Abstract
Genetic variants of cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase (VKORC1) are known to influence warfarin dose, but the effect of other genes has not been fully elucidated. We genotyped 183 polymorphisms in 29 candidate genes in 1496 Swedish patients starting warfarin treatment, and tested for association with response. CYP2C9*2 and *3 explained 12% (P = 6.63 × 10−34) of the variation in warfarin dose, while a single VKORC1 SNP explained 30% (P = 9.82 × 10−100). No SNP outside the CYP2C gene cluster and VKORC1 regions was significantly associated with dose after correction for multiple testing. During initiation of therapy, homozygosity for CYP2C9 and VKORC1 variant alleles increased the risk of over-anticoagulation, hazard ratios 21.84 (95% CI 9.46; 50.42) and 4.56 (95% CI 2.85; 7.30), respectively. One of 8 patients with CYP2C9*3/*3 (12.5%) experienced severe bleeding during the first month compared with 0.27% of other patients (P = .066). A multiple regression model using the predictors CYP2C9, VKORC1, age, sex, and druginteractions explained 59% of the variance in warfarin dose, and 53% in an independent sample of 181 Swedish individuals. In conclusion, CYP2C9 and VKORC1 significantly influenced warfarin dose and predicted individuals predisposed to unstable anticoagulation. Our results strongly support that initiation of warfarin guided by pharmacogenetics would improve clinical outcome.
Introduction
Warfarin is an oral anticoagulant widely used for the prevention of thrombotic events and to treat a confirmed episode of venous thrombosis. Effectiveness and safety of warfarin therapy are routinely monitored by the international normalized ratio (INR), which is the ratio of the time required for a patient's blood to coagulate relative to that of a reference sample. Although highly effective, warfarin's usability is limited by a narrow therapeutic range combined with a pronounced interindividual variability in the dose required for adequate anticoagulation.1 Clinical use of warfarin is further complicated by a substantial risk for hemorrhagic side effects, which is increased in patients with low-dose requirements.2 A minor part of the variability in dose requirement has been ascribed to effects of demographic and clinical factors such as age, drug interactions, concomitant diseases, and vitamin K intake,3 while a major part is ascribed to genetic factors.4
The first gene documented to influence warfarin dose requirement was CYP2C9.5 It encodes the enzyme cytochrome P450 2C9 that mediates the metabolism of the enantiomer S-warfarin, which is more potent than R-warfarin.6 CYP2C9 is situated in the CYP2C cluster, a region of high linkage disequilibrium (LD).7 The most common CYP2C9 variant alleles in whites are *2 and *3. The clearance of S-warfarin is reduced in individuals homozygous for CYP2C9*2 (rs1799853, R144C),5 and is still lower in those homozygous for CYP2C9*3 (rs1057910, I359L).8 Recent studies have focused on warfarin's target of action, vitamin K epoxide reductase,9 which is encoded by VKORC1.10,11 Several common noncoding VKORC1 polymorphisms have been studied, for example, rs9923231 that is located upstream of VKORC1 (−1639 G>A) and associated with low expression, and the intronic rs2359612 (2255 C>T).12,13 In different populations, CYP2C9 explains on average 12% (range, 4%-20%) of the variability in dose requirements, while VKORC1 explains 27% (range, 15%-34%).14–19 There is relatively little information about the effect of CYP2C9 and VKORC1 during initiation of warfarin,20 but the risk of overanticoagulation and bleeding has been associated with CYP2C9 variants in a few studies.2,21–25 An interim analysis of the 219 first patients in the Swedish Warfarin Genetics (WARG) cohort showed that CYP2C9*2 and *3 alleles increased the risk of supratherapeutic INR (> 3) during the first week of treatment, RR = 2.8 (95% CI 1.2; 6.7) and RR = 6.1 (95% CI 2.7; 13.6), respectively.26 The overall incidence of serious bleeding in the WARG cohort was 2.6 per 100 patient years (95% CI 1.7; 3.5), and the risk was increased in patients with potentially interacting medication and in males.27
Several genes in the warfarin interactive pathways apart from CYP2C9 and VKORC1 have been tested for association with interindividual differences in warfarin dose requirement. In our independent pilot study, 201 Swedish patients were genotyped for single nucleotide polymorphisms (SNPs) in 29 candidate genes (Figure S1; available on the Blood website; see the Supplemental Materials link at the top of the online article).28 SNPs in the genes CYP2C18, CYP2C19, PROC, APOE, EPHX1, CALU, GGCX, and ORM1 were nominally associated (P < .05) with warfarin dose, and so were haplotypes of CYP2C19, PROC, GGCX, ORM1-2, PROZ, F7, F9, and NR112.28 These genes have previously rendered inconsistent results in a number of studies that were underpowered to accurately detect genes with a modest effect.4 In the pilot cohort, APOE *E4 (rs429358) was associated with higher warfarin dose requirements,29 the opposite was shown in a British sample,30 and no effect was seen in Italians.31 A GGCX SNP (rs699664) was associated with lower dose in the Japanese, but not in the Israeli or European Americans.16,32,33 In 3 studies warfarin dose increased with the number of microsatellite repeats in GGCX,34–36 while a fourth found the opposite.33 No firm conclusions can be drawn from these small studies.
This study aimed to produce reliable results from 29 candidate genes by genotyping functional SNPs and tagSNPs in the hitherto largest prospectively collected warfarin cohort.27 We also tested for association with response to warfarin during initiation of warfarin, and generated a dosing algorithm for warfarin using demographic, clinical and genetic factors.
Methods
Patients
The WARG study (http://www.druggene.org/) is a prospective multicenter study of warfarin bleeding complications and predictors of response to warfarin.37 Between 2001 and 2005, patients starting warfarin (Waran; Nycomed, Stockholm, Sweden) were collected at 40 outpatient clinics distributed throughout Sweden. The majority of these centers were specialized at anticoagulation, the remainder were primary health care centers. The patients were subject to treatment according to standard care at each center, without specific warfarin dosing algorithms. Other analyses in this patient cohort have been presented in 2 previous publications.26,27
Study size was calculated based on the expected number of bleeding events (see “Statistical analysis”). In total, 1523 first-time warfarin users aged 18 to 92 years were recruited. The majority were of Swedish origin, but ethnicity was not registered. Patients were recruited regardless of indication for treatment, and apart from established contraindications for warfarin, the only exclusion criteria were previous exposure to warfarin and age younger than 18 years. There were no restrictions regarding target INR, planned treatment duration or comorbidities. The study was approved by Karolinska Institutet Ethics Committee and regional ethics committees concerned; informed consent was obtained in accordance with the Declaration of Helsinki.
