 62.0 | 59.0-65.0 | Referent |
| 1-49% | 34 | 64.5 | 61.3-67.6 | ||
| >50% | 43 | 66.0 | 63.6-68.5 | 0.04 | |
| Mucinous component (in MSI-L/MSS tumors) | 0% | 405 | 59.4 | 58.5-60.3 | Referent |
| 1-49% | 153 | 60.5 | 58.9-62.1 | ||
| >50% | 72 | 62.9 | 60.5-65.3 | 0.008 | |
| Signet ring cells | 0% | 648 | 59.9 | 59.2-60.7 | Referent |
| 1-49% | 48 | 64.0 | 60.8-67.2 | 0.02 | |
| >50% | 15 | 68.3 | 63.5-73.1 | 0.002 | |
| Signet ring cells (in CIMP-high tumors) | 0% | 85 | 63.6 | 62.0-65.3 | |
| 1-49% | 20 | 68.1 | 63.6-72.7 | ||
| >50% | 3 | 67.8 | 51.4-84.1 | ||
| Signet ring cells (in CIMP-low/0 tumors) | 0% | 563 | 59.4 | 58.6-60.2 | Referent |
| 1-49% | 28 | 61.1 | 56.8-65.4 | ||
| >50% | 12 | 68.5 | 62.5-74.4 | 0.006 |
Only significant p values are shown.
CI, confidence interval; CIMP, CpG island methylator phenotype; LOH, loss of heterozygosity; MSI-H, microsatellite instability-high; MSI-L, microsatellite instability-low; MSS, microsatellite stable; TIL, tumor infiltrating lymphocyte.
DISCUSSION
DNA methylation physiologically plays an important role in X-chromosome inactivation, imprinting and repression of retrotransposons and endogenous retroviruses.1 On the other hand, promoter CpG islands of tumor suppressor genes are usually unmethylated, allowing active transcription. In cancer cells, however, genome-wide hypomethylation and/or hypermethylation of specific CpG islands often occur.1 We conducted this study to determine the relation between these two abnormal epigenomic phenomena in colorectal cancer. To estimate genome-wide methylation level, we measured LINE-1 methylation, which has been shown to be correlated with global DNA methylation level.28, 29, 31 We have shown that both MSI and CIMP are inversely correlated with LINE-1 hypomethylation, suggesting that MSI/CIMP phenomena and genome-wide hypomethylation may represent different pathways to colorectal cancer.
Detection of epigenetic aberrations in cancer and precancerous lesions is important in cancer research.2, 20, 41-43 We utilized quantitative DNA methylation assays - Pyrosequencing for LINE-1 methylation and MethyLight real-time PCR for specific CpG islands, both of which are robust and can reproducibly differentiate low-level methylation from high-level methylation.28, 29, 37 MethyLight has high analytical sensitivity with 1-5% methylation level easily detectable.37 In contrast, Pyrosequencing has superior precision and accuracy at higher methylation levels as observed in LINE-1.28, 29, 37
Previous studies have examined a relationship between MSI, CIMP and LINE-1 methylation in colorectal cancer. One study did not show a correlation of MSI or CIMP status with LINE-1 methylation level in colorectal cancer.27 A study using LINE-1 Pyrosequencing assay showed a correlation of LINE-1 methylation with MSI, but not with CIMP,30 possibly due to the classic methylation marker panel used. Thus, the authors suggested that MSI, rather than CIMP, was the main molecular alteration associated with lack of LINE-1 hypomethylation.30 In contrast, our findings clearly indicate that both CIMP and MSI are associated with lack of LINE-1 hypomethylation. Our CIMP-specific marker panel can distinguish CIMP-high that is associated with BRAF mutation from CIMP-low that is associated with KRAS mutation.34, 39 On the other hand, CIMP+ tumors determined by the classic CIMP panel (including MINT markers) are associated with both BRAF and KRAS mutations,14, 44 implying biological heterogeneity among the CIMP+ tumors determined by the classic CIMP markers. Our resource of a large number of colorectal cancers (derived from two large prospective cohorts) has provided us with a sufficient power to demonstrate the correlation of LINE-1 methylation with both MSI and CIMP. Nonetheless, a higher LINE-1 methylation level in normal colon tissue than tumor (shown by the study by Estecio et al.30) is in complete agreement with our current data.
