Summary
In Drosophila embryos and larvae, a small number of identified motor neurons innervate body wall muscles in a highly stereotyped pattern. Although genetic screens have identified many proteins that are required for axon guidance and synaptogenesis in this system, little is known about the mechanisms by which muscle fibers are defined as targets for specific motor axons. To identify potential target labels, we screened 410 genes encoding cell-surface and secreted proteins, searching for those whose overexpression on all muscle fibers causes motor axons to make targeting errors. Thirty such genes were identified, and a number of these were members of a large gene family encoding proteins whose extracellular domains contain leucine-rich repeat (LRR) sequences, which are protein interaction modules. By manipulating gene expression in muscle 12, we showed that four LRR proteins participate in the selection of this muscle as the appropriate synaptic target for the RP5 motor neuron.
Keywords: synapse, neuromuscular junction, cell adhesion molecule, genetic redundancy, gain-of-function screen, EP element, Tartan, Capricious, CG14351, Hattifattener, CG8561, Als
Introduction
Genetic screens in Drosophila and C. elegans have identified cell-surface and secreted (CSS) proteins involved in axon guidance, synaptic target selection, and presynaptic differentiation. In some cases these steps involve distinct sets of CSS proteins, while in others overlapping sets of proteins are involved in multiple steps. Mutations affecting immunoglobulin (Ig)-domain cell adhesion molecules (CAMs), cadherins, receptor tyrosine phosphatases, and Wnt proteins can produce phenotypes in which axons reach their targets but then fail to form normal synapses (Inaki et al., 2007; Klassen and Shen, 2007) (reviewed by (Johnson and Van Vactor, 2003; Shen, 2004).
One of the most accessible systems for examination of target selection is the Drosophila larval neuromuscular system. This is a simpler subset of the central nervous system (CNS), with only 36 identified motor neurons per abdominal hemisegment (reviewed by (Ruiz-Canada and Budnik, 2006)). These neurons innervate a set of 30 body wall muscle fibers in an invariant pattern.
Motor axons leave the CNS in three nerve roots: the ISN, SN, and TN. One of the ISN branches, ISNb (also called SNb) initially follows the ISN pathway, leaving it at the "exit junction" to enter the ventrolateral muscle (VLM) field. ISNb innervates seven muscles: 6, 7, 12, and 13 (internal layer), and 14, 30, and 28 (external layer). The SNa branch bifurcates at the dorsal edge of the VLMs; its ventral (or posterior) branch innervates muscles 5 and 8, which are immediately dorsal to the VLMs. The two TN axons fasciculate with the axon of the peripheral LBD neuron to form the TN tract. Every axon and synapse in the neuromuscular system can be visualized in dissected "fillet" preparations of embryos or larvae stained with antibodies such as monoclonal antibody (mAb) 1D4 (Vactor et al., 1993).
Neuromuscular junctions (NMJs) increase greatly in size during the three stages (instars) of larval life in order to keep pace with the growth of the muscle fibers. Each NMJ arbor has a stereotyped pattern of outgrowth. However, NMJ growth is not autonomously determined within the motor neuron, but is driven by a homeostatic relationship between the neuron and its postsynaptic target (reviewed by (Davis, 2006)). Some of the signaling mechanisms involved in homeostasis resemble those used for synaptic development and plasticity in mammalian systems. Furthermore, the NMJ is a glutamatergic synapse that uses orthologs of vertebrate AMPA receptors. These features make the Drosophila NMJ a useful genetic model system for excitatory synapses in the mammalian brain.
Many proteins involved in guidance of motor neuron growth cones have been identified. However, we know relatively little about how individual fibers within a muscle field are selected for innervation by specific motor axons. Two models, which are not mutually exclusive, have been proposed. In the relative balance model, each fiber is defined as a target by a mixture of generally expressed attractive and repulsive axon guidance cues that distinguish it from its neighbors (Winberg et al., 1998). In the lock-and-key model, individual muscle fibers are specified by molecular labels that are recognized by receptors expressed by the innervating neurons (Hoang and Chiba, 1999).
The relative balance model was evaluated by altering expression of Netrin B (NetB), Semaphorin II (SemaII), and Fasciclin II (FasII) proteins on the VLMs. Netrins and Semaphorins regulate axon guidance in multiple contexts, while FasII is a homophilic Ig-CAM involved in axonal fasciculation. SemaII and FasII are expressed by all muscles, while NetB is expressed by muscles 6 and 7, which are innervated by the RP3 neuron. Removal of the attractive NetB cue reduces innervation of the 6/7 cleft by RP3, as does increasing muscle expression of the repulsive SemaII cue.
Reducing SemaII levels causes the TN to make ectopic contacts on the VLMs, and this phenotype is strengthened by increasing FasII expression on muscles. Thus, VLM innervation can be controlled by adjusting the relative levels of a few attractive, repulsive, and adhesive cues. However, these cues are not essential for targeting: in a SemaII NetB double mutant lacking both attraction and repulsion, or in a FasII mutant, RP3 innervates the 6/7 cleft and the ISNb has a relatively normal morphology (Winberg et al., 1998).
Investigations of the lock-and-key model have focused on CSS proteins that are expressed on individual VLM fibers. Fasciclin III (FasIII) is a homophilic Ig-CAM that is expressed on the RP3 growth cone and at the 6/7 cleft. Its adhesive properties and expression pattern make it an ideal candidate for an RP3 target label; but in a FasIII null mutant, RP3 still innervates muscles 6 and 7. When FasIII is expressed on the surrounding muscles in a wild-type background, however, RP3 mistargets to these muscles. Removing FasIII from the neuron suppresses mistargeting (Chiba et al., 1995; Kose et al., 1997). These results show that FasIII-mediated homophilic adhesion can divert RP3 to the wrong muscles, but is not required for RP3's recognition of its normal target.
Capricious (Caps) is a leucine-rich repeat (LRR) protein that is expressed on muscle 12, as well as on ventral muscles and a subset of dorsal muscles. When Caps is ectopically expressed on all muscles, a strong larval mistargeting phenotype ("12→13 loopback") is observed in which the muscle 12 NMJ sends a collateral branch back onto muscle 13. caps null mutants have low-penetrance loopback phenotypes involving only a few boutons(Shishido et al., 1998).
mRNA encoding the secreted protein Wnt4 is enriched in muscle 13. In Wnt4 LOF mutant embryos, RP5 axons, which normally target to muscle 12, transiently form synapses on muscle 13, suggesting that Wnt4 is a repulsive cue that normally prevents RP5 from synapsing on muscle 13 (Inaki et al., 2007).
The data summarized above show that although alterations in CSS protein expression can influence target selection in the neuromuscular system, none of the CSS cues that have been identified thus far are required for normal targeting. This may be partially due to genetic redundancy. Each muscle fiber might express several different cues, any of which can be used for target recognition by innervating growth cones. Consistent with this interpretation, the only identified gene for which LOF mutations produce strong targeting phenotypes is abrupt (ab), which encodes a transcription factor expressed in muscles. In ab embryos, ISNb nerves have a variety of different abnormal morphologies, suggesting that axons cannot recognize any of the VLMs as targets. Perhaps Ab regulates a battery of ISNb muscle targeting cues, and all of these cues are absent or misregulated in ab mutants (Hu et al., 1995).
The results on FasIII and Caps suggest that new target labels could be identified by performing a gain-of-function (GOF) screen for genes whose overexpression on all muscles cause axonal mistargeting phenotypes without affecting the structures of the muscles themselves. Overexpression/misexpression screens are commonly done using "EP"-like transposable elements, which contain "UAS" sequences recognized by the yeast transcriptional activator GAL4 upstream of a basal promoter (Rorth et al., 1998). Like other P elements, EPs tend to transpose into the 5' ends of genes. When an EP line with a 5' insertion is crossed to a "driver" line expressing GAL4 in a particular cell type, the gene downstream of the EP will be expressed in that cell type in the F1 progeny of the cross (Brand and Perrimon, 1993). In one screen of this type, 500 random EP insertions were crossed to the pan-muscle 24B-GAL4 driver (Luo et al., 1994) and F1 larvae examined for mistargeting defects (Umemiya et al., 2002).
In our screen, we planned to examine individual larvae by confocal microscopy in order to ensure that we could detect subtle phenotypes. This meant that we could not analyze thousands of lines, as would be required in order to identify genes encoding CSS proteins in a random EP screen. We thus decided to conduct a "directed" screen of genes encoding CSS proteins that are likely to be involved in cell recognition events. To do this, we constructed a database of Drosophila genes that encode such proteins, and found EP-like insertions that could drive 410 of these genes. The EP lines were crossed to 24B-GAL4 and the resulting F1 larvae examined for mistargeting and synaptic phenotypes.
Our screen cannot assay all CSS proteins encoded in the genome. However, we hoped that identification of all the mistargeting genes within the ~40% of the CSS repertoire accessible through EP lines would inform us as to which protein families are likely to include target labels. We may have succeeded in this effort, because our screen identified 16 CSS LRR proteins that either cause axonal mistargeting or affect NMJ arbor structure when they are expressed on all muscles. Here we present evidence that four LRR proteins actually function as synaptic targeting cues.
Results
A database of cell-surface and secreted proteins
To create a database of Drosophila CSS proteins potentially involved in cell recognition, we performed BLAST searches with extracellular (XC) domain sequences from a variety of species and collated published information (see Supplemental Text for details). The database (Supplementary Table 1) contains 976 proteins and more than 80 domain types. We then searched through all existing collections of UAS-containing EP-like lines and obtained those with insertions 5' to CSS database genes that could be used to drive their expression. These insertions should drive 410 of the 976 genes in the database, or over 40% of the cell recognition repertoire (Supplementary Table 1 and Supplementary Table 2). 53 of these 410 genes encode proteins with LRRs.
Targeting events examined in the screen
The ISNb contains the axons of seven type I (glutamatergic) neurons, including RP1, 3, 4, and 5, which are 1b (big boutons) neurons (Hoang and Chiba, 2001; Landgraf et al., 1997). After entering the VLM field, the ISNb axon bundle extends dorsally, traveling between the external (14 and 28) and internal (6 and 7) muscle layers (diagram in Figure 1B). The RP3 axon leaves the bundle and forms an NMJ in the cleft between muscles 6 and 7. The remaining axons then traverse to the internal face of muscle 13, where RP1 and 4 form synapses. The RP5 axon grows past muscle 13 to the ISNb termination point on the internal face of muscle 12. In early stage 17 embryos, the nascent RP5 synapses are at the ventral edge of muscle 12, but by 1st instar RP5 has formed an NMJ that extends across the muscle surface. By 3rd instar, all of the VLMs are also innervated by a 1s (small boutons) neuron.