One hundred eighty-one previously genotyped individuals from the pilot cohort28 were used to validate the algorithm derived from the WARG cohort. The pilot cohort was not part of the WARG cohort.
Data collection
Patients were monitored from the start of warfarin treatment until warfarin was withdrawn or until the data collection phase was terminated on August 12, 2005, whichever came first. To facilitate data collection and real-time online monitoring of the study, an internet-based data collection system was developed, as described by Lindh et al.37 The time between visits was not standardized. Throughout the study, complete data on warfarin doses and INRs were registered by the participating centers. Data on age, sex, and indication for treatment were collected, but not on body weight and height. All concomitant medications were entered, and medications potentially interacting with warfarin were identified (Table S1).
Deaths and bleeding events occurring during the study period were reported. A bleeding event was assessed as serious if it fulfilled the World Health Organization (WHO) criteria for a serious adverse drug reaction, that is, if it was lethal, life-threatening, permanently disabling, or lead to hospital admission or prolongation of hospital stay.38 The hospital study database was matched with the national hospital discharge registry to identify any overlooked cases of serious bleeding.27
Main outcome measures
Primary outcome measures were INR and warfarin dose (mg/week). Target INR was estimated from the individual target INR range using linear interpolation. Therapeutic INR was defined as INR between 2 and 3 irrespective of the individual target INR range. A minimum of 3 consecutive therapeutic INR measurements denoted stable anticoagulation. Maintenance dose was defined as the mean of all doses given to a patient during stable anticoagulation. Stable maintenance dose was calculated from all weekly doses that were unchanged over a minimum of 3 consecutive visits during therapeutic INR. Secondary outcome measures were time to the first INR in a series of 3 therapeutic INRs (stable anticoagulation), fraction of the study time spent within INR range 2 to 3 (TIR), overanticoagulation (INR > 4.0), and the incidence of bleeding.
Genotyping
Patients were genotyped retrospectively using DNA samples collected at the start of therapy. All genotyping was performed at the Wellcome Trust Sanger Institute, Hinxton, United Kingdom.
DNA was extracted from whole blood using the MagnaPure LC method (MagnaPure DNA Isolation Kit-Large Volume; Roche Diagnostics, Mannheim, Germany). An LD map of the Swedish population was constructed and tagSNPs were determined by Tagger; r2 = 0.8 of pair-wise tagging.39 Functional polymorphisms from the literature were added. SNP assays were designed with the AssayDESIGNER v3.0.
All SNPs but 2 were genotyped with Homogeneous Mass Extend and iPLEX assays (Sequenom, Hamburg, Germany). Primer sequences are available on request. MassEXTEND PCR amplification was performed as described elsewhere.28,40 For iPLEX assays (multiplexed up to 29 SNPs per well), 8 ng genomic DNA in 2 μL deionized water, 1 μL 500 nM primer mix, 2 μL PCR mix including 0.1 μL QIAGEN Hotstar Taq polymerase, 0.1 μL 25 mM dNTP, 0.325 μL 25 mM MgSO4, 0.625 μL QIAGEN PCR buffer, and 0.85 μL deionized water were mixed. The 384-well PCR plates were heated to 94°C for 15 minutes, followed by 45 cycles of 94°C for 20 seconds, 56°C for 30 seconds, 72°C for 1 minute, and a final step at 72°C for 3 minutes. For the iPLEX mass extension, 2 μL extension mix was added to the 384-well plate, which included 0.041 μL iPLEX enzyme, 0.2 μL 10X iPLEX buffer, 0.2 μL iPLEX termination mix, 0.559 μL deionized water, and 1 μL primer mix of 5.5 μM for low mass extension primers, and 1.1 μM for extension primers with higher mass. PCR conditions were 94°C for 30 seconds, followed by 45 cycles of 94°C for 5 seconds, 52°C for 5 seconds, 80°C for 5 seconds, and a final step of 72°C for 3 minutes. Sample clean-up of Mass EXTEND/iPLEX assays and MALDI-TOF mass spectrometry analyses were performed as described elsewhere.40
Data quality was assessed by 16 duplicates (8 DNAs) and 16 negative controls (water) per 384-well plate. SNPs with more than one discrepant call or showing self priming in the negative control were removed. Finally, we removed nonpolymorphic SNPs, SNPs with call rate below 70%, and markers that departed from the Hardy Weinberg equilibrium (HWE P < .001).
Genotyping of 2 functional CYP2C9 polymorphisms was performed using the ABI PRISM 7500 Sequence Detection System and TaqMan Pre-Developed Assay Reagent kits for Allelic Discrimination (Applied Biosystems, CA). The part numbers are 4312559 for rs1799853 (*2) and 4312560 for rs1057910 (*3).
Statistical analysis
The WARG study was powered to detect an effect of CYP2C9 variant alleles on the risk of bleeding. Assuming that bleeding occurs in 10% of all treated patients and that the prevalence of CYP2C9 variant alleles is 36%, it was calculated that 1470 patients (allowing for a dropout rate of 10%) would give the study a power of 0.8 at a significance level of 0.05 (2-sided) to detect a doubled risk of bleeding in carriers of these alleles.
R version 2.5.1 (R foundation for statistical computing, Vienna, Austria) and SAS version 9.1.3 (SAS Institute, Cary, NC) were used for statistical analyses. Univariate and multivariable analyses of predictor impact on the square root of warfarin dose and time in range were calculated using linear regression analyses. To account for partial dependence among tests of SNPs in LD, we applied a Bonferroni correction for multiple testing based on the effective number of independent tests (Meff) calculated by a spectral decomposition method.41–43 Meff was 172 in our study and hence a P value of less than .001 was required for significance. To avoid losing power to detect true signals because of loss of data, comparisons were focused on maintenance dose rather than on stable maintenance dose.
LD was visualized using the HaploView software.44 The QTPHASE component of UNPHASED software was used to test for association of haplotypes with warfarin dose.45 Pair-wise LD was quantified by the standard r2 measure.46 The coefficient of determination, R2, was used to measure the proportion of explained variance. Association with time to stable INR, overanticoagulation, and bleeding were evaluated with the log- rank test. Hazard ratios were estimated with Cox regression analyses. Risk of bleeding was compared with the Fisher exact test and the Pearson test. The prediction models were based on verified findings and only significant variables (P < .05) were allowed in the final model. Performance of the final prediction model was evaluated with cross validation to achieve unbiased estimates. The training data set was randomly selected as 70% of the data and the procedure was repeated 10 000 times. The median and 2.5/97.5 percentiles of the resulting distribution of the R2 values were calculated. The final prediction model was validated in a separate cohort.
Results
Baseline characteristics
Between December 2001 and August 2005, 1542 warfarin naive patients were included at 40 centers. Sixteen patients were subsequently excluded because of failure to meet the inclusion criteria and 3 because of withdrawal of consent. In total, 1523 patients were included in the WARG study. Genotyping results were obtained from 1496 patients aged 18 to 92 years. In 1457 patients (97.4%), target INR was between 2.4 and 2.6. Fourteen patients (0.9%) had a target INR below 2.4. Thirty-four patients (2.3%) had a prosthetic heart valve, and 25 of them (1.7%) had a target above 2.6. The most common comorbidities were hypertension, diabetes, malignancy, cardiac failure, and previous stroke. Baseline characteristics of the genotyped patients are presented in Table 1.
Table 1.
Baseline characteristics of the replication cohort (WARG)
| Total number of genotyped patients | 1496 |
|---|---|
| Age, y | 66 (57;74) |
| Sex, male/female | 947 (63%)/549 (37%) |
| Indication for warfarin treatment | |
| Atrial fibrillation | 762 (51%) |
| Deep venous thrombosis | 378 (25%) |
| Pulmonary embolism | 188 (13%) |
| Heart valve transplant, artificial | 34 (2.3%) |
| Cerebral infarction/transient ischemic attack | 28 (1.9%) |
| Heart valve transplant, biologic | 25 (1.7%) |
| Cardiomyopathy/cardiac failure | 23 (1.5%) |
| Other indications* | 58 (3.9%) |
| Planned treatment duration | |
| Less than 3 mo | 13 (0.9%) |
| 3-5 mo | 214 (14%) |
| 6-12 mo | 387 (26%) |
| More than 12 mo, specified | 14 (0.9%) |
| Infinite or not predefined | 868 (58%) |
| Warfarin induction, mg | |
| First dose | 10 (1.25;15) |
| Second dose | 7.5 (1.25;15) |
| Third dose | 5.0 (1.25;10) |
| INR, international normalized ratio | |
| Lower limit of therapeutic interval | 2.1 (1.5;3.1) |
| Target value | 2.5 (1.8;3.0) |
| Upper limit of therapeutic interval | 3.0 (2.0;3.6) |
| Percent of time within therapeutic INR interval | 63.8 (50.9;73.5) |
| Patients experiencing bleeding events | |
| Serious bleeding | 28 (1.9%) |
| All bleeding | 146 (9.8%) |
| Comedication, number of drugs | 2 (0;18) |
| Patients using drugs that interact with warfarin† | |
| No interacting drug | 781 (52%) |
| Drugs decreasing warfarin effect | 56 (3.7%) |
| Drugs potentiating warfarin effect (reflected in INR) | 691 (46%) |
| Genotypes of VKORC1‡ and CYP2C9§ | |
| VKORC1 rs9923231 (−1639) G/G | 529 (36%) |
| VKORC1 rs9923231 (−1639) A/G | 714 (49%) |
| VKORC1 rs9923231 (−1639) A/A | 218 (15%) |
| CYP2C9*1/*1 | 988 (66%) |
| CYP2C9*1/*2 | 277 (19%) |
| CYP2C9*1/*3 | 177 (12%) |
| CYP2C9*2/*2 | 21 (1.4%) |
| CYP2C9*2/*3 | 19 (1.3%) |
| CYP2C9*3/*3 | 8 (0.5%) |
Age and time within therapeutic INR interval are presented as median (interquartile range). Other parameters are presented as median (range) or number of patients (percent).
Includes carotid dissection and stenosis, unspecified cardiac embolism, myocardial infarction, peripheral artery disease, sinus thrombosis, and cardiac arrhythmias other than atrial fibrillation.
Drugs that interact without affecting INR are not presented here. Some patients have drugs that both increase and decrease INR.
VKORC1 rs9923231 genotypes are missing in 35 patients.
CYP2C9 genotyping results from *2 and *3 alleles are missing in 6 patients.
Warfarin was given once daily, usually in the evening. A wide variety of warfarin induction strategies were used (Table 1). The most common 3 day combination was 10 mg – 7.5 mg – 5 mg (average 9.7 mg – 7.2 mg – 5.5 mg). Specifically, 48% received 10 mg the first day, 29% received 10 mg – 7.5 mg the first 2 days and 13% were prescribed exactly 10 mg – 7.5 mg – 5 mg the first 3 days. Other first-day doses were 7.5 mg (given to 15% of the patients), 12.5 mg (13%), 15 mg (11%), 5 mg (8%) and less than 5 mg (5%). Dose adjustments were according to local routines.
INR was checked in all patients before starting warfarin, and in 14% INR was also checked the day after the first dose was given. INR was measured in 38% of the patients after 2 doses, in 52% after 3 doses, in 33% after 4 doses, in 30% after 5 doses, in 36% after 6 doses, and in 38% after 7 doses. INR was usually measured twice during the second week, weekly during weeks 3 through 5 and less frequently once stable anticoagulation dose was achieved. On average, each patient measured INR 25.6 times per treatment year, and the overall mean time between visits was 12.75 days. Most patients were monitored for at least 3 months. The median follow-up time was 6 months, and the aggregated follow-up time 1214 warfarin-exposed patient-years.
During the whole observation period 1324 patients (88%) reached stable anticoagulation defined as INR 2 to 3 over 3 consecutive visits. Their mean maintenance doses while on stable anticoagulation varied between 6.0 and 130 mg per week. Only 850 patients (57%) reached stable maintenance dose defined as stable anticoagulation and unchanged dose over 3 consecutive visits. Patients were on 100 other drugs that potentially interact with warfarin. A complete list of these drugs is presented in Table S1.
Univariate association with warfarin dose
We successfully genotyped 183 selected functional and tagSNPs in 29 candidate genes across 1496 patients. Table S2 shows the percent of maintenance dose variance (R2) explained by univariate analysis of all SNPs, and if available corresponding results from the pilot study.28
The strongest association with dose was observed for VKORC1 (Table 2, Figure S2A). Two nearly perfectly concordant VKORC1 SNPs, rs2359612 and rs9923231, each explained 29.8% (P = 9.82 × 10−100) and 29.3% (1.03 × 10−97) of the variance in maintenance dose. In the subset of patients with stable maintenance dose, VKORC1 rs2359612 explained 33.3% (P = 5.78 × 10−73) and rs9923231 explained 32.8% (P = 3.97 × 10−72) of the variance. Univariate association with warfarin dose was nearly identical for rs2359612 and rs9923231, whose genotypes are almost completely concordant with each other. Henceforth VKORC1 results will focus on the possibly functional rs9923231.12,13 Due to the high LD, haplotypes did not explain more of the variance in dose than the individual VKORC1 polymorphisms rs2359612 or rs9923231 did.