The relation between global methylation level and prevalence of hypermethylation in individual CpG islands (i.e., CIMP status) is intuitively plausible. In addition, our results suggest that global methylation level is also correlated with MSI independent of CIMP; specifically, MSI-H non-CIMP-high tumors demonstrate high levels of LINE-1 methylation similar to CIMP-high tumors. Since separation of non-CIMP-high from CIMP-high in MSI-H tumors is very clear cut using our CIMP marker panel,34 high LINE-1 methylation level cannot be explained by misclassification of MSI-H CIMP-high tumors as MSI-H CIMP-low/0 tumors. Further studies are necessary to elucidate the mechanism of high LINE-1 methylation level in MSI-H tumors.
We have also shown that, in non-MSI-H tumors, 18q loss of heterozygosity (LOH) is correlated with low LINE-1 methylation level, supporting the link between genome-wide hypomethylation and chromosomal instability (CIN). CIN has been inversely associated with overall CIMP status.10, 15 Recent data indicate that different patterns of chromosomal aberrations are associated with methylation in different sets of promoters.45 In addition, the relation between global hypomethylation and chromosomal aberrations has been repeatedly shown.21-23, 46-48 A study using ApcMin/+ mice has shown that hypomorphic alleles of the Dnmt1 gene cause DNA hypomethylation in intestinal cells leading to increased microadenoma formation through LOH at the Apc locus,23 suggesting a pathogenetic link between DNA hypomethylation and LOH. Genome-wide DNA demethylation has been correlated with chromosomal instability and genomic copy number changes.22, 46 Genetic disruption of DNA methyltransferase or treatment with 5-aza-deoxycytidine results in chromosomal translocations or rearrangements.47, 48 In another study using LINE-1 COBRA assay,21 the presence of LOH at 5q, 8p or 17p was correlated with low LINE-1 methylation level in colorectal cancer, and in particular, the association between 18q LOH and LINE-1 hypomethylation was stronger than the associations between LINE-1 hypomethylation and the other LOH markers.21 These data collectively support the link between global DNA hypomethylation and CIN.
In summary, both MSI and CIMP are inversely correlated with LINE-1 hypomethylation, supporting the presence of distinct pathways to colorectal cancer - MSI/CIMP and global DNA hypomethylation. Our data also support the link between global DNA hypomethylation and chromosomal instability.
ACKNOWLEDGEMENTS
We deeply thank the Nurses' Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires; hospitals and pathology departments throughout the U.S. for providing us with tumor tissue materials; Walter Willett, Sue Hankinson, and many other staff members who implemented and have maintained the cohort studies; and Jean-Pierre Issa, Lanlan Shen and Liying Yan for their assistance in the development of the LINE-1 assay.
Funding: This work was supported by National Institute of Health (NIH)/National Cancer Institute (NCI) grants P01 CA87969, P01 CA55075K07 and K07 CA122826 (to S.O.), and in part by grants from the Bennett Family Fund and from the Entertainment Industry Foundation (EIF) through the EIF National Colorectal Cancer Research Alliance (NCCRA). M.O. was supported by a fellowship grant from the Japanese Foundation for Multidisciplinary Treatment of Cancer. The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH.