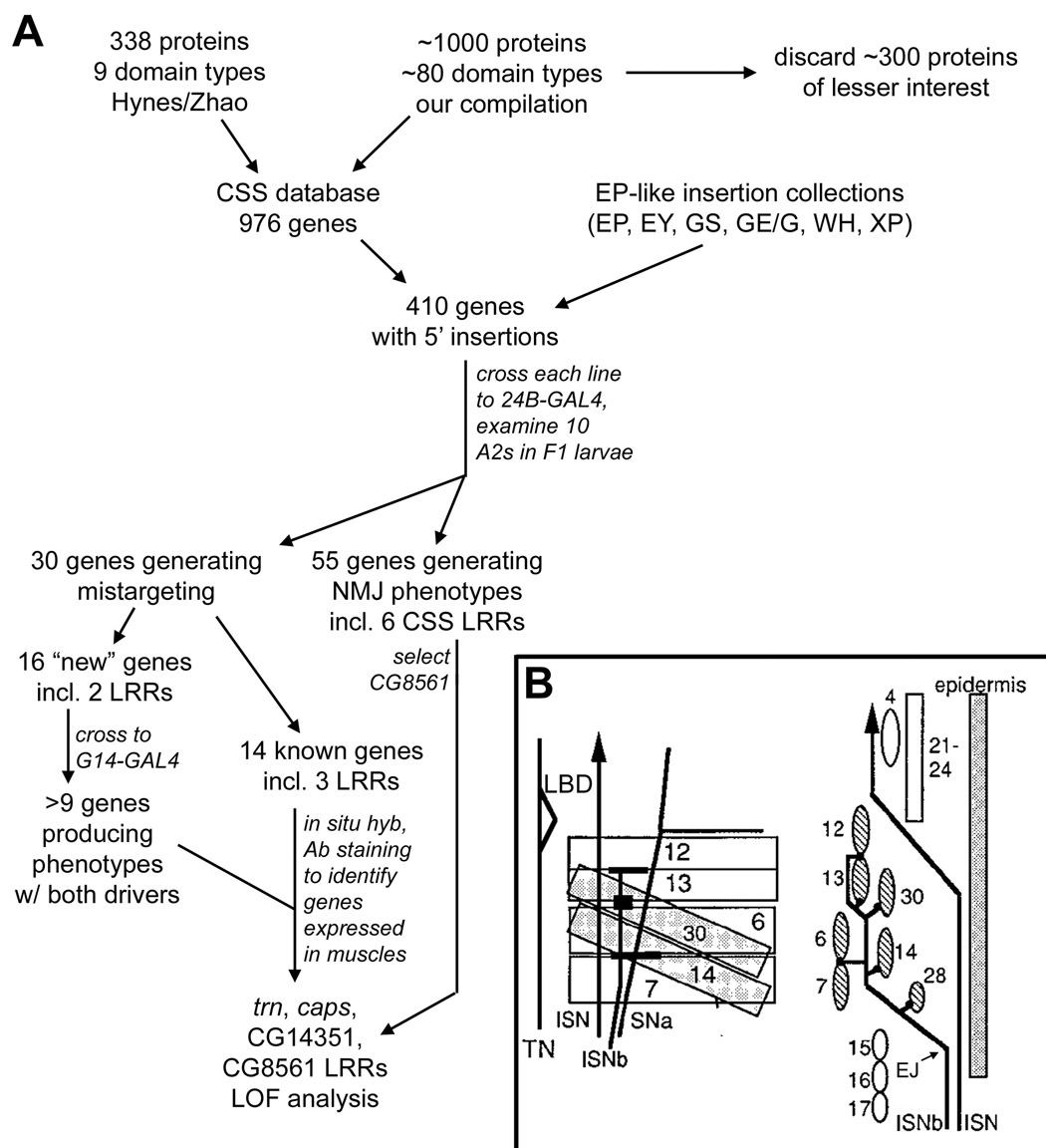
Figure 1. Flowchart of the screen.
(A) Outline of the steps in the screen. (B) Diagram of the pathways taken by ISNb axons within the VLM field. Left, face-on view of the VLMs. The LBD (triangle shape) is indicated. Underlying muscles 14 and 30 are shaded. Right, side view, with the interior of the embryo to the left. EJ, exit junction. The seven VLM fibers are shaded.
The internal surfaces of muscles 13 and 12 lie in a plane, and the RP5 growth cone that will innervate muscle 12 has full access to both muscles when it reaches the embryonic ISNb termination point. This may account for the observation that the muscle 12 vs. 13 decision is often perturbed in ectopic expression experiments (Shishido et al., 1998; Umemiya et al., 2002).
Execution of the screen
We screened the CSS insertion collection for genes conferring ISNb mistargeting and/or presynaptic terminal phenotypes when overexpressed in postsynaptic cells. To do this, 462 insertion lines representing 410 genes (Supplementary Table 2) were crossed to the 24B-GAL4 driver, which confers high-level expression in all somatic muscles from stage 12 onward, persisting into 3rd instar (Luo et al., 1994). F1 3rd instar larvae from these crosses were stained with mAb 1D4 to visualize axons and NMJs. Muscles were visualized with UAS-GFP driven by 24B-GAL4 or by Alexa-488-phalloidin staining. For the initial screen of each cross, we examined the VLM regions of 10 A2 hemisegments (five larvae) using confocal microscopy. Genes producing mistargeting were then tested with another early pan-muscle driver (G14-GAL4) and a pan-neuronal driver (Elav-GAL4; see screen flowchart in Figure 1A). The normal pattern of innervation is shown in Figure 2A and depicted schematically in Figure 8. The 30 genes conferring mistargeting with ≥30% penetrance are listed in Table 1, and Figure 2 shows examples of phenotypes.
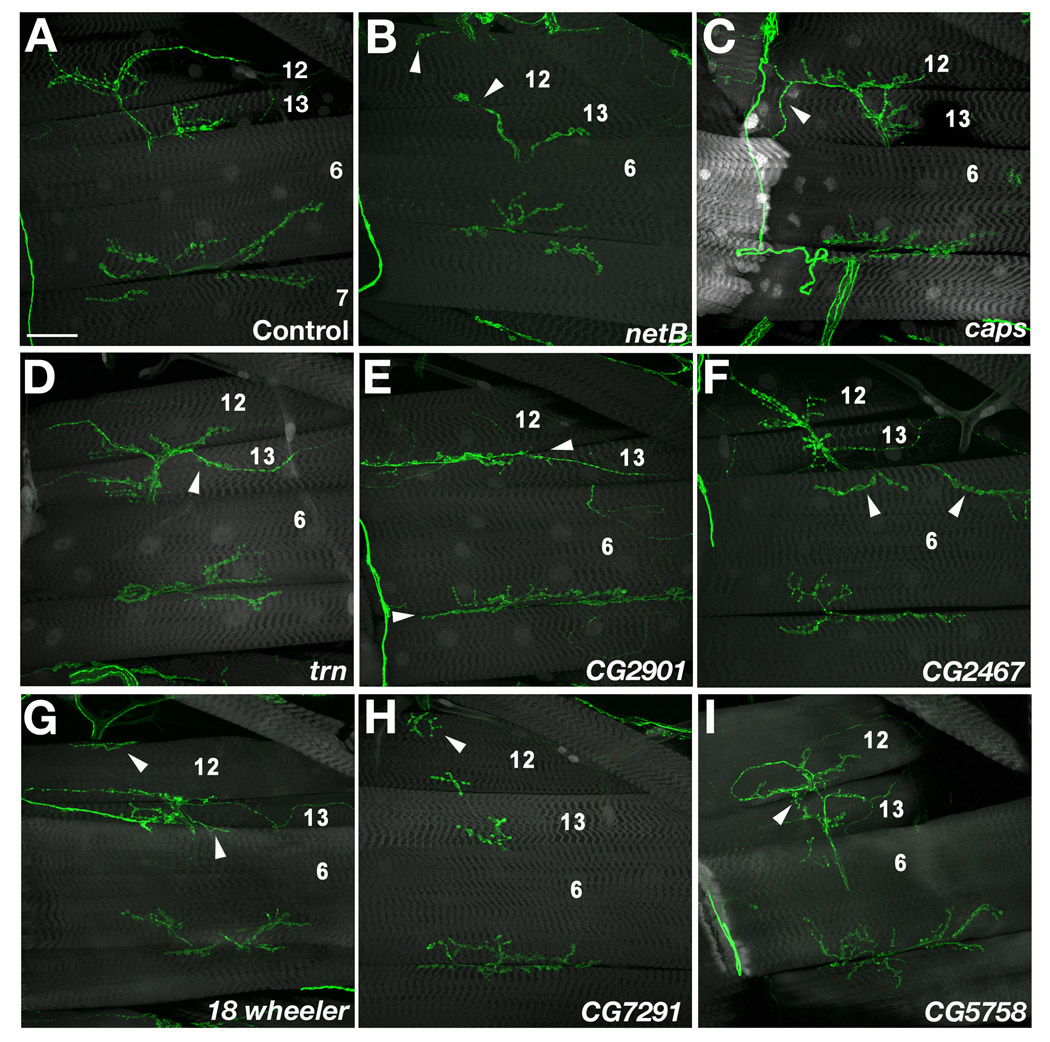
Figure 2. Examples of mistargeting phenotypes.
Confocal z-stack images of 3rd instar F1 larvae stained with 1D4 (green) and Alexa-488-phalloidin or anti-GFP (gray). (A) Control (UAS-GFP, 24B-GAL4×w). The NMJs on muscle (m) 6/7, 13, and 12 are evident. (B) NetB. Top arrowhead, an NMJ from an unidentified neuron grows onto m12 from below (Type 3 phenotype; see Figure 4A for phenotypic diagrams). Bottom arrowhead, truncated normal m12 NMJ. (C) caps. (D) trn. Arrowheads in (C) and (D), 12→13 loopback phenotype (Type 1). (E) CG2901. Arrowhead, the entire NMJ remains at the 12/13 junction and does not grow onto m12 (Type 2). (F) pot/CG2467. Arrowheads, ectopic NMJs (probably from m13) innervating m6 (Type 7). (G) 18w. Arrowheads, Type 3 (top) and Type 7 (bottom) ectopic NMJs. (H) CG7291/NPC2. Arrowhead, Type 3 ectopic NMJ. (I) CG5758. Arrowhead, 12→13 loopback phenotype (Type 1); note the complex multilooped structure of this NMJ. Bar in (A), 50µm; applies to all panels.
Figure 8. Effects of muscle 12-specific overexpression and knockdown of LRR genes.
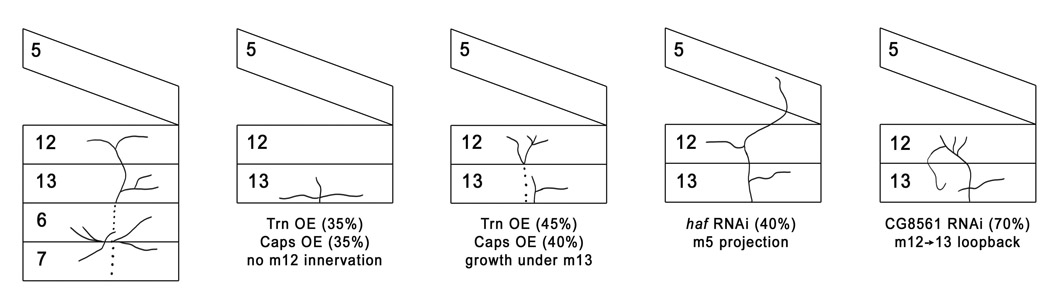
Representative phenotypes caused by m12-specific perturbations (genes or RNAi driven by 5053A-GAL4) are illustrated. Dotted lines indicate that axons travel under (external to) m13, m6, and m7. OE, overexpression.
Table 1. Mistargeting genes identified in the screen.
