Abstract
Mammalian cell membranes provide an interface between the intracellular and extracellular compartments. It is currently thought that cytoplasmic signaling adapter proteins play no functional role within the extracellular tumor environment. Here, by selecting combinatorial random peptide libraries in tumor-bearing mice, we uncovered a direct, specific, and functional interaction between CRKL, an adapter protein [with Src homology 2 (SH2)- and SH3-containing domains], and the plexin-semaphorin-integrin domain of β1 integrin in the extracellular milieu. Through assays in vitro, in cellulo, and in vivo, we show that this unconventional and as yet unrecognized protein–protein interaction between a regulatory integrin domain (rather than a ligand-binding one) and an intracellular adapter (acting outside of the cells) triggers an alternative integrin-mediated cascade for cell growth and survival. Based on these data, here we propose that a secreted form of the SH3/SH2 adaptor protein CRKL may act as a growth-promoting factor driving tumorigenesis and may lead to the development of cancer therapeutics targeting secreted CRKL.
Keywords: cancer, CrkL, integrin, phage display
Cell membranes have evolved as a tight and compartmentalized, but dynamic, interface between intracellular and extracellular contents (1, 2). To maintain such homeostasis, transmembrane receptors mediate bidirectional signaling across the cell surface through a complex spatial and temporal organization (3, 4). Thus, protein location in signal transduction is central to specificity of cellular responses (2, 5, 6). For example, cell surface receptors such as integrins undergo conformational changes elicited through ligand binding to enable a cross-talk with signal transduction cascades such as the MAPK pathways; conventional integrin ligands include ECM proteins that recognize integrin extracellular domains and cytoskeletal proteins that interact with intracellular domains (3–4, 7–9).
To gain insight into signal transduction across cell membranes in cancer, we set out to identify functional protein interactions in a tumor xenograft model; we reasoned that a combinatorial approach (10–14) in vivo might provide clues by emulating ligand–receptor binding in the context of the tumor microenvironment. Here, we show a specific interaction between the intracellular signaling protein CRKL and a regulatory (rather than ligand-binding) β1 integrin extracellular domain. Surprisingly, we found that CRKL targets the plexin-semaphorin-integrin (PSI) domain of β1 integrin chain located outside of the cell and promotes cell growth and survival. These results indicate an unrecognized integrin-mediated outside-in function for intracellular mediators, such as Src homology 2 (SH2)- and SH3-containing proteins, in activating proliferative pathways.
Results
Combinatorial Selection in Vivo Yields Tumor-Homing Peptides.
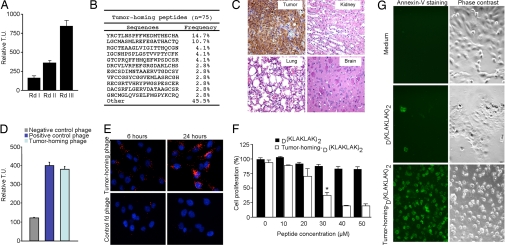
We administered a phage library (12, 14, 15) i.v. into nu/nu (nude) mice bearing human DU145-derived prostate cancer xenografts and recovered tumors after 24-h circulation. We recovered an enriched population of tumor-targeting phage (Fig. 1A) and individual phage clones (Fig. 1B). The dominant peptide (YRCTLNSPFFWEDMTHECHA) was functionally characterized; we evaluated its tumor-targeting specificity in vivo in tumor-bearing mice. After i.v. administration of YRCTLNSPFFWEDMTHECHA-phage, we observed marked homing to tumors (insertless phage served as a negative control) with barely detectable phage localization in several control organs (Fig. 1C). We also in vitro-targeted DU145 cells with an aqueous-to-organic phase separation assay (16) and a phage-based immunofluorescence assay (Kaposi Sarcoma cells; KS1767); consistently, YRCTLNSPFFWEDMTHECHA-phage bound to tumor cell surfaces to a greater extent than negative control phage (Fig. 1 D and E). We next evaluated the internalization of the peptide by fusing a proapoptotic motif (13, 17–19) to the tumor-homing sequence. We found targeted cell death relative to controls (Fig. 1 F and G), indicating that YRCTLNSPFFWEDMTHECHA mediates ligand-directed internalization. These results show that YRCTLNSPFFWEDMTHECHA targets tumor cells and enables internalization.
Fig. 1.
