Abstract
The selective RNA-binding protein QKI play a key role in advancing oligodendrocyte-dependent myelination, which is essential for the function and development of the CNS. The emerging evidence that QKI abnormalities are associated with schizophrenia and may underlie myelin impairment in this devastating disease has greatly increased interest in understanding the function of QKI. Despite the discovery of the biochemical basis for QKI-RNA interaction, a comprehensive model is currently missing regarding how QKI regulates its mRNA ligands to promote normal myelinogenesis and how deficiency of the QKI pathway is involved in the pathogenesis of human diseases that affect CNS myelin. In this review, we will focus on the role of QKI in regulating distinct mRNA targets at critical developmental steps to promote oligodendrocyte differentiation and myelin formation. In addition, we will discuss molecular mechanisms that control QKI expression and activity during normal myelinogenesis as well as the pathological impact of QKI deficiency in dysmyelination mutant animals and in human myelin disorders.
Keywords: CNS myelination, myelin disorders, oligodendrocytes, post-transcriptional regulation, QKI, RNA-binding protein, schizophrenia
The selective RNA-binding protein quaking (QKI), encoded by the qkI gene, governs the cellular behavior of the downstream mRNA ligands at various post-transcriptional steps (for recent reviews, see [1–3]). QKI is broadly expressed in multiple tissues from early embryonic development [4]. Several QKI protein iso-forms that share the same RNA-binding domain and activity are derived from alternative splicing at the C-terminal exons. However, these isoforms exhibit distinct patterns of subcellular localization and developmentally programmed expression [5,6], hence exerting sophisticated, sometimes even opposing functional influence, on target mRNAs and cell function [7,8]. More than a thousand mRNA species contain the QKI recognition element (QRE) that can directly bind QKI in vitro [9,10]. Many of these bioinformatically predicted QKI target mRNAs encode key components in cell-fate specification, growth/differentiation, migration and cell–cell communication [9]. Indeed, QKI has been shown to impact cell cycle progression and differentiation, and play essential roles in the establishment of various cell lineages during development [11–13]. Furthermore, QKI is the founding member of a protein family named signal transduction activator of RNA, owing to its amino acid domains characteristic of interaction with both RNA and signal-transduction molecules [14]. The phosphorylation-dependent regulation of QKI’s RNA-binding activity [15] supports the idea that QKI may act as a pivotal factor connecting posttranscriptional regulation with developmental signals [14].
Much attention has been delivered to the role of QKI in myelin development in the CNS, because a genetic deletion at the mouse qkI locus causes severe CNS dysmyelination in the well-known quakingviable (qkv)-mutant mice [13]. In addition, more recent studies demon strate that QKI is necessary and sufficient for promoting rodent oligodendrocyte (OL) differentiation [3,16]. Although the function of the QKI protein in human OLs has not been characterized, the highly conserved genes driving CNS myelination and the identical amino acid sequences in rodent and human QKI proteins suggest that QKI most likely play an important role in regulating human myelination as well. It has been known for decades that OLs are solely responsible for CNS myelination. Neuronal axons are ensheathed by specialized OL membrane lamellae composed of highly abundant myelin structural proteins and lipids, which maintain saltatory conductance and allow for rapid information flow between distant brain areas. Thus, the recent discovery of OL and myelin impairment in schizophrenia (SCZ) offers a plausible explanation for the deficits in long-range connectivity, the characteristic clinical phenotype of this mental morbidity (for recent reviews see [17–20]). In particular, the emerging evidence that reduced QKI expression and genetic alterations in the human QKI gene are associated with SCZ patient brains [21–24] raises an intriguing possibility that QKI deficiency may be an underlying factor for the myelin etiology in SCZ. However, no comprehensive model has been proposed to describe how QKI regulates distinct mRNA ligands during OL development to promote myelinogenesis, and how deficits in the QKI pathway lead to myelin impairment. Moreover, how QKI expression and activity are regulated during normal myelin development and how misregulation of QKI may occur in human diseases that affect CNS myelin, represented by SCZ and multiple sclerosis (MS), remains poorly understood. In this review, we will focus on the function of QKI isoforms in regulating OL-specific targets at key steps of myelin development, the mechanisms that regulate QKI expression and activity, and the potential pathologic impact of QKI in myelin-related human diseases. Readers are referred to other recent publications for the full capacity of information regarding the general RNA-binding properties of QKI, the bioinformatically identified QKI mRNA ligands and the function of QKI orthologs in nonmyelin tissues and invertebrates [1,10,25–27].
Essential roles of QKI in advancing OL development & CNS myelination
To date, most of the studies regarding QKI function have been conducted in mouse models. The first clue that QKI may be important for CNS myelination is suggested by the severe dysmyelination phenotype found in the mutant mice homozygous for a spontaneous recessive allele named quaking [28]. The mutant has a normal life span, and hence is referred to as ‘qkv’ later to contrast the spontaneous dysmyelination mutations that affect longevity [29], as well as the N-ethyl-N-nitrosourea (ENU)-induced mutations in the qkI locus that result in recessive embryonic lethality [30]. Homozygous qkv mice (qkv/qkv) form only 5–10% of the normal amount of myelin in the CNS, and the limited amount of myelin usually fails to compact [13,29]. Consequently, qkv/qkv mice suffer from vigorous tremors beginning at around postnatal day 10, when myelin function becomes critical. The severity of the phenotype progressively increases with age and is often accompanied by clonic–tonic seizures. Many key myelin structural proteins are markedly reduced. However, no mutations were found in any myelin structural genes, suggesting that the qkv mutation must affect a regulatory gene essential for myelination. The qkv lesion, identified in 1996 by the Artz group [4], encompasses a genomic deletion greater than 1 Mbp on mouse chromosome 17. qkv deletes the protein coding sequence of parkin and parkin coregulated (pacrg) and extends to approximately 900 bp upstream to the transcription start site of qkI, which is predicted to contain an OL-specific enhancer for qkI transcription [31,32]. As a result, expression of the qkI gene is specifically reduced in qkv/qkv OLs [5,33].
Unlike in humans, where loss-of-function mutations in parkin lead to autosomal juvenile Parkinson’s disease, the deletion of parkin and pacrg in qkv/qkv mice does not result in Parkinson-like, dopaminergic neuropathology [32]. Moreover, ENU-induced missense mutations in qkI fail to complement the dysmyelination in compound qkv heterozygous mice, indicating that neither parkin nor pacrg is responsible for the qkv dysmyelination phenotype [30]. These early studies suggest that deficiency in qkI gene expression in OLs is the cause for the dysmyelination phenotype. Consistent with this view, a new ENU-induced mutation (qke5), which maps to the qkI locus yet is outside the qkI coding region, causes diminished QKI expression in OLs and severe dysmyelination similar to that in the qkv/qkv mutant [34].
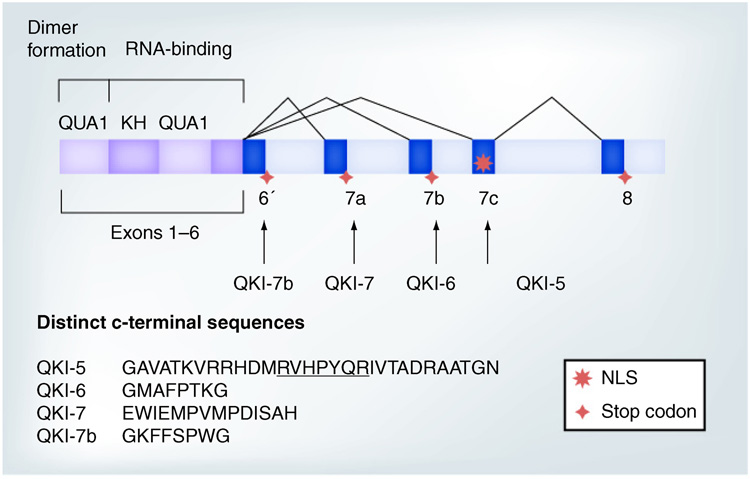
The 3’ coding exons in human and mouse QKI primary transcripts are subjected to extensive alternative splicing, which generates four mRNA isoforms, each giving rise to a QKI protein isoform with distinct amino acids in the very C-terminus (Figure 1) [4,31]. QKI protein isoforms are named QKI-5, -6, -7 and -7b [31], based on the length (kb) of the corresponding mRNAs. Little is known regarding the expression and function of QKI-7b, which was discovered more recently. Interestingly, QKI isoforms are differentially affected in the qkv/qkv mutant. QKI-6 and -7 are more severely reduced than QKI-5 [5,33,35], which cannot be a direct consequence of the transcriptional misregulation of qkI due to the qkv deletion. Therefore, QKI expression must also be affected by undefined post-transcriptional and/or post-translational mechanisms in the qkv/qkv mutant.
Figure 1. QKI mRNA isoforms, including QKI-5, QKI-6, QKI-7 and QKI-7b, are derived from alternative splicing of the 3’exons.
This highlights the extended RNA-binding domain, composed of the QUA1, KH and QUA2 regions that mediate dimerization and RNA-binding, respectively. The alternatively spliced exons and unique C-terminal amino acids in human QKI proteins are illustrated. Note, a NLS is encoded by the unique exon 7c in QKI-5, which is underlined in the QKI-5 C-terminal amino acids. The amino acids in QKI-5, -6 and -7 are 100% identical in mouse and human. The mouse QKI-7b sequence is GKYDSCTM, referred to as QKI-G previously [8].
NLS: Nuclear localization signal.
The direct evidence demonstrating that QKI is indeed required for CNS myelination is provided by a recent report, in which OL-specific expression of QKI-6, the most abundant QKI isoform in the brain, can rescue the dysmyelination and tremor phenotype in the qkv/qkv mice [35]. At the molecular level, deficits in major myelin structural proteins are completely restored at the peak of myelination. However, in earlier postnatal days, the QKI-6 transgene only selectively rescues the expression of its high-affinity mRNA ligands, represented by myelin basic protein (MBP) mRNA. This suggests that QKI-6 alone may not be sufficient for early myelinogenesis. Other QKI isoforms must also be necessary for advancing OL/myelin development.
Consistent with the idea that QKI is also required for promoting early OL development before myelin formation, elimination of QKI by RNAi in OL progenitor cells (OLPs) abrogates morphologic differentiation and delays OL maturation [16]. The blockade of differentiation can only be partially rescued by individual QKI isoforms, suggesting that more than one QKI isoform is needed to achieve full activity for OL differentiation. Reciprocally, overexpression of QKI isoforms in OLPs enhances OL differentiation [3,16]. These observations indicate that QKI is necessary and sufficient for advancing OLP differentiation, in addition to its functional necessity in actual myelin synthesis.
QKI regulates distinct mRNA targets at different stages of OL development & myelination
In various examples, QKI has been shown to govern the splicing, stability, translation and subcellular localization of its bound RNA ligands in rodent OLs and model cell lines [7,15,26,36– 38]. The RNA-binding activity of QKI depends on the hnRNP K homologous domain and the immediately adjacent QUA2 domain shared in all QKI isoforms (Figure 1) [1,39]. The N-terminal QUA1 domain mediates dimerization between QKI isoforms, which is also important for binding RNA [40,41]. Purified recombinant QKI directly interacts with the QRE, which contains a core sequence of ACUAAY sufficient for binding QKI [9,10,39], often accompanied by UAAY located within 1–20 bp to form a bipartite motif [9]. Bioinformatic analysis identified 1433 mouse mRNA species that harbor one or more QREs, among which a small group of mRNAs has been confirmed to associate with QKI in the developing brain [9]. In addition, almost all functionally validated QKI target mRNAs carry QREs in their 3’-untranslated region (3’UTR). Interestingly, many of the QRE-containing QKI ligand mRNAs are conserved between mouse and human. However, no direct evidence has been reported to demonstrate the influence of QKI to any putative human mRNA targets, except the association of QKI deficiency with altered expression of some QRE-containing OL mRNAs in the SCZ brains [21]. Thus, the functional influence of QKI on rodent mRNA targets discussed below may or may not always apply to the corresponding human genes.
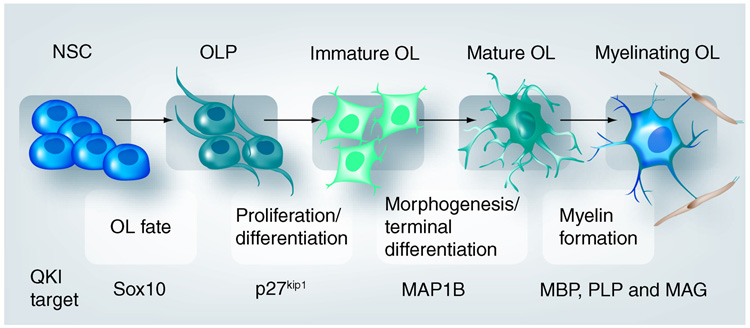
Unfortunately, the bioinformatically defined QRE does not predict OL-specific functional targets of QKI that play critical roles at different stages of OL development. As illustrated in Figure 2, OL development can be divided into several key steps based on the drastic morphological changes before the ultimate formation of myelin on axons. Tightly regulated expression of distinct genes, in response to various extracellular and intrinsic developmental signals, marks cells at various stages in the OL lineage and governs the progression of myelinogenesis [42,43]. A prevailing issue in understanding how QKI advances OL and myelin development is to delineate the function of each QKI isoform in regulating d evelopmental stage-specific mRNA ligands.
Figure 2. Hallmark steps in oligodendroglia lineage development with drastic morphological changes before ultimate myelin formation.
Representative QKI mRNA targets that are critical for OL and myelin development are listed below the relevant steps. Among these QKI targets, the functional influence of QKI is predicted for human Sox10, PLP, MBP and MAG based on postmortem studies of schizophrenia brain. All the indicated QKI targets, except Sox10, are functionally validated in mouse models. The full name of the QKI target mRNAs and their function are described in detail in the text.
MAG: Myelin associated glycoprotein; MBP: Myelin basic protein; NSC: Neural stem cells; OL: Oligodendrocyte; OLP: Oligodendroglia progenitor cells; PLP: Proteolipid protein.
Oligodendrocytes are derived from pluripotent neural stem cells in which QKI-5 is detected [44]. During neuron–glia fate specification, QKI expression is silenced in neurons but maintained in various glia types [44]. Thus, QKI is recognized as one of the earliest glial markers [45] and is implicated in neuron–glia fate determination [44]. Consistent with this notion, the Sox10 mRNA, which encodes a basic helix–loop–helix (bHLH) transcription factor known to play key roles in OL fate decision and early OL lineage establishment [46], harbors a putative QRE in the 3’UTR that is conserved in human and mouse [21]. Furthermore, deficiency of QKI in SCZ brains is found to associate with reduced expression of the Sox10 mRNA [21], suggesting that Sox10 is one of the functional targets for QKI in early OL lineage establishment. More interestingly, mouse mRNAs encoding several other Sox family members, including Sox2, Sox21 and Sox30, are also predicted to contain QREs [9]. However, whether these mRNAs are indeed bound and regulated by QKI has not been validated.
Upon entering the OL lineage, the bipolar-shaped OLPs and the immature OLs with simple processes (Figure 2) continue to proliferate until they become terminally differentiated. QKI-5, -6 and -7 are all present in OLPs before they differentiate and migrate away from the subventricular zone, with QKI-5 being the most abundant [44]. The transition from proliferation to differentiation is a key step in OL development, in which the cyclin E/cdk2 inhibitor p27kip1 plays a critical role in cell cycle arrest [45,47,48]. Mouse p27kip1 mRNA harbors a QRE in its 3’UTR that mediates QKI binding [3]. Moreover, forced expression of QKI stabilizes p27kip1 mRNA, increases p27kip1 protein expression and enhances cell cycle exit and differentiation of primary cultured OLPs. Although these observations indicate p27kip1 is a functional target for QKI to control OLP proliferation, overexpression of p27kip1 alone is not sufficient to drive OLP differentiation, despite the success in cell cycle arrest [49]. Loss of p27kip1 did not alter the timing for OLP differentiation in vivo [45]. In addition, siRNA-mediated QKI knockdown abrogates the differentiation of an OLP cell line without affecting proliferation [16]. Therefore, QKI must also act on additional t argets to advance OLP differentiation.
One such QKI target is the microtubule associated protein 1B (MAP1B), which promotes microtubule assembly and morphogenesis of OLPs before myelin synthesis occurs [50,51]. Multiple copies of the bipartite QRE are clustered in a region within the rodent MAP1B 3’UTR, which mediates in vitro binding of QKI [52]. In cultured OLPs, elimination of QKI results in reduced stability of the MAP1B mRNA. Reciprocally, forced expression of QKI increases MAP1B expression. Moreover, in the developing mouse brain, MAP1B protein and QKI-7 are co-upregulated in OLs upon their physical interaction with neuronal axons [53]. These results indicate that MAP1B mRNA is a representative target of QKI during OL morphogenic differentiation and the initial phase of myelin ensheathment.
The final stage of OL development is myelin formation. Numerous myelin structural proteins are rapidly synthesized and accumulated to very high levels specifically in OLs, which is essential for the formation of the specialized myelin lamellae [29,54]. At least three of the most abundant myelin structural protein mRNAs, which encode the MBP, proteolipid protein (PLP) and myelin-associated glycoprotein (MAG), are demonstrated as functional targets of QKI in the mouse. MBP is essential for myelin compaction, and is the only major myelin structural protein whose absence causes failure in CNS myelination [29]. PLP is responsible for the formation of intraperiod lines in the compact myelin and MAG is important for axon–glia interaction and the formation of the nodes of Ranvier. The MBP mRNA is the best-characterized QKI mRNA ligand [7,39], and binds QKI with higher efficiency in vivo than other myelin mRNAs [35,37]. QKI deficiency in the qkv/qkv mutant causes destabilization of MBP and PLP mRNAs [37], which can be rescued by the QKI-6 transgene driven by the OL-specific PLP promoter [35]. Importantly, when the QKI transgene is expressed at low levels during early development, MBP expression, but not PLP expression, is selectively rescued in the qkv/qkv mutant [35], suggesting that MBP mRNA is a preferential t arget for QKI in mouse myelinogenesis.
Besides the functional impact on mRNA stability, QKI isoforms also control mRNA splicing and subcellular localization during myelination [7,38]. Importantly, QKI isoforms do not always localize to the same subcellular compartment. The unique C-terminus of QKI-5 contains a nuclear localization signal (Figure 1), which allows QKI-5 to be predominantly localized to the nucleus at steady state, despite its ability of nuclear–cytoplasmic shuttling [5,6]. By contrast, the rest of the QKI isoforms are largely detected in the cytoplasm, and can be transported to the distal processes of mature OLs [5,53]. As a consequence, QKI isoforms exert a distinct influence on their mRNA targets. During normal OL development, the MBP mRNA is transported into the distal region of OL processes and compact myelin to support local translation of MBP protein [55]. Despite the essential function of QKI in stabilizing the MBP mRNA and accelerated production of the MBP protein, overexpression of QKI-5 causes nuclear retention of the MBP mRNA in rodent OLs, which negatively affects MBP protein synthesis [7]. In addition, the less severe reduction of QKI-5 in the qkv mutant results in unbalanced QKI isoforms [5,7,33], which may explain the aberrant accumulation of MBP mRNA in the nucleus and the perikaryon [7,55], as well as the failure in targeting the MBP mRNA into compact myelin [37].
Unlike MBP mRNA, in which the 3’UTR mediates QKI-binding [9,37,39], a 53-nucleotide intronic segment in the mouse MAG pre-mRNA binds QKI-5 and mediates QKI-5-dependent alternative splicing of MAG [38]. Whether this interaction is nuclear-specific, perhaps facilitated by other splicing factors, remains elusive. Nonetheless, the small isoform of MAG, predominantly expressed in normal adults, becomes aberrantly elevated during early development with concurrent reduction of the alternatively spliced large MAG in qkv/qkv mutant mice [38,56], as well as in SCZ brains [21]. In both cases, the cytoplasmic QKI-7 is preferentially reduced [5,21,33]. Thus, whether cytoplasmic QKI isoforms may coordinate with QKI-5 in regulating splicing is an intriguing question to be addressed by future studies.
Taken together, the above evidence indicates that QKI acts on distinct mRNA targets at different stages of OL development. Regardless of the significant progress made in understanding QKI–RNA interaction and the functional requirement of QKI in myelinogenesis in animal models, the entire pool of QKI mRNA targets at each critical stage of OL development still remains elusive. Identification of QKI targets that govern different steps of OL development, possibly by microarray analysis of OL mRNAs co-immunoprecipitated with QKI at different developmental windows, will greatly improve our knowledge regarding how the QKI pathway advances OL development and how QKI deficiency impacts myelin disorders.
Developmental regulation of QKI isoforms: a double play of transcription & splicing
Since the qkv deletion is located upstream of the qkI transcription start site and causes severe reduction of all the QKI mRNAs isoforms in myelinating OLs and Schwann cells [57], it is believed that qkv must affect a cell type-specific enhancer that tightly controls qkI transcription in myelinating cells. However, such an enhancer still remains to be identified. In fact, no published studies have specifically examined qkI transcription in OLs during development. The previously reported northern blot ana lysis, using total brain RNA, revealed increased QKI-6 and -7 mRNAs during the active phase of myelinogenesis with a concurrent decline of QKI-5 mRNA [5], but could not definitively address how qkI transcription is regulated. Nonetheless, since qkv does not affect QKI expression in non-myelinating tissues [57], the DNA sequence downstream of qkv must harbor sufficient promoter activity.
Sequence analysis revealed that the mouse qkI promoter is GC-rich and lacks a TATA box [31]. Several potential binding sites for the Sp1 zinc finger transcription factor can be predicted surrounding the qkI transcription start site [31]. Sp1 is expressed in OLs and known to be upregulated during OL differentiation owing to p27kip1-dependent stabilization of the Sp1 protein [58]. Whether Sp1 indeed binds to the qkI promoter and whether the developmentally programmed upregulation of Sp1 enhances qkI transcription during OL differentiation still remain to be determined. Nonetheless, since Sp1 is broadly expressed in nearly all cell types, it is unlikely to be responsible for OL-specific transcription of qkI. Whether OL-specific transcription factors, represented by the bHLH family that controls OL fate determination and differentiation [59,60], may also regulate qkI transcription is an intriguing possibility to be explored. Considering the key roles of histone acetylation-dependent chromatin modulation in regulating transcription during OL development [61], qkI transcription may also be modulated by histone acetylation, which in turn alters the accessibility of t ranscription factors at the qkI promoter.
Obviously, transcription is not the only mechanism that controls QKI expression. The reciprocal changes in the developmental profile of mRNA isoforms derived from qkI [5] suggest regulation of alternative splicing of the pre-mRNA. This idea is further supported by the fact that mRNA isoforms encoding various QKI protein isoforms are differentially affected in the qkv/qkv mutant as well as in SCZ patients [21,33]. This is not surprising, considering the fact that most myelin-specific gene transcripts are subjected to extensive alternative splicing and the isoform patterns are vigorously regulated during development [2,13,29]. The alterations in the expression levels of splicing factors during OL development, as represented by the reduction of hnRNP A1, H and L [62,63], likely contributes to the regulation of alternative splicing of qkI.
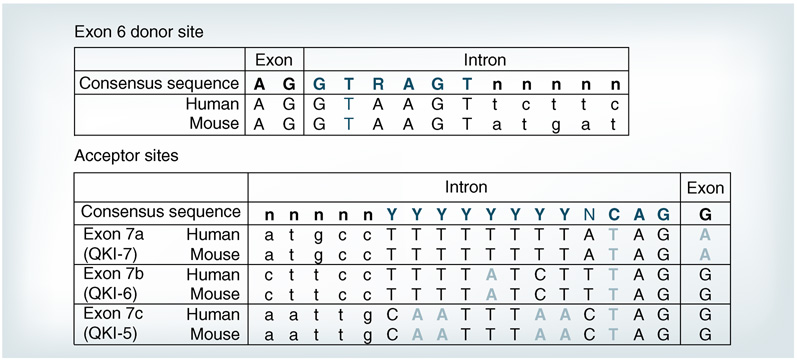
Sequence analysis of the 3’ intron–exon boundaries of qkI reveals several interesting features that may provide a molecular basis for the extensive alternative splicing of qkI (Figure 3). The alternative splicing of downstream exons exclusively occurs at the 3’ end of exon 6, the last common exon in each QKI isoform (Figure 1). The 3’-splice site of exon 6 displays perfect homology with the consensus donor sequence (Figure 3, top panel). All the alternatively spliced qkI introns are flanked by GU---AG, which satisfies the primary requirement for splicing [64,65]. However, none of the acceptor sites at the intron–exon boundaries downstream of exon 6 matches with the consensus sequence (Figure 3, bottom panel). Multiple mismatched bases indicated by bolded letters are found in these acceptor sites in all the vertebrate species examined, suggesting these unusual splice acceptor sites are evolutionarily conserved. Presumably, these imperfect acceptor sequences could result in weak binding by the spliceosome complex, and hence they may need help from specific splicing factors to generate the QKI isoforms during OL development.
Figure 3. The donor and acceptor sites for alternative splicing of QKI primary transcripts in mouse and human are highly conserved.
The donor site at exon 6 is illustrated in the top panel, which matches perfectly with the consensus sequence for splicing listed at top. The bottom panel depicts acceptor sites for the alternatively spliced exons in human and mouse. Note the diverse mismatches (light gray) in the QKI 3’ exon acceptor sites in comparison to the consensus sequence listed on top.
It is peculiar that the QKI-5-specific exon 7c is at the most 3’-end yet appears to be the default splice site, since QKI-5 is the predominant isoform during embryonic development and in many non-glia adult tissues [5,33]. In addition, the acceptor site in exon 7c contains more mismatched bases compared with the upstream acceptor sites (Figure 3, bottom panel). Thus, a plausible hypothesis predicts that splicing suppressors must block acceptor sites upstream of exon 7c in order to allow the formation of the QKI-5 transcript. Conceivably, developmentally programmed downregulation of these suppressors may lead to increased production of QKI-7 and -6. The decline of several hnRNP splicing suppressors during OL differentiation [63] is consistent with this idea, although which factor is involved in regulating qkI splicing remains elusive.
An alternative possibility for the differential expression of QKI isoforms concerns the differential stability of these mRNAs. All QKI mRNAs harbor lengthy 3’UTRs of 4–6 kb. The 3’UTR in QKI-5 is unique whereas the 3’UTRs in QKI-6 and -7 are largely overlapping and contain numerous AT-rich elements that may mediate the degradation of QKI mRNAs via the canonical mRNA decay mechanism [66,67]. In addition, QKI may regulate its own expression, since predicted QREs are also found in the 3’UTRs of both human and mouse QKI mRNA isoforms [9]. Whether QKI mRNA isoforms indeed display differential stability during OL development needs to be determined in future studies.
Phosphorylation of QKI: linking developmental signals to mRNA cellular behavior
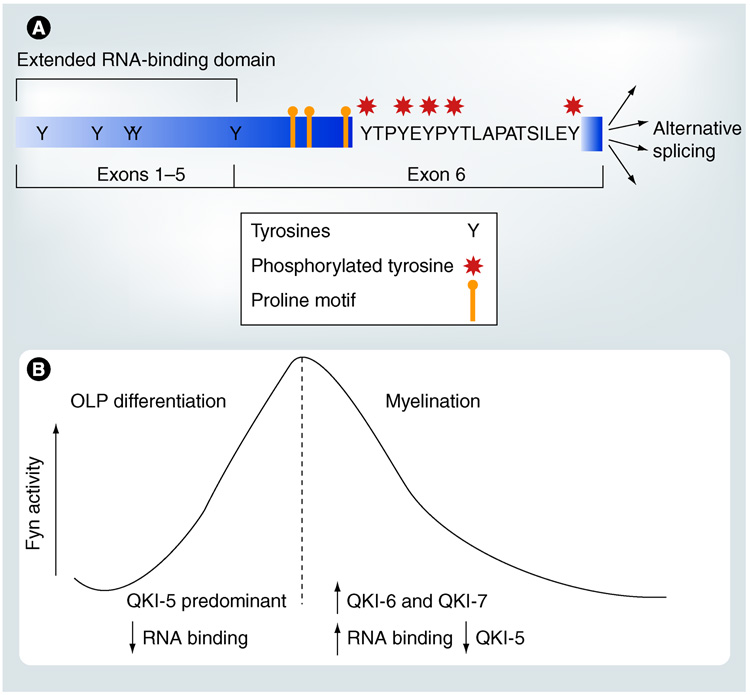
Many signal-transduction proteins harbor Srchomology 3 (SH3) domains [68,69]. The presence of proline-rich putative SH3-binding motifs following the RNA-binding domain in QKI (Figure 4A) leads to the hypothesis that QKI may bind SH3 domain-containing proteins in the signal transduction pathway besides its mRNA ligands, thus connecting developmental signals to mRNA homeostasis [14]. In support of this hypothesis, more recent studies demonstrated that the C-terminal tyrosine cluster of QKI, but not the N-terminal tyrosine residues (Figure 4A), are phosphorylated by the Src family protein tyrosine kinases (Src-PTKs) in transfected cells, brain lysates, and isolated compact myelin membranes [15]. Although QKI does not form a stable complex with Src-PTKs, mutating one of the predicted SH3-binding motifs in QKI significantly reduced Src-dependent phosphorylation of QKI [15]. More importantly, Src-PTK-mediated phosphorylation of QKI negatively regulates the ability of QKI to interact with the MBP mRNA, one of the best characterized QKI mRNA ligands [15]. Whether such regulation occurs with other QKI mRNA ligands still awaits future analysis. Considering the vigorous regulation of Src-PTK activity during OL and myelin development [70–72], Src-PTK-mediated phosphorylation of QKI offers a practical mechanism to regulate the cellular behavior of QKI mRNA ligands at various post-transcriptional steps.
Figure 4. Src-PTK-dependent phosphorylation of QKI and a working hypothesis for the function of developmentally programmed changes in Fyn activity in relation to QKI isoform expression during oligodendrocyte and myelin development.
(A) The cluster of tyrosines encoded by exon 6 (marked by asterisks) are phosphorylated by the Src-PTK member Fyn in oligodendrocytes, likely mediated by the proline-rich putative SH3-binding motifs. Although there are tyrosines in the N-terminus of the protein, they are not phosphorylated by Src-PTK. (B) Fyn activity is increased during OLP differentiation when QKI–5 is the predominant isoform, which presumably reduces the RNA-binding activity of QKI–5. This may offer a means to release nuclear retention of QKI’s target mRNA. By contrast, the developmentally programmed decline of Fyn activity during accelerated myelinogenesis correlates with increased expression of cytoplasmic QKI isoforms. This conceivably may increase QKI’s RNA-binding activity, which in turn may protect QKI’s mRNA targets from degradation represented by accelerated accumulation of the myelin basic protein mRNA.
OLP: Oligodendroglia progenitor cells.
In the developing OLs, Fyn is the major Src- PTK member [71]. A convincing set of data demon strates the essential function of Fyn in OL and myelin development. Upon induced differentiation of primary cultured OLPs, the activity of Fyn, but not other Src-PTKs, is drastically increased [71]. In addition, inhibition of Fyn activity prevents OL differentiation. During in vivo myelin d evelopment, Fyn is regulated bidirectionally (Figure 4B). The activity of Fyn is gradually increased in the initial phase of myelinogenesis, followed by a developmentally programmed decline in the second postnatal week [70,72]. fyn−/− mice suffer from impaired OL differentiation, abnormal MBP expression/phosphorylation, and deficits in myelin synthesis, especially abnormal formation/maintenance of the compact myelin [57,70,73–76]. It is important to note that the myelin defects in fyn−/− brain exist despite the upregulation of s everal other Src-PTKs as a compensatory response to the lack of Fyn [77]. Thus, the function of Fyn in myelinogenesis cannot be substituted by other Src-PTKs.
Despite the critical role of Fyn in early OL differentiation [71], fyn−/− brain does not show MBP deficiency and hypomyelination until accelerated myelinogenesis starts in wild-type brain during the second postnatal week [33]. The level of MBP accumulates more slowly in the fyn−/− forebrain in the most vigorous phase of myelin synthesis and never reaches the level of wild-type controls. Importantly, the onset of hypomyelination in fyn−/− brain occurs within the developmental window when Fyn activity declines in wild-type brain [72,75], suggesting that reduced Fyn activity may release a repression mechanism for myelination during normal development. The negative regulation of QKI’s RNA-binding activity by Src-PTKs is a candidate for such regulation. The decline of Fyn activity upon maturation of OLs and the initiation of myelin formation should facilitate the interaction between QKI and MBP mRNA. This, in combination with the increased QKI-6 and -7 expression in the cytoplasm and decreased expression of the nuclear QKI-5 (Figure 4B), presumably should increase the cytoplasmic stability of the MBP mRNA and the expression of MBP protein. Indeed, the decline of Src-PTK-dependent tyrosine phosphorylation of QKI in isolated myelin correlates with the rapid accumulation of MBP protein [15]. By this means, QKI conceivably converts the developmentally programmed downregulation of Fyn into a positive signal in post-transcriptional r egulation of MBP accumulation and a ccelerated myelin synthesis.
Whether and how Fyn and QKI may also functionally coordinate in early OL differentiation before active myelinogenesis remains unknown. It is particularly puzzling that both QKI and Fyn are upregulated and play important roles in advancing OL differentiation, yet Fyn negatively regulates QKI’s RNA-binding activity. Importantly, QKI-5 is the predominant isoform during early OL differentiation when Fyn activity is upregulated (Figure 4B). By contrast, the cytoplasmic QKI isoforms are expressed at low levels before initiation of myelinogenesis [44,53]. Despite the ability for QKI-5 to shuttle between nucleus and cytoplasm [6], at steady state QKI-5 is predominantly localized in the nucleus, which is known to cause nuclear retention of its mRNA ligands [7]. Thus, although QKI-5 may stabilize mRNAs that are important for OL differentiation, represented by the p27kip1 mRNA and the MAP1B mRNA [3,52], these mRNAs may need to dissociate from QKI-5 in order to exit the nucleus and to allow translation of the corresponding proteins in the cytoplasm. Therefore, an intriguing hypothesis can be postulated that Fyn-dependent phosphorylation of QKI-5 and the negative effect on QKI’s RNA-binding activity may serve as a means for releasing nuclear retention of mRNAs during early OL differentiation, an idea that deserves investigation by future studies.
Besides Fyn, other kinases have also been shown to play important roles in promoting OL development and myelination. Recent discoveries revealed that Cdk-5, a serine/threonine kinase that is key for brain development [78,79], is activated upon OL differentiation and plays an essential role in supporting the development of OL processes [80]. In addition, transgenic mice harboring forced Akt signaling exhibit overproduction of myelin without affecting OL proliferation [81]. Furthermore, MAPK pathways have also been demonstrated to play important roles in OLP migration, proliferation, survival and differentiation [82,83]. Nonetheless, whether these signal transduction pathways are connected with QKI function still remains elusive. It is also worth mentioning that in addition to phosphorylation, QKI can also be arginine-methylated [84]. Whether and how other post-translational modifications on QKI also modulate its RNA-binding activity still remains to be addressed by future studies.
QKI & myelin disorders: potential roles of QKI in schizophrenia & myelin repair in multiple sclerosis
Although little is known about the function of human QKI, the essential role of QKI in mouse myelin development [35], the similarity in the organization of mouse qkI gene and human QKI gene, and the identical amino acid sequence of QKI in human and rodents [31,85] together suggest that QKI most likely plays key roles in myelination of the human CNS. Growing evidence from neuroimaging and neuropathological studies has revealed that impairment of OL and myelin development is a common morbidity in major psychiatric disorders represented by SCZ [17,20]. In particular, QKI deficiency is implicated in the etiology of myelin defect in SCZ [21,23].
The human QKI gene is located at chromosome 6q26 [85]. Interestingly, 6q25-27 has long been recognized to contain susceptibility loci that associate with SCZ [86,87]. Most recently, one locus was mapped to a 0.5 Mb region in which QKI is the only known gene [22]. It is particularly interesting that two SNPs located in intron 3 and 7b of the QKI gene, respectively, are shown to cosegregate with the majority of SCZ individuals in a large pedigree [22]. In addition, several independent studies demonstrate reduced QKI mRNA expression in multiple regions of SCZ patient brains [22–24,88], suggesting the functional involvement of QKI in SCZ etiology. Currently, molecular mechanisms that lead to the reduced QKI expression in the SZC brain remain elusive. Since QKI proteins are also expressed in non-myelinating glia, the reported reduction of QKI mRNAs in SCZ brains likely underestimates the quantitative deficiency of QKI in OLs. Among QKI mRNA isoforms, QKI-7 and -7b are preferentially reduced in SCZ brains [21], indicating post-transcriptional misregulation of QKI and suggesting the preferential importance of these QKI isoforms in SCZ etiology. Interestingly, we found that the SCZ-associated SNP in intron 7b potentially ablates a splicing enhancer site, which may alter binding of some splicing factors, based on sequence prediction using the RESCUE method, which is commonly applied in the splicing field [89,90]. Whether this is an underlying factor for the differential reduction of QKI isoform mRNAs in SCZ remains undetermined at this point.
More importantly, QKI deficiency in SCZ is associated with reduced expression of mRNAs critical for OL and myelin development [21,91]. Noticeably, several of these mRNAs harbor predicted QREs, and therefore are likely direct targets of QKI [21]. The best examples include the mRNAs for MBP, MAG and PLP, which are functionally validated QKI targets in mice. Hence, QKI deficiency may serve as an underlying factor for the impairment of OL and myelin development in SCZ. Despite the reduced QKI expression in SCZ in general, patients treated with typical antipsychotics harbor relatively higher levels of QKI mRNA as compared with patients treated with atypical antipsychotics [21]. This observation suggests that some antipsychotics may also modulate myelination in addition to regulating dopaminergic signaling in neurons. Future studies are needed to investigate how antipsychotics may regulate QKI expression, and whether this may help to improve the deficiency in myelin development in SCZ patients, especially considering the fact that the first episode of SCZ is often diagnosed in young adults when myelination is not completed in the frontal cortex [92,93].
The most prevalent human demyelination disorder is MS, which affects more than two million people in the world [94]. This devastating disease results from repetitive autoimmune attacks that destroy CNS myelin, often leading to permanent neurological disabilities due to the damage of neuronal axons as a result of the loss of protective myelin [95,96]. Efficient repair of the damaged myelin is the ultimate solution for the treatment of MS. Unfortunately, no current therapies enhance remyelination. In fact, molecular and cellular mechanisms that control myelin repair in the adult CNS remain largely unknown, which is an obvious hurdle in developing strategies against MS. The generation/proliferation of OLPs, migration of OLPs to the lesion, differentiation to mature OLs and the ensheathment of demyelinated axons most likely recapitulates the process in neonatal myelin development (Figure 2). Considering the essential roles of QKI in OL lineage development and myelinogenesis, the functional importance of QKI can be predicted for myelin repair. However, QKI expression declines to low levels in the adult brain after the completion of CNS myelination, which likely functions to maintain the integrity of formed myelin sheaths and axonal conductance [15]. Whether and how QKI is upregulated in adult OLPs to advance migration/differentiation of OLPs and repair the damaged myelin in CNS lesions still remain elusive. Whether increased QKI expression, for instance in OLs of the Flag-QKI-transgenic mice, may enhance myelination and repair is an important issue to be addressed by future studies. In addition, the recent discovery that the g chain of immunogloblin Fc receptor– Fyn signaling is critical for myelin repair [97] raises an intriguing possibility for the involvement of Src-PTK-dependent phosphorylation of QKI in myelin repair.
Conclusion
A convincing set of evidence derived from molecular, cellular and animal studies has demonstrated that post-transcriptional regulation by the selective RNA-binding protein QKI is necessary and sufficient for CNS myelin development, which in turn governs the rapid conduction of nerve impulses in the brain. Although the entire pool of OL-specific QKI mRNA targets have not been identified, at various key points of development QKI appears to regulate distinct mRNA targets in OLs to promote myelinogenesis (Figure 2). A combination of regulations at the level of transcription and alternative splicing generates multiple QKI isoform proteins at different stages of OL development, which display distinct subcellular distribution. These QKI isoforms act at different post-transcriptional steps and exert differential influences, sometimes even opposing effects, to regulate the homeostasis and subcellular localization of their mRNA targets. The vigorous regulation of QKI isoform expression, together with the phosphorylation-dependent modulation of QKI’s RNA-binding activity, suggests that QKI is a pivotal factor that mediates extracellular and developmental signals to regulate the cellular behavior of a broad spectrum of mRNA species during myelin development. More importantly, recent genetic and molecular discoveries identify QKI as a susceptibility factor for the etiology of myelin impairment in psychiatric disorders represented by SCZ, thus extending the significance of QKI-related research beyond the scope of myelin biology.
Executive summary
Introduction
The selective RNA-binding protein QKI plays important roles in post-transcriptional regulation of its bound mRNA ligands.
Many bioinformatically identified QKI mRNA targets encode key proteins that govern normal cell growth and development.
Accumulating evidence indicates the functional requirement of QKI, particularly in oligodendrocyte (OL) development and CNS myelination.
Recent discoveries suggest that QKI is a potential susceptibility factor for schizophrenia (SCZ) and QKI deficiency may contribute to the myelin defect in SCZ and multiple sclerosis (MS).
Essential roles of QKI in advancing oligodendrocyte development & CNS myelination
Identification of the qkv spontaneous dysmyelination mutation and reduced expression of QKI in qkv-mutant OLs is the first indication for the functional importance of QKI in CNS myelination.
Expression of a QKI transgene specifically in OLs can rescue the dysmyelination and tremor phenotype of qkv mice, indicating that QKI is necessary for CNS myelinogenesis.
Manipulating QKI expression demonstrates that QKI is also necessary and sufficient for promoting OL progenitor cell differentiation before actual myelination.
QKI regulates distinct mRNA targets at different stages of oligodendrocyte development & myelination
The QKI recognition elements located in the 3’-untranslated region mediate direct interaction of QKI with its mRNA targets.
Alternatively spliced QKI isoforms are differentially expressed during OL and myelin development, which display distinct subcellular distribution and sometimes even opposing influence to the QKI mRNA ligands.
QKI regulates distinct mRNA targets at different stages of OL and myelin development.
QKI isoforms act at different post-transcriptional steps to control mRNA homeostasis and subcellular localization in OL and myelin development.
Developmental regulation of QKI isoforms: a double play of transcription & splicing
The qkv mutation affects an OL-specific transcriptional mechanism of qkI, likely due to the loss of an undefined OL-specific enhancer.
The QKI primary transcript is subjected to extensive alternative splicing. The diverse mismatches from the consensus sequence at the predicted weak 3’ splice acceptor sites of QKI suggest regulation by splicing factors.
The expression levels of many splicing factors are vigorously regulated during OL differentiation, which may control the developmentally programmed splicing of QKI.
Phosphorylation of QKI: linking signal pathways to mRNA behavior
QKI can be phosphorylated by the Src family protein tyrosine kinases (Src-PTKs), which negatively regulate QKI-RNA interaction.
Fyn is the major Src-PTK in developing OLs, and Fyn deficiency results in post-transcriptional abnormality of QKI mRNA ligands and CNS hypomyelination.
Fyn activity is increased during OLP differentiation but declines in the most active phase of myelin synthesis.
A working hypothesis is proposed regarding the functional significance of Fyn-mediated negative regulation of QKI’s RNA-binding activity in early OLP differentiation and accelerated myelinogenesis.
QKI & myelin disorders: potential roles of QKI in schizophrenia & myelin repair
Chromosome 6 contains susceptibility loci associated with SCZ, including the human QKI locus at 6q26.
QKI isoforms are differentially reduced in the postmortem brain of SCZ patients, which correlates with decreased QKI mRNA ligands in myelination.
QKI is upregulated in SCZ patients treated with typical antipsychotics.
The essential function of QKI in bona fide neonatal myelin development suggests that it may also play critical roles in myelin repair in demyelination diseases represented by MS.
Conclusion
QKI is necessary and sufficient for OL and CNS myelin development.
Tight regulation of QKI isoform expression and activity allows QKI to exert differential influences on distinct mRNA targets at various key steps of OL development, which forms a pivotal pathway to advance myelinogenesis.
The genetic alterations in the human QKI locus and deficiencies in QKI expression associated with SCZ suggest the involvement of QKI in the etiology of SCZ, possibly as an underlying mechanism for myelin impairment.
Future perspective
Further delineating the molecular interaction between QKI isoforms and their RNA ligands and identification of QKI’s functional targets specifically in OLs should provide important insights in understanding the function of QKI in normal myelinogenesis as well as the pathological impact of the QKI pathway in myelin defects in SCZ.
The potential role of QKI in myelin repair after demyelination is an intriguing question to be explored, which may help to develop novel strategies against MS.
Future perspective
Several prevailing questions in understanding the function of QKI still remain to be answered. Despite the extensive ana lysis of QRE for direct interaction with QKI in vitro, how the functional domains in QKI interact with RNA requires the solution of the 3D structure of QKI. Regarding the in vivo situation, whether RNA sequences adjacent to the QRE may modulate QKI binding, perhaps by recruiting other RNA-binding proteins specifically expressed in OLs, is unknown. In addition, the full capacity of developmental stage-specific mRNA targets of QKI in OLs still remains elusive. Furthermore, molecular mechanisms that control QKI isoform expression and activity, which can be predicted to include OL-specific transcription factors, splicing factors, as well as signaling pathways that govern phosphorylation and/or other post-translational modifications of QKI, are poorly understood. Considering the pathological involvement of QKI, how QKI expression is affected in SCZ and whether malfunction of the QKI-pathway indeed underlies the myelin defects in SCZ need to be determined. Moreover, despite the essential role of QKI in the de novo myelinogenesis during neonatal development, whether and how QKI may also promote myelin repair after demyelination in adults is an intriguing possibility that may lead to identification of a new therapeutic target against MS. It is important to note that QKI defects are not limited to myelin disorders. Genetic deletions in the QKI locus have also been associated with human glioblastoma [85,98] and some cases of autism [99]. QKI function is also implicated in ataxia in one of the ENU mutant mouse lines, suggesting a possible connection to human ataxic diseases [1,34,100]. Therefore, further investigations are warranted to delineate the molecular mechanisms by which the QKI pathway governs normal brain development and function and how QKI malfunction impacts various human CNS diseases.
Acknowledgments
Financial & competing interests disclosure
This work is supported by NIH grant NS39551, NMSS grant RG 4010-A-2 and NASRAD Independent Investigator Award to Yue Feng. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Katrina Bockbrader, Department of Pharmacology, Emory University School of Medicine, 1510 Clifton Road, Atlanta, GA 30322, USA, Tel.: +1 404 727 0351, Fax: +1 404 727 0365, kbockbr@emory.edu.
Yue Feng, Department of Pharmacology, Emory University School of Medicine, 1510 Clifton Road, Atlanta, GA 30322, USA, Tel.: +1 404 727 0351, Fax: +1 404 727 0365, yfeng@emory.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪ ▪ of considerable interest
- 1.Chenard CA, Richard S. New implications for the QUAKING RNA binding protein in human disease. J. Neurosci. Res. 2008;86:233–242. doi: 10.1002/jnr.21485. [DOI] [PubMed] [Google Scholar]
- 2.McInnes LA, Lauriat TL. RNA metabolism and dysmyelination in schizophrenia. Neurosci. Biobehav. Rev. 2006;30:551–561. doi: 10.1016/j.neubiorev.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 3. Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S. Protection of p27kip1 mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat. Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. First evidence suggesting that QKI can promote oligodendrocyte progenitor cells from proliferation to differentiation by regulating the p27kip1 mRNA.
- 4. Ebersole TA, Chen Q, Justice MJ, Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. Identification of the qkv mutation and the mouse qkI gene, first prediction of the alternatively spliced QKI isoforms.
- 5. Hardy RJ, Loushin CL, Friedrich VL, Jr, et al. Neural cell type-specific expression of QKI proteins is altered in quakingviable mutant mice. J. Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. First evidence that QKI is expressed in glia but not in neurons and QKI isoforms display distinct subcellular localization. Characterization of differential deficiency of QKI isoforms specifically in oligodendrocytes of the qkv/qkv-mutant mice, suggesting that QKI is important in myelination.
- 6.Wu J, Zhou L, Tonissen K, Tee R, Artzt K. The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 1999;274:29202–29210. doi: 10.1074/jbc.274.41.29202. [DOI] [PubMed] [Google Scholar]
- 7. Larocque D, Pilotte J, Chen T, et al. Nuclear retention of MBP mRNAs in the quaking viable mice. Neuron. 2002;36:815–829. doi: 10.1016/s0896-6273(02)01055-3. First evidence that QKI isoforms can exert opposing effects on myelination.
- 8.Pilotte J, Larocque D, Richard S. Nuclear translocation controlled by alternatively spliced isoforms inactivates the QUAKING apoptotic inducer. Genes Dev. 2001;15:845–858. doi: 10.1101/gad.860301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005;12:691–698. doi: 10.1038/nsmb963. Identification of QKI recognition element and bioinformatical prediction of putative target mRNAs for QKI.
- 10.Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 2004;11:20–28. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- 11.Noveroske JK, Lai L, Gaussin V, et al. Quaking is essential for blood vessel development. Genesis. 2002;32:218–230. doi: 10.1002/gene.10060. [DOI] [PubMed] [Google Scholar]
- 12.Dawson MR, Polito A, Levine JM, Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol. Cell. Neurosci. 2003;24:476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 13.Hardy RJ. Molecular defects in the dysmyelinating mutant quaking. J. Neurosci. Res. 1998;51:417–422. doi: 10.1002/(SICI)1097-4547(19980215)51:4<417::AID-JNR1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 14.Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Y, Lu Z, Ku L, Chen Y, Wang H, Feng Y. Tyrosine phosphorylation of QKI mediates developmental signals to regulate mRNA metabolism. EMBO J. 2003;22:1801–1810. doi: 10.1093/emboj/cdg171. First report demonstrating regulation of the RNA-binding activity of QKI by Src-PTK-dependent tyrosine phosphorylation.
- 16. Chen Y, Tian D, Ku L, Osterhout DJ, Feng Y. The selective RNA-binding protein quaking I (QKI) is necessary and sufficient for promoting oligodendroglia differentiation. J. Biol. Chem. 2007;282:23553–23560. doi: 10.1074/jbc.M702045200. QKI is necessary and sufficient for oligodendrocyte progenitor cell development and differentiation, involving cell cycle-independent mechanisms.
- 17.Sokolov BP. Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse Comorbidity, shared traits, or molecular phenocopies? Int. J. Neuropsychopharmacol. 2007;10:547–555. doi: 10.1017/S1461145706007322. [DOI] [PubMed] [Google Scholar]
- 18.Karoutzou G, Emrich HM, Dietrich DE. The myelin-pathogenesis puzzle in schizophrenia: a literature review. Mol. Psychiatry. 2008;13:245–260. doi: 10.1038/sj.mp.4002096. [DOI] [PubMed] [Google Scholar]
- 19.Segal D, Koschnick JR, Slegers LH, Hof PR. Oligodendrocyte pathophysiology: a new view of schizophrenia. Int. J. Neuropsychopharmacol. 2007;10:503–511. doi: 10.1017/S146114570600722X. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y. Convergence and divergence in the etiology of myelin impairment in psychiatric disorders and drug addiction. Neurochem. Res. 2008;33(10):1940–1949. doi: 10.1007/s11064-008-9693-x. [DOI] [PubMed] [Google Scholar]
- 21. Aberg K, Saetrex P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc. Natl Acad. Sci. USA. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. Linking QKI deficiency and QKI target mRNA reduction to myelin impairment in schizophrenia; suggesting the potential impact of QKI deficiency in myelin etiology in schizophrenia.
- 22. Aberg K, Saetre P, Lindholm E, et al. Human QKI, a new candidate gene for schizophrenia involved in myelination. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2006;141:84–90. doi: 10.1002/ajmg.b.30243. First indication that deficiencies of QKI isoform expression are correlated with schizophrenia.
- 23.Haroutunian V, Katsel P, Dracheva S, Davis KL. The human homolog of the QKI gene affected in the severe dysmyelination ‘quaking’ mouse phenotype: downregulated in multiple brain regions in schizophrenia. Am. J. Psychiatry. 2006;163:1834–1837. doi: 10.1176/ajp.2006.163.10.1834. [DOI] [PubMed] [Google Scholar]
- 24.Lauriat TL, Shiue L, Haroutunian V, et al. Developmental expression profile of quaking, a candidate gene for schizophrenia, and its target genes in human prefrontal cortex and hippocampus shows regional specificity. J. Neurosci. Res. 2008;86:785–796. doi: 10.1002/jnr.21534. [DOI] [PubMed] [Google Scholar]
- 25.Larocque D, Richard S. QUAKING KH domain proteins as regulators of glial cell fate and myelination. RNA Biol. 2005;2:37–40. doi: 10.4161/rna.2.2.1603. [DOI] [PubMed] [Google Scholar]
- 26.Lakiza O, Frater L, Yoo Y, et al. STAR proteins quaking-6 and GLD-1 regulate translation of the homologues GLI1 and tra-1 through a conserved RNA 3’UTR-based mechanism. Dev. Biol. 2005;287:98–110. doi: 10.1016/j.ydbio.2005.08.038. [DOI] [PubMed] [Google Scholar]
- 27.Volk T, Israeli D, Nir R, Toledano-Katchalski H. Tissue development and RNA control: ‘HOW’ is it coordinated? Trends Genet. 2008;24:94–101. doi: 10.1016/j.tig.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Sidman S, Dickie M, Appel S. Mutant mice (Quaking and Jimpy) with deficient myelination in the central nervous system. Science. 1964;144:309–311. doi: 10.1126/science.144.3616.309. [DOI] [PubMed] [Google Scholar]
- 29.Campagnoni AT. Molecular biology of myelin proteins from the central nervous system. J. Neurochem. 1988;51:1–4. doi: 10.1111/j.1471-4159.1988.tb04827.x. [DOI] [PubMed] [Google Scholar]
- 30.Cox RD, Hugill A, Shedlovsky A, et al. Contrasting effects of ENU induced embryonic lethal mutations of the quaking gene. Genomics. 1999;57:333–341. doi: 10.1006/geno.1999.5804. [DOI] [PubMed] [Google Scholar]
- 31.Kondo T, Furuta T, Mitsunaga K, et al. Genomic organization and expression analysis of the mouse qkI locus. Mamm. Genome. 1999;10:662–669. doi: 10.1007/s003359901068. [DOI] [PubMed] [Google Scholar]
- 32.Lorenzetti D, Antalffy B, Vogel H, Noveroske J, Armstrong D, Justice M. The neurological mutant quaking(viable) is Parkin deficient. Mamm. Genome. 2004;15:210–217. doi: 10.1007/s00335-003-2333-5. [DOI] [PubMed] [Google Scholar]
- 33.Lu Z, Zhang Y, Ku L, Wang H, Ahmadian A, Feng Y. The quakingviable mutation affects qkI mRNA expression specifically in myelin-producing cells of the nervous system. Nucleic Acids Res. 2003;31:4616–4624. doi: 10.1093/nar/gkg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noveroske JK, Hardy R, Dapper JD, Vogel H, Justice MJ. A new ENU-induced allele of mouse quaking causes severe CNS dysmyelination. Mamm. Genome. 2005;16:672–682. doi: 10.1007/s00335-005-0035-x. [DOI] [PubMed] [Google Scholar]
- 35. Zhao L, Tian D, Xia M, Macklin WB, Feng Y. Rescuing qkV dysmyelination by a single isoform of the selective RNA-binding protein QKI. J. Neurosci. 2006;26:11278–11286. doi: 10.1523/JNEUROSCI.2677-06.2006. Oligodendrocyte-specific expression of a QKI transgene rescues the qkv dyemyelination phenotype and restores normal myelin. First direct/ functional evidence that QKI is essential for myelination.
- 36.Saccomanno L, Loushin C, Jan E, Punkay E, Artzt K, Goodwin EB. The STAR protein QKI-6 is a translational repressor. Proc. Natl Acad. Sci. USA. 1999;96:12605–12610. doi: 10.1073/pnas.96.22.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Zhang Y, Li D, Feng Y. Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J. Neurosci. 2000;20:4944–4953. doi: 10.1523/JNEUROSCI.20-13-04944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu JI, Reed RB, Grabowski PJ, Artzt K. Function of quaking in myelination: regulation of alternative splicing. Proc. Natl Acad. Sci. USA. 2002;99:4233–4238. doi: 10.1073/pnas.072090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryder SP, Williamson JR. Specificity of the STAR/GSG domain protein QKI: implications for the regulation of myelination. RNA. 2004;10:1449–1458. doi: 10.1261/rna.7780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T, Damaj BB, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and QKI: role of the KH domain. Mol. Cell. Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen T, Richard S. Structure-function analysis of QKI: a lethal point mutation in mouse quaking prevents homodimerization. Mol. Cell. Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67:2072–2080. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- 43.Rosenberg SS, Powell BL, Chan JR. Receiving mixed signals: uncoupling oligodendrocyte differentiation and myelination. Cell. Mol. Life Sci. 2007;64:3059–3068. doi: 10.1007/s00018-007-7265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardy RJ. QKI expression is regulated during neuron-glial cell fate decisions. J. Neurosci. Res. 1998;54:46–57. doi: 10.1002/(SICI)1097-4547(19981001)54:1<46::AID-JNR6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 45.Casaccia-Bonnefil P, Hardy RJ, Teng KK, Levine JM, Koff A, Chao MV. Loss of p27kip1 function results in increased proliferative capacity of oligodendrocyte progenitors but unaltered timing of differentiation. Development. 1999;126:4027–4037. doi: 10.1242/dev.126.18.4027. [DOI] [PubMed] [Google Scholar]
- 46.Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 47.Durand B, Fero ML, Roberts JM, Raff MC. p27kip1 alters the response of cells to mitogen and is part of a cell-intrinsic timer that arrests the cell cycle and initiates differentiation. Curr. Biol. 1998;8:431–440. doi: 10.1016/s0960-9822(98)70177-0. [DOI] [PubMed] [Google Scholar]
- 48.Tikoo R, Osterhout DJ, Casaccia-Bonnefil P, Seth P, Koff A, Chao MV. Ectopic expression of p27kip1 in oligodendrocyte progenitor cells results in cell-cycle growth arrest. J. Neurobiol. 1998;36:431–440. [PubMed] [Google Scholar]
- 49.Tang XM, Beesley JS, Grinspan JB, Seth P, Kamholz J, Cambi F. Cell cycle arrest induced by ectopic expression of p27 is not sufficient to promote oligodendrocyte differentiation. J. Cell Biochem. 1999;76:270–279. doi: 10.1002/(sici)1097-4644(20000201)76:2<270::aid-jcb10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 50.Vouyiouklis DA, Brophy PJ. Microtubule-associated protein MAP1B expression precedes the morphological differentiation of oligodendrocytes. J. Neurosci. Res. 1993;35:257–276. doi: 10.1002/jnr.490350305. [DOI] [PubMed] [Google Scholar]
- 51.Fischer I, Konola J, Cochary E. Microtubule associated protein (MAP1B) is present in cultured oligodendrocytes and co-localizes with tubulin. J. Neurosci. Res. 1990;27:112–124. doi: 10.1002/jnr.490270117. [DOI] [PubMed] [Google Scholar]
- 52.Zhao L, Ku L, Chen Y, Xia M, LoPresti P, Feng Y. QKI binds MAP1B mRNA and enhances MAP1B expression during oligodendrocyte development. Mol. Biol. Cell. 2006;17:4179–4186. doi: 10.1091/mbc.E06-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu HY, Dawson MR, Reynolds R, Hardy RJ. Expression of QKI proteins and MAP1B identifies actively myelinating oligodendrocytes in adult rat brain. Mol. Cell. Neurosci. 2001;17:292–302. doi: 10.1006/mcne.2000.0941. [DOI] [PubMed] [Google Scholar]
- 54.Deber CM, Reynolds SJ. Central nervous system myelin: structure, function, and pathology. Clin. Biochem. 1991;24:113–134. doi: 10.1016/0009-9120(91)90421-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbarese E. Spatial distribution of myelin basic protein mRNA and polypeptide in quaking oligodendrocytes in culture. J. Neurosci. Res. 1991;29:271–281. doi: 10.1002/jnr.490290302. [DOI] [PubMed] [Google Scholar]
- 56.Fujita N, Sato S, Kurihara T, Inuzuka T, Takahashi Y, Miyatake T. Developmentally regulated alternative splicing of brain myelin-associated glycoprotein mRNA is lacking in the quaking mouse. FEBS Lett. 1988;232:323–327. doi: 10.1016/0014-5793(88)80762-2. [DOI] [PubMed] [Google Scholar]
- 57.Lu Z, Ku L, Chen Y, Feng Y. Developmental abnormalities of myelin basic protein expression in fyn knock-out brain reveal a role of Fyn in posttranscriptional regulation. J. Biol. Chem. 2005;280:389–395. doi: 10.1074/jbc.M405973200. [DOI] [PubMed] [Google Scholar]
- 58.Wei Q, Miskimins WK, Miskimins R. The Sp1 family of transcription factors is involved in p27kip1-mediated activation of myelin basic protein gene expression. Mol. Cell. Biol. 2003;23:4035–4045. doi: 10.1128/MCB.23.12.4035-4045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marquardt T, Pfaff SL. Cracking the transcriptional code for cell specification in the neural tube. Cell. 2001;106:651–654. doi: 10.1016/s0092-8674(01)00499-8. [DOI] [PubMed] [Google Scholar]
- 60.Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J. Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubicki M, McCarley RW, Shenton ME. Evidence for white matter abnormalities in schizophrenia. Curr. Opin Psychiatry. 2005;18:121–134. doi: 10.1097/00001504-200503000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Havlioglu N, Wang J, Fushimi K, et al. An intronic signal for alternative splicing in the human genome. PLoS ONE. 2007;2:E1246. doi: 10.1371/journal.pone.0001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang E, Dimova N, Cambi F. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007;35:4164–4178. doi: 10.1093/nar/gkm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang MQ. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 66.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 67.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 68.Schlessinger J. SH2/SH3 signaling proteins. Curr. Opin. Genet. Dev. 1994;4:25–30. doi: 10.1016/0959-437x(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 69.Gmeiner WH, Horita DA. Implications of SH3 domain structure and dynamics for protein regulation and drug design. Cell Biochem. Biophys. 2001;35:127–140. doi: 10.1385/CBB:35:2:127. [DOI] [PubMed] [Google Scholar]
- 70.Umemori H, Sato S, Yagi T, Aizawa S, Yamamoto T. Initial events of myelination involve Fyn tyrosine kinase signalling. Nature. 1994;367:572–576. doi: 10.1038/367572a0. [DOI] [PubMed] [Google Scholar]
- 71.Osterhout DJ, Wolven A, Wolf RM, Resh MD, Chao MV. Morphological differentiation of oligodendrocytes requires activation of Fyn tyrosine kinase. J. Cell Biol. 1999;145:1209–1218. doi: 10.1083/jcb.145.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kramer EM, Klein C, Koch T, Boytinck M, Trotter J. Compartmentation of Fyn kinase with glycosylphosphatidylinositol-anchored molecules in oligodendrocytes facilitates kinase activation during myelination. J. Biol. Chem. 1999;274:29042–29049. doi: 10.1074/jbc.274.41.29042. [DOI] [PubMed] [Google Scholar]
- 73.Umemori H, Kadowaki Y, Hirosawa K, et al. Stimulation of myelin basic protein gene transcription by Fyn tyrosine kinase for myelination. J. Neurosci. 1999;19:1393–1397. doi: 10.1523/JNEUROSCI.19-04-01393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seiwa C, Sugiyama I, Yagi T, Iguchi T, Asou H. Fyn tyrosine kinase participates in the compact myelin sheath formation in the central nervous system. Neurosci. Res. 2000;37:21–31. doi: 10.1016/s0168-0102(00)00100-0. [DOI] [PubMed] [Google Scholar]
- 75.Sperber BR, Boyle-Walsh EA, Engleka MJ, et al. A unique role for Fyn in CNS myelination. J. Neurosci. 2001;21:2039–2047. doi: 10.1523/JNEUROSCI.21-06-02039.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goto J, Tezuka T, Nakazawa T, Sagara H, Yamamoto T. Loss of Fyn tyrosine kinase on the C57BL/6 genetic background causes hydrocephalus with defects in oligodendrocyte development. Mol. Cell. Neurosci. 2008;38:203–212. doi: 10.1016/j.mcn.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 77.Grant SG, Karl KA, Kiebler MA, Kandel ER. Focal adhesion kinase in the brain: novel subcellular localization and specific regulation by Fyn tyrosine kinase in mutant mice. Genes Dev. 1995;9:1909–1921. doi: 10.1101/gad.9.15.1909. [DOI] [PubMed] [Google Scholar]
- 78.Lew J, Huang QQ, Qi Z, et al. A brain-specific activator of cyclin-dependent kinase 5. Nature. 1994;371:423–426. doi: 10.1038/371423a0. [DOI] [PubMed] [Google Scholar]
- 79.Tsai LH, Delalle I, Caviness VS, Jr, Chae T, Harlow E. p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature. 1994;371:419–423. doi: 10.1038/371419a0. [DOI] [PubMed] [Google Scholar]
- 80.Miyamoto Y, Yamauchi J, Chan JR, et al. Cdk5 regulates differentiation of oligodendrocyte precursor cells through the direct phosphorylation of paxillin. J. Cell Sci. 2007;120:4355–4366. doi: 10.1242/jcs.018218. [DOI] [PubMed] [Google Scholar]
- 81.Flores AI, Narayanan SP, Morse EN, et al. Constitutively active Akt induces enhanced myelination in the CNS. J. Neurosci. 2008;28:7174–7183. doi: 10.1523/JNEUROSCI.0150-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fragoso G, Haines JD, Roberston J, Pedraza L, Mushynski WE, Almazan G. p38 mitogen-activated protein kinase is required for central nervous system myelination. Glia. 2007;55:1531–1541. doi: 10.1002/glia.20567. [DOI] [PubMed] [Google Scholar]
- 83.Frost EE, Zhou Z, Krasnesky K, Armstrong RC. Initiation of oligodendrocyte progenitor cell migration by a PDGF-A activated extracellular regulated kinase (ERK) signaling pathway. Neurochem. Res. 2008 doi: 10.1007/s11064-008-9748-z. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cote J, Boisvert FM, Boulanger MC, Bedford MT, Richard S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell. 2003;14:274–287. doi: 10.1091/mbc.E02-08-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li ZZ, Kondo T, Murata T, et al. Expression of Hqk encoding a KH RNA binding protein is altered in human glioma. Jpn J. Cancer Res. 2002;93:167–177. doi: 10.1111/j.1349-7006.2002.tb01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lindholm E, Ekholm B, Shaw S, et al. A schizophrenia-susceptibility locus at 6q25, in one of the world’s largest reported pedigrees. Am. J. Hum. Genet. 2001;69:96–105. doi: 10.1086/321288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tastemir D, Demirhan O, Sertdemir Y. Chromosomal fragile site expression in Turkish psychiatric patients. Psychiatry Res. 2006;144:197–203. doi: 10.1016/j.psychres.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 88.McCullumsmith RE, Gupta D, Beneyto M, et al. Expression of transcripts for myelination-related genes in the anterior cingulate cortex in schizophrenia. Schizophr. Res. 2007;90:15–27. doi: 10.1016/j.schres.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 91.Katsel P, Davis KL, Haroutunian V. Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: a gene ontology study. Schizophr. Res. 2005;79:157–173. doi: 10.1016/j.schres.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 92.Hafner H. Onset and course of the first schizophrenic episode. Kaohsiung J. Med. Sci. 1998;14:413–431. [PubMed] [Google Scholar]
- 93.Steen RG, Mull C, McClure R, Hamer RM, Lieberman JA. Brain volume in first-episode schizophrenia: systematic review and meta-analysis of magnetic resonance imaging studies. Br. J. Psychiatry. 2006;188:510–518. doi: 10.1192/bjp.188.6.510. [DOI] [PubMed] [Google Scholar]
- 94.Steinman L, Martin R, Bernard C, Conlon P, Oksenberg JR. Multiple sclerosis: deeper understanding of its pathogenesis reveals new targets for therapy. Annu. Rev. Neurosci. 2002;25:491–505. doi: 10.1146/annurev.neuro.25.112701.142913. [DOI] [PubMed] [Google Scholar]
- 95.Howe CL. Immunological aspects of axon injury in multiple sclerosis. Curr. Top. Microbiol. Immunol. 2008;318:93–131. doi: 10.1007/978-3-540-73677-6_5. [DOI] [PubMed] [Google Scholar]
- 96.Vanderlocht J, Hellings N, Hendriks JJ, Stinissen P. Current trends in multiple sclerosis research: an update on pathogenic concepts. Acta Neurol. Belg. 2006;106:180–190. [PubMed] [Google Scholar]
- 97.Seiwa C, Yamamoto M, Tanaka K, et al. Restoration of FcRγ/Fyn signaling repairs central nervous system demyelination. J. Neurosci. Res. 2007;85:954–966. doi: 10.1002/jnr.21196. [DOI] [PubMed] [Google Scholar]
- 98.Ichimura K, Mungall AJ, Fiegler H, et al. Small regions of overlapping deletions on 6q26 in human astrocytic tumours identified using chromosome 6 tile path array-CGH. Oncogene. 2006;25:1261–1271. doi: 10.1038/sj.onc.1209156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sukumar S, Wang S, Hoang K, et al. Subtle overlapping deletions in the terminal region of chromosome 6q24.2-q26: three cases studied using FISH. Am. J. Med. Genet. 1999;87:17–22. [PubMed] [Google Scholar]
- 100.Lim J, Hao T, Shaw C, et al. A protein- protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]