Abbreviations and HUGO Gene Nomenclature Committee (HGNC)-approved official gene symbols
- CACNA1G
calcium channel, voltage-dependent, T type alpha-1G subunit
- CDKN2A
cyclin-dependent kinase inhibitor 2A (p16/INK4A)
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- CIN
chromosomal instability
- CRABP1
cellular retinoic acid binding protein 1
- IGF2
insulin-like growth factor 2
- LINE-1
long interspersed nucleotide element-1
- LOH
loss of heterozygosity
- MSI
microsatellite instability
- MSI-H
microsatellite instability-high
- MSI-L
microsatellite instability-low
- MSS
microsatellite stable
- NEUROG1
neurogenin 1
- PMR
percentage of methylated reference (degree of DNA methylation)
- RUNX3
runt-related transcription factor 3
- SOCS1
suppressor of cytokine signaling 1
- TIL
tumor infiltrating lymphocyte
Footnotes
Competing interests: none declared
The novelty and impact of this paper:
To our knowledge, this paper is the first to provide evidence for an inverse association between CIMP and genomic hypomethylation, independent of MSI.
Our findings suggest that MSI/CIMP and genomic hypomethylation may represent different pathways to colorectal cancer.
REFERENCES
- 1.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 2.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–31. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. The necessity of a human epigenome project. Carcinogenesis. 2006;27:1121–5. doi: 10.1093/carcin/bgl033. [DOI] [PubMed] [Google Scholar]
- 4.Grady WM. CIMP and colon cancer gets more complicated. Gut. 2007;56:1498–500. doi: 10.1136/gut.2007.125732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–87. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 7.van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51:797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitehall VL, Wynter CV, Walsh MD, Simms LA, Purdie D, Pandeya N, Young J, Meltzer SJ, Leggett BA, Jass JR. Morphological and molecular heterogeneity within nonmicrosatellite instability-high colorectal cancer. Cancer Res. 2002;62:6011–4. [PubMed] [Google Scholar]
- 9.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 10.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, Boland CR. The CpG Island Methylator Phenotype and Chromosomal Instability Are Inversely Correlated in Sporadic Colorectal Cancer. Gastroenterology. 2007;132:127–38. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka H, Deng G, Matsuzaki K, Kakar S, Kim GE, Miura S, Sleisenger MH, Kim YS. BRAF mutation, CpG island methylator phenotype and microsatellite instability occur more frequently and concordantly in mucinous than non-mucinous colorectal cancer. Int J Cancer. 2006;118:2765–71. doi: 10.1002/ijc.21701. [DOI] [PubMed] [Google Scholar]
- 12.Goel A, Li MS, Nagasaka T, Shin SK, Fuerst F, Ricciardiello L, Wasserman L, Boland CR. Association of JC Virus T-Antigen Expression With the Methylator Phenotype in Sporadic Colorectal Cancers. Gastroenterology. 2006;130:1950–61. doi: 10.1053/j.gastro.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 13.Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner G, Weisenberger DJ, Campan M, Laird PW, Loda M, Fuchs CS. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–6. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samowitz W, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–45. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Ogino S, Kawasaki T, Kirkner GJ, Ohnishi M, Fuchs CS. 18q loss of heterozygosity in microsatellite stable colorectal cancer is correlated with CpG island methylator phenotype-negative (CIMP-0) and inversely with CIMP-low and CIMP-high. BMC Cancer. 2007;7:72. doi: 10.1186/1471-2407-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–30. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 17.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–92. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 18.Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, 3rd, Jaenisch R. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell. 2005;8:275–85. doi: 10.1016/j.ccr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, Voravud N, Sriuranpong V, Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–6. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 20.Deng G, Nguyen A, Tanaka H, Matsuzaki K, Bell I, Mehta KR, Terdiman JP, Waldman FM, Kakar S, Gum J, Crawley S, Sleisenger MH, et al. Regional hypermethylation and global hypomethylation are associated with altered chromatin conformation and histone acetylation in colorectal cancer. Int J Cancer. 2006;118:2999–3005. doi: 10.1002/ijc.21740. [DOI] [PubMed] [Google Scholar]
- 21.Matsuzaki K, Deng G, Tanaka H, Kakar S, Miura S, Kim YS. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11:8564–9. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci U S A. 2005;102:13580–5. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frigola J, Sole X, Paz MF, Moreno V, Esteller M, Capella G, Peinado MA. Differential DNA hypermethylation and hypomethylation signatures in colorectal cancer. Hum Mol Genet. 2005;14:319–26. doi: 10.1093/hmg/ddi028. [DOI] [PubMed] [Google Scholar]
- 25.Ehrlich M, Woods CB, Yu MC, Dubeau L, Yang F, Campan M, Weisenberger DJ, Long T, Youn B, Fiala ES, Laird PW. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene. 2006;25:2636–45. doi: 10.1038/sj.onc.1209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bariol C, Suter C, Cheong K, Ku SL, Meagher A, Hawkins N, Ward R. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol. 2003;162:1361–71. doi: 10.1016/S0002-9440(10)63932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iacopetta B, Grieu F, Phillips M, Ruszkiewicz A, Moore J, Minamoto T, Kawakami K. Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa. Cancer Sci. 2007;98:1454–60. doi: 10.1111/j.1349-7006.2007.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, Luna R, Rashid A, Shen L, Estecio MR, Kantarjian HM, Garcia-Manero G, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 29.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, Tajara EH, Issa JP. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colditz GA, Hankinson SE. The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–96. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 33.Chan AT, Ogino S, Fuchs CS. Aspirin and the Risk of Colorectal Cancer in Relation to the Expression of COX-2. New Engl J Med. 2007;356:2131–42. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 34.Ogino S, kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S, Odze RD, kawasaki T, Brahmandam M, Kirkner GJ, Laird PW, Loda M, Fuchs CS. Correlation of pathologic features with CpG island methylator phenotype (CIMP) by quantitative DNA methylation analysis in colorectal carcinoma. Am J Surg Pathol. 2006;30:1175–83. doi: 10.1097/01.pas.0000213266.84725.d0. [DOI] [PubMed] [Google Scholar]
- 36.Dietmaier W, Hartmann A, Wallinger S, Heinmoller E, Kerner T, Endl E, Jauch KW, Hofstadter F, Ruschoff J. Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am J Pathol. 1999;154:83–95. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino S, kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, Fuchs CS. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–17. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotypelow (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–8. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 41.Iacopetta B, Grieu F, Li W, Ruszkiewicz A, Caruso M, Moore J, Watanabe G, Kawakami K. APC gene methylation is inversely correlated with features of the CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2006;119:2272–8. doi: 10.1002/ijc.22237. [DOI] [PubMed] [Google Scholar]
- 42.Slattery ML, Curtin K, Sweeney C, Levin TR, Potter J, Wolff RK, Albertsen H, Samowitz WS. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120:656–63. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 43.Okochi-Takada E, Nakazawa K, Wakabayashi M, Mori A, Ichimura S, Yasugi T, Ushijima T. Silencing of the UCHL1 gene in human colorectal and ovarian cancers. Int J Cancer. 2006;119:1338–44. doi: 10.1002/ijc.22025. [DOI] [PubMed] [Google Scholar]
- 44.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–5. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Derks S, Postma C, Carvalho B, van den Bosch SM, Moerkerk PT, Herman JG, Weijenberg MP, de Bruine AP, Meijer GA, van Engeland M. Integrated analysis of chromosomal, microsatellite and epigenetic instability in colorectal cancer identifies specific associations between promoter methylation of pivotal tumour suppressor and DNA repair genes and specific chromosomal alterations. Carcinogenesis. 2007 doi: 10.1093/carcin/bgm270. (published online) [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capella G, Ribas M, Peinado MA. Chromosomal Instability Correlates with Genome-wide DNA Demethylation in Human Primary Colorectal Cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 47.Karpf AR, Matsui S. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005;65:8635–9. doi: 10.1158/0008-5472.CAN-05-1961. [DOI] [PubMed] [Google Scholar]
- 48.Ji W, Hernandez R, Zhang XY, Qu GZ, Frady A, Varela M, Ehrlich M. DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res. 1997;379:33–41. doi: 10.1016/s0027-5107(97)00088-2. [DOI] [PubMed] [Google Scholar]