Targeting tumor xenografts reveal a β1 integrin mimic peptide. (A) Isolation of tumor-homing phage by screening of a random cyclic X2CX14CX2 phage library on DU145-derived prostate cancer tumor-bearing mice. Bars represent mean ± SD from triplicate plating. (B) Peptide sequence analysis from the third round of in vivo selection. Seventy-five peptide sequences were recovered and analyzed. Frequency reflects the number of phage from the total recovered pool. (C) Targeting specificity for the phage displaying the peptide YRCTLNSPFFWEDMTHECHA in the prostate tumor-bearing mice. Antiphage staining by peroxidase reactivity was found in the tumor but not in control organs. Representative images are shown. (D) The YRCTLNSPFFWEDMTHECHA-displaying phage binds to the cell surface on DU145 cells. A phage clone displaying the sequence RGD-4C (12) served as a positive control, and fd-tet (insertless) was a negative control. Bars represent mean ± SD from triplicate plating. (E) Immunolocalization of tumor-homing phage on the cell surface of nonpermeabilized KS1767 cells. (F and G) Synthetic peptide YRCTLNSPFFWEDMTHECHAGG-D(KLAKLAK)2 (30 μM) enables internalization by DU145 cells. Cell viability with WST-1 reagent (F) and an anti-annexin-V FITC-labeled antibody (G) confirmed targeted cell death by the tumor-homing peptide internalization. Representative images are shown. *, P < 0.001 at 30 μM vs. D(KLAKLAK)2 alone (Student's t test). (Magnifications: C, ×200; E, ×400; G, ×200.)
A Tumor-Homing Peptide Sequence Mimics a Regulatory Integrin Extracellular Domain.
To determine whether the peptide sequence mimics a native protein, we performed a similarity search of YRCTLNSPFFWEDMTHECHA and other selected peptide sequences. By using BLAST followed by protein alignment, we found that the peptides resembled sequences present on β1 integrin. Unexpectedly, the dominant sequence YRCTLNSPFFWEDMTHECHA had similarity to the PSI domain (residues 26–78) of the β1 integrin; moreover, we found that other selected peptides also appeared within the same region. We then asked whether the similarity of the peptide YRCTLNSPFFWEDMTHECHA was specific for the PSI domain of the β1 integrin or common to other known integrin β chains. After fit analysis and molecular modeling, we concluded that the homology between YRCTLNSPFFWEDMTHECHA and the PSI domain of β1 integrin was indeed the best alignment (Fig. S1 A–C).
A Cytoplasmic Adapter Serves as a Receptor for the PSI Domain-Like Tumor-Homing Peptide.
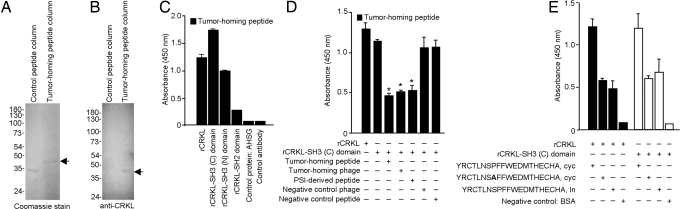
The integrin PSI domain has been characterized for regulatory activity (20–22). Given our results, we hypothesized that this domain might also function as a ligand-receptor binding site in β1 integrins and set out to purify extracellular proteins that might bind to this region. We used affinity chromatography to identify binding partners to YRCTLNSPFFWEDMTHECHA (Fig. 2 and Fig. S2). We precleared a DU145-derived cell extract in a control peptide column, passed the precleared extract through the YRCTLNSPFFWEDMTHECHA column, and performed an acidic elution to detect a specific gel band corresponding to an ≈40-kDa protein (Fig. 2A). Mass spectrometry identified such protein as CRKL (ref. 23 and Fig. S1D); we validated the purified protein by immunoblotting (IB) with an anti-CRKL antibody (Fig. 2B). Next, we constructed recombinant His-tag proteins (rCRKL) and 3 corresponding domains [rCRKL-SH2, rCRKL-SH3 (N), and rCRKL-SH3 (C)] for binding assays and found that the peptide bound to rCRKL preferentially through the 2 SH3 domains of the protein. In contrast, little to no binding was detected through the SH2 domain or to control proteins (Fig. 2C); several controls with unrelated sequences had no binding. We also performed binding studies with a synthetic peptide of the β1 integrin PSI domain (NSTFLQEGMPTSA; residues 50–62), which overlaps the selected sequences in the native PSI (Fig. S1 A–C); again, NSTFLQEGMPTSA bound to rCRKL, rCRKL-SH3 (N), and rCRKL-SH3 (C) (Fig. S2A). Moreover, we generated cyclic and linear peptides from the PSI domain and found that disulfide bonds are not essential for binding to CRKL (Fig. S3A). Finally, we showed that the interaction between SH3 (C) and the tumor-homing peptide is specifically inhibited by the synthetic peptide and by the phage clone itself (Fig. 2D). These findings show that the tumor-homing peptide and β1 integrin-specific PSI domain-mimic, YRCTLNSPFFWEDMTHECHA, targets SH3 domains of CRKL. Other studies have shown that CRKL-SH3 domains homodimerize (24). The tumor homing peptide targets the monomeric and dimeric forms of CRKL-SH3 (data not shown), suggesting that the peptide-binding site is not involved in CRKL dimerization; however, the exact mechanism of peptide interaction with SH3 domains and competition with the PSI domain warrant additional study. Furthermore, SH3 domains bind to PXXP and non-PXXP motifs in addition to motifs with a single Pro (25–29). Because YRCTLNSPFFWEDMTHECHA does not contain any such motifs, we used site-directed mutagenesis to evaluate the binding attributes of the tumor-homing peptide. Binding to the CRKL-SH3 (C) domain depended on the Pro and the cyclic Cys-Cys bridge in the peptide (Fig. 2E); insertless phage and unrelated peptides were controls. We also performed mutational analysis of CRKL SH3 (C) and found the binding region to be between Gly-236 and Trp-277 (Fig. S3B). Both the tumor-homing phage and the PSI-derived phage (CNSTFLQEGMPTSAC) bind to rCRKL (Fig. S2B). Additional CRKL SH3 domain mutants (including domains that cannot bind PXXP or non-PXXP motifs) might yield insights into the binding of the tumor-homing peptide in future studies. By affinity chromatography, we established that CRKL can be purified from DU145 serum-free conditioned medium and that a control column with a mutant tumor-homing peptide no longer bound to CRKL at detectable levels (Fig. S2C).
Fig. 2.
Receptor identification and validation. (A) Receptor purification by peptide YRCTLNSPFFWEDMTHECHA affinity chromatography. A 40-kDa band was detected in the Coomassie blue-stained gel and was excised for protein sequencing. (B) An anti-CRKL antibody confirmed the identity of the 40-kDa band by immunoblot. (C) The recombinant His-tag CRKL (rCRKL), rCRKL-SH3 (N) domain, and rCRKL-SH3 (C) domain bind to the tumor-homing peptide YRCTLNSPFFWEDMTHECHA. P < 0.001 vs. controls (Student's t test). (D) The binding activity of the tumor-homing peptide to rCRKL-SH3 (C) domain is inhibited by the tumor-homing peptide YRCTLNSPFFWEDMTHECHA, the PSI-derived (NSTFLQEGMPTSA) peptide, or the tumor-homing phage displaying YRCTLNSPFFWEDMTHECHA. Bars represent mean ± SD from triplicate wells. *, P < 0.001 vs. rCRKL alone or rCRKL-SH3(C) (Student's t test). (E) Binding properties of the rCRKL-SH3 (C) to the tumor-homing peptide. A Pro → Ala mutant is shown in bold. cyc, cyclic; ln, linear. P < 0.02 vs. Pro → Ala cyc peptide (Student's t test).
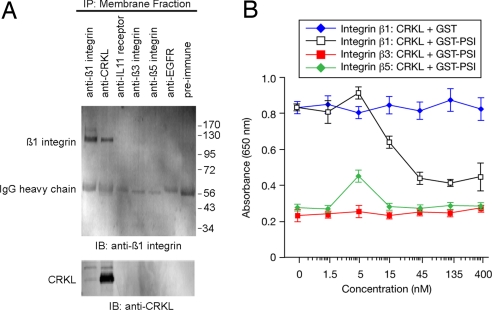
By reciprocal coimmunoprecipitation (IP) assays with membrane fractions, we showed that CRKL and β1 integrin form a complex at the membrane; in contrast, control antibodies raised against membrane receptors (anti-IL11R, anti-EGFR, anti-β3 or -β5 integrins) had no association with CRKL or β1 integrin (Fig. 3A). Finally, the interaction between CRKL and β1 integrin is inhibited (concentration-dependent) by a recombinant PSI domain (IC50 = 20 nM; Fig. 3B); consistent with the co-IP, control integrins showed no binding (Fig. 3A).
Fig. 3.
Interaction between CRKL and β1 integrin. (A) The association of CRKL with β1 integrin by reciprocal coIP from a DU145 membrane fraction with either anti-CRKL antibody or anti-β1 integrin antibody. The following unrelated antibodies served as negative controls: anti-IL11R, anti-EGFR, anti-β3, anti-β5, and preimmune serum. (B) A concentration-dependent inhibition of CRKL binding to β1 integrin by recombinant GST-PSI protein. The integrins αvβ3 and αvβ5 were used as controls. Mean ± SD from triplicate wells are shown.
CRKL Can Localize Outside of Cells.
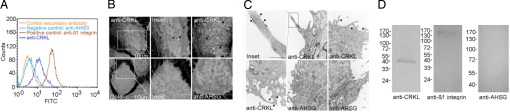
Molecular imaging showed colocalization of CRKL and β1 integrin (Fig. 4 and Fig. S4), indicating a specific molecular interaction at the cell surface. Because the tumor-homing peptide binds DU145 cell surfaces, we tested the possibility that CRKL may also exist outside of the cell membrane. FACS analysis of nonpermeable DU145 cells showed surface labeling with an anti-CRKL antibody (Fig. 4A). We found cell-surface labeling by immunofluorescence and confocal imaging in cells treated similarly (Fig. S4A). We also evaluated ultrastructural localization by scanning and transmission electron microscopy (TEM) with intact membranes. These imaging approaches yielded only modest CRKL cell-surface labeling but remain consistent with nonenhanced fluorescence detection methods (Fig. 4 B and C). Finally, we used biochemical approaches to confirm cell membrane localization of CRKL: cell surface labeling by biotinylation (Fig. 4D) and detergent membrane fractionation (Fig. S4B). By either method, we found CRKL present on the cell membrane (Fig. 4D and S4B). Antibodies against unrelated intracellular proteins served as controls (Fig. S4B). These results show that CRKL, in addition to its cytoplasmic location, is present on the surface.
Fig. 4.
CRKL is localized at the cell surface. (A) Flow cytometry analysis of CRKL on DU145 cells. Immunolabeling was performed with monoclonal anti-CRKL, anti-β1 integrin, and anti-AHSG antibodies. (B) Scanning electron microscopy of CRKL localization. Scale bar, 5 μm unless specified (arrows indicate CRKL-gold conjugated labeling). (C) TEM of CRKL showing individual CRKL-gold particles on the cell surface (arrowheads). The DU145 cells used in B–D were fixed under conditions in which cell plasma membranes were retained intact. Scale bar 500 nm. Representative images are shown. (D) (Left) Cell-surface biotinylated DU145 cells exhibit a surface location of CRKL. (Negative controls Center and Right). An anti-CRKL polyclonal antibody was used in (B–D).
Functional Studies and Transport Mechanisms.
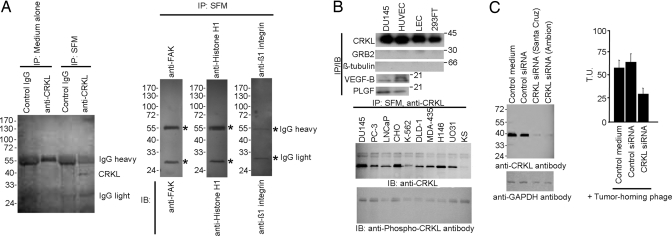
There are two potential explanations for finding extracellular CRKL: active transport by a secretory mechanism and/or passive release of intracellular contents caused by cell death. Because CRKL lacks a classic transmembrane domain, we evaluated whether CRKL is secreted from tumor cells (Fig. 5 and Fig. S5); we found that DU145 cells cultured in a serum-free medium do secrete unphosphorylated CRKL. In contrast, CRKL was not detected in controls as shown by IP either with anti-CRKL or control antibodies (Fig. 5A). To assess the generality of these data, we examined a panel of tumor cell lines in serum-free media and found that they also secrete unphosphorylated CRKL (Fig. 5B), indicating that this phenomenon is not cell type specific and highly depends on the tumor microenvironment for surface binding. Next, we sought to determine whether CRKL secretion had detectable effects on cell proliferation and migration. To show specificity, we added an anti-CRKL-neutralizing antibody to the serum-free medium of DU145 cells. We found that the anti-CRKL antibody does neutralize extracellular CRKL and reduced cell proliferation and cell migration; preimmune serum or control antibodies did not yield detectable effects on cell proliferation or migration (Fig. S6 A and B). To further understand the role of CRKL in tumor cells, we silenced CRKL with siRNA; again, we found reductions in cell proliferation, adhesion, and migration with CRKL reduction (Fig. S6 A–C). As an additional control, we showed that the decrease in proliferation is rescued by exogenous CRKL (Fig. S6D); only background apoptosis (≈1%) was detected in CRKL siRNA-treated cells or cells in serum-free medium. Finally, we found, in the CRKL siRNA-treated cells, reduced binding of the tumor-homing phage to the cells, indicating that tumor-homing phage might bind through secreted CRKL (Fig. S5C). Given that CRKL lacks a hydrophobic N-terminus for secretion via the endoplasmic reticulum and Golgi-dependent pathway (30), we tested whether secretory inhibitors might prevent cell release of CRKL. We showed that the inhibitors brefeldin A and thapsigargin do not inhibit CRKL secretion (Fig. 5F); in contrast, we found that glybenclamide, an inhibitor of ABC transporters, prevents CRKL release (Fig. S5C). There are at least 4 processes through which a protein can be secreted without a classic signal peptide (31–33). Our data are consistent with CRKL secretion by a nonclassical export pathway via ABC transporters. Other growth factors use ABC transporters (34, 35), suggesting a plausible working hypothesis. These results do not exclude the possibility that cell death caused by clonal selection or postcytotoxics could also generate extracellular CRKL in the tumor microenvironment. We thus designed binding assays to detail the biochemical interactions among CRKL, β1 integrins, and the PSI-mimetic. With rCRKL (GST and His-tag), our results suggest that CRKL can oligomerize (Fig. S7), perhaps through the SH3 domains (24, 36, 37).
Fig. 5.
CRKL secretion and cell signaling. (A) CRKL is secreted by DU145 cells cultured in serum-free medium (SFM) after 24 h as shown by detergent-free IP with an anti-CRKL antibody. Control antibodies (anti-FAK, anti-histone H1, and anti-β1 integrin) do not pull down their corresponding proteins in the cultured SFM. (B) (Upper) CRKL is secreted in several cell lines. IP and IB for GRB2 and β-tubulin served as negative controls; Placental growth factor (PLGF) and VEGF-B served as positive controls. (Lower) Other cell types cultured in SFM appeared to secrete only the unphosphorylated form of CRKL. (C) siRNA silencing of CRKL reduced tumor-homing phage binding to DU145 cells. SEM from triplicate wells is shown.
Furthermore, addition of exogenous CRKL leads to the phosphorylation of proteins in the MAPK and integrin-mediated signaling cascades, suggesting direct activation of migratory and proliferative pathways (Fig. S8).
CRKL-Mediated Ligand-Directed Tumor Targeting.
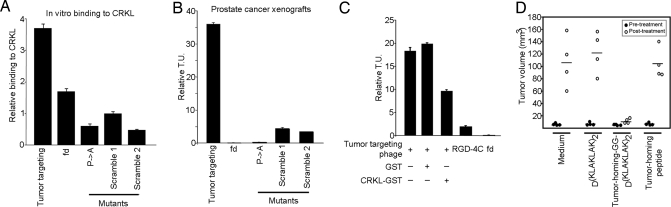
We next evaluated the targeting of CRKL-binding phage in vitro (Fig. 6A) and in tumor-bearing mice. First, we produced constructs displaying the selected CRKL-binding peptide or control (mutant or scrambled) peptides. Targeting was tested in human tumor xenografts [DU145 (Fig. 6B) and KS1767 (Fig. S9A)] and an isogenic mouse tumor [EF43-FGF4 (Fig. S9B)]. We observed marked and specific tumor homing after i.v. administration of CRKL-binding phage; in contrast, controls showed no tumor localization (Fig. 6 and Fig. S9). Further, we showed that homing is inhibited whether CRKL-binding phage is preincubated with rCRKL before administration to tumor-bearing mice (Fig. 6C). Notably, the tumor targeting of CRKL-binding phage was at least ≈10-fold higher relative to that of RGD-4C phage (10, 12, 15–17, 38), used as positive control. We then conducted a pilot preclinical trial with 4 size-matched cohorts of tumor-bearing mice (Fig. 6D). Animals received the following: (i) synthetic PSI domain-mimic peptide, (ii) synthetic PSI domain-mimic peptide fused to a proapoptotic motif (13, 17–19) to induce targeted apoptosis upon receptor-mediated internalization, (iii) synthetic proapoptotic motif, or (iv) vehicle. Peptides were administered at equimolar concentrations. The PSI domain-mimic peptide had no detectable effect on tumor growth, whereas the targeted proapoptotic motif inhibited tumor growth.
Fig. 6.
Tumor targeting. (A) In vitro phage binding of tumor-homing or controls (insertless, mutant, or scrambled) phage on rCRKL. Results are expressed as mean ± SEM of triplicate wells. (B) In vivo homing of targeted or control phage constructs in mice bearing prostate tumor xenografts. Tumor-homing phage localized to tumors preferentially compared with controls. Representative data from 2 independent experiments are shown. Bars represent mean ± SD. Tumor targeting, YRCTLNSPFFWEDMTHECHA; fd, fd-tet phage (negative control); P->A, YRCTLNSAFFWEDMTHECHA; Scramble 1, YRFCTSPFHEWHLENTDMCA; Scramble 2, YRECTDSPHEFHLWNTMCAF (bold indicates residue mutations). (C) Targeting inhibition in tumor-bearing mice. The tumor-homing phage was preincubated with control GST or recombinant GST-CRKL before administering to nude mice bearing size-matched human tumors (DU145-derived). Inhibition was observed in the pretreated tumor-homing phage by recombinant CRKL. Results from 2 independent experiments are shown. (D) Tumor treatment with a synthetic tumor-homing proapoptotic peptidomimetic. Cohorts of size-matched nude mice bearing human prostate cancer xenografts (DU145-derived) were used. Markedly reduced tumor growth was observed in tumor-bearing mice treated with synthetic tumor-homing proapoptotic peptidomimetic YRCTLNSPFFWEDMTHECHA-GG-D(KLAKLAK)2. Equimolar amounts of YRCTLNSPFFWEDMTHECHA or D(KLAKLAK)2 showed no differences in tumor volume compared with untreated animals. P < 0.001 (Student's t test).
Discussion
Consistent with the data presented here, it is likely that extracellular CRKL plays an as-yet-unrecognized role in the tumor microenvironment by triggering cell proliferation and migration. Because intracellular CRKL is also phosphorylated after addition of exogenous rCRKL, one might speculate that extracellular CRKL can function as an autocrine or paracrine factor within tumors. Our results establish an unusual connection between signaling and cell adhesion molecules, in which their relationship at the cell surface can trigger signaling events from the extracellular milieu. Based on the “switchblade” structural model for integrin activation (39) we propose a working model in which extracellular CRKL activates integrins through binding to the β1 integrin PSI domain (Fig. S10). We show that intracellular unphosphorylated CRKL is secreted by a nonclassical active transport and/or released through cell death into the microenvironment (step 1), where its SH3 domains bind specifically to the PSI domain of the β1 integrin on the tumor cell surface (step 2). Upon binding, the β1 integrin changes conformation from a bent to extended (active) form and thereby triggers downstream phosphorylation of target proteins in the integrin-mediated pathway (steps 3 and 4) and/or MAPK pathway (steps 5–7), affecting tumor cell migration and proliferation (step 8). Precedent for intracellular molecules such as nuclear proteins (40, 41), transcription factors (42), and stress-response chaperones (13, 43, 44) on the cell surface has been reported. Ligand–receptor interactions between signaling molecules and surface receptors within the extracellular environment may have general biological significance.
Materials and Methods
Reagents.
Anti-CRKL (Santa Cruz, Cell Signaling,Epitomics, or Upstate Biotechnology), antiphospho-CRKL (Cell Signaling), anti-β1 integrin (Chemicon or BD), anti-IL11R (Santa Cruz), anti-β3 and anti-β5 integrins (45), anti-EGFR (46), anti-grb2 (Santa Cruz), anti-α6 integrin (Chemicon), anti-fetuin A/α2-Heremans-Schmid glycoprotein (AHSG; R&D Systems), preimmune serum (Jackson), anti-His (Santa Cruz), anti-GST (Santa Cruz), and anti-GAPDH (Ambion) were used. Peptides were synthesized to our specifications (AnaSpec). Nude mice were purchased (Harlan), and tumors were generated as described (13, 47). The Institutional Animal Care and Use Committee at the University of Texas M. D. Anderson Cancer Center approved animal experiments.
Cell Culture.
Cell lines were purchased (ATCC) unless otherwise specified. Human lung epithelial cells and human umbilical vein endothelial cells were purchased and cultured in EGM-2 (Cambrex). Tumor cells were maintained in DMEM-high glucose (GIBCO) supplemented with 10% FBS, penicillin, streptomycin, and 4 mM glutamine (ICN) in a humidified atmosphere of 95% air and 5% CO2 at 37 °C.
Phage Display Library Selection.
Screenings were performed as described (12–15). A random phage library displaying an insert with the general arrangement X2CX12CX2 (C, cysteine; X, any residue) was administered (tail vein) into tumor-bearing nude mice and allowed to circulate for 24 h. Mice were placed under deep anesthesia, tumors were excised and weighed, and the bound phage population was recovered and processed.
Affinity Chromatography and Mass Spectrometry.
Peptide affinity columns were made by 1-ethyl-3-(3-dimethylaminopropyl)carboiimide and diaminodipropylamine immobilization resin (Pierce). Cell extracts were prepared and first passed through a nonspecific control peptide column followed by the tumor-homing peptide column. Columns were washed, eluted with Gly (pH 2.2), analyzed by SDS/PAGE, and stained with Coomassie blue. Affinity purification of CRKL from serum-free conditioned medium was performed and confirmed with recombinant GST-tag fusion protein expressing tumor-homing or mutant control peptide (Pro → Ala). Serum-free conditioned medium (200 mL at 48-h culture) was concentrated for the affinity purification. Fusion proteins were coupled to GST-resin beads and poured into a column. The concentrated serum-free conditioned medium was added to the columns for an overnight incubation. After several washes, the bound CRKL was eluted for Western blot analysis. The blot was probed with anti-GST and anti-CRKL antibodies.
Cell Surface and Membrane Localization.
Cell surface labeling with biotin (42), phage binding assays (16), and membrane fractionation (44) were performed as described. Immunofluorescence (IF), FACS, and TEM analyses were performed through standard protocols. For details, see SI Text.
Mutagenesis.
Primers are summarized in Table S1. For details on recombinant proteins and scrambled and mutant tumor-homing phage, see SI Text.
siRNA.
CRKL (mRNA accession no. NM_005207), β1 integrin (mRNA accession no. NM_002211), and control siRNAs were purchased from Santa Cruz, Ambion, and Dharmacon. siRNA oligonucleotides are summarized in Table S2. Oligofectamine (Invitrogen) or DharmaFect (Dharmacon) were used to transfect siRNAs into DU145 cells (1–2 × 105 cells per well). Transfected cells were incubated for 48–72 h before processing, recovered, and lysed in the presence of protease inhibitors. Tumor-homing phage binding activity was examined in CRKL-silenced cells. For rescue experiments, up to 1.5 μg of recombinant CRKL was added to CRKL siRNA-transfected cells. Exogenous His-tag recombinant CRKL (400 ng/mL) or controls (EGF, 200 ng/mL; migration inhibitory factor, 300 ng/mL; phorbol 12-myristate 13-acetate, 300 ng/ml) were used in β1 integrin-silenced experiments. Equal amounts of protein were loaded and resolved by SDS/PAGE followed by Western blot analysis with the appropriate antibodies. Cell proliferation assays were performed with WST-1 (Roche). Cell migration assays were performed in a Boyden chamber assay (Corning).
IP.
Cells were synchronized for 24–48 h in RPMI serum-free 1640 medium, followed by centrifugation and sterile filtration and preabsorption to control antibodies. An equal volume of PBS was added to the recovered supernatant before IP with the appropriate antibodies. Brefeldin A, thapsigargin, and glybenclamide were used. Cells were incubated with the compounds in serum-free medium for 9 h before IP with the appropriate antibodies. No detergents were used in the IP. This protocol favors the finding of CRKL in a free soluble state rather than in vesicles.
Peptide Binding and Internalization.
Peptides were coated on microtiter plates followed by blocking and washing. Mixtures were incubated, washed, and labeled with the appropriate antibodies. HRP-conjugated secondary antibodies were added followed by 3,3′–5,5′–tetramethylbenzidine (TMB) substrate (Calbiochem), and complexes were analyzed in an ELISA reader. To determine the inhibitory activity of the tumor-homing peptide, the PSI-derived peptide or the phage clone displaying the tumor-homing peptide were incubated with the rCRKL-SH3 (C) domain. Unrelated peptide sequences and insertless phage served as negative controls. Mixtures were incubated and added to wells coated with the tumor-homing peptide. After incubation, the wells were washed and labeled with the appropriate antibodies. For details and recombinant protein information, see SI Text.
Phage and Protein Binding Assays.
Phage binding assays on purified proteins were carried out as described (19, 48). For a detailed description, see SI Text.
In Vivo Tumor Targeting and Inhibition.
In vivo targeting experiments with phage were performed as described (10, 12, 13, 47). For details, see SI Text.
Therapy in Tumor-Bearing Mice.
Tumor-bearing mice were size-matched and divided into individual cohorts (n = 4 mice per group). The tumor-homing peptide was synthesized fused with the proapoptotic motif D(KLAKLAK)2. Unconjugated peptide YRCTLNSPFFWEDMTHECHA or D(KLAKLAK)2 served as controls. Peptides were administered i.v. at 300 μg per mouse per week, and tumor volumes were measured (12, 13).
Supplementary Material
Acknowledgments.
We thank Drs. Tony Hunter, Claudio Joazeiro, Helene Sage, and Richard Sidman for helpful discussions. This work was supported by grants from the National Institutes of Health (to W.A. and R.P.), the Department of Defense (to P.J.M., W.A., and R.P.) and awards from the V Foundation, the Marcus Foundation, and the Gillson-Longenbaugh Foundation (to R.P. and W.A.). M.C.V. is a Fellow from the Susan G. Komen Breast Cancer Foundation. R.R. and G.R.S. are Scholars from the Odyssey Program of the University of Texas M. D. Anderson Cancer Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807543105/DCSupplemental.
References
- 1.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 2.Cho W, Stahelin RV. Membrane–protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 3.Martin KH, Slack JK, Boerner SA, Martin CC, Parsons JT. Integrin connections map: To infinity and beyond. Science. 2002;296:1652–1653. doi: 10.1126/science.296.5573.1652. [DOI] [PubMed] [Google Scholar]
- 4.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 5.Mochly-Rosen D. Localization of protein kinases by anchoring proteins: A theme in signal transduction. Science. 1995;268:247–251. doi: 10.1126/science.7716516. [DOI] [PubMed] [Google Scholar]
- 6.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 7.Hunter T. The age of cross-talk: Phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 9.Blume-Jensen P, Hunter T. Oncogenic kinase signaling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 10.Hajitou A, et al. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 11.Arap W, et al. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 12.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 13.Arap MA, et al. Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell. 2004;6:275–284. doi: 10.1016/j.ccr.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 15.Pasqualini R, Arap W, Rajotte D, Ruoslahti E. In: Phage Display: A Laboratory Manual. Barbas CF III, Burton DR, Scott JK, Silverman GJ, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. pp. 22.1–22.24. [Google Scholar]
- 16.Giordano RJ, Cardó-Vila M, Lahdenranta J, Pasqualini R, Arap W. Biopanning and rapid analysis of selective interactive ligands. Nat Med. 2001;7:1249–1253. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]
- 17.Ellerby HM, et al. Anticancer activity of targeted proapoptotic peptides. Nat Med. 1999;5:1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 18.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med. 2004;10:625–632. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
- 19.Zurita AJ, et al. Combinatorial screenings in patients: The interleukin-11 receptor α as a candidate target in the progression of human prostate cancer. Cancer Res. 2004;64:435–439. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
- 20.Shi M, et al. The crystal structure of the plexin-semaphorin-integrin domain/hybrid domain/I-EGF1 segment from the human integrin β2 subunit at 1.8-Å resolution. J Biol Chem. 2005;280:30586–30593. doi: 10.1074/jbc.M502525200. [DOI] [PubMed] [Google Scholar]
- 21.Mould AP, et al. Evidence that monoclonal antibodies directed against the integrin β subunit plexin/semaphorin/integrin domain stimulate function by inducing receptor extension. J Biol Chem. 2005;280:4238–4246. doi: 10.1074/jbc.M412240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnaout MA, Mahalingam B, Xiong JP. Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- 23.ten Hoeve J, Morris C, Heisterkamp N, Groffen J. Isolation and chromosomal localization of CRKL, a human crk-like gene. Oncogene. 1993;8:2469–2474. [PubMed] [Google Scholar]
- 24.Harkiolaki M, Gilbert RJ, Jones EY, Feller SM. The C-terminal SH3 domain of CRKL as a dynamic dimerization module transiently exposing a nuclear export signal. Structure (London) 2006;14:1741–1753. doi: 10.1016/j.str.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Mayer BJ. SH3 domains: Complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 26.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 27.Kato M, Miyazawa K, Kitamura N. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J Biol Chem. 2000;275:37481–37487. doi: 10.1074/jbc.M007251200. [DOI] [PubMed] [Google Scholar]
- 28.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, et al. Structural basis for specific binding of the Gads SH3 domain to an RxxK motif-containing SLP-76 peptide: A novel mode of peptide recognition. Mol Cell. 2003;11:471–481. doi: 10.1016/s1097-2765(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 30.Walter P, Gilmore R, Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984;38:5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- 31.Prudovsky I, et al. The nonclassical export routes: FGF1 and IL-1α point the way. J Cell Sci. 2003;116:4871–4881. doi: 10.1242/jcs.00872. [DOI] [PubMed] [Google Scholar]
- 32.Reynwar BJ, et al. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- 33.McNiven MA, Thompson HM. Vesicle formation at the plasma membrane and trans-Golgi network: The same but different. Science. 2006;313:1591–1594. doi: 10.1126/science.1118133. [DOI] [PubMed] [Google Scholar]
- 34.Flieger O, et al. Regulated secretion of macrophage migration inhibitory factor is mediated by a nonclassical pathway involving an ABC transporter. FEBS Lett. 2003;551:78–86. doi: 10.1016/s0014-5793(03)00900-1. [DOI] [PubMed] [Google Scholar]
- 35.Hamon Y, et al. Interleukin-1β secretion is impaired by inhibitors of the ATP binding cassette transporter, ABC1. Blood. 1997;90:2911–2915. [PubMed] [Google Scholar]
- 36.Kishan KV, Scita G, Wong WT, Di Fiore PP, Newcomer ME. The SH3 domain of Eps8 exists as a novel intertwined dimer. Nat Struct Biol. 1997;4:739–743. doi: 10.1038/nsb0997-739. [DOI] [PubMed] [Google Scholar]
- 37.Kristensen O, et al. A unique set of SH3–SH3 interactions controls IB1 homodimerization. EMBO J. 2006;22:785–797. doi: 10.1038/sj.emboj.7600982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasqualini R, Koivunen E, Ruoslahti E. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- 39.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 40.Sinclair JF, O'Brien AD. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-γ of enterohemorrhagic Escherichia coli O157:H7. J Biol Chem. 2002;277:2876–2885. doi: 10.1074/jbc.M110230200. [DOI] [PubMed] [Google Scholar]
- 41.Hovanessian AG, et al. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res. 2000;261:312–328. doi: 10.1006/excr.2000.5071. [DOI] [PubMed] [Google Scholar]
- 42.Monferran S, Paupert J, Dauvillier S, Salles B, Muller C. The membrane form of the DNA repair protein Ku interacts at the cell surface with metalloproteinase 9. EMBO J. 2004;23:3758–3768. doi: 10.1038/sj.emboj.7600403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin BK, et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J Biol Chem. 2003;278:7607–7616. doi: 10.1074/jbc.M210455200. [DOI] [PubMed] [Google Scholar]
- 44.Mintz PJ, et al. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol. 2003;21:57–63. doi: 10.1038/nbt774. [DOI] [PubMed] [Google Scholar]
- 45.Barker TH, et al. SPARC regulates extracellular matrix organization through its modulation of integrin-linked kinase activity. J Biol Chem. 2005;280:36483–36493. doi: 10.1074/jbc.M504663200. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein NI, Prewett M, Zuklys K, Rockwell P, Mendelsohn J. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res. 1995;11:1311–1318. [PubMed] [Google Scholar]
- 47.Marchiò S, et al. Aminopeptidase A is a functional target in angiogenic blood vessels. Cancer Cell. 2004;5:151–162. doi: 10.1016/s1535-6108(04)00025-x. [DOI] [PubMed] [Google Scholar]
- 48.Smith GP, Scott JK. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.