Table 2.
Replication of factors that showed a strong relationship with dose in the pilot cohort28
| Genetic variant or other factor | Pilot cohort |
Replication cohort |
|||||
|---|---|---|---|---|---|---|---|
| N | R2 | P | N | R2 | P | ||
| VKORC1 | rs9923231 | 181 | 31.7% | 1.91 × 10−15 | 1293 | 29.3% | 1.03 × 10−97 |
| VKORC1 | rs2359612 | 200 | 29.0% | 2.30 × 10−15 | 1292 | 29.8% | 9.82 × 10−100 |
| CYP2C9 | *2 and *3 | 201 | 15.9% | 2.29 × 10−6 | 1321 | 11.8% | 6.63 × 10−34 |
| CYP2C9 | rs4917639 | 197 | 11.8% | 4.94 × 10−6 | 1092 | 11.7% | 4.61 × 10−30 |
| PROC | rs2069919 | 182 | 9.0% | 2.16 × 10−4 | 1294 | 0.2% | 2.07 × 10−1 |
| EPHX1 | rs4653436 | 196 | 4.8% | 8.48 × 10−3 | 1207 | 0.1% | 4.25 × 10−1 |
| GGCX* | rs12714145/rs7568458 | 198 | 3.4% | 3.32 × 10−2 | 1252 | 0.1% | 4.97 × 10−1 |
| ORM1 | rs1687390 | 149 | 2.6% | 4.96 × 10−2 | 1310 | 0% | 9.14 × 10−1 |
| Age | 201 | 9.2% | 1.20 × 10−5 | 1324 | 14.5% | 8.00 × 10−47 | |
| Sex | 201 | 2.6% | 2.29 × 10−2 | 1324 | 1.2% | 6.89 × 10−5 | |
| Interaction (increased INR) | 201 | 3.6% | 6.67 × 10−2 | 1324 | 1.5% | 8.00 × 10−6 | |
Univariate R2 values (percentage of dose variance explained) and P values from the pilot cohort and the replication (WARG) cohort are shown. In the replication cohort, 1324 patients with a minimum of 3 consecutive therapeutic INRs are taken into account. All genotyping results are used in the pilot cohort, which is separate from the replication cohort. N represents the number of patients with data for the factor in each study.
In the replication study GGCX rs7568458 was genotyped instead of rs12714145. They are in high LD.
The second most significant gene was CYP2C9 (Table 2; Figure S2B). In univariate analysis, CYP2C9*2 explained 4.1% of the variance in maintenance dose (P = 1.34 × 10−12), while CYP2C9*3 explained 6.3% (P = 1.82 × 10−19). The alleles *2 and *3 together explained 11.8% (P = 6.63 × 10−34) of the variance in maintenance dose (Table 2). The intronic CYP2C9 SNP rs4917639 was associated with a composite minor allele formed by aggregating *2 and *3 (r2 = 0.836),28 and explained 11.7% of the variance (P = 4.61 × 10−30). In the subset of patients with stable maintenance dose, *2 and *3 together explained 12.3% of the variance (P = 3.67 × 10−22), while rs4917639 explained 12.1% (P = 1.99 × 10−20). Haplotypes did not explain more of the variance in dose than the combination of CYP2C9 *2 and *3 did.
The CYP2C cluster genes CYP2C8 and CYP2C19 contained SNPs with association P values below the significance threshold (P < .001), but these associations were almost entirely attributed to LD with CYP2C9 (see “Multiple models of dose” and Table S2). Polymorphisms in CYP1A1, CYP2C18, CYP3A4, EPHX1, F10, ORM1, and PROS1 were nominally significant (P < .05), but failed correction for multiple testing. We searched for haplotypes outside VKORC1 and the CYP2C gene cluster that might influence warfarin dose, but none of the tested haplotypes exhibited association below Bonferroni Meff correction (P < .001).
Age, sex, and drugs that interact with warfarin by increasing INR were all associated with maintenance dose (Table 2). Thirty-four patients (2.3%) in WARG had prosthetic heart valves, but only 11 had a target INR of 3. There were too few prosthetic heart valve patients with a high target INR and stable anticoagulation to detect a significant effect of indication.
Multiple models of dose
Given the lack of evidence for additional dose prediction by other genetic variants or haplotypes, our multiple models focused on the substantial genetic prediction provided by single polymorphisms in VKORC1 and CYP2C9. Multiple regression models of each SNP together with VKORC1 rs9923231 plus CYP2C9*2 and *3 revealed one significant CYP2C SNP that gave evidence of being independent. This CYP2C19 SNP (rs3814637; P = 2.2 × 10−6) predicted 0.7% of the variance in dose after accounting for rs9923231, *2 and *3.
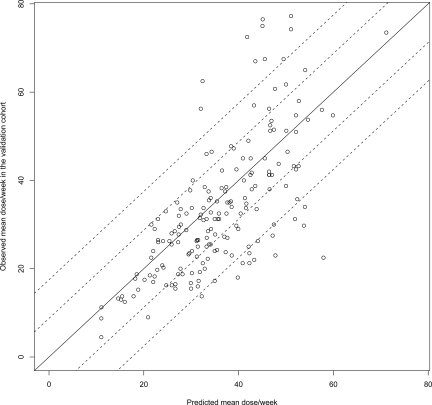
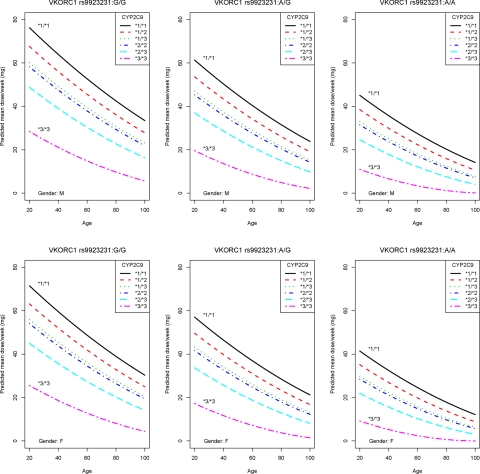
A multiple regression model using the predictors VKORC1 rs9923231, CYP2C9*2 and *3, age, sex, and drug interactions explained 58.7% of the variance in warfarin dose (Table 3). Cross- validation in the same cohort gave a median R2 of 58.1% (95% CI 52.4%-63.4%) from 10 000 runs. When validated in 181 genotyped individuals from the pilot cohort, the model predicted 52.8% of the variance in dose (Figure 1). The predicted weekly dose was calculated by the algorithm [9.46 832 − (0.90 112 for rs9923231 A/G) – (2.01 863 for rs9923231 A/A) – (0.50 836 for *1/*2) – (0.97 546 for *1/*3) – (1.10 204 for *2/*2) – (1.74 761 for *2/*3) – (3.40 061 for *3/*3) – (0.03 686 × age) – (0.27 698 for females) – (0.06 992 × number of drugs that increase INR)]2. The algorithm is illustrated by nomograms (Figure 2) that show predicted dose for different genotypes, sexes, and ages in patients without interacting drugs.