Orange, black, and green genes are "new"; blue and red genes were previously characterized using genetics. %, phenotypic penetrance. "mRNA OE?" indicates whether we showed that the mRNA is overexpressed by in situ hybridization.
| gene name/CG | insertion | protein domains | SP, TM domains | 24B % | G14 % | elav % | mRNA OE? | LOF phenotype | mRNA/prot in muscles? | in other cells contacted by motor axons? |
|---|---|---|---|---|---|---|---|---|---|---|
| CG14351/haf | EY11244 | LRR-8 | TM-1 | 30% | 30% | 0% | yes | none known | yes^ | |
| CG3413/windpipe | GE13015 | LRR-2 | SP, TM-1 | 30% | 30% | 30% | yes | none known | no | tracheae* |
| CG2901 | GE3026 | SPX, EXS | TM-4? | 100% | 70% | 0% | yes | none known | yes^ | tracheae, TN^ |
| CG9342 | G3187 | lipoprotein N-terminal | SP | 30% | 20% | 0% | yes | none known | yes (mesoderm)^ | |
| CG8668 | G2829 | glycosyltransferase | SP | 30% | 40% | 0% | yes | none known | ? | ectoderm (BDGP) |
| CG11372/galectin | GS13007 | ConA | SP | 40% | 40% | 30% | nt | none known | yes (BDGP) | |
| CG7291/NPC2 | GE12107 | ML | SP | 30% | 20% | 0% | yes | none known | no signal^ | |
| CG12004 | GS16151 | DUF300 | TM-7 | 30% | 30% | 0% | nt | none known | ? | |
| CG5758 | GS22939 | Fasciclin I | SP | 50% | nt | nt | nt | none known | ? | |
| CG32373 | GS14433 | Sushi/CCP(EGF-like) | SP | 30% | nt | nt | nt | none known | ? | |
| CG7179 | GS13316 | CUB | SP | 30% | nt | nt | nt | none known | yes? (BDGP) | |
| CG7447 | EY7651 | EGF-like | SP | 30% | 0% | 10% | yes | none known | muscle attachment | sites^ |
| CG14469/Dpr12 | WH7539 | Ig | SP | 30% | 50% | nt | yes | none known | no^ | no (CNS only)^ |
| CG2578/ten-a | GE1914 | EGF-like | TM-1 | 40% | nt | nt | nt | none known | no* | no (CNS only)* |
| CG5634/distracted | EY05482 | CUB | (SP), TM-1 | 40% | 0% | 0% | yes | none known | no signal^ | |
| CG10772/fur-1 | EP3499 | Furin | TM-2 | 30% | 40% | nt | nt | none known | no^ | CNS*^ |
| tartan | GS10885 | LRR-10 | SP, TM-1 | 60% | 40% | nt | nt | CNS,PNS,tracheae,discs | yes* | |
| 18-wheeler | GE14005 | LRR-20 | SP, TM-1 | 40% | 20% | 0% | yes | segmentation, defense | no* | tracheae, ectoderm* |
| capricious | GS10839 | LRR-11 | SP, TM-1 | 60% | nt | nt | nt | mistargeting (GOF) | yes | |
| NetB | GS1029 | Netrin | SP | 80% | nt | nt | nt | mistargeting (GOF), CNS | yes | |
| papillote/CG2467 | G1385 | ZP/PAN | SP, TM-1 | 50% | 70% | 10% | yes | epidermis | no | ventral epidermis* |
| neuromusculin | GS9446 | Ig9 | TM-1 | 30% | nt | nt | nt | CNS axons | yes* | |
| hikaru genki | EY03336 | Sushi/CCP(EGF-like) | SP | 40% | nt | nt | nt | locomotor defects | no | secreted into clefts* |
| robo2 | EP2582 | Ig/FN3 | (SP), TM-1 | 40% | nt | nt | yes* | CNS axon guidance | no* | tracheae, epidermis* |
| pvf3/CG34378 | f04177 | PDGF | SP | 30% | 10% | 0% | yes | blood cell migration | no?(foregut, MT)* | |
| shifted | GE946 | WIF | SP | 30% | nt | nt | nt | Hh signaling | ubiquitous, st. 14* | |
| turtle | GS9718 | Ig/FN3 | TM-1 | 40% | nt | nt | nt | locomotor defects | no (CNS only)* | no (CNS only)* |
| neurexin IV | EP604 | EGF-like | SP | 30% | 30% | 30% | yes | septate junctions | not reported* | glia* |
| Gliotactin | GE13413 | acetylcholinesterase | (SP), TM-1 | 30% | 30% | nt | nt | blood-nerve barrier | no | |
| outstretched | G17133 | none | SP | 30% | 20% | 0% | yes | many phenotypes | ubiquitous* |
For protein/mRNA expression, ^ indicates that the source is our unpublished data, and * indicates that the information is published. These data and references can be found in FlyBase (http://flybase.bio.indiana.edu/).
We also found 55 other genes whose overexpression in postsynaptic muscles caused major alterations in the morphologies of NMJ presynaptic terminals without affecting the structures of the muscles themselves (Supplementary Figure 1, Supplementary Table 3). The screen is described in detail in the Supplemental Text.
Selection of mistargeting genes
To identify mistargeting genes that are normally expressed in cells that would be contacted by ISNb axons during their outgrowth through the VLM field, and therefore might encode genuine targeting molecules, we examined their expression in wild-type embryos using in situ hybridization. The cells of interest include the VLMs themselves, ventral peripheral nervous system (PNS) neurons, ventral tracheal branches, ventral epidermis, and peripheral glia. Six "new" genes (those not previously characterized using genetics; orange in Table 1) were found to be expressed in some of these cells, as were six previously characterized genes (blue in Table 1). We noted that five of these 12 (orange+blue) genes encode cell-surface LRR proteins: Caps, Tartan (Trn), 18-wheeler (18w), CG14351, and CG3413/Windpipe. All other domain types (Ig, ZP/PAN, Netrin, ConA, and others) are represented by only a single gene within this group.
Within the LRR set, we initially focused on Trn, Caps and CG14351 because they are normally expressed by muscles (Artero et al., 2003; Shishido et al., 1998) (Supplementary Figure 2). We also selected CG8561, an LRR gene identified as producing NMJ phenotypes (Figure 1A, Supplementary Table 3), for further investigation. CG8561 generates mistargeting with 20% penetrance and is expressed in muscles (Supplementary Figure 3).
Tartan and Capricious regulate motor axon guidance and targeting in the embryo
The XC domains of Trn and Caps are 65% identical (Chang et al., 1993; Shishido et al., 1998). Studies of Trn and Caps function in imaginal discs suggest that the two proteins can interact with a common receptor (Milan et al., 2005). If so, Trn and Caps might function in a redundant manner to regulate axon guidance and to label muscles as axonal targets. caps LOF mutants have very weak mistargeting phenotypes (Shishido et al., 1998), so Caps alone is not necessary for targeting.
We could not assess trn LOF phenotypes in larvae, because trn mutants die before 3rd instar. We made trn RNAi lines from our own inverted repeat constructs, but these did not produce larval phenotypes when crossed to 24B-GAL4 or other drivers. Thus, to define the trn phenotype, and to ascertain whether Trn and Caps have redundant functions, we examined trn, caps, and trn caps phenotypes in embryos.
We evaluated phenotypes for two trn alleles: trn28.4, which deletes coding sequence and is likely to be a null allele, and trns064117, an embryonic lethal insertion mutation that does not interrupt the coding region. We also examined caps65.2, a null excision allele, and the double mutant trns064117 caps65.2. We quantitated motor axon phenotypes in late stage 16/early stage 17 embryo fillets by staining with mAb 1D4 and scoring segments A2–A7 (Figure 3H).
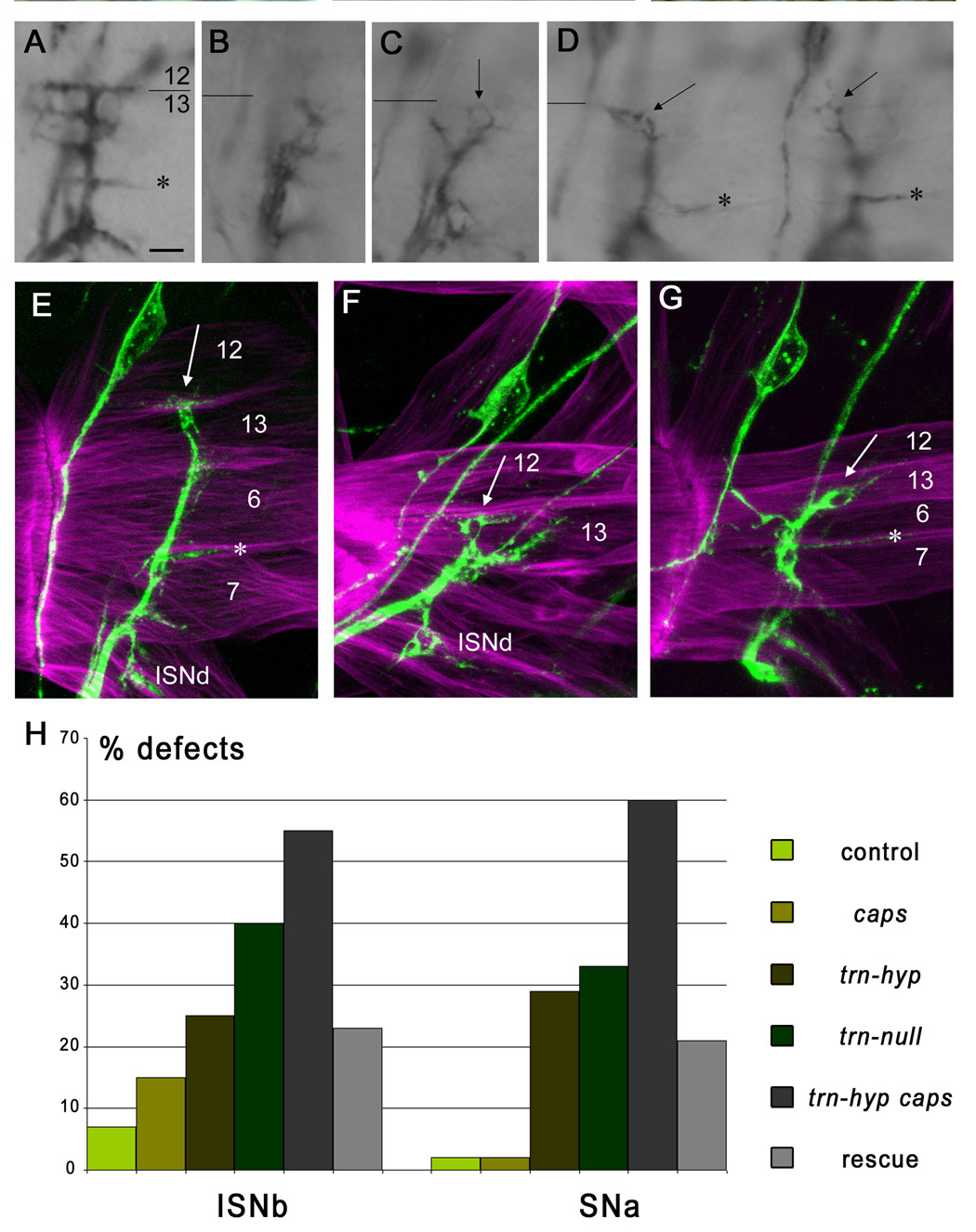
Figure 3. Looped ISNb nerves in trn caps mutants.
(A–C) ISNbs in early stage 17 embryos stained with mAb 1D4 using HRP immunohistochemistry; these are brightfield images, as the loops are too faint to see with DIC optics. (A) is a control hemisegment. Loops indicated by arrows in (B–D); note that all are at the 12/13 border or on m13. The 12/13 border is indicated by a line in each panel. NMJs at the 6/7 cleft indicated by asterisks (*). (E–G) Loops visualized by confocal microscopy. 1D4 is green, Alexa-phalloidin is magenta. (E) is a control hemisegment, and (F) and (G) are trn caps. The control hemisegment was stretched more during dissection so the muscles are wider. The hemisegment in (F) is an A7, so the ventral muscles have a different morphology from (E) and (G). Arrow in (E), muscle 12 NMJ. Arrows in (F) and (G), terminal loops. Stars (*), muscle 6/7 NMJs. ISNd is indicated in (E) and (F). (H) Bar graph of total phenotypic penetrances (% defects) for ISNb and SNa in control (TM3-GFP/+; n=468 hemisegments for ISNb, n=432 for SNa), caps65.2 (n=202 for ISNb, n=152 for SNa), trns064117 (labeled as trn-hyp; n=197 for ISNb, n=143 for SNa), trn28.4 (labeled as trn-null; n=216 for ISNb, n=160 for SNa), trns064117 caps65.2(labeled as trn-hyp caps; n=196 for ISNb, n=140 for SNa), and Elav-GAL4, UAS-Trn, trns064117 caps65.2 (rescue of trn-hyp caps by neuronal Trn; labeled as "Rescue"; n=202 for ISNb, n=170 for SNa). p<.001 (Chi-square test) for differences between trn-hyp and trn-hyp caps, and between trn-hyp caps and Rescue. Bar in (A), 10 µm for (A–D), 5 µm for (E–G).
trn28.4 homozygotes had a mild CNS phenotype and some muscle patterning defects (see also (Artero et al., 2003)). We quantitated phenotypes in hemisegments with normal muscles, and found that they had strong motor axon guidance phenotypes that primarily affected the ISNb and SNa nerves (Figure 3H). These included "stall" phenotypes in which the ISNb was truncated ventral to muscle 13, suggesting that the axons stopped prematurely, "bypass" phenotypes in which the ISNb failed to leave the ISN pathway at the exit junction, and SNa bifurcation failures (see Figure 1B for normal embryonic ISNb pathways).
trns064117 mutants had no visible CNS phenotypes, and only rare muscle patterning defects. They displayed a similar spectrum of motor axon guidance phenotypes to trn28.4 embryos, but with a lower penetrance (25% ISNb, 29% SNa defects for trns064117, vs. 40% ISNb, 33% SNa defects for trn28.4; Figure 3H). caps65.2 embryos had weak ISNb phenotypes and no SNa phenotypes (15% total ISNb defects and 2% SNa defects were observed in caps mutants, while the background ISNb defect penetrance was 7% for TM3armGFP/+ (balancer control) embryos; the background SNa defect penetrance was 2%). Ectopic ISNb projections to the TN were reported in an earlier study of caps mutant embryos, but the penetrance was not quantitated (Abrell and Jackle, 2001). We occasionally saw such phenotypes (12%), but they were seen with a similar frequency in balancer controls.
Double mutants (trns064117 caps65.2) also had no CNS phenotypes, and <7% of hemisegments had a missing muscle. The penetrances of the ISNb and SNa phenotypes (55% and 60%, respectively) in double mutant embryos were roughly doubled relative to trns064117 single mutants, and were higher than those observed in trn-null mutants (Figure 3H). The differences in penetrance between trn and trn caps are statistically significant (p<001, Chi-square test). This shows that the two genes interact.
The trn caps phenotype was rescued to ~20% penetrance for both ISNb and SNa phenotypes (Figure 3H) by neuronal expression of Trn driven by Elav-GAL4. Rescue is statistically significant (see Figure 3 legend), and the ISNb phenotypic penetrance in neuronal rescue embryos is only slightly higher than in caps single mutants, suggesting that loss of Trn from neurons accounts for the ISNb axon guidance errors. For SNa, the penetrance in rescued embryos is still considerably higher than for caps single mutants, so loss of Trn from muscles may also contribute to this phenotype. We did not examine rescue by GAL4-driven expression of Trn in muscles, since this produces the dominant phenotypes that allowed us to identify trn as a gene of interest in our screen.
Double mutant ISNbs that passed the center of muscle 13 sometimes exhibited a striking "terminal loop" phenotype at early stage 17 that has not been described for any other mutant. It was never observed in trn or caps single mutants (n=197 hemisegments for trn, n=202 for caps). Most ISNb nerves with the loop phenotype had formed synapses at the muscle 6/7 cleft, showing that overall ISNb development is relatively normal. We also confirmed that the loop hemisegments all had normal muscle patterning. The distal edge of the terminal loop was at the muscle 12/13 junction or on the surface of muscle 13 (Figures 3A–G).
The terminal loop phenotype is reminiscent of the 12→13 loopback phenotypes observed in the larval screen (Figures 2B, C), and suggests that the RP5 axon destined for muscle 12 did not recognize it as the preferred target and turned back onto muscle 13. We estimate that >20% of ISNbs in double mutants that reach the center of muscle 13 have these terminal loops. However, the exact penetrance of the loop phenotype is difficult to determine, because the loop appears to involve only one axon and is thus hard to visualize using immunohistochemistry. Also, the phenotype would only be detectable if the loop is big enough to distinguish. An axon that doubled back on itself might represent the same phenotype, but we would not be able to see this unless it was connected to a loop (e.g. Figure 3B). Note also that we cannot unequivocally determine if the loops are axons or if they are parts of growth cones, although growth cones are smaller and are not normally seen at this stage of development. A structure with a similar appearance might also be produced by two axons that separate and then rejoin.
Trn overexpression on muscle 12 alters targeting
The results described above suggest that loss of both Trn and Caps can cause the RP5 axon to fail to recognize muscle 12, so that it turns back onto muscle 13 and forms a loop. Caps is normally expressed on muscle 12, and has been proposed to be an attractive cue for RP5, based on the fact that when Caps is overexpressed on all muscles, the 3rd instar muscle 12 NMJ often sends loopback collaterals onto muscle 13 (Shishido et al., 1998) (Figure 2C).
To evaluate Trn's function in targeting and compare it to that of Caps, we first defined the expression patterns of Trn protein and a trn enhancer trap. trn mRNA is expressed in the ectoderm, mesoderm, tracheae, CNS, and PNS at various stages, and its expression pattern changes rapidly throughout development (Chang et al., 1993; Krause et al., 2006). Examination of a trn-nuclear lacZ enhancer trap shows that trn is transcribed in all of the VLMs at stages 14 through early 16 (Supplementary Figure 4).
Trn protein is expressed in a subset of muscle founder cells at stage 12 (Artero et al., 2003), but its expression in later embryos has not been described. We stained stage 14–16 embryos with the rabbit anti-Trn antibody described by (Chang et al., 1993). During late stage 14 through early stage 16, the ventral and lateral patches of Trn expression are separated by a gap that corresponds to the region containing muscles 12 and 13 (Supplementary Figure 5), suggesting that Trn protein is expressed at lower levels on these muscles than on muscles 6 and 7. By late stage 16, Trn protein is no longer detected on the VLMs. In 3rd instar larvae, there is no detectable expression of Trn protein in the VLM region (data not shown).
To evaluate the consequences of Trn muscle overexpression, we drove the GS10885 (trn EP) line with three different GAL4 drivers and examined 3rd instar larvae. These are 24B, H94, and 5053A. H94 drives expression in muscles 13, 6, and 4, and at low levels in 12; it turns off by the end of embryogenesis and is weaker than 24B. 5053A is selective for muscle 12 only, and drives high-level expression in both embryos and larvae (Supplementary Figure 6).
All three drivers generated mistargeting at high frequencies (37–60%; Figure 4H). 24B-GAL4 × GS10885 (abbreviated as 24B::Trn) and H94::Trn both produced a spectrum of mistargeting defects. The most common 24B::Trn phenotypes (see bar graph; Figure 4I) were innervation of muscle 12 from the dorsal side by unknown axons (Type 3; Figure 4C) and 12→13 loopbacks (Type 1) (see phenotype diagram in Figure 4A). H94::Trn produced Type 1, Type 2 (reduced innervation of 12), and Type 8 (no innervation of 13; Figure 4E). These phenotypes are hard to interpret mechanistically, because ISNb axons traverse multiple Trn-overexpressing muscles during outgrowth, and the mistargeting axons sometimes cannot be identified.
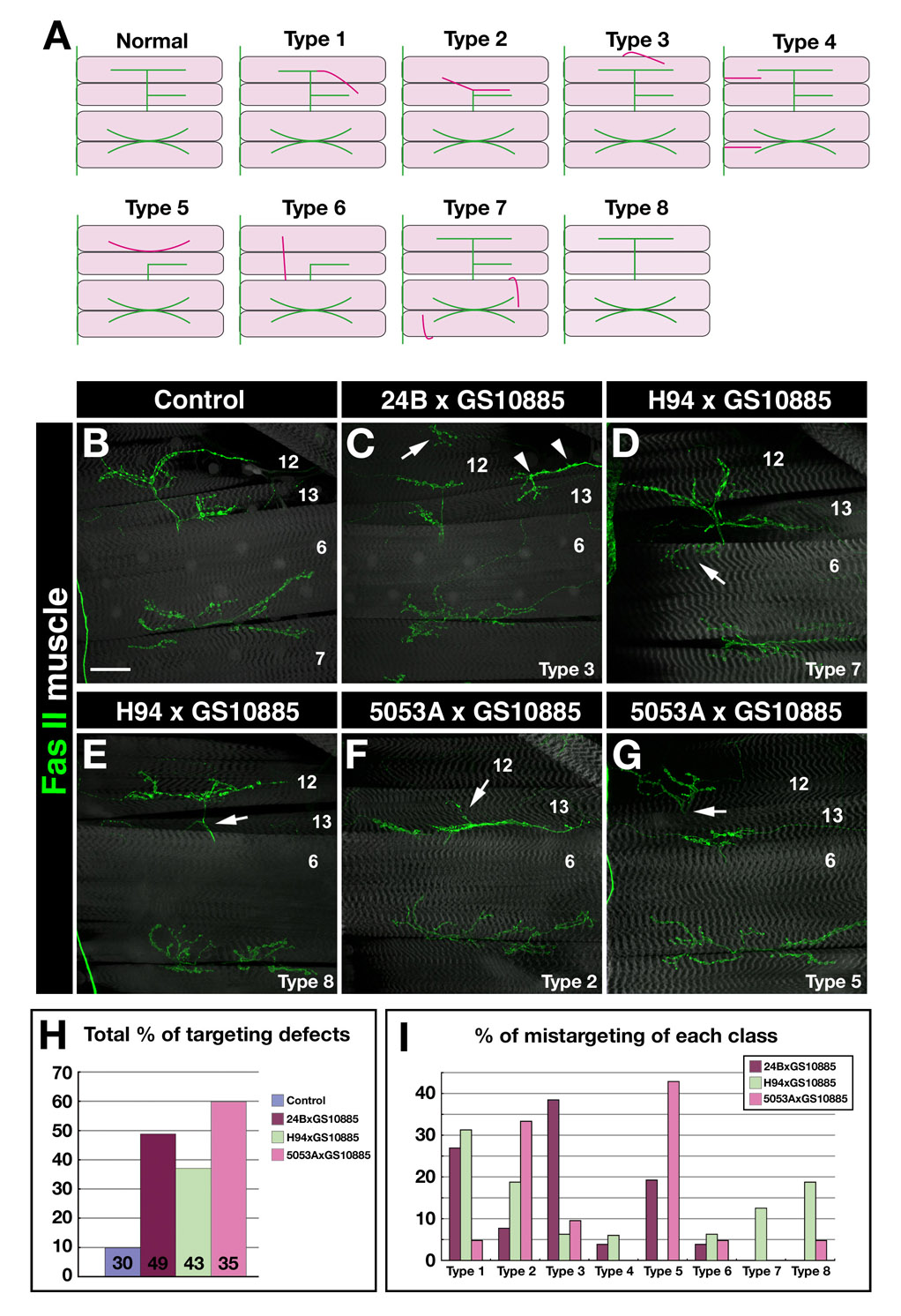
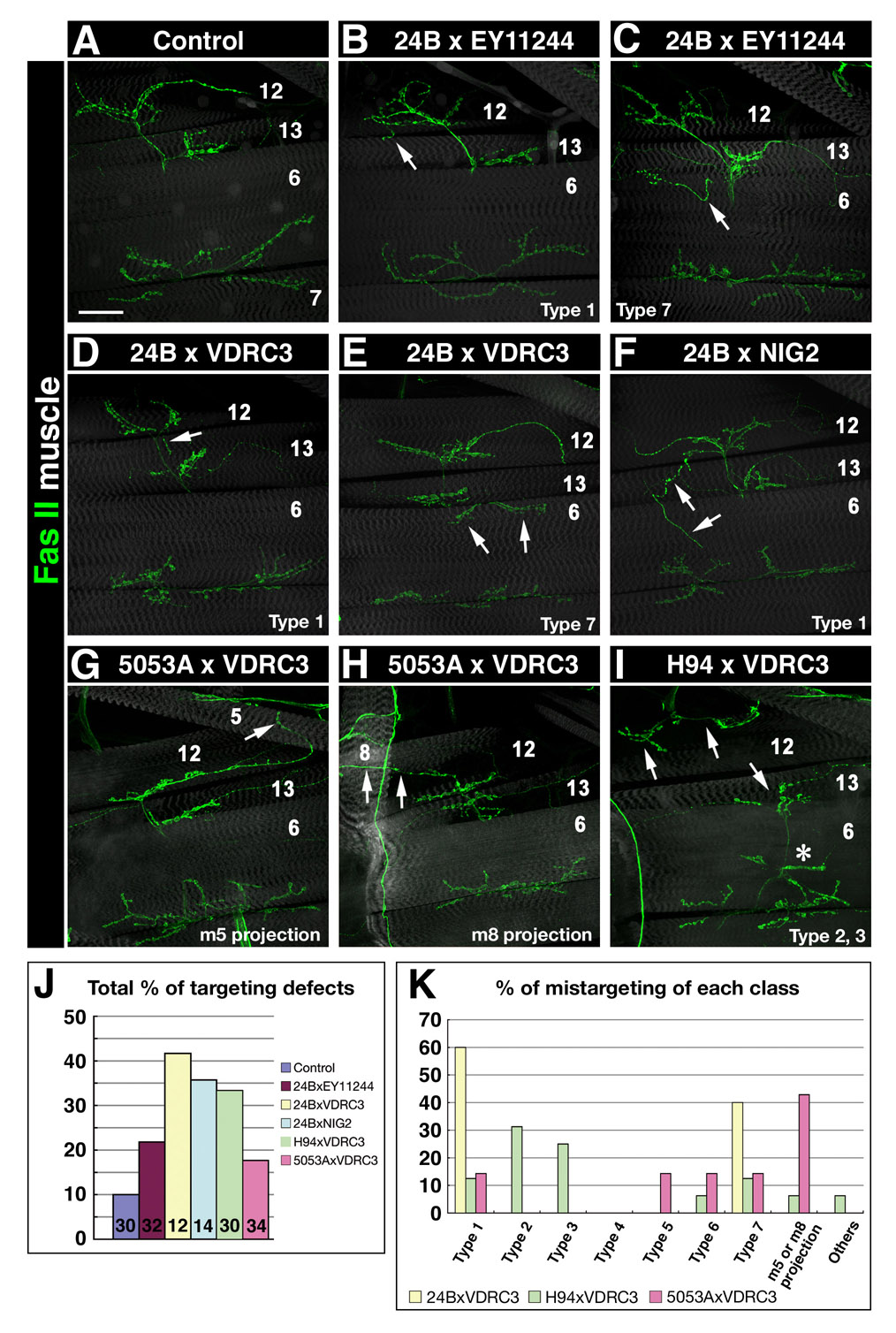
Figure 4. Trn overexpression in muscle subsets causes mistargeting.
(A) Classification of observed mistargeting phenotypes. Green lines indicate normally patterned NMJs on muscles 6,7, 12, and 13. The TN is indicated at the left. Red lines indicate ectopic (mistargeted) NMJs. (B–G) Confocal z-stack images of the VLM regions of 3rd instar F1 larvae stained with 1D4 (anti-FasII; green). Muscles (gray) were visualized by UAS-GFP driven by 24B-GAL4 (B,C) or by phalloidin staining (D–G). (B) Control (UAS-GFP, 24B-GAL4 × w). (C) Overexpression of intact Trn (in GS10885 × UAS-GFP, 24B-GAL4). Arrow in (C), ectopic NMJ wrapping over the dorsal edge of m12 (Type 3). Arrowheads in (C), ectopic NMJ on m12/13, presumably split off from the normal NMJ. (D–E) Trn overexpression on muscles 13 and 6 (in GS10885 × H94-GAL4). Arrow in (D), ectopic innervation of m6 (Type 7); arrow in (E) indicates the absence of innervation on m13 (only an axon traversing m13 is observed; Type 8). (F–G) Trn overexpression on m12 only (in GS10885 × 5053A-GAL4). Arrow in (F), a branch of the m13 NMJ extends dorsally but there is no innervation of m12 (Type 2); arrow in (G), the NMJ on m12 emerges from under m13, and there is no axon crossing over m13 (Type 5). (H) Bar graph of total phenotypic percentages. Numbers of A2 hemisegments examined indicated on bars. (I) Bar graph showing the distribution of phenotypes among the categories illustrated in (A). Bar in (B), 50µm: applies also to (C–G).
By contrast, when muscle 12 innervation is examined in crosses where the muscle 12-specific 5053A-GAL4 is used to drive perturbing agents, the results can be more easily interpreted, because the axons are known and only the last stage of their targeting should be affected. In 5053A::Trn larvae, the most prevalent phenotypes (Figure 4I) were Type 2 (reduced innervation of 12; Figure 4F) and Type 5 (muscle 12-innervating axons grow under 13 to reach 12 rather than over it; Figure 4G). The Type 2 phenotype could be interpreted as repulsion from muscle 12 when it expresses Trn, but the Type 5 phenotype is not readily explained in the context of a simple attractive or repulsive model, since Trn on muscle 12 seems to be affecting RP5 axonal growth past the adjacent muscle 13 (a non-autonomous effect).
To better understand the effects of driving targeting cues on muscle 12, we expressed Caps from 24B, H94, and 5053A. Consistent with published data (Shishido et al., 1998), the phenotypes observed in crosses of UAS-Caps to 24B-GAL4 were primarily Type 1 (12→13 loopbacks; 70%); this was also observed for H94-driven expression. If Caps is an attractive cue for muscle 12 innervation by RP5, as suggested by these results, then driving excess expression on muscle 12 might be expected to have no effect. However, we found that overexpressing Caps on muscle 12 produced primarily Type 2 and Type 5 phenotypes, exactly as observed for Trn (Supplementary Figure 7). This indicates that Caps cannot be understood in simple terms as an attractive muscle 12 cue. Rather, muscle 12 expression of either Caps and Trn produces complex changes in RP5 targeting, possibly as a consequence of increased occupancy of the putative neuronal Caps/Trn receptor (see below for further discussion). In addition, our embryonic data (Fig. 3) and published results (Taniguchi et al., 2000) show that neuronal Caps and Trn are also important for axon guidance. These neuronal functions might involve interactions with a different receptor(s).
CG14351/Haf is a targeting cue required for innervation of all ventrolateral muscles
We initially characterized CG14351 expression and LOF phenotypes in embryos. There are two GAL4 enhancer trap lines just upstream of the 5' end of the sequenced CG14351 Drosophila Gold Collection (DGC) cDNA, with insertion sites separated by 130 bp. One has an expression pattern (http://flymap.lab.nig.ac.jp/~dclust/getdb.html) described as "muscle subset plus salivary glands", and the other as "CNS, gut subset, salivary glands". The "muscle subset" insertion line, P(GawB)NP0212, drives lacZ expression at highest levels in the VLMs and ventral muscles during part of stage 16 (Supplementary Figure 2).
To examine protein expression, we made a mouse antibody against a fragment of the XC domain expressed in E. coli. We confirmed that the antibody recognizes the protein by showing that it stained the appropriate striped pattern in germ-band extended embryos in which CG14351 was driven by engrailed-GAL4 (data not shown). In wild-type embryos, the antibody stains muscles, the CNS, the PNS, and the salivary glands (Supplementary Figure 2). Thus, motor axon phenotypes caused by CG14351 LOF mutations could arise from loss of expression in neurons, muscles, or both.
We were primarily interested in whether CG14351 functions as a muscle targeting cue, so we examined both conventional mutations and CG14351 RNAi constructs expressed in muscles. The RNAi lines were generated by the Vienna Drosophila RNAi Center (VDRC) (Dietzl et al., 2007) and the National Institute of Genetics, Japan (NIG). As described below, three insertion mutations and two independent RNAi constructs crossed to pan-muscle drivers all produced strong ISNb phenotypes and generated similar kinds of targeting errors.
The CG14351 gene spans 53 kb, and has a 43 kb second intron that contains coding exons for two other genes of unknown function. The CG14351 coding sequence is all 3' to this intron. We examined two PiggyBac splice-trap insertions from the Exelixis collection within the second intron, RBe04649 and RBe02960. These are unidirectional splice-traps, and are >7 kb away from the transcribed regions for the two embedded genes (CG10869, CG31935), both of which are transcribed in the opposite direction from CG14351. (See http://flybase.bio.indiana.edu/cgi-bin/gbrowse/dmel/?Search=1;name=FBgn0031349 for the transcript map).
We also obtained a line, LL01240, with an insertion of a new splice-trap PiggyBac vector (Schuldiner et al., 2008) at position −903 relative to the 5' end of the DGC cDNA. Based on the LL01240 phenotype (see below), we speculate that there may be additional 5' untranslated exon(s) in this gene, so that LL01240 would perturb splicing. The LL01240 insertion should only affect the CG14351 gene, as it is >30 kb from any other gene. To confirm that the LL01240 phenotype is not due to other insertions on the chromosome, we examined it in transheterozygous combinations with two different deficiency (Df) mutations that remove the CG14351 gene, as well as in transheterozygotes with RBe02960.
We obtained three UAS-driven RNAi lines for CG14351 from the VDRC and NIG collections. The sequences used for the UAS-RNAi constructs differ between VDRC and NIG lines. All three produced lethality when crossed to a strong pancellular driver, tubulin (tub)-GAL4. These pancellular RNAi embryos also had CNS defects. Knockdown of neuronal CG14351 by crossing Elav-GAL4 to the strongest RNAi line, VDRC2, also produced alterations in the CNS axon ladder, suggesting that neuronal CG14351 is involved in axon guidance (data not shown). However, the insertion mutations did not produce CNS defects.
All three insertion mutants and the two strongest RNAi lines crossed to muscle drivers produced a similar spectrum of embryonic ISNb phenotypes, as assayed by staining with mAb 1D4. These are ordered by phenotypic strength as follows: VDRC2 × 24B-GAL4 (60% of hemisegments have defects) > LL01240 (homozygotes or over Dfs; 50%) > RBe04649, RBe02960, NIG2 × 24B-GAL4, and VDRC2 × G14-GAL4 (30–40%). Driver-alone and balancer/+ controls were all <10% (Figure 5K).
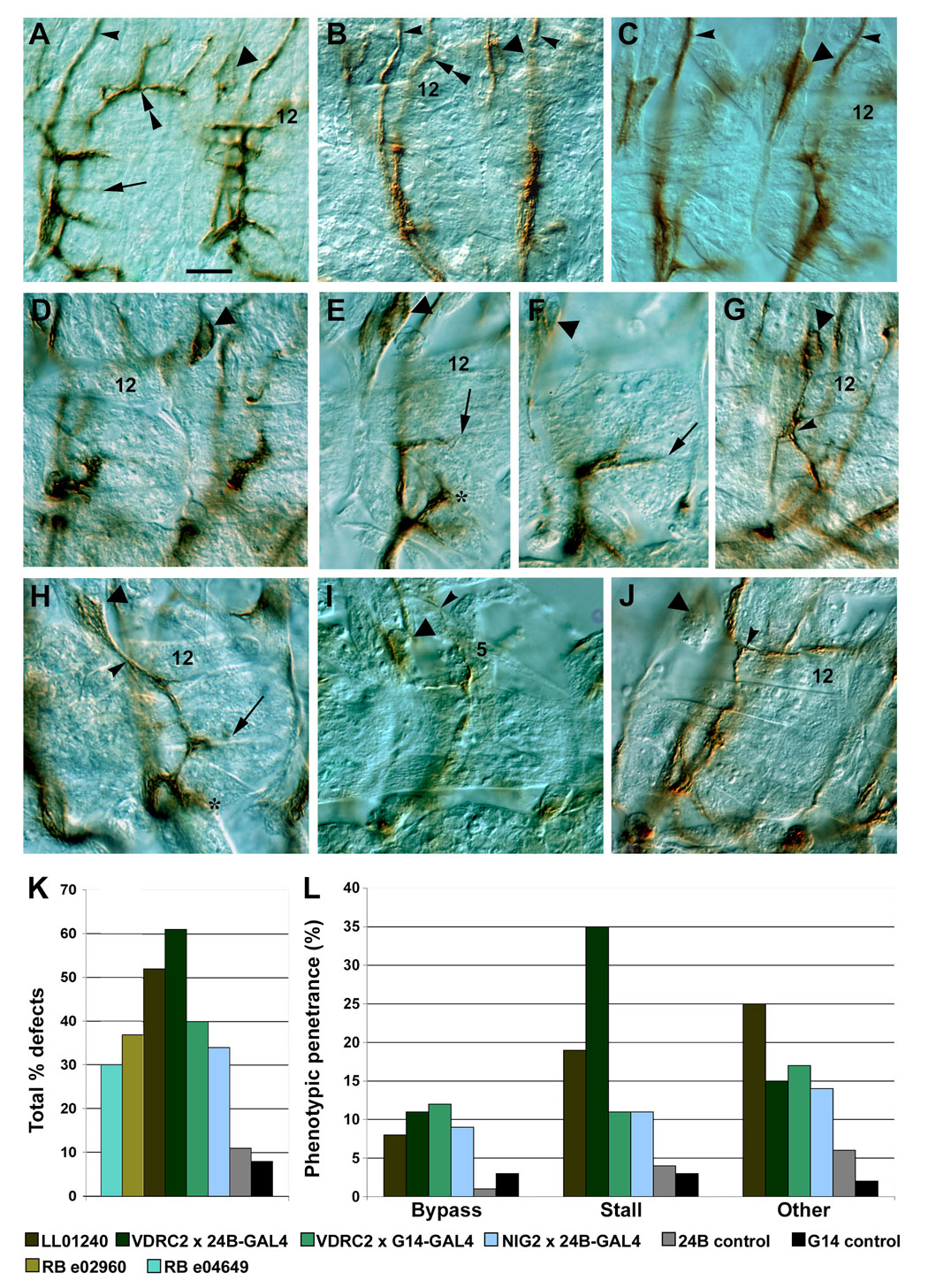
Figure 5. haf (CG14351) mutant and RNAi phenotypes in embryos.
(A–J) ISNb nerves (brown) in late stage 16/early stage 17 embryos stained with mAb 1D4, visualized with HRP immunohistochemistry and DIC optics. m12 is labeled for reference in many panels; arrows indicate m6/7 NMJs, triangles indicate LBD cells (part of the TN), and double arrowheads indicate the SNa. Single arrowheads in (A–C): ISN. The allele in all images of mutants is LL01240. (A) Early stage 17 control. (B,C) Bypass phenotypes, in the haf mutant. In the left hemisegment in (B) the ISNb extends along the SNa, while in the right hemisegment in (B) and the left hemisegment in (C) it extends along the ISN. In the right hemisegment in (C) the ISNb appears to initially follow the SNa, then leave it and extend onto the VLMs. (D–F) Various kinds of stall phenotypes. (D,E) VDRC2 haf RNAi × 24B-GAL4. The ISNb is truncated and curled, ending at the 6/7 cleft in (D); in (E) the ISNb is split at the exit junction; one part stops at the 6/7 cleft, and the other forms an abnormal branch to m14 (asterisk). (F) VDRC2 haf RNAi × G14-GAL4. The ISNb stops at the 6/7 cleft and forms a forked NMJ. (G–J) "Other" phenotypes. (G) VDRC2 haf RNAi × 24B-GAL4. The ISNb emerges into focus, grows across the internal surfaces of the VLMs, and joins the TN at the point marked by the arrowhead. (H) VDRC2 haf RNAi × G14-GAL4. The ISNb splits, forming a short ventral branch (*); the remainder of the nerve grows over the VLMs, forming NMJs at m6/7 and m12/13, then joins the TN (arrowhead), which has abnormally crossed over the ISN. (I,J) haf mutant. In (I) the ISNb grows underneath the VLMs, splits at the dorsal edge of m12, and sends one branch to the LBD, while the other branch (arrowhead) contacts the lateral muscle m5. In (J) the ISNb grows on top of the VLMs and splits (arrowhead) at the dorsal edge of m12. One branch extends along the muscle edge, while the other follows the ISN pathway. (K) Bar graph of phenotypic penetrances for the mutant and RNAi lines. n=135 hemisegments for the RBe04649 mutant, n=203 for RBe02960, n=418 for LL01240, n=226 for VDRC2 RNAi × 24B-GAL4, n=243 for VDRC2 RNAi × G14-GAL4, n=193 for NIG RNAi × 24B-GAL4, n=231 for 24B-GAL4 × w control, n=129 for G14-GAL4 × w control. Penetrances of other genotypes not indicated on bar graph: LL01240/Df(2L) dp-79b, 51% (n=218), LL01240/Df (2L) ast2, 57% (n=267), LL01240/RBe02960, 32% (n=264). Differences between mutants/RNAi and controls are significant (p<.001, Chi-square test). (L) Distribution of phenotypes in selected haf mutant and RNAi genotypes. Bar in (A), 20 µm; applies to all panels.
A wide variety of phenotypes were seen, including: 1) bypass phenotypes in which the ISNb failed to enter the VLM field and instead followed the ISN or SNa pathway (Figures 5B,C); 2) stall phenotypes in which the ISNb ended ventral to muscle 13, often following a incorrect trajectory to the stall point (Figures 5D–F); 3) ISNbs with several different kinds of abnormal trajectories (classified as "other"; phenotypic distribution in Figure 5L). Among these were: growth onto the TN (Figures 5F,G), growth past the VLMs to contact lateral muscle 5 (Figure 5I), growth to the dorsal edge of the VLMs followed by splitting (Figure 5J), and splitting at or near the exit junction (Figures 5E,H).
In summary, our data indicate that CG14351 is required for normal motor axon targeting into and within the VLM field. No consistent guidance errors are produced by loss of muscle CG14351. Rather, ISNbs appear to make a variety of different abnormal decisions, suggesting that they are unable to choose a trajectory. The phenotypes are specific for VLM and ventral muscle fields, as few errors (<10% penetrance) were observed for SNa (lateral muscles) and the ISN (lateral and dorsal muscles).
To further analyze CG14351 function, we drove the VDRC3 and NIG2 RNAi constructs with 24B, H94 (13,6,4), and 5053A (12 only), and examined the consequences in 3rd instar larvae (larvae do not survive when the strongest construct, VDRC2, is crossed to 24B-GAL4). All three drivers produced mistargeting phenotypes, with 24B-driven VDRC3 RNAi generating the highest penetrance (>40% of A2 hemisegments) (Figure 6J). A variety of different phenotypes were seen with 24B and H94, including Type 1 (12→13 loopback; Figure 6D), Type 7 (innervation of m6 from the dorsal side; Figure 6E), Type 2 (reduced innervation of 12; Figure 6I) and Type 3 (innervation of 12 from the dorsal side; Figure 6I). CG14531 overexpression (EY11244 × 24B) produced a different spectrum of phenotypes, with Type 1 (12→13 loopback; Figure 6B) and Type 7 (innervation of 6/7 from the dorsal side; Figure 6C) being most common. Since CG14351 is normally expressed on the VLMs, these phenotypes are either due to excess signaling through muscle CG14351 or to an alteration in the relative amounts of CG14351 on the VLMs versus other muscle groups.
Figure 6. Expressing haf (CG14351) RNAi in muscle subsets causes mistargeting.
(A–I) Confocal z-stack images of 3rd instar F1 larvae stained with 1D4 (green) and Alexa-phalloidin or UAS-GFP expression (gray). (A) Control (UAS-GFP, 24B-GAL4 × w). (B,C) Haf muscle overexpression (in EY11244 × UAS-GFP, 24B-GAL4). Arrow in (B), loopback collateral from m12 onto m13 (Type 1). Arrow in (C), ectopic innervation of m6 (Type 7). (D, E) VDRC3 haf RNAi × UAS-GFP, 24B- GAL4. Arrow in (D), 12→13 loopback (Type 1). Arrow in (E), ectopic m6 innervation (Type 7). (F) NIG2 haf RNAi × UAS-GFP, 24B-GAL4. Arrow, a long loopback collateral from m12 to m6. (G, H) VDRC3 haf RNAi × 5053A-GAL4 (muscle 12 only). Arrows, axons not only make normal NMJs on m12, but also arborize on m5 (arrow in G) or m8 (arrows in H). (I) VDRC3 haf RNAi × H94-GAL4 (muscles 13, 6). Arrows, ectopic NMJ wrapping over the dorsal edge of m12 (Type 3); arrowhead, abnormal ending on m13 (extreme Type 2). Asterisk, normal NMJ on m30/14. (J) Bar graph of mistargeting penetrances in control, overexpression, and RNAi larvae with pan-muscle or muscle subset drivers. Number of A2 hemisegments examined indicated on bars. (K) Bar graph showing the distribution of mistargeting phenotypes among the categories illustrated in Figure 4A. Bar in (A), 50µm: applies also to (B–I).
Driving CG14351 RNAi with 5053A-GAL4 produced a unique phenotype (<40% of affected hemisegments) in which the muscle 12-innervating axons sent ectopic branches to lateral muscles 5 or 8 (Figures 6G, H). This phenotype was also seen in the embryo (Figure 5I).
Based on these findings, and on the fact that the other two mutations in this phenotypic category (inability to find VLM targets, with generation of varied ISNb phenotypes) are named clueless (an allele of ab) and walkabout (wako) (Hu et al., 1995; Vactor et al., 1993), we named this gene hattifattener (haf), an English translation of a word invented for the Moomin stories by the Finnish author Tove Jansson. Hattifatteners are worm-like creatures that float randomly around the world in little boats. In Japan, they are called Nyoro-nyoro (see http://en.wikipedia.org/wiki/Hattifattener).
Muscle knockdown of CG8561 produces larval mistargeting phenotypes
24B-GAL4-driven expression of CG8561 from two different EP-like lines (GS10548 and GE12785) produced synaptic bouton phenotypes and mistargeting (Figures 7B, H, J; Supplementary Table 3). We could not detect CG8561 mRNA in embryos using in situ hybridization. To examine protein expression, we generated a mouse antiserum against a fusion protein made in E. coli and a rabbit antiserum against a synthetic peptide. Both antisera were verified as being able to recognize ectopically expressed CG8561 as described above for anti-Haf. Staining with either antiserum showed that in wild-type embryos CG8561 is expressed in muscles, tracheae, and CNS axons. It is also localized to the NMJ in 3rd instar larvae (Supplementary Figure 3).
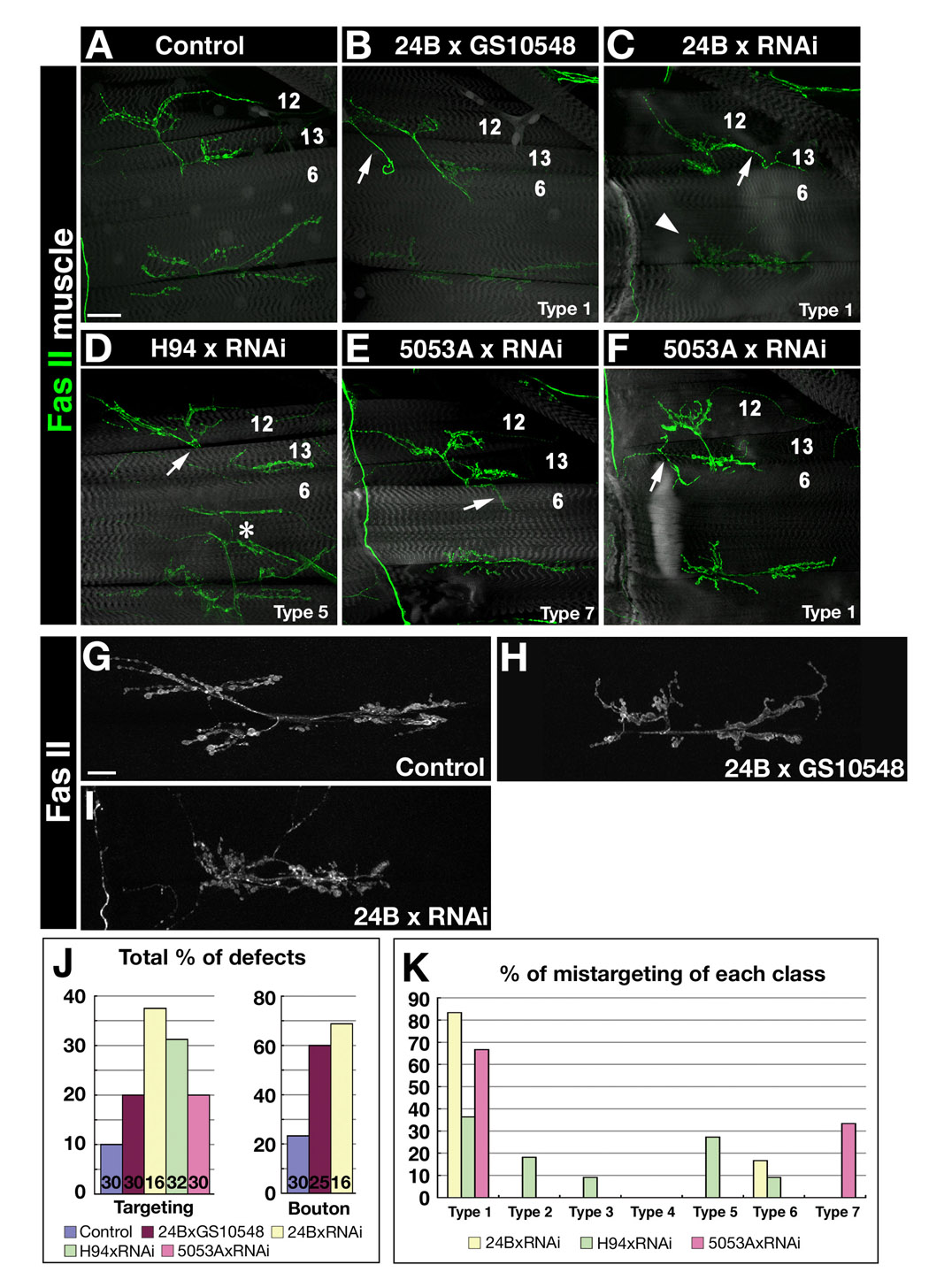
Figure 7. Expressing CG8561 RNAi in muscles causes mistargeting and synaptic phenotypes.
(A)–(C) Confocal z-stack images of 3rd instar F1 larvae stained with 1D4 (green) and Alexa-phalloidin or UAS-GFP expression (gray). (A) Control (UAS-GFP, 24B-GAL4 × w). (B) CG8561 muscle overexpression (in GS10548 × UAS-GFP, 24B-GAL4). Arrow, long loopback collateral from m12 to m6 (Type 1). (C) CG8561 RNAi × 24B-GAL4. Arrow, m12→m13 loopback (Type 1). Arrowhead, tangled NMJ arbor. (D) CG8561 RNAi × H94-GAL4 (muscles 13, 6). Arrow, abnormal NMJ on m12 emerging from under m13 (Type 5). Asterisk, normal NMJ on m30/14. (E, F) CG8561 RNAi × 5053A-GAL4 (muscle 12 only). Arrow in (E), ectopic innervation of m6 (Type 7); arrow in (F), m12→m13 loopback (Type 1). (G–I) Confocal z-stacks showing higher-magnification views of the 6/7 NMJ in the indicated genotypes. In (H), boutons are fused. In (H) and (I), the arbor is tangled. (J) Bar graph of penetrances for mistargeting and synaptic bouton phenotypes in control, overexpression, and RNAi larvae. Numbers of A2 hemisegments examined indicated on bars. (K) Bar graph showing the distribution of mistargeting phenotypes among the categories illustrated in Figure 4A. Bar in (A), 50µm; applies also to (B–F); in (G), 20µm; applies also to (H–I).
There are no insertion mutations in the CG8561 gene. We obtained an RNAi line for CG8561 from the VDRC, and crossed this to a variety of drivers. When crossed to tub-GAL4, CG8561 RNAi produced embryonic/early larval lethality, and the embryos had an uncondensed ventral nerve cord (Supplementary Figure 8). No phenotypes were produced by crossing CG8561 RNAi to the pan-neuronal driver Elav-GAL4.
CG8561 RNAi × 24B-GAL4 embryos did not have motor axon phenotypes. However, 3rd instar larvae expressing CG8561 RNAi in muscles displayed mistargeting and synaptic bouton phenotypes, and these were stronger than the overexpression phenotypes we initially identified in the screen (Figures 7B, H). 38% of A2 muscle RNAi hemisegments had mistargeting phenotypes, 80% of which were Type 1 (12→13 loopback; Figure 7C), while 69% had bouton phenotypes, characterized by tangled arbors that failed to extend normally along the 6/7 cleft (Figure 7I).
As with Haf, we also drove CG8561 RNAi in muscle subsets with H94 and 5053A. We obtained a clear result with 5053A (12 only): >65% of affected hemisegments (see bar graph of Figure 7K) had 12→13 loopback phenotypes (Type 1; Figure 7F).
Discussion
We assembled a database of 976 Drosophila CSS proteins that are likely to be involved in cell recognition events during development (Supplementary Table 1). We found EP lines that allowed us to express 410 of these genes in muscles (Supplementary Table 2), and defined all the genes that alter presynaptic NMJ terminal patterning and structure without visibly affecting the muscles themselves (screen summarized in Figure 1A). The screen identified 30 genes that cause mistargeting of axons within the VLM field with a penetrance of ≥30% (Table 1, Figure 2), and 55 genes that produce major alterations in synaptic boutons or the structures of NMJ arbors with ≥60% penetrance (Supplementary Figure 1, Supplementary Table 3).
LRR genes represented five of the 12 mistargeting genes of interest (orange+blue genes in Table 1). To evaluate the roles of the LRR genes in synaptic targeting, we examined LOF mutants, and also knocked down or overexpressed the genes using both pan-muscle drivers and drivers expressed only in specific muscle fibers. For mechanistic analysis, we focus here primarily on results obtained by driving targeting genes or RNAi constructs in muscle 12 only (driver pattern in Supplementary Figure 6), and examining the consequences for innervation of muscle 12 by RP5 and the 1s neuron. These results are interpretable because the axons are known and only the last stage of their targeting should be affected. By contrast, mistargeting phenotypes observed with pan-muscle expression could result from errors at any point along the axonal trajectory, and the axons that mistargeted cannot be identified in many cases. There may also be targeting errors that we cannot detect using mAb 1D4, because it labels all motor axons. We can only see NMJs that display morphological abnormalities, and thus could miss phenotypes in which an axon from one motor neuron is replaced by another one, if it forms a similar NMJ. Ideally, these experiments should be performed using reagents that label single identified motor axons, but these are not yet available.
Tartan and Capricious have redundant functions in targeting
LRRs are ~24 aa protein domains that can be found outside the cell or in cytoplasmic proteins. A chain of LRRs forms a concave binding surface that is used for interactions with other proteins. The LRR domains of Trn and Caps are interchangeable, suggesting that they can interact with a common receptor (Milan et al., 2005). Trn and Caps are involved in cell-cell interactions in tracheae and imaginal discs, and Caps regulates layer-specific targeting in the optic lobe (Krause et al., 2006; Milan et al., 2001; Shinza-Kameda et al., 2006).
Within the VLM field, Caps is expressed on muscle 12 (Shishido et al., 1998), while Trn appears to be expressed on all VLMs, but with higher levels on muscles 6 and 7 (Figure 3; Supplementary Figure 4, Supplementary Figure 5). trn caps double mutant embryos have stronger motor axon phenotypes than trn single mutants, and they exhibit ISNb terminal loop phenotypes that are suggestive of RP5 mistargeting (Figure 3).
Because muscle 12 NMJs send loopback branches onto muscle 13 when Caps is expressed in all muscles, it was proposed to be an attractive cue which facilitates targeting of RP5 to muscle 12 (Shishido et al., 1998). However, the true situation may be more complex, because selective overexpression of either Caps or Trn on muscle 12 produces phenotypes in which muscle 12-destined axons either stall on muscle 13 (Type 2), so that muscle 12 remains uninnervated, or grow under muscle 13 rather than over it to reach muscle 12 (Type 5; Figure4, Figure8, Supplementary Figure 7). These apparently non-autonomous effects (alteration of axonal extension over an adjacent muscle) might be explained by Trn- or Caps-induced alterations in the pattern of myopodia, projections from the muscle that reach out to contact innervating axons and direct their growth. Myopodia can extend over distances similar to the width of a muscle fiber (Ritzenthaler and Chiba, 2003; Ritzenthaler et al., 2000). Perhaps when Trn or Caps is expressed on muscle 12, the myopodia extend under muscle 13 rather than over it. If RP5 axons contact these aberrant myopodia, they may grow under 13 to reach 12; if they fail to contact them, they may stall on muscle 13.
Haf/CG14351 is necessary for axonal targeting to all ventrolateral muscles
CG14351, which we denoted as Haf, is a large protein (1316 aa), and LRRs occupy only aa ~100–350 of the XC domain. Haf has a signal sequence, a single transmembrane region, and a large cytoplasmic domain (~500 aa). It appears to be expressed by all VLMs (Supplementary Figure 2).
The embryonic motor axon phenotypes observed in the haf insertion mutant and in haf RNAi × pan-muscle-GAL4 embryos indicate that ISNb cannot innervate any of the VLMs in a normal manner if Haf is not expressed in muscles. Only 40–50% of ISNbs have a normal morphology. The remainder bypass onto the ISN or SNa or follow abnormal trajectories within the VLM field, sometimes contacting inappropriate targets (Figure 5). The phenotypes are highly variable, suggesting that many different kinds of errors are produced by loss of Haf.
We suggest that in the embryo Haf is a permissive muscle factor that is required for target selection by all muscles within the VLM field, but does not define the identities of specific fibers. This model is also consistent with the larval phenotypes that result from knocking down Haf expression on muscle 12 only. In this case, the RP5 and 1s axons seem to reach muscle 12 and form NMJs in a normal manner, but they also extend further and form ectopic synapses on lateral muscles 5 or 8 (Figure 6, Figure 8). This phenotype suggests that a stable NMJ on muscle 12 sometimes cannot form when Haf is knocked down, and in these cases the axons (or NMJ branches) continue to grow until they reach the lateral muscles.
CG8561 mediates synaptic targeting and arbor growth
CG8561 is a 1092 aa protein that has a signal sequence but lacks a transmembrane region. Its C-terminal sequence is characteristic of proteins that are attached to membranes by glycosyl-phosphatidylinositol anchors. It appears to be expressed by all muscles (Supplementary Figure 3).
CG8561 muscle RNAi and muscle overexpression produce mistargeting and NMJ arbor phenotypes in larvae. When CG8561 RNAi is expressed in muscle 12 only, the muscle 12 NMJ sends loopback branches to muscle 13 in 70% of affected hemisegments (Figure 7, Figure 8). This implies that CG8561 confers a preference for the RP5 and 1s axons to choose muscle 12, and in its absence these axons do not strongly prefer muscle 12 to the adjacent muscle 13.
CG8561 has a vertebrate ortholog, the acid-labile subunit (Als) of the IGF-1 binding complex. CG8561 mRNA is expressed in the larval fat body (FB) and in a group of neurosecretory cells (NSCs) that express insulin-like peptides. Starvation causes downregulation of the mRNA in the FB and NSCs. These data suggest that CG8561 (dAls) may be involved in insulin/IGF-1 signaling (Colombani et al., 2003). Interestingly, the single fly insulin/IGF-1 receptor, InR, is expressed in neurons and is required for guidance of photoreceptor axons into the optic lobe (Song et al., 2003).
The future of target selection in the neuromuscular system
Our identification of 30 mistargeting genes among the 410 CSS genes we screened suggests that there may be ~70 mistargeting genes in the entire cell-recognition database, and perhaps twice that many if genes with lower mistargeting percentages are included. If we assume that the screen is capable of identifying all CSS proteins involved in targeting, our results seem inconsistent with a simple version of the lock-and-key model discussed in the Introduction, because we did not find mRNAs or proteins within the mistargeting set that are expressed in small subsets of muscles. Also, the four LRR proteins examined in this paper have complex effects on targeting that are not explainable by a simple model (Figure 8).
We screened 53 LRR proteins, and 16 of these produced mistargeting, NMJ phenotypes, or both (Supplementary Table 3). If the failure of an LRR protein to produce a phenotype when overexpressed in muscles in the course of our screen indicates that it is not involved in targeting or synapse development, then there are 48 signal sequence-containing LRR proteins that remain to be examined for expression patterns, GOF phenotypes, and LOF phenotypes. A comprehensive analysis of this large family may help to define mechanisms involved in target selection and synaptic growth.
Experimental Procedures
Analysis of larval and embryonic phenotypes
For 3rd instar larval preparations, dissected fillets were fixed for 30 min with 4% paraformaldehyde at room temperature. Washing was done with 0.3% TritonX-100 in PBS. Blocking was done with 0.3% TritonX-100+0.1% BSA+5% normal goat serum in PBS. Incubation with primary antibody was done overnight at 4 °C. Labeled fillets were mounted with anti-fade Vectashield medium (Vector Laboratories, Inc., Burlingame, CA USA). Confocal images were captured with an LSM510 instrument. Images were processed using Adobe Photoshop. Staining of whole-mount embryos was done as described by (Patel, 1994). Staining of live-dissected embryos was done as in (Fox and Zinn, 2005). The following primary antibodies were used: mouse anti-FasII mAb 1D4 (DSHB), diluted 1:5; mouse anti-Futsch mAb 22C10 (DSHB) diluted 1:4; rabbit anti-Trn (Chang et al., 1993), diluted 1:400; mouse anti-Haf diluted 1:200; rabbit anti-CG8561 diluted 1:1000; mouse anti-CG8561 diluted 1:200; rat anti-Troponin T1 diluted 1:500 (Abcam Inc., Cambridge, MA, USA); goat anti-horseradish peroxidase (HRP) (Jackson ImmunoResearch, West Grove, PA, USA). Alexa Fluor-conjugated phalloidin was diluted 1:40 (Invitrogen-Molecular Probes, Carlsbad, CA, USA ). Cy3-conjugated or HRP-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA) were used at dilutions of 1: 400 (Cy3) or 1:200 (HRP).
Antibodies against Haf were generated using a GST fusion protein containing aa 543–689. Antibodies against CG8561 were made using a GST fusion protein containing aa 683–1019, or against a synthetic peptide (aa 1030–1046; VSRDSDGNTRKWFSGQC). Protein was purified from inclusion bodies in E. coli and injected into mice at the Caltech Monoclonal Antibody Facility, or into rabbits at a commercial facility (Medial & Biological Laboratories Co., Ltd).
Supplementary Material
Acknowledgements
We thank members of the Zinn and Suzuki groups, Akinao Nose, and Liqun Luo for helpful discussions, Violana Nesterova for technical assistance, figure preparation, and the gene name, Susan Ou for antibody work, Liqun Luo for the LL01240 insertion mutant, Stephen Cohen for the trn caps double mutant, Akinao Nose for UAS-Caps and anti-Caps antibody, and Allen Laughon for anti-Trn antibody. We also thank Flybase, the Bloomington, Szeged, Kyoto, and Harvard stock centers, GenExel, Inc., the VDRC, and the National Institute of Genetics for information and fly stocks. This work was supported by an NIH RO1 grant to K.Z., NS28182, by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Naito Foundation to M. Kurusu, and by a grant from the Mitsubishi Foundation to E. Suzuki. M. Kurusu was supported as a postdoc at Caltech by a Postdoctoral Fellowship for Research Abroad of the Japan Society for the Promotion of Science for Young Scientists. Confocal analysis was performed at the Caltech Biological Imaging Facility. Mouse antisera were generated by the Caltech Monoclonal Antibody Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrell S, Jackle H. Axon guidance of Drosophila SNb motoneurons depends on the cooperative action of muscular Kruppel and neuronal capricious activities. Mech Dev. 2001;109:3–12. doi: 10.1016/s0925-4773(01)00511-1. [DOI] [PubMed] [Google Scholar]
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M. Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development. 2003;130:6257–6272. doi: 10.1242/dev.00843. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chang Z, Price BD, Bockheim S, Boedigheimer MJ, Smith R, Laughon A. Molecular and genetic characterization of the Drosophila tartan gene. Dev Biol. 1993;160:315–332. doi: 10.1006/dbio.1993.1310. [DOI] [PubMed] [Google Scholar]
- Chiba A, Snow P, Keshishian H, Hotta Y. Fasciclin III as a synaptic target recognition molecule in Drosophila. Nature. 1995;374:166–168. doi: 10.1038/374166a0. [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Davis GW. Homeostatic control of neural activity: from phenomenology to molecular design. Annu Rev Neurosci. 2006;29:307–323. doi: 10.1146/annurev.neuro.28.061604.135751. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Fox AN, Zinn K. The heparan sulfate proteoglycan syndecan is an in vivo ligand for the Drosophila LAR receptor tyrosine phosphatase. Curr Biol. 2005;15:1701–1711. doi: 10.1016/j.cub.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Hoang B, Chiba A. 'Identify' and 'lock in': molecular integration during synaptic target recognition. Cell Mol Life Sci. 1999;55:1399–1406. doi: 10.1007/s000180050380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang B, Chiba A. Single-cell analysis of Drosophila larval neuromuscular synapses. Dev Biol. 2001;229:55–70. doi: 10.1006/dbio.2000.9983. [DOI] [PubMed] [Google Scholar]
- Hu S, Fambrough D, Atashi JR, Goodman CS, Crews ST. The Drosophila abrupt gene encodes a BTB-zinc finger regulatory protein that controls the specificity of neuromuscular connections. Genes Dev. 1995;9:2936–2948. doi: 10.1101/gad.9.23.2936. [DOI] [PubMed] [Google Scholar]
- Inaki M, Yoshikawa S, Thomas JB, Aburatani H, Nose A. Wnt4 Is a Local Repulsive Cue that Determines Synaptic Target Specificity. Curr Biol. 2007 doi: 10.1016/j.cub.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Johnson KG, Van Vactor D. Receptor protein tyrosine phosphatases in nervous system development. Physiol Rev. 2003;83:1–24. doi: 10.1152/physrev.00016.2002. [DOI] [PubMed] [Google Scholar]
- Klassen MP, Shen K. Wnt signaling positions neuromuscular connectivity by inhibiting synapse formation in C. elegans. Cell. 2007;130:704–716. doi: 10.1016/j.cell.2007.06.046. [DOI] [PubMed] [Google Scholar]
- Kose H, Rose D, Zhu X, Chiba A. Homophilic synaptic target recognition mediated by immunoglobulin-like cell adhesion molecule Fasciclin III. Development. 1997;124:4143–4152. doi: 10.1242/dev.124.20.4143. [DOI] [PubMed] [Google Scholar]
- Krause C, Wolf C, Hemphala J, Samakovlis C, Schuh R. Distinct functions of the leucine-rich repeat transmembrane proteins capricious and tartan in the Drosophila tracheal morphogenesis. Dev Biol. 2006;296:253–264. doi: 10.1016/j.ydbio.2006.04.462. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Bossing T, Technau GM, Bate M. The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J Neurosci. 1997;17:9642–9655. doi: 10.1523/JNEUROSCI.17-24-09642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Milan M, Perez L, Cohen SM. Boundary formation in the Drosophila wing: functional dissection of Capricious and Tartan. Dev Dyn. 2005;233:804–810. doi: 10.1002/dvdy.20384. [DOI] [PubMed] [Google Scholar]
- Milan M, Weihe U, Perez L, Cohen SM. The LRR proteins capricious and Tartan mediate cell interactions during DV boundary formation in the Drosophila wing. Cell. 2001;106:785–794. doi: 10.1016/s0092-8674(01)00489-5. [DOI] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler S, Chiba A. Myopodia (postsynaptic filopodia) participate in synaptic target recognition. J Neurobiol. 2003;55:31–40. doi: 10.1002/neu.10180. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler S, Suzuki E, Chiba A. Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat Neurosci. 2000;3:1012–1017. doi: 10.1038/79833. [DOI] [PubMed] [Google Scholar]
- Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, Cohen SM. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Budnik V. Introduction on the use of the Drosophila embryonic/larval neuromuscular junction as a model system to study synapse development and function, and a brief summary of pathfinding and target recognition. Int Rev Neurobiol. 2006;75:1–31. doi: 10.1016/S0074-7742(06)75001-2. [DOI] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K. Molecular mechanisms of target specificity during synapse formation. Curr Opin Neurobiol. 2004;14:83–88. doi: 10.1016/j.conb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Shinza-Kameda M, Takasu E, Sakurai K, Hayashi S, Nose A. Regulation of layer-specific targeting by reciprocal expression of a cell adhesion molecule, capricious. Neuron. 2006;49:205–213. doi: 10.1016/j.neuron.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Shishido E, Takeichi M, Nose A. Drosophila synapse formation: regulation by transmembrane protein with Leu-rich repeats, CAPRICIOUS. Science. 1998;280:2118–2121. doi: 10.1126/science.280.5372.2118. [DOI] [PubMed] [Google Scholar]
- Song J, Wu L, Chen Z, Kohanski RA, Pick L. Axons guided by insulin receptor in Drosophila visual system. Science. 2003;300:502–505. doi: 10.1126/science.1081203. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Shishido E, Takeichi M, Nose A. Functional dissection of drosophila capricious: its novel roles in neuronal pathfinding and selective synapse formation. J Neurobiol. 2000;42:104–116. doi: 10.1002/(sici)1097-4695(200001)42:1<104::aid-neu10>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Umemiya T, Takasu E, Takeichi M, Aigaki T, Nose A. Forked end: a novel transmembrane protein involved in neuromuscular specificity in drosophila identified by gain-of-function screening. J Neurobiol. 2002;51:205–214. doi: 10.1002/neu.10063. [DOI] