Abstract
The antitumor antibiotic and cancer chemotherapeutic agent mitomycin C (MC) alkylates and cross-links DNA, forming six major MC-deoxyguanosine adducts of known structures in vitro and in vivo. Two of these adducts are derived from 2,7-diaminomitosene (2,7-DAM), a non-toxic reductive metabolite of MC formed in cells in situ. Several methods have been used for analysis of MC-DNA adducts in the past; however, a need exists for a safer, more comprehensive and direct assay of the six-adduct complex. Development of an assay, based on mass spectrometry is described. DNA from EMT6 mouse mammary tumor cells, Fanconi Anemia –A fibroblasts, normal human fibroblasts, and MCF-7 human breast cancer cells was isolated after MC or DMC treatment of the cells, digested to nucleosides and submitted to liquid chromatography electrospray-tandem mass spectrometry. Two fragments of each parent ion were monitored (“multiple reaction monitoring”; MRM). Identification and quantitative analysis was based on a standard mixture of six adducts, the preparation of which is described here in detail. The lower limit of detection of adducts is estimated as 0.25 picomol. Three initial applications of this method are reported: (i) differential kinetics of adduct repair in EMT6 cells; (ii) analysis of adducts in MC- or DMC-treated Fanconi Anemia cells; and (iii) comparison of the adducts generated by treatment of MCF-7 breast cancer cells with MC and DMC. Notable results are the following: repair removal of the DNA interstrand cross-link and of the two adducts of 2,7-DAM is relatively slow; both MC and DMC generate DNA interstrand cross-links in human fibroblasts, Fanconi Anemia-A fibroblasts and MCF-7 cells as well as EMT6 cells; DMC shows a stereochemical preference of linkage to the guanine-2-amino group opposite from that of MC.
Introduction
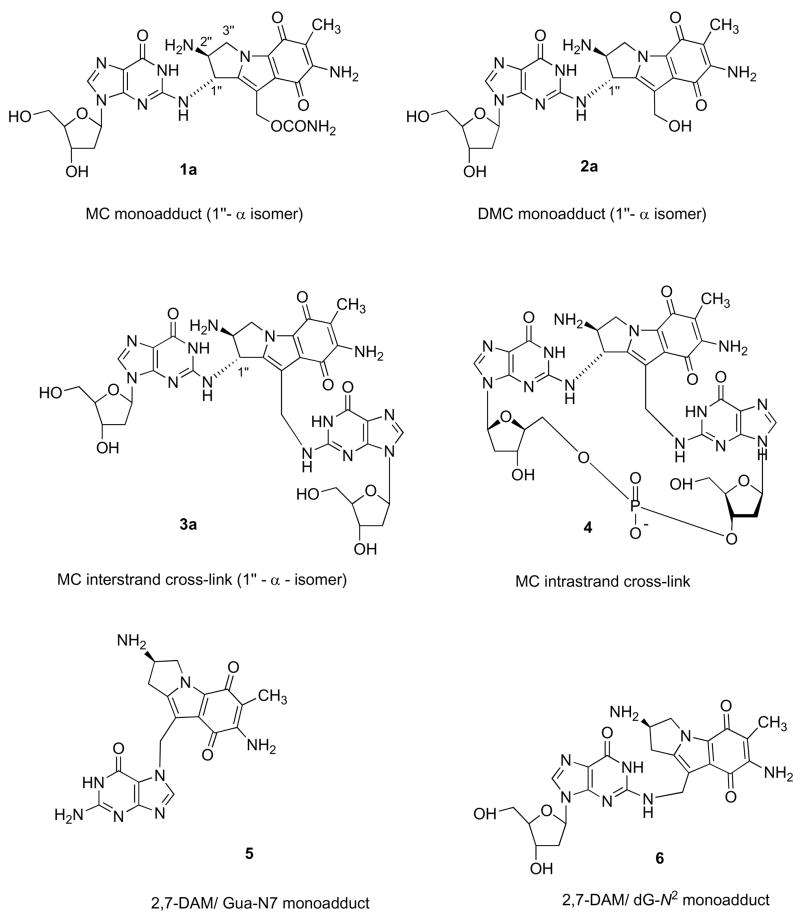
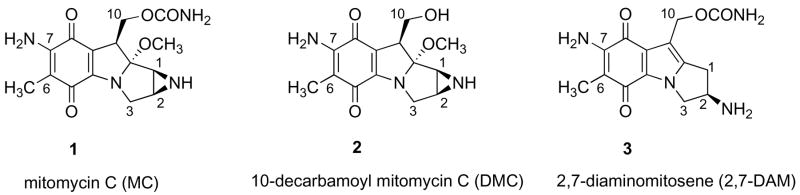
Mitomycin C (1; MC1), an antitumor antibiotic is used in clinical cancer chemotherapy (1). It has been shown to alkylate DNA monofunctionally and bifunctionally, resulting in covalent DNA monoadducts and DNA interstrand and intrastrand cross-links (2). These DNA lesions are thought to constitute the molecular basis of the cytotoxicity of MC to tumor cells; in particular, the observed frequencies of the DNA interstrand cross-links (ICLs) have been shown to correlate with the cytotoxicities of MC and its derivatives 10-decarbamoyl mitomycin C (2; DMC) and 2,7-diaminomitosene (3; 2,7-DAM) in various tumor cells (3, 4, 15). Treatment of EMT6 mouse mammary tumor cells resulted in the formation of six major DNA adducts as were shown using HPLC and [3H]-labelled MC of high specific activity for analysis of the DNA adducts in digests of DNA isolated after drug treatment (4). Each of the six adducts has also been isolated from biomimetic reactions of MC with DNA in vitro, enabling elucidation of their structures by microscale spectroscopic methods (Figure 2; 2, 5–7).
Figure 2.
Chemical structures of major adducts of DMC
MC adducts have been detected in several in vivo systems by 32P-postlabeling methodology (8). Organ tissues of rats (9), human colon cancer cells (10), chick embryos (11), human breast cancer cells, breast tumor biopsies, and xenografted tumors in mice (12) contained MC adducts in their DNA after administration of the drug. Assignments of specific structures (Figure 2) to the 32P-labelled spots separated on TLC were limited by the lack of availability of authentic standards to the investigators (9), with the exception of the ICL adduct 3a (10–12). Typically, 10−6–10−8 MC adducts per DNA nucleotide have been detectable in these studies (9–12). Specific detection of MC-induced cross-links in DNA in vivo at similar sensitivities has been reported by a different approach, based on the altered denaturation-renaturation behavior of cross-linked DNA relative to native DNA upon alkaline denaturation, seen using agarose gel electrophoresis (13) or “alkaline elution” (14, 15).
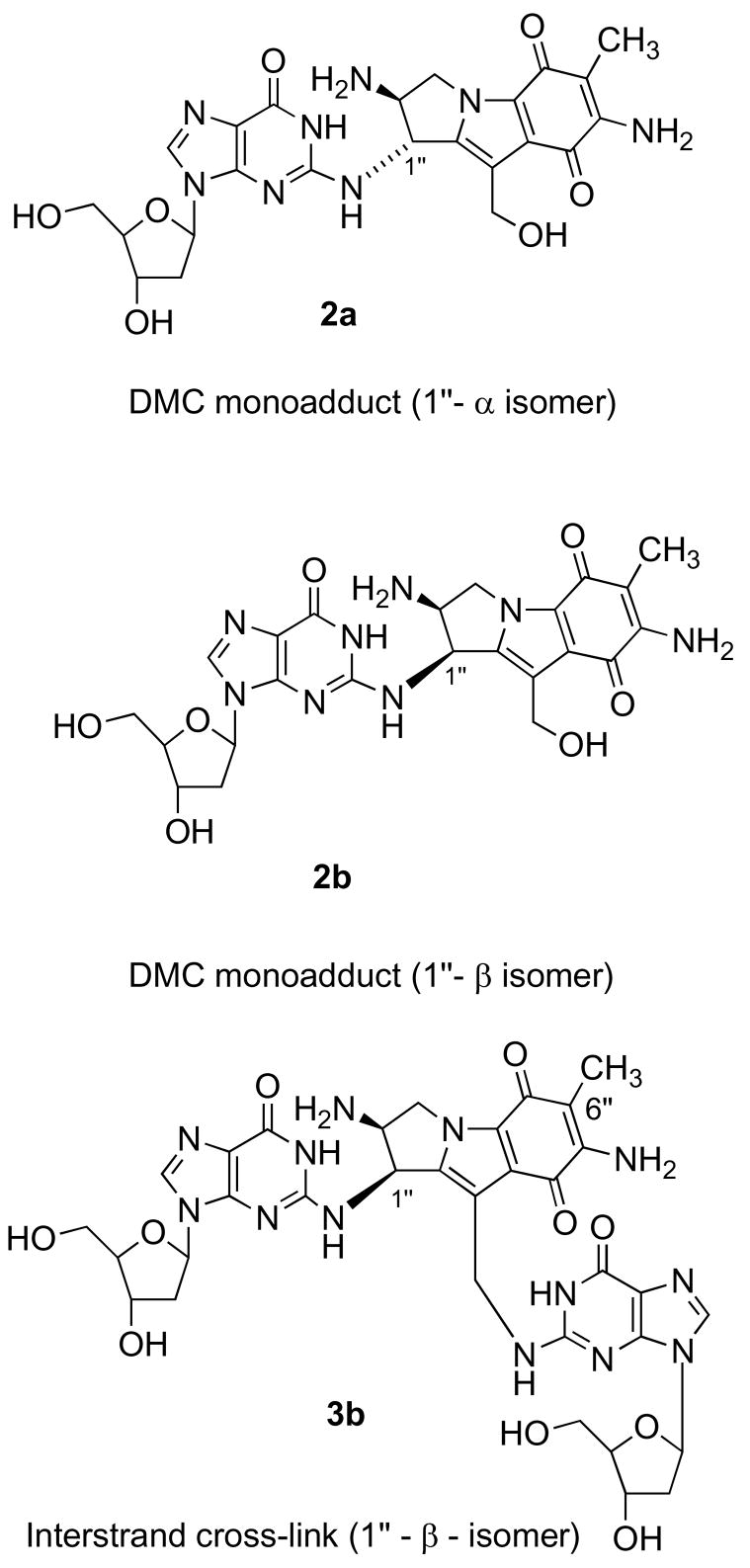
Recently, a more direct, safe, comprehensive and sensitive general method of assay of DNA adducts has emerged, employing liquid chromatography-electrospray tandem mass spectrometry (LC-ESI MS/MS; 16–20). In view of the continued interest in the biological and chemotherapeutic responses to DNA cross-linkers and alkylating agents, as typified by MC and its analogs (21–24), we explored this methodology for the first time with respect to DNA adducts of MC, DMC, and 2,7-DAM. This was especially timely, due to the continued trend toward non-radioactive assays and the fact that the [3H]-labeled MC used in our previous analyses (25) is no longer available. We report the development of an MS-based assay of MC adducts and the application of the assay to several research objectives: examination of the differential repair kinetics of MC adducts in EMT6 cells, identification of the adducts formed in normal human fibroblasts in comparison with FA-A cells after MC and DMC treatments, and assay of the adducts formed in MC- and DMC-treated MCF-7 human breast cancer cells. These last two objectives are of particular interest because treatment of EMT6 cells with DMC resulted in cross-link adducts (3), contrary to the previously held notion that DMC is the monofunctional counterpart of MC; furthermore, the major adducts of DMC had 1″-β stereochemistry of their linkage to DNA (3; Figure 3), which is opposite of that of the major adducts of MC (Figure 2). Using the MS method, we obtained similar results for three additional human cell lines, corroborating the previous results by confirming the presence of cross-links and demonstrating 1″-β adducts with DMC.
Figure 3.
Chemical structures of six major adducts of reductively activated MC
After completion of this work, development of a method utilizing the LC-MS/MS technique for DNA ICLs formed in human skin melanoma cells treated with 8-methoxypsoralen and UVA light was reported (26). Digestion with nuclease P1 alone generated cross-linked tetranucleotides, which were detected in negative-ion ESI-MS/MS mode, in contrast to the positive-ion mode used in the present work.
An essential part of this work was preparation of the adducts of MC and DMC used as standards in the LC-MS/MS assay. A comprehensive description of the preparation of six major adducts for this purpose is given here for the first time.
Experimental Section
Materials
Mitomycin C (1) was obtained from Bristol-Myers Squibb Co., Wallingford, CT and from Kyowa Hakko Kogyo Co. Ltd., Japan. 10-Decarbamoyl mitomycin C (3) was synthesized from mitomycin C by a published procedure (25). Phosphodiesterase I (snake venom diesterase, Crotalus adamanteus venom, EC 3.1.4.1) and alkaline phosphatase (Escherichia coli, EC 3.1.3.1) were obtained from Worthington Biochemical Corp., Freehold, NJ. and nuclease P1 (Penicillium citrinum, EC 3.1.30.1) from Sigma Life Sciences, St. Louis, MO. Sep-Pak C-18 cartridges (“Classic Short Body”) were purchased from Waters Corp., Milford, MA.
Cell lines, treatment of cells with MC or DMC and isolation of nuclear DNA from drug-treated cells
EMT6 mouse mammary tumor cells were cultured and maintained as exponentially growing monolayers in Waymouth’s medium supplemented with 15% Fetal Clone (Hyclone) and antibiotics, as described previously (28). Approximately 3×108 cells were suspended and treated in suspension culture with 2 μM MC for 1h under hypoxia as described previously (28). Incubation under hypoxia was used because our previous studies showed that treatment under severe hypoxia induces greater cytotoxicity, more adducts and a higher proportion of cross-links than treatment under air (3, 4, 7, 28). Cells were washed twice to remove the drug and the cell suspension was divided into 3 equal volumes, designated as the 0-h sample, 16-h sample and 24-h sample, respectively. The 16-h and 24-h samples were incubated in fresh medium for 16 and 24 hours, respectively to allow DNA repair, while the 0-h sample was processed immediately. Cells were counted before the isolation of nuclear DNA from each of the samples. Nuclear DNA was isolated using a Qiagen blood and cell culture DNA maxi kit following the manufacturer’s instructions (Qiagen, Valencia, CA). DNA was resuspended in TE buffer.
Immortalized cell lines derived from normal human fibroblasts (PD751) and fibroblasts from Fanconi Anemia patient from complementation group A (FA-A; PD220) were obtained from the Fanconi Anemia depository at Oregon Health Sciences Institute. Cells were propagated in alfaMEM (GIBCO) supplemented with 10% Bovine Growth Serum (Hyclone) and antibiotics. For experiments, cells were suspended and approximately 1×108 cells were treated with 2 μM MC or 2 μM DMC for 1 h under hypoxia. Nuclear DNA was isolated from the cells after drug treatment, as described above.
MCF-7 human breast cancer cells, obtained from American Type Culture Collection, were grown in Dulbecco’s modified Eagle’s medium (Mediatech) supplemented with 10% fetal bovine serum (Gemini) and penicillin (50 units/mL)-streptomycin (50 μg/mL) solution (Mediatech). Cells were seeded at 80 % confluence and incubated overnight prior to addition of drug. A total of approximately 2.7 × 107 cells were treated with 10 μM MC or 10 μM DMC for 24 h at 37°C in an atmosphere of 95% air/5% CO2. At the end of the incubation period, cells were harvested, washed 2x in ice-cold PBS, and spun down after each wash at × 300g. DNA was extracted from the cell pellet as described above. The isolated DNA was suspended in TE buffer.
Enzymatic digestion of DNA from drug-treated cells and isolation of the adduct fraction by Sep-Pak chromatography
The lyophilized DNA, typically 20–30 A260 units, obtained from 108 cells was digested to the nucleoside level by the following protocol: Nuclease P1 (1.0 unit/A260 unit of DNA) was added to the DNA in dilute aqueous acetic acid at pH 5.0 (2.5 A260 units/mL), followed by incubation for 4 h at 37 °C. The pH was adjusted to 8.2 by addition of 0.5 M Tris, and MgCl2 was added to a concentration of 1.0 mM. Addition of snake venom phosphodiesterase (2.25 units/A260 unit of DNA) and a 2-h incubation at 37 °C were followed by the addition of alkaline phosphatase (1.6 units/A260 unit of DNA) and incubation overnight at 37 °C. The digest was then heated at 90 °C for 1 h to hydrolyze guanine-N7 adducts. The solution was concentrated to 1 mL and applied to a Sep-Pak C-18 cartridge, previously washed consecutively with 10 mL acetonitrile, 10 mL H2O and 2 mL of 10 mM ammonium acetate. The unmodified nucleosides were eluted with 120 mL H2O, then 10 mL 5% methanol in H2O, followed by quantitative elution of the adducts with 60% methanol–water (20 mL)5. This last fraction was lyophilized, and submitted to analysis by mass spectrometry, described below.
Synthesis of adducts of MC and DMC for use as authentic standards for mass spectrometry
MC monoadduct 1a
The 14-mer self-complementary oligonucleotide 5′-ATATA(CG)2TATAT (60 A260 units; 6 μmol nucleotide units) was annealed in 3 mL 0.1 M potassium phosphate, pH 7.5, 90°C for 5 min followed by slow cooling to ice bath temperature. MC (24 μmol in 2.7 mL of the same buffer) and 0.3 mL of a deaerated solution of 40 mM Na2S2O4 in H2O (12 μmol) were added, and the mixture was stirred for 60 min in the ice bath, open to air, followed by applying it onto a 2.5 × 56 cm Sephadex G-25 column, and eluting it with 0.02 M NH4HCO3. The void volume fraction was lyophilized. The residue was dissolved in 15 mL 0.1 M Tris, pH 8.6 - 5 mM MgCl2 and digested by adding 159 units of SVD and 85 units of alkaline phosphatase, followed by overnight incubation at 45°C. The digestion mixture was lyophilized to dryness and redissolved in 1 mL H2O. The adduct was isolated in several batches by HPLC chromatography, using a Varian column (C18 300 Å, 4.6 × 250 mm). Elution conditions were as follows: Buffer A, 0.02 M ammonium acetate; buffer B, 70% buffer A-30% acetonitrile, υ/υ; linear gradient of 20–60% B in 60 min, at a flow-rate of 1 mL/min. The elution time of 1a was 16.8 min. The collected and pooled fractions (1.5 A260 units of 1a) were lyophilized.
MC interstrand cross-link 3a
The same amount of the 14-mer oligo was alkylated by a procedure similar to that above, except conducted anaerobically under Argon and with 5-fold total amount of Na2S2O4, added in five portions at 5-min intervals. The oligo product was digested and adduct 3a was isolated by the same protocol as above. The elution time of 3a was 19–20 min; the yield was 1.4 A260 units.
DMC monoadduct 2a can be conveniently prepared by quantitative hydrolysis of 1a at 90 °C for 4–6 hours in an aqueous solution.
DMC monoadduct 2b, isomeric with 2a, was obtained from 1b, the 1″-β isomer of the 1″-α MC monoadduct 1a in Figure 2, in an analogous procedure; 1b was synthesized previously by reductive alkylation of dG by MC (29). Elution times of 2a and 2b in the above HPLC system: 14 and 28 min, respectively.
Monoadduct 5, derived from 2,7-DAM (3) was synthesized by reductive alkylation of duplex oligo 5′-ATCTATGGTACC. 5′-GTACCTAT by 2,7-DAM, followed by heating of the crude product to release adduct 5 by depurination (30). HPLC elution time: 23 min.
Monoadduct 6, the major DNA alkylation product of 2,7-DAM in vivo, could not be prepared by the above biomimetic route in sufficient yields. Instead, it was obtained from organic synthesis, according to our recent procedure (31).
Two additional adducts 3b and 4, observed previously in DNA treated with the drugs in vivo, were not prepared as standards in the present work; nevertheless, both were characterized earlier by LC-ESI MS and other methods (3, 4; Tomasz, unpublished results).
Molar concentration of adduct stock solutions
Individual adducts were obtained by elution from HPLC, then were lyophilized and dissolved in H2O. Their absorbances were determined at 260 and 254 nm. Molar concentrations were calculated using the following molar extinction coefficients [ε(H2O, 254 nm)]: 1a, 2a, 2b, 24,000; 3a, 30,000 (32); 5, 13,8006a; 6, 24,70006b.
Mass spectrometry
LC-ESI MS/MS
An Applied Biosystems ABI-5000 tandem mass spectrometer (Concord, ONT, Canada) equipped with a Turbo V ion source was interfaced directly with an Agilent Model 1100 liquid chromatograph (Santa Clara, CA) equipped with a degasser, autosampler, column oven and diode array detector. The column used in this work was Phenomenex Luna 3 μ, C8 (2), 100 Å, 150 × 2mm. The LC column was maintained at 35 °C. Mobile phase conditions: mobile phase A, 95/5 water/acetonitrile, containing 0.1% formic acid, 5 mM ammonium formate; mobile phase B, 5/95 water/acetonitrile, containing the same modifiers. Elution program: 100 % A for 15 min, 0–30% B in 30 min, 30 % B for 15 min. The flow rate was 0.2 mL/min. The diode array detector recorded UV spectra from 190 to 400 nm.
The eluent from the LC column is introduced into the ABI Turbo V ion source where it is entrained in a nebulizing gas (GS1) flow of 50 mL/min and electrosprayed at a potential of 5000 V, resulting in nebulization of the eluent. Dry nitrogen gas (GS2) heated to 650°C impinges on the spray at 40 mL/min, speeding evaporation of the droplets before they reach the entrance to the mass spectrometer. A modest flow (15 mL/min) of dry nitrogen gas out of the entrance of the mass spectrometer, referred to as curtain gas (CUR), minimizes chemical background noise in the instrument. The MS inlet heater plate was ON and at its fixed value of 100 °C. Pressure of the nitrogen gas in the collision cell (CAD) was 6.0 and the CAD entrance potential was 10 V.
The parent ions and their fragment pairs were monitored sequentially throughout the course of the run, dwelling on each parent-fragment pair for 50 msec before moving onto the next. The cycle was repeated at approximately every 1.5 sec. For each adduct, the parent ion and its two fragment ions are listed in Table 1, along with the corresponding experimental variables.
Table 1.
Various parameters of the LC/MS runs
| Adduct(s) | Q1 m/z (parent ion) | Q3 m/z (fragment ion pair) | DPa | CEb | CxPc |
|---|---|---|---|---|---|
| 2a, 2b | 528 | 411, 393 | 90 | 15 | 40 |
| 1a | 571 | 552, 376 | 90 | 15, 41 | 50, 20 |
| 3a, 3b3 | 776 | 660, 544 | 100 | 19 | 38 |
| 5 | 395.5 | 214, 227 | 71 | 2.7, 40 | 24, 32 |
| 6 | 511 | 395, 244 | 90 | 17, 33 | 38, 36 |
Declustering potential, V
Collision energy, V
Collision cell exit potential, V
Quantitative analysis
Separate calibration curves of each adduct in the 6-adduct standard mixture were constructed by 2- to 100-fold range dilutions of the standard adduct mixture. The undiluted mixture contained 5 nmol/1.0 mL of each adduct. Two fragment ions were monitored for a particular adduct, resulting in two calibration curves, one for each MRM fragment pairs. The adduct samples were analyzed in duplicate injections and their quantities were calculated separately from each of the two different fragment calibration curves. The four values thus obtained for each adduct were averaged and standard errors were calculated. All calibration curves were linear, with correlation coefficients in the range of 0.98–0.99 (Figures S3A and S3B). DNA adduct frequencies7 of the samples were calculated from the formula:
where “μmol DNA digested” was calculated from the A260 values of the DNA samples before digestion, using 6,500/mol−1cm−1 as the molar extinction coefficient A260 of mammalian DNA (4).
Results
Synthesis of six mitomycin-deoxyguanosine adducts for use as standards for MS/MS spectrometry
This is the first comprehensive account of the practical preparation of five major adducts of MC and one of DMC. Adducts 1a, 3a and 5 (Figure 2) were obtained by biomimetic syntheses, i.e. reductive alkylation of duplex oligonucleotides which contained target guanines in a sequence context optimal for their alkylation, as determined in previous work (2,37). The 1″-isomer pair of decarbamoyl monoadducts 2a and 2b (Figure 3) were obtained from 1a and 1b, simply by aqueous hydrolysis of the 10″-carbamate groups. The biomimetic route was impractical for the synthesis of the 2,7-DAM adduct 6, therefore 6 was prepared by organic synthesis (31). Each adduct was synthesized in yields of approximately 1 to 2 A254 units, corresponding to 30–40 nanomoles depending on their 254 nm extinction coefficients. Attempts to prepare two major adducts, the 1″-β isomer of the interstrand cross-link 3b and the intrastrand cross-link 4 were unsuccessful. Nevertheless, 3b was detected and quantitated in the samples by selecting for the same MS/MS fragmentations as those for 3a (Table 1), and using the calibration curves of 3a. Adduct 4 was detected only in the form of the parent ion.
MRM-LC-ESI MS/MS spectrometry
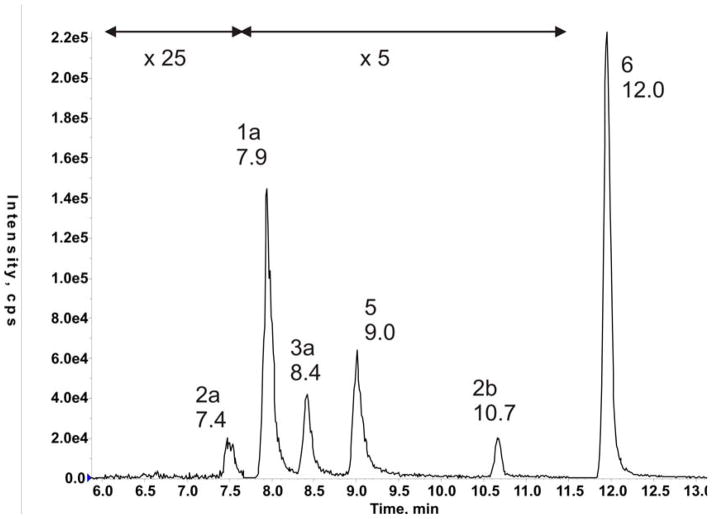
Each adduct was monitored by two fragments of the parent ion (Table 1). The full MS/MS spectrum of adduct 3a is shown in Figure 7 as a representative example. A plot of the signals of the various MS/MS pairs in the standard six-adduct mixture as function of the LC retention time is shown in Figure 8. The six adducts are well separated and the order of elution corresponds to that observed in HPLC systems used previously (3, 4). The intrastrand cross-link adduct 4, for which no standard was available, was observed as the parent ion (m/z 838), and only in one sample. (Supplementary Information, Figure S2).
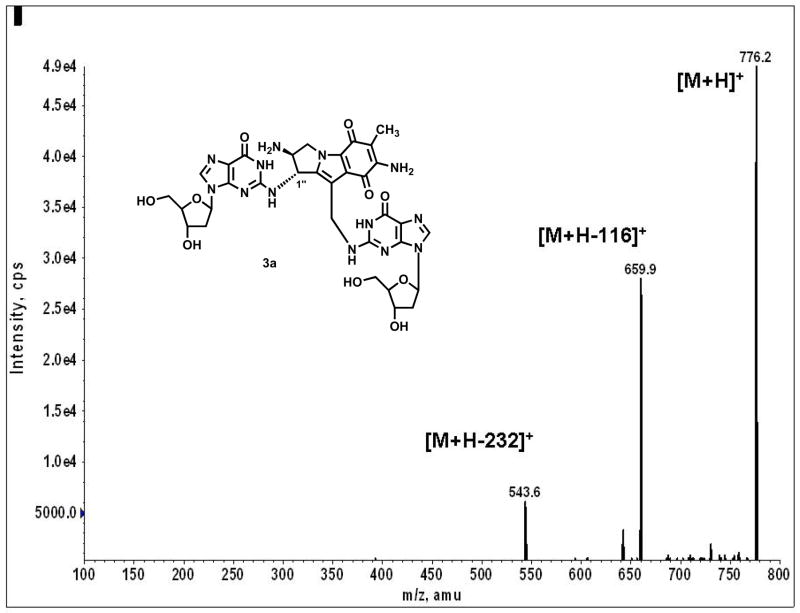
Figure 7.
MS/MS spectrum of the ICL adduct 3a. The two major fragments m/z 659.9 and m/z 543 represent loss of one and two deoxyribose units, respectively, from the parent ion, m/z 776.4.
Figure 8.
LC elution pattern of the standard six-adduct mixture (0.25 picomol of each adduct). Adducts were monitored by two MS/MS fragment signals (Table 1) and the sum of the two signal intensities are plotted against elution time for qualitative illustration of the separation of the adducts in the LC system. The numbers above the peaks specify the particular adduct and its retention time in minutes.
Treatment of EMT6 cells with MC, followed by 0, 16 and 24 h post-incubation for potential adduct repair
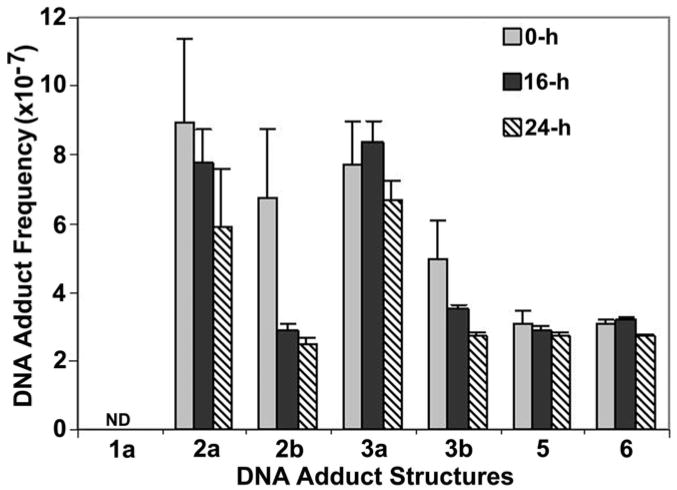
The treatment (2 μM MC, 1 h, hypoxia) reduces cell survival to approximately 10% of the control level, as assayed previously by a clonogenic assay (3, 4). There was no significant increase in cell number in the the drug-treated cultures during the 24-h post-incubation period, suggesting that cell proliferation was minimal during this period, as expected for these confluent cultures (data not shown). The individual DNA adduct frequencies in the 0-, 16- and 24-h post-incubation samples are shown side-by-side for comparison in Figure 4. After 24 hours of post-incubation significantly decreased frequencies were observed for three adducts 2a (34 %), 2b (63 %), and 3b (45 %). In contrast, the frequencies of adducts 3a, 5, and 6 decreased only by 9 %, 13 %, and 13 %, respectively. Although there is a relatively large experimental error associated with some of the adduct frequencies7 in the first group (up to 28 %), it is clear that the second group of adducts disappears more slowly, if at all. This leads to the qualitative conclusion that the kinetics of removal of the major cross-link adduct of MC, 3a, and the two monoadducts 5 and 6 of the MC metabolite 2,7-DAM (2; Figure 1) formed in situ is slower than that of the other adducts.
Figure 4.
Change of frequencies of DNA adducts (adduct/DNA-nucleotide) in MC-treated EMT6 cells as a result of post-treatment incubation in drug-free medium. EMT6 cells were treated with 2 μM for 1 h under hypoxia. Cells were lysed and their DNA was isolated immediately after treatment (“0-h” samples), or were incubated in drug-free medium for (i) 16 hours (“16-h” samples) or (ii) for 24 hours (“24-h” samples), before isolation of their DNA. Error limits are shown in % relative standard deviation units (see Experimental Section).
Figure 1.
Chemical structures of MC, DMC and 2,7-DAM
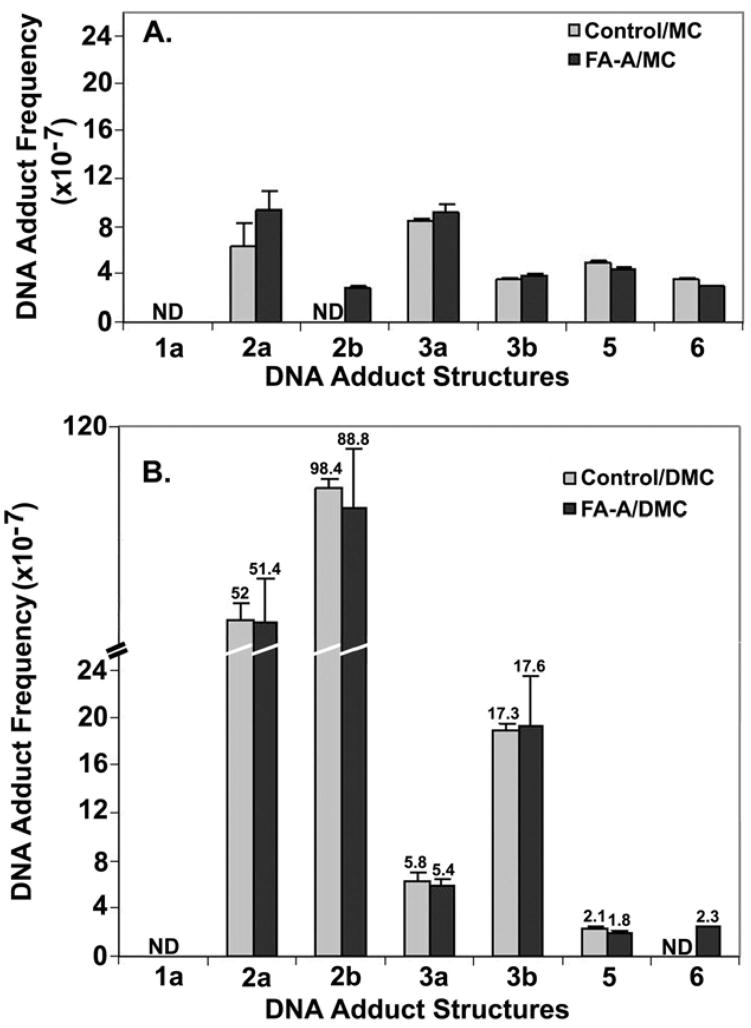
DNA adducts generated in normal human fibroblasts and FA-A fibroblasts treated with MC or DMC
The cultures were treated with 2 μM drug (MC or DMC) for 1 h under hypoxia and nuclear DNA was isolated immediately after cell treatment. The DNA adduct frequencies7 for normal fibroblasts and FA-A cells treated with MC are shown in Figure 5A, those from the same pair of cells treated with DMC are shown in Figure 5B. Two major conclusions may be drawn from these results: (i) There is very little difference between the DNA adduct frequencies of normal and FA-A cells, for both MC and DMC treatments. (ii) DMC treatment generates certain adducts at frequencies 5–10-fold greater than those resulting from MC treatment, in good agreement with our previous results for EMT6 cells (3).
Figure 5.
Frequencies of DNA adducts formed in immortalized normal human fibroblasts (“Control”) and FA-A fibroblasts, treated with (A) 2 μM MC, for 1 h, under hypoxia, or (B) 2 μM DMC, for 1 h, under hypoxia. For clarity, in (B) the frequency values of the bars are indicated above the bar error limits. Error limits are shown in % relative standard deviation units.
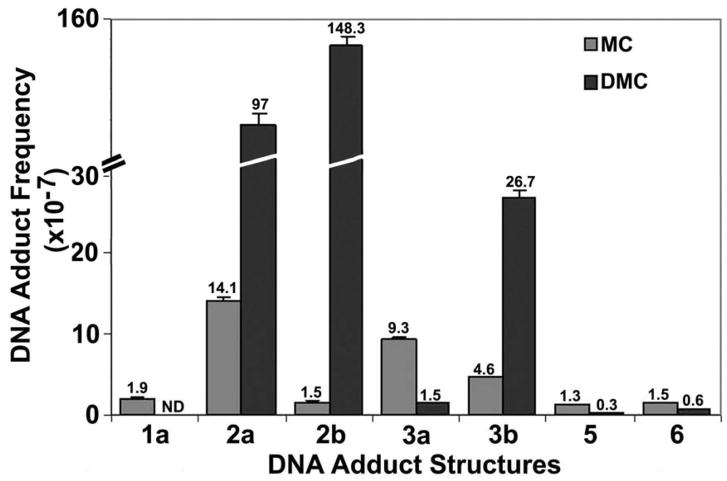
Comparison between the DNA adducts generated in MCF-7 human breast cancer cells by treatment with MC or DMC
The MCF-7 cells were treated with 10 μM MC or DMC for 24 h under aerobic conditions. This DMC treatment was shown previously to result in a 50% reduction in mitochondrial activity in either presence or absence of p53 protein while the MC treatment reduced the mitochondrial activity by only 10 % under similar conditions (33). The DNA adduct frequencies generated by these treatments are shown in Figure 6. Again, treatment with DMC generates 5–10-fold greater frequencies of adducts 2a, 2b and 3b than similar treatment with MC, a result strikingly consistent with those observed with the Fanconi cell/normal cell pairs shown in Figure 5 above, as well as with our earlier results obtained in EMT6 cells treated with the two drugs (3).
Figure 6.
Frequencies of DNA adducts formed in MCF-7 human breast cancer cells treated with 10 μM MC (gray bars) or 10 μM DMC (black bars) for 24 hours under aerobic conditions. The individual frequency values of the bars are indicated above the bar error limits. Error limits are shown in % relative standard deviation units.
This consistency across four different cell lines validates the present MRM/MS results. Their validity is also supported by the fact that the adduct distribution, determined previously in the EMT6 cells after treatment with 2 μM MC for 1 h under hypoxia in our laboratory, using the more cumbersome and error-prone [3H]-labeled methodology, indicated the same adduct pattern, (4).
Discussion
In spite of a long-standing interest and extensive investigations, important aspects of the chemical and biological mechanism of the cytotoxic activity of MC are still not completely understood. In particular, the mechanism(s) of DNA cross-link repair (24), mechanism of hypersensitivity to MC and other cross-linking agents in cells from Fanconi Anemia (FA) patients (40), structure-function relationships of the six major DNA adducts of MC with respect to mutagenesis, DNA repair and cytotoxicity (e.g. 30, 41), and other relevant processes are subject to continuing inquiry. Analysis of the DNA damage induced by MC and its derivatives on the chemical level can be a useful tool in these investigations. The subject of this report is development of an LC-ESI tandem mass spectrometric analysis of MC adducts formed in cells, and several initial applications of this method of analysis.
The LC-ESI MS/MS analysis is shown here to be equally sensitive to the previously reported radiolabel analysis using [3H]-labeled MC (4), estimating, that perhaps as a lower limit, 0.25 pmol of an adduct (2a) in the standard mixture (Figure 8) gave a detectable signal in the mass spectrometer. The overall validity of this method is reflected by the similarity of the results to those obtained using other analytical methods (3, 4), as discussed in the Results section above.
The first application addressed the possibility of differential kinetics of adduct repair in MC-treated EMT6 mouse mammary tumor cells. The experimental design consisted of post-treatment incubations in the absence of MC for 0, 16 or 24 hours, and analyzing DNA adducts in the three cultures. In order to correct for potential new DNA synthesis and cell division during the post-treatment incubations cells were counted immediately after treatment and in the 16-h and 24 h post-incubation cultures. As expected, no significant change in cell counts was observed in these confluent drug-treated cultures. The results of the analyses (Figure 4) suggest that 33% of 2a, 54% of 2b and 44% of 3b are removed after 24 hours of post-incubation, while the frequencies7 of the interstrand cross-link 3a and the 2,7-DAM adducts 5 and 6 change little during this period. Repair kinetics of the MC interstrand cross-links have been reported in the literature (13–16, 34, 35). In view of the diverse origin of the cells and techniques used in these investigations it is difficult to compare them with present results, except with those of ref. 15, in which EMT6 cells showed no cross-link repair up to 17 h post-incubation, after which a large loss of cross-links appeared by 24 hours, as assayed by alkaline elution. No MC cross-link removal at all was observed in wild-type and mutant Chinese Hamster cells up to 48 hours of post incubation (35). Our preliminary conclusion is that 3a is repaired slowly, if at all, in our system. The 2,7-DAM adducts 5 and 6 seem to be persistent as well; this is interesting, since the in situ metabolite of MC, 2,7-DAM, and its adducts are not appreciably cytotoxic (7, 28, 37). Perhaps the 2,7-DAM adducts originate from regions of DNA insensitive to replication inhibitors or perhaps these adducts do not hamper the processivity of polymerases (30).
Cross-link repair kinetics has been studied by Hanawalt and coworkers (36). In CHO cells psoralen cross-links, the major groove counterparts of the MC cross-link, were removed in the first 24 hours somewhat faster than the psoralen monoadducts (40% and 35% removal, respectively) from the bulk genome. However, the rate of transcription-coupled repair of this cross-link was much faster than that of the monoadducts. Our results reflect only global genome repair of the mitomycin damage; further investigations are needed to confirm and extend the present findings.
Analysis of adducts in Fanconi Anemia cells
FA cells have been postulated to have a repair deficiency that selectively affects the repair of DNA interstrand cross-links but not monoadducts (44). This assumption is based mostly on the fact that various FA cells are hypersensitive to MC but have normal sensitivity to DMC, which was once thought to be a strictly monofunctional analog of MC (38, 42). However, recent studies in our laboratory have clearly shown that under appropriate conditions DMC forms DNA interstrand cross-links with purified DNA in vitro and also in DMC-treated EMT6 cells (3). When EMT6 cells were treated with either MC or DMC at the same concentration, the two drugs produced cross-link adduct 3a in a 1.3:1 ratio, respectively, and in addition, DMC also formed a new stereoisomeric cross-link adduct 3b (Figure 3) which was approximately 10-times more abundant than 3a (Table 2). This new cross-link was undetectably low in the MC-treated cells. Furthermore, DMC formed approximately 20-fold more total adducts overall, than MC. These results explained why DMC was more cytotoxic than MC to EMT6 cells and to repair –proficient and repair –deficient Chinese hamster cell lines (3, 43). The present analysis of the adducts of MC and DMC formed in FA-A and normal human fibroblast cells (Figures 5A, 5B) confirms our earlier study of adducts in EMT6 cells: DMC treatment generates the same cross-link adduct 3a as seen with MC, and, in addition, forms stereoisomeric cross-link 3b (Figure 5A, 5B). Similar experiments with a third cell line, MCF-7, derived from human breast carcinoma cells, yielded the same results (Figure 6).
Table 2.
Ratio of stereoisomeric adducts in MC- and DMC-treated cells
| Cell Line | MC treatment | DMC treatmentb | ||
|---|---|---|---|---|
| 2a:2b | 3a:3b | 2a:2b | 3a:3b | |
| EMT6 | ∞a, c | ∞a, c | 0.25d | 0.1+0.01e |
| EMT6, 0-h | 1.34 | 1.57 | - | - |
| MCF-7 | 9.4 | 2.0 | 0.65 | 0.06 |
| Human Fibroblast | ∞c | 2.4 | 0.55 | 0.34 |
| FA-A | 3.4 | 2.4 | 0.58 | 0.28 |
ref. 4
ref. 3
No 2b and 3b detected
10 μM DMC
20 μM DMC
Mapping of the DMC-generated adducts in the earlier (3) as well as in the present investigation leads to the conclusion that DMC is a DNA cross-linking agent, rather than a strictly monofunctional analog of MC as was previously believed. Furthermore, DMC shows the opposite stereochemical preference of alkylation of the guanine-2-amino group from that of MC in those cell lines (Table 2).
In our past studies the 1″-β-linked adducts 2b and 3b (Figure 3) have not been detected in MC-treated cells or with in vitro alkylations by MC. They were now detected, indicating that the MRM-LC/MS/MS method is uniquely sensitive and suitable to identify known low-abundance components of complex mixtures3.
Regardless of its different stereochemical preference of alkylation, it is clear that DMC forms the same interstrand cross-link adduct 3a as MC, although at slightly lower (60–70%) frequency. We have recently found that Fanconi Anemia cells are more sensitive to DMC than previously assessed4, which is consistent with this finding.
The biological significance, if any, of 1″-α vs. 1″-β stereochemistry of mitomycin adducts remains unknown. However, our group reported that MC and DMC treatments can result in induction of the p53 pathway and apoptosis, while DMC has more efficacy to induce p53-independent cell death signaling in human cancer cell lines (33, 37). It was speculated (37) based on our earlier report (3), that the large proportion of the 1″-β stereoisomeric adducts generated by DMC may play a role in these phenomena. Another explanation, simply that DMC produces more cross-links and more monoadducts overall than MC, cannot be excluded, however. The present work confirms the striking difference between the DNA adduct patterns of MC and DMC in three cell lines, thus opening the way to investigate the basis of differential modes of cell death induced by the two drugs.
Supplementary Material
Figure S1. Relative intensity of the MS/MS signals (a), and diode array UV signals (b), in the LC run of 10 picomol of each adduct in the standard 6-adduct mixture in superimposed plots.
Figure S2. MS chromatogram of the MCF-7/MC adducts sample, illustrating detection of the intrastrand cross-link adduct 4 by parent ion signal only, and of the isomeric ICL adduct 3b by monitoring the same two MS/MS fragments as for the detection of ICL adduct 3a.
Figure S3. Representative external calibration curves of adducts in the 6-standard adduct mixture. A: One of the two standard calibration curves of adduct 1a, monitoring the m/z=552 MS/MS fragment ion. B: One of the two standard calibration curves of ICL adduct 3a, monitoring the m/z=660 MS/MS fragment ion.
Table 3.
Frequencies of 1″-β isomer adducts in MC- and DMC-treated cellsa
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants CA28681 (M.T.), S06 GM60654 (J.B.), CA28681 and CA129186 (S.R.), a National Center for Research Resources award RR03037 to support the research infrastructure at Hunter College, and a Minority Biomedical Research Initiative in Science Enhancement grant GM60665 (E.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or NCRR. This work was also supported by a PSC-CUNY Research Award 60035-3738 (to E.C.).
Footnotes
Abbreviations: MC, mitomycin C; DMC, 10-decarbamoyl mitomycin C; 2,7-DAM, 2,7-diaminomitosene; ICL, interstrand cross-link; LC-ESI MS/MS, liquid chromatography-electrospray tandem mass spectrometry; FA, Fanconi Anemia; FA-A, Fanconi Anemia complementation group A; SVD, snake venom diesterase; LC, liquid chromatography; MRM, multiple reaction monitoring;
This stereochemical preference is not seen in cell-free systems of alkylation using purified DNA, DMC and a reducing agent (3, 35).
Adduct 3b was not present in the standard adduct mixture. Its quantitation is based on the two calibration curves of the 3a fragments. This does not affect the relative amounts of 3b in MC- vs. DMC-treated cells and most likely it does not affect its absolute amount either.
S. Rockwell, Y. Liu, M. Gilmore-Herbert, and M. Tomasz, “Response of Aerobic and Hypoxic Fanconi Anemia Cells to Mitomycin C and Decarbamoyl Mitomycin C”, presented at the Radiation Research Society 52nd Annual Meeting, Denver, CO, October, 2005.
In control experiments HPLC of the Sep-Pak fractions showed that only a relatively small amount of unmodified dA (estimated as 10 % or less of the total dA) elutes with the adduct fraction. Recovery of the adducts from the Sep-Pak is likely to be quantitative as indicated in earlier work: A 100 % methanol wash was applied after the 60 % methanol elution of the adducts in a large scale control adduct preparation. HPLC of this 100% methanol wash showed no UV signals corresponding to any adduct (S. Ladwa, unpublished).
Calculated as the sum of E254 of 2,7-DAM and 7-ethylguanine.
Calculated as the sum of E254 of 2,7-DAM and 2′-deoxyguanosine.
DNA adduct frequency is defined here, as in our previous publications, as mol DNA adduct/mol DNA-nucleotide (or simply, adduct/DNA base).
This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Verweij J, den Hartigh J, Pinedo HM. Antitumor Antibiotics. J. B. Lippincott Company; Philadelphia, PA: 1990. [Google Scholar]
- 2.Tomasz M, Palom Y. The mitomycin bioreductive antitumor agents: cross-linking and alkylation of DNA as the molecular basis of their activity. Pharmacol Ther. 1997;76:73–87. doi: 10.1016/s0163-7258(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 3.Palom Y, Suresh Kumar G, Tang LQ, Paz MM, Musser SM, Rockwell S, Tomasz M. Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbamoyl mitomycin C and mitomycin C. Chem Res Toxicol. 2002;15:1398–1406. doi: 10.1021/tx020044g. [DOI] [PubMed] [Google Scholar]
- 4.Bizanek R, Chowdary D, Arai H, Kasai M, Hughes CS, Sartorelli AC, Rockwell S, Tomasz M. Adducts of mitomycin C and DNA in EMT6 mouse mammary tumor cells: effects of hypoxia and dicumarol on adduct patterns. Cancer Res. 1993;53:5127–5134. [PubMed] [Google Scholar]
- 5.Tomasz M, Chowdary D, Lipman R, Shimotakahara S, Veiro D, Walker V, Verdine GL. Reaction of DNA with chemically or enzymatically activated mitomycin C: isolation and structure of the major covalent adduct. Proc Natl Acad Sci USA. 1986;83:6702–6706. doi: 10.1073/pnas.83.18.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomasz M, Lipman R, Chowdary D, Pawlak J, Verdine GL, Nakanishi K. Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA. Science. 1987;235:1204–1208. doi: 10.1126/science.3103215. [DOI] [PubMed] [Google Scholar]
- 7.Palom Y, Belcourt MF, Musser SM, Sartorelli AC, Rockwell S, Tomasz M. Structure of adduct X, the last unknown of the six major DNA adducts of mitomycin C formed in EMT6 mouse mammary tumor cells. Chem Res Toxicol. 2000;13:479–488. doi: 10.1021/tx000024j. [DOI] [PubMed] [Google Scholar]
- 8.Randerath K, Reddy MV, Gupta RC. 32P-labeling test for DNA damage. Proc Natl Acad Sci U S A. 1981;78:6126–6129. doi: 10.1073/pnas.78.10.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy MV, Randerath K. 32P-analysis of DNA adducts in somatic and reproductive tissues of rats treated with the anticancer antibiotic, mitomycin C. Mutat Res. 1987;179:75–88. doi: 10.1016/0027-5107(87)90043-1. [DOI] [PubMed] [Google Scholar]
- 10.Pan SS, Yu F, Hipsher C. Enzymatic and pH modulation of mitomycin C-induced DNA damage in mitomycin C-resistant HCT 116 human colon cancer cells. Mol Pharmacol. 1993;43:870–877. [PubMed] [Google Scholar]
- 11.Warren AJ, Maccubbin AE, Hamilton JW. Detection of mitomycin C-DNA adducts in vivo by 32P-postlabeling: time course for formation and removal of adducts and biochemical modulation. Cancer Res. 1998;58:453–461. [PubMed] [Google Scholar]
- 12.Warren AJ, Mustra DJ, Hamilton JW. Detection of mitomycin C-DNA adducts in human breast cancer cells grown in culture, as xenografted tumors in nude mice, and in biopsies of human breast cancer patient tumors as determined by 32P-postlabeling. Clin Cancer Res. 2001;7:1033–1042. [PubMed] [Google Scholar]
- 13.Matsumoto A, Vos JMH, Hanawalt PC. Repair analysis of mitomycin C-induced DNA crosslinking in ribosomal RNA genes in lymphoblastoid cells from Fanconi’s anemia patients. Mutat Res. 1989;217:185–192. doi: 10.1016/0921-8777(89)90070-0. [DOI] [PubMed] [Google Scholar]
- 14.Dorr RT, Bowden GT, Alberts DS, Liddil JD. Interactions of mitomycin C with mammalian DNA detected by alkaline elution. Cancer Res. 1985;45:3510–3516. [PubMed] [Google Scholar]
- 15.Fracasso PM, Sartorelli AC. Cytotoxicity and DNA lesions produced by mitomycin C and porfiromycin in hypoxic and aerobic EMT6 and Chinese hamster ovary cells. Cancer Res. 1986;46:3939–3944. [PubMed] [Google Scholar]
- 16.Giese RW, Vouros P. Methods development toward the measurement of polyaromatic hydrocarbon-DNA adducts by mass spectrometry. Res Rep Health Eff Inst. 1993:1–25. [PubMed] [Google Scholar]
- 17.Doerge DR, Churchwell MI, Marques MM, Beland FA. Quantitative analysis of 4-aminobiphenyl-C8-deoxyguanosyl DNA adducts produced in vitro and in vivo using HPLC-ES-MS. Carcinogenesis. 1999;20:1055–1061. doi: 10.1093/carcin/20.6.1055. [DOI] [PubMed] [Google Scholar]
- 18.Andrews CL, Vouros P, Harsch A. Analysis of DNA adducts using high-performance separation techniques coupled to electrospray ionization mass spectrometry. J Chromatogr A. 1999;856:515–526. doi: 10.1016/s0021-9673(99)00779-7. [DOI] [PubMed] [Google Scholar]
- 19.Singh G, Gutierrez A, Xu K, Blair IA. Liquid chromatography/electron capture atmospheric pressure chemical ionization/mass spectrometry: analysis of pentafluorobenzyl derivatives of biomolecules and drugs in the attomole range. Anal Chem. 2000;72:3007–3013. doi: 10.1021/ac000374a. [DOI] [PubMed] [Google Scholar]
- 20.Paehler A, Richoz J, Soglia J, Vouros P, Turesky RJ. Analysis and quantification of DNA adducts of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in liver of rats by liquid chromatography/electrospray tandem mass spectrometry. Chem Res Toxicol. 2002;15:551–561. doi: 10.1021/tx010178e. [DOI] [PubMed] [Google Scholar]
- 21.Takeiri A, Mishima M, Tanaka K, Shioda A, Harada A, Watanabe K, Masumura KI, Nohmi T. A newly established GDL1 cell line from gpt delta mice well reflects the in vivo mutation spectra induced by mitomycin C. Mutat Res. 2006;609:102–115. doi: 10.1016/j.mrgentox.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Budach W, Paulsen F, Welz S, Classen J, Scheithauer H, Marini P, Belka C, Bamberg M. Mitomycin C in combination with radiotherapy as a potent inhibitor of tumour cell repopulation in a human squamous cell carcinoma. Br J Cancer. 2002;86:470–476. doi: 10.1038/sj.bjc.6600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montie JE. Intravesical therapy for bladder cancer: empiricism at the helm. J Natl Cancer Inst. 2001;93:572–573. doi: 10.1093/jnci/93.8.572. [DOI] [PubMed] [Google Scholar]
- 24.Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arai H, Kasi M. Synthesis of [C6-CH3-14C] and C6-CH3-3H] mitomycin C. J Labelled Compd Radiopharm. 1991;28:903–908. [Google Scholar]
- 26.Cao H, Hearst JE, Corash L, Wang Y. LC-MS/MS for the detection of DNA interstrand cross-links formed by 8-methoxypsoralen and UVA irradiation in human cells. Anal Chem. 2008;80:2932–2938. doi: 10.1021/ac7023969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinoshita S, Uzu K, Nakano K, Takahashi T. Mitomycin derivatives. 2 Derivatives of decarbamoylmitosane and decarbamoylmitosene. J Med Chem. 1971;14:109–112. doi: 10.1021/jm00284a006. [DOI] [PubMed] [Google Scholar]
- 28.Palom Y, Belcourt MF, Kumar GS, Arai H, Kasai M, Sartorelli AC, Rockwell S, Tomasz M. Formation of a major DNA adduct of the mitomycin metabolite 2,7-diaminomitosene in EMT6 mouse mammary tumor cells treated with mitomycin C. Oncol Res. 1998;10:509–521. [PubMed] [Google Scholar]
- 29.Tomasz M, Lipman R, Verdine GL, Nakanishi K. Reassignment of the guanine-binding mode of reduced mitomycin C. Biochemistry. 1986;25:4337–4344. doi: 10.1021/bi00363a024. [DOI] [PubMed] [Google Scholar]
- 30.Utzat CD, Clement CC, Ramos LA, Das A, Tomasz M, Basu AK. DNA adduct of the mitomycin C metabolite 2,7-diaminomitosene is a nontoxic and nonmutagenic DNA lesion in vitro and in vivo. Chem Res Toxicol. 2005;18:213–223. doi: 10.1021/tx049813h. [DOI] [PubMed] [Google Scholar]
- 31.Champeil E, Paz MM, Ladwa S, Clement CC, Zatorski A, Tomasz M. Synthesis of an oligodeoxyribonucleotide adduct of mitomycin C by the postoligomerization method, via a triamino mitosene (2008) . J Am Chem Soc. 2008;130:9556–9565. doi: 10.1021/ja802118p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suresh Kumar G, Lipman R, Cummings J, Tomasz M. Mitomycin C-DNA adducts generated by DT-diaphorase. Revised mechanism of the reductive activation of mitomycin C. Biochemistry. 1997;36:14128–14136. doi: 10.1021/bi971394i. [DOI] [PubMed] [Google Scholar]
- 33.Boamah EK, White DE, Talbott KE, Arva NC, Berman D, Tomasz M, Bargonetti J. Mitomycin-DNA adducts induce p53-dependent and p53-independent cell death pathways. ACS Chemical Biology. 2007;2:399–407. doi: 10.1021/cb700060t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyn RE, Jenkins SF, Thompson LH. Defective removal of DNA cross-links in a repair-deficient mutant of Chinese hamster cells. Cancer Res. 1982;42:3106–3110. [PubMed] [Google Scholar]
- 35.Larminat F, Cambois G, Zdzienicka MZ, Defais M. Lack of correlation between repair of DNA interstrand cross-links and hypersensitivity of hamster cells towards mitomycin C and cisplatin. FEBS Lett. 1998;437:97–100. doi: 10.1016/s0014-5793(98)01209-5. [DOI] [PubMed] [Google Scholar]
- 36.Islas AL, Vos JM, Hanawalt PC. Differential introduction and repair of psoralen photoadducts to DNA in specific human genes. Cancer Res. 1991;51:2867–2873. [PubMed] [Google Scholar]
- 37.Abbas T, Olivier M, Lopez J, Houser S, Xiao G, Kumar GS, Tomasz M, Bargonetti J. Differential activation of p53 by the various adducts of mitomycin C. J Biol Chem. 2002;277:40513–40519. doi: 10.1074/jbc.M205495200. [DOI] [PubMed] [Google Scholar]
- 38.Tomasz M, Lipman R, Mcguinness BF, Nakanishi K. Isolation and Characterization of a Major Adduct between Mitomycin-C and DNA. J Am Chem Soc. 1988;110:5892–5896. [Google Scholar]
- 39.Subramaniam G, Paz MM, Suresh Kumar G, Das A, Palom Y, Clement CC, Patel DJ, Tomasz M. Solution structure of a guanine-N7-linked complex of the mitomycin C metabolite 2,7-diaminomitosene and DNA. Basis of sequence selectivity. Biochemistry. 2001;40:10473–10484. doi: 10.1021/bi010965a. [DOI] [PubMed] [Google Scholar]
- 40.Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, Ishiai M, Yamamoto K, Takata M, Arakawa H, Buerstedde JM, Yamazoe M, Kawamoto T, Araki K, Takahashi JA, Hashimoto N, Takeda S, Sonoda E. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- 41.Maccubbin AE, Mudipalli A, Nadadur SS, Ersing N, Gurtoo HL. Mutations induced in a shuttle vector plasmid exposed to monofunctionally activated mitomycin C. Environ Mol Mutagen. 1997;29:143–151. [PubMed] [Google Scholar]
- 42.Carrano AV, Thompson LH, Stretka DG, Minkler JL, Mazrimas JA, Fong S. DNA crosslinking, sister chromatid exchange and specific locus mutations. Mutat Res. 1979;63:175–188. doi: 10.1016/0027-5107(79)90114-3. [DOI] [PubMed] [Google Scholar]
- 43.Kim SY, Rockwell S. Cytotoxic potential of monoalkylation products between mitomycins and DNA: studies of decarbamoyl mitomycin C in wild-type and repair-deficient cell lines. Oncol Res. 1995;7:39–47. [PubMed] [Google Scholar]
- 44.Fujiwara Y, Tatsumi M. Cross-link repair in human cells and its possible defect in Fanconi’s anemia cells. J Mol Biol. 1977;113:635–649. doi: 10.1016/0022-2836(77)90227-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Relative intensity of the MS/MS signals (a), and diode array UV signals (b), in the LC run of 10 picomol of each adduct in the standard 6-adduct mixture in superimposed plots.
Figure S2. MS chromatogram of the MCF-7/MC adducts sample, illustrating detection of the intrastrand cross-link adduct 4 by parent ion signal only, and of the isomeric ICL adduct 3b by monitoring the same two MS/MS fragments as for the detection of ICL adduct 3a.
Figure S3. Representative external calibration curves of adducts in the 6-standard adduct mixture. A: One of the two standard calibration curves of adduct 1a, monitoring the m/z=552 MS/MS fragment ion. B: One of the two standard calibration curves of ICL adduct 3a, monitoring the m/z=660 MS/MS fragment ion.