Table 3.
The WARG warfarin dose algorithm
| Coefficients | Estimate | Standard error | P |
|---|---|---|---|
| Intercept | 9.46832 | 0.11867 | < 2 × 10−16 |
| VKORC1 rs9923231 G/G | 0 | ||
| VKORC1 rs9923231 A/G | −0.90112 | 0.04959 | < 2 × 10−16 |
| rs9923231 A/A | −2.01863 | 0.06799 | < 2 × 10−16 |
| CYP2C9*1/*1 | 0 | ||
| *1/*2 | −0.50836 | 0.05811 | < 2 × 10−16 |
| *1/*3 | −0.97546 | 0.07077 | < 2 × 10−16 |
| *2/*2 | −1.10204 | 0.19767 | 3.0 × 10−8 |
| *2/*3 | −1.74761 | 0.20391 | < 2 × 10−16 |
| *3/*3 | −3.40061 | 0.33091 | < 2 × 10−16 |
| Age, y | −0.03686 | 0.00172 | < 2 × 10−16 |
| Male sex* | 0 | ||
| Female sex* | −0.27698 | 0.04682 | 4.2 × 10−9 |
| Interaction × number of drugs that increase INR† | −0.06992 | 0.01867 | .001 |
VKORC1 G/G and CYP2C9*1/*1 were used as the reference genotypes in the algorithm. The model predicts the square root of dose; hence dose (mg/week) is obtained by squaring the result. Example: A 50-year-old male with no interacting drugs is genotyped as VKORC1 A/G and CYP2C9*1/*3. The resulting square root of weekly dose is: 9.46832 – (0.90112 for A/G) – (0.97546 for *1/*3) – (0.03686 × 50 y) = 5.74874. To obtain his predicted weekly dose, transform by squaring the result: 5.748742 ≈ 33 mg/week.
Sex is a substitute for body weight and height, which are not available in the WARG cohort.
Take the number of interacting drugs that potentially increase INR and multiply by −0.06992. Drugs that decrease the effect of warfarin also have a large impact on the individual, but are not included in the algorithm, since too few were present in the cohort to get reliable results.
Figure 1.
Validation of the multiple regression model. The solid line illustrates perfect prediction in the pilot cohort and the dashed reference lines show deviation from the prediction of plus or minus 1.25 mg (½, pill) and plus or minus 2.5 mg (1 pill) per day. Predictors: VKORC1 rs9923231, CYP2C9 *2 and *3, age, sex, and drug interactions. rs9923231 is used in this algorithm since it is possibly functional.
Figure 2.
The dose algorithm illustrated by nomograms. The nomograms show predicted maintenance dose for VKORC1 and CYP2C9 genotypes, sexes, and ages with the number of interacting drugs increasing warfarin effect set as 0, hence they do not account for drug interactions (Table 3).
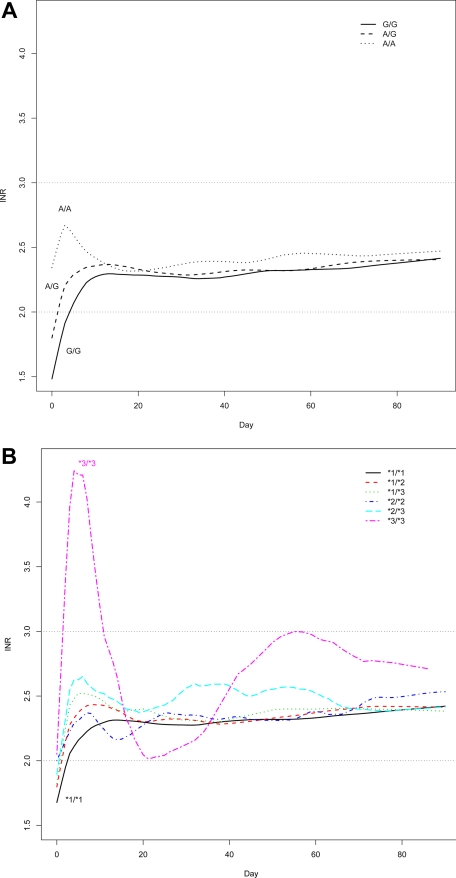
Time to stable anticoagulation
Carriers of VKORC1 variant alleles reached the first INR above 2 more rapidly than others, as illustrated by Figure 3A. Anticoagulation appeared more unstable in the 8 individuals homozygous for CYP2C9*3, which is visualized in Figure 3B. However, none of the 29 genes significantly affected time to stable anticoagulation defined as the first therapeutic INR in a row of at least 3 INRs between 2 and 3. The median time to stable anticoagulation was 16 days in the 1324 patients who reached it.
Figure 3.
Time to stable anticoagulation. (A) Lowess smoothed plot of INR values over time as a function of VKORC1 rs9923231 (−1639 G>A) genotype. (B) Lowess smoothed plot of INR values over time as a function of CYP2C9 *2 and *3 genotype.
Time in range
During the first 3 months, the median time in range, 2 to 3 (TIR), was 67% (average, 65%). VKORC1 rs9923231 G>A had a nominal effect on TIR during the first 3 months (P = .003, and for rs2359612 C>T P = .002). Average TIR was the highest (70%) in A/A individuals (in LD with rs2359612 T/T) and the lowest (64%) in G/G individuals (in LD with rs2359612 C/C). CYP2C9*2 and *3 also nominally affected this outcome (P = .041). TIR was the lowest in *3/*3 individuals (average, 53%), and relatively high in *2/*3 heterozygotes (average 67% to 72%). After correction for multiple testing (P < .001), no candidate gene was significantly associated with TIR during the first 3 months.
The median time in range 2 to 3 during the entire treatment period was 64% (average, 61%). When the 39 patients with a target INR outside 2.4 to 2.6 were removed, the overall average TIR increased by 0.1%. VKORC1 rs9923231 was significantly associated with overall time in range (P = 1.68 × 10−4, and for rs2359612 P = 1.47 × 10−4). As above, average TIR was the highest (65%) in A/A individuals (rs2359612 T/T) and the lowest (60%) in rs9923231 G/G individuals (rs2359612 C/C). NR1I3 SNP rs3003596 was almost significantly associated with overall TIR after correction for multiple testing (P = .001). CYP2C9 was not associated with overall TIR.
Overanticoagulation
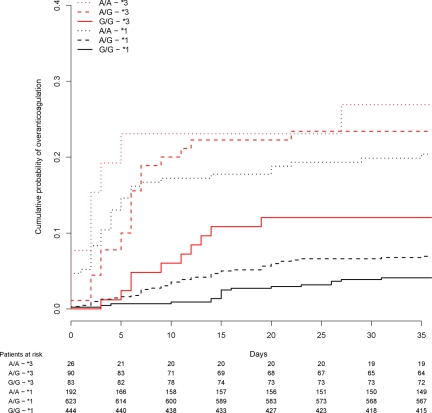
CYP2C9*3 was strongly associated with supra-therapeutic anticoagulation (INR peaks more than 4) during the first 5 weeks of treatment (P < 1.11 × 10−16). The risk was especially pronounced in CYP2C9*3 homozygotes, hazard ratio 21.84 (95% CI 9.46;50.42, P = 5.1 × 10−13). VKORC1 rs9923231 was also associated with INR > 4 within 5 weeks (P = 6.56 × 10−12, and forrs2359612 P = 4.19 × 10−12). Patients carrying rs9923231 A/A had an increased risk of INR > 4 within 5 weeks, hazard ratio 4.56 (95% CI 2.85;7.30, P = 2.4 × 10−10). Twenty-seven percent of patients with at least one CYP2C9*3 allele and VKORC1 rs9923231 A/A had an INR peak higher than 4 during the first 5 weeks of therapy (Figure 4). In comparison, only 6% of patients with no *3 alleles and rs9923231 G/G had INR higher than 4 during the first 5 weeks.
Figure 4.
Survival (Kaplan-Meier) curve showing time to first INR peak greater than 4. VKORC1 rs9923231 (−1639 G>A) and CYP2C9*3 are presented. *3 indicates a carrier of at least 1 CYP2C9*3 allele, while *1 in this case means that *3 is not present. The table beneath the graph shows the number of individuals that had not reached INR > 4 at certain time points.
Bleeding during warfarin treatment occurred in 146 patients (9.8%) and serious bleeding in 28 (1.9%) patients (Table 1). There were tentative, but nonsignificant, differences between CYP2C9 genotypes in the incidence of bleeding and serious bleeding within 5 weeks (both Fisher exact test P = .066). One of 8 patients (12.5%) with CYP2C9*3/*3 experienced a serious bleeding event during the first 5 weeks, compared with 0.27% of patients with other genotypes. There were no significant differences between CYP2C9 genotypes concerning the incidence of bleeding or serious bleeding when the whole observation period was taken into account (Fisher exact test P = .323 and P = .079, respectively). There were no differences between VKORC1 genotypes concerning the incidence of bleeding or serious bleeding within 5 weeks (both Pearson test P = .639). Nor were there any differences between VKORC1 genotypes in the incidence of bleeding or serious bleeding during the whole observation period (Pearson test P = .535 and P = .640, respectively).
Discussion
Anticoagulant response to warfarin therapy is largely influenced by demographic, clinical, and genetic factors.4,47 At least 2 genes are closely associated with warfarin dose requirements: warfarin's target gene, VKORC1, and the gene of its main metabolising enzyme, CYP2C9.12–20 A number of algorithms for warfarin maintenance dose prediction using these 2 genes together with demographic and clinical factors have been reported.48 In 2007, the US Food and Drug Administration (FDA) Center for Drug Evaluation and Research updated the label of warfarin to include information on genetic testing.47 The FDA encourages personalized dosing in patients with VKORC1 and CYP2C9 variant alleles, and randomized clinical trials of pharmacogenetic dosing are now in progress in Europe and the United States. However, 3 important issues need to be considered before starting clinical trials of personalized warfarin therapy: (1) whether other genes influence warfarin dose, (2) the optimal dosing algorithm for warfarin maintenance dose, and (3) how to calculate loading dose from predicted maintenance dose.
The Swedish WARG study is the first comprehensive study with a sample size adequately powered to detect smaller effects among warfarin candidate genes. To preserve statistical power, analyses were made using observations of all doses during stable anticoagulation, rather than only unchanged doses over a minimum of 3 consecutive visits. A disadvantage of using all doses is that temporary dose adjustments caused by external factors such as acute illness and change in comedications are included. On the other hand, observations from many more patients can be used, making this definition more reliable. In fact, the 2 different dose definitions gave almost identical statistical results, but P values were less significant when fewer observations were used. Our findings, using both definitions of dose, underscore the importance of VKORC1 and CYP2C9 as the primary genetic factors for warfarin dose prediction. The 2 best predictors of dose, VKORC1 rs9923231 and rs2359612, were in very high LD (R2 > 0.99) in our population. From a statistical perspective either, or possibly both, of these SNPs could be causative. According to Rieder et al, minor alleles of rs9923231 and rs2359612 are present in the low-dose haplotype A, while wild-type alleles characterize the high-dose haplotype B.12 Separating the influence of the 2 SNPs is difficult in European populations where they are in almost complete concordance. However, their respective influence may be more readily separated in other populations (eg, African Americans) with a more diverse distribution of VKORC1 haplotypes.12 Other significant associations we found in the CYP2C cluster were almost entirely attributed to LD with CYP2C9, since the implicated SNPs were either nonsignificant or contributed less than 1% of dose variance (CYP2C19 rs3814637) in multiple regression models that contained the *2 and *3 alleles. Several genes that showed a small effect on warfarin dose in our pilot study28 were not replicated in the WARG cohort. This demonstrates the need of large samples to identify true associations with genetic variants having a small effect in candidate gene studies and especially in whole genome association studies of warfarin pharmacogenetics.
Contrary to most published pharmacogenetic warfarin studies, our patients were followed prospectively from the start of therapy and were drawn from a diverse and dispersed set of anticoagulation clinics that represent a broad, real-world collection of contributors. Limitations of the WARG study are that only one ethnic group is represented and that data on bodyweight and height were not collected. A multiple regression model using the predictors VKORC1, CYP2C9, age, sex, and drug interactions explained 59% of the variance in warfarin maintenance dose in the WARG patients and predicted 53% when tested in an independent sample. Both this and other available warfarin algorithms48 rely on SNPs that increase sensitivity to warfarin and lack SNPs that cause resistance. Although rare genetic variants that cause warfarin resistance have been found, common resistance SNPs that can be incorporated into an algorithm are as yet unknown.11 This makes current models fit poorly at high doses; however, the most important task is to discover low-dose patients to avoid early overdosing.
WARG's large sample size and prospective nature make it possible to study anticoagulation over time as a function of genotypes. In agreement with Limdi et al,25 we did not find an association between VKORC1 and risk of bleeding, but homozygosity for VKORC1 variant alleles increased the risk of early INR peaks over 4. A prospective warfarin study by Schwarz et al comprising 297 patients reported a similar effect of common VKORC1 variant alleles on INR response.20 CYP2C9 variant alleles are associated with difficulties in achieving stable anticoagulation and in some studies an increased risk of early bleeding.2,21–26 It is also known that the CYP2C9*3 variant causes a major reduction in warfarin metabolism,8 while the more common *2 has a minor effect.5 In our study, homozygosity for *3 greatly increased the risk of INR peaks over 4 and tended to increase the risk of bleeding during the first month of treatment. Contrary to this, Schwarz et al claimed that CYP2C9 has a limited influence on initial variability in sensitivity to warfarin.20 In their study, all 11 patients genotyped as CYP2C9 poor metabolizers (*2/*2, *2/*3, and *3/*3) were grouped together for statistical analysis, and the number of individuals per genotype was not revealed. The minor allele frequency for *3 was 4.8% making the expected number of *3/*3 individuals less than 1 (0.23%). By grouping a singular *3/*3 individual together with less detrimental genotypes, the effect of *3/*3 could easily be obscured. Our results do not support the Schwarz study conclusion that initial variability in the response to warfarin is more strongly associated with genetic variability in VKORC1 than with CYP2C9.20
A wide variety of warfarin induction strategies were used in our study. In 2 small prospective clinical trials, warfarin induction using predicted warfarin dose has been compared with standard warfarin loading dose.49,50 In the trial by Caraco et al, the control group received a standard loading dose of 5 mg the first day.49 The pharmacogenetic doses were adjusted according to CYP2C9 genotype. In Anderson's clinical trial, patients randomized to standard therapy got 10 mg/day the first 2 days.50 Patients dosed according to VKORC1 and CYP2C9 received double the predicted maintenance dose. Both studies claim that pharmacogenetic dosing increased the efficiency of warfarin initiation. However, the different standard loading doses used in these studies, as well as in ours, illustrate the lack of consensus concerning warfarin induction. Furthermore, little is known about how to predict loading doses from a warfarin maintenance dose model.
The WARG study indicates that genetic forecasting would improve anticoagulant therapy and has partial answers to 3 issues that need to be discussed before clinical trials are initiated: (1) none of the other candidate genes substantially influence response to warfarin, but results from large whole genome association studies remain to be seen; (2) the Swedish WARG dose algorithm is robust, but should be adjusted for body size; and (3) “standard” warfarin induction is by no way standard, and the optimal way to predict a loading dose from a pharmacogenetic model has not been established.
In conclusion, we have shown that in a large prospective study, VKORC1 and CYP2C9 predict warfarin dose and individuals predisposed to unstable anticoagulation during initiation of therapy. We anticipate that adequately sized clinical trials of patients randomized to pharmacogenetic versus conventional dosing will demonstrate that warfarin dose prediction improves the safety and cost effectiveness of oral anticoagulant treatment.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the late Stefan Lundgren for Biobanking and counseling, and the medical staff and participants of the study for their valuable contribution.
This work was supported by the Wellcome Trust and the following Swedish funding agencies: The Science Council (Medicine), the Heart and Lung Foundation, Torsten and Ragnar Söderberg, Thuréus and Selander Foundations, the Society of Medicine, the Foundation for Strategic Research, and The Clinical Research Support (ALF) at Karolinska Institutet and Uppsala University.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.W., L.Y.C., J.D.L., A.R., and P.D. contributed to study concept and design; J.D.L., L.H., and A.R. acquired patient data; L.Y.C., M.J.R.G., S.B., and P.D. were responsible for methods and genotyping; M.W., L.Y.C., J.D.L., and N.E. interpreted data; N.E. and R.M. did statistical analysis; M.W., L.Y.C., J.D.L., and N.E. drafted the manuscript; M.W., R.M., A.R., and P.D. critically revised the manuscript; and M.W., A.R., and P.D. obtained funding.
Conflict-of-interest disclosure: The study was partly supported by research funding from Nycomed Ltd (Stockholm) to A.R. The sponsors had no role in the writing of this paper. The remaining authors declare no competing financial interests.
Correspondence: Anders Rane, Division of Clinical Pharmacology, Karolinska Institutet, SE-141 86 Stockholm, Sweden; e-mail: anders.rane@ki.se.
References
- 1.Landefeld C, Beyth R. Anticoagulant-related bleeding: clinical epidemiology, prediction and prevention. Am J Med. 1993;95:315–328. doi: 10.1016/0002-9343(93)90285-w. [DOI] [PubMed] [Google Scholar]
- 2.Aithal G, Day C, Kesteven P, Daly A. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 3.Loebstein R, Yonath H, Peleg D, et al. Interindividual variability in sensitivity to warfarin- Nature or nurture? Clin Pharmacol Ther. 2001;70:159–164. doi: 10.1067/mcp.2001.117444. [DOI] [PubMed] [Google Scholar]
- 4.Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2007;7:99–111. doi: 10.1038/sj.tpj.6500417. [DOI] [PubMed] [Google Scholar]
- 5.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Kaminsky L, Zhang Z. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73:67–74. doi: 10.1016/s0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- 7.Walton R, Kimber M, Rockett K, Trafford C, Kwiatkowski D, Sirugo G. Haplotype block structure of the cytochrome P450 CYP2C gene cluster on chromosome 10. Nat Genet. 2005;37:915–916. doi: 10.1038/ng0905-915. [DOI] [PubMed] [Google Scholar]
- 8.Steward DJ, Haining RL, Henne KR, et al. Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics. 1997;7:361–367. doi: 10.1097/00008571-199710000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Bell RG, Sadowski JA, Matschiner JT. Mechanism of action of warfarin. Warfarin and metabolism of vitamin K 1. Biochemistry. 1972;11:1959–1961. doi: 10.1021/bi00760a034. [DOI] [PubMed] [Google Scholar]
- 10.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 11.Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 12.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 13.Yuan HY, Chen JJ, Lee MT, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 14.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 15.Veenstra DL, You JH, Rieder MJ, et al. Association of Vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2005;15:687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- 16.Vecsler M, Loebstein R, Almog S, et al. Combined genetic profiles of components and regulators of the vitamin K-dependent gamma-carboxylation system affect individual sensitivity to warfarin. Thromb Haemost. 2006;95:205–211. doi: 10.1160/TH05-06-0446. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi H, Wilkinson GR, Nutescu EA, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics. 2006;16:101–110. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI] [PubMed] [Google Scholar]
- 18.Aquilante CL, Langaee TY, Lopez LM, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz UI, Ritchie MD, Bradford Y, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higashi M, Veenstra D, Kondo L, et al. Association between CYP 2C9 genetic variants and anticoagulation-related outcomes during warfarin treatment. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 22.Peyvandi F, Spreafico M, Siboni SM, Moia M, Mannucci PM. CYP2C9 genotypes and dose requirements during the induction phase of oral anticoagulant therapy. Clin Pharmacol Ther. 2004;75:198–203. doi: 10.1016/j.clpt.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Joffe HV, Xu R, Johnson FB, Longtine J, Kucher N, Goldhaber SZ. Warfarin dosing and cytochrome P450 2C9 polymorphisms. Thromb Haemost. 2004;91:1123–1128. doi: 10.1160/TH04-02-0083. [DOI] [PubMed] [Google Scholar]
- 24.Margaglione M, Colaizzo D, D'Andrea G, et al. Genetic modulation of oral anticoagulation with warfarin. Thromb Haemost. 2000;84:775–778. [PubMed] [Google Scholar]
- 25.Limdi NA, McGwin G, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83:312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindh JD, Lundgren S, Holm L, Alfredsson L, Rane A. Several-fold increase in risk of overanticoagulation by CYP2C9 mutations. Clin Pharmacol Ther. 2005;78:540–550. doi: 10.1016/j.clpt.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Lindh JD, Holm L, Dahl ML, Alfredsson L, Rane A. Incidence and predictors of severe bleeding during warfarin treatment. J Thromb Thrombolysis. 2008;25:151–159. doi: 10.1007/s11239-007-0048-2. [DOI] [PubMed] [Google Scholar]
- 28.Wadelius M, Chen LY, Eriksson N, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. doi: 10.1007/s00439-006-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohnke H, Sörlin K, Granath G, Wadelius M. Warfarin dose related to apolipoprotein E (APOE) genotype. Eur J Clin Pharmacol. 2005;61:381–388. doi: 10.1007/s00228-005-0936-3. [DOI] [PubMed] [Google Scholar]
- 30.Sconce EA, Daly AK, Khan TI, Wynne HA, Kamali F. APOE genotype makes a small contribution to warfarin dose requirements. Pharmacogenet Genomics. 2006;16:609–611. doi: 10.1097/01.fpc.0000220567.98089.b5. [DOI] [PubMed] [Google Scholar]
- 31.Kohnke H, Scordo MG, Pengo V, Padrini R, Wadelius M. Apolipoprotein E (APOE) and warfarin dosing in an Italian population. Eur J Clin Pharmacol. 2005;61:781–783. doi: 10.1007/s00228-005-0982-x. [DOI] [PubMed] [Google Scholar]
- 32.Kimura R, Miyashita K, Kokubo Y, et al. Genotypes of vitamin K epoxide reductase, gamma-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thromb Res. 2007;120:181–186. doi: 10.1016/j.thromres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Rieder MJ, Reiner AP, Rettie AE. gamma-Glutamyl carboxylase (GGCX) tagSNPs have limited utility for predicting warfarin maintenance dose. J Thromb Haemost. 2007;5:2227–2234. doi: 10.1111/j.1538-7836.2007.02744.x. [DOI] [PubMed] [Google Scholar]
- 34.Shikata E, Ieiri I, Ishiguro S, et al. Association of pharmacokinetic (CYP2C9) and pharmacodynamic (factors II, VII, IX, and X; proteins S and C; and gamma-glutamyl carboxylase) gene variants with warfarin sensitivity. Blood. 2004;103:2630–2635. doi: 10.1182/blood-2003-09-3043. [DOI] [PubMed] [Google Scholar]
- 35.Chen LY, Eriksson N, Gwilliam R, Bentley D, Deloukas P, Wadelius M. Gamma-glutamyl carboxylase (GGCX) microsatellite and warfarin dosing. Blood. 2005;106:3673–3674. doi: 10.1182/blood-2005-04-1711. [DOI] [PubMed] [Google Scholar]
- 36.Herman D, Peternel P, Stegnar M, Breskvar K, Dolzan V. The influence of sequence variations in factor VII, gamma-glutamyl carboxylase and vitamin K epoxide reductase complex genes on warfarin dose requirement. Thromb Haemost. 2006;95:782–787. [PubMed] [Google Scholar]
- 37.Lindh JD, Kublickas M, Westgren M, Rane A. Internet based clinical trial protocols—as applied to a study of warfarin pharmacogenetics. Br J Clin Pharmacol. 2004;58:482–487. doi: 10.1111/j.1365-2125.2004.02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.WHO. Glossary of terms related to patient and medication safety. Vol. 2008: Council of Europe: Committee of Experts on Management of Safety and Quality in Health Care (SP-SQS) Expert Group on Safe Medication Practices; 2005.
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 40.Whittaker P, Bumpstead S, Downes K, Ghori J, Deloukas P. Single nucleotide polymorphism analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. In: Celis J, editor. Cell Biology: A Laboratory Handbook. 3rd ed. Elsevier; 2006. [Google Scholar]
- 41.Cheverud JM. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87:52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- 42.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H. A permutation procedure for the haplotype method for identification of disease-predisposing variants. Ann Hum Genet. 2001;65:189–196. doi: 10.1017/S0003480001008491. [DOI] [PubMed] [Google Scholar]
- 44.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 45.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 46.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gage BF, Lesko LJ. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb. 2008;25:45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- 48.Wu AH. Use of genetic and nongenetic factors in warfarin dosing algorithms. Pharmacogenomics. 2007;8:851–861. doi: 10.2217/14622416.8.7.851. [DOI] [PubMed] [Google Scholar]
- 49.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 50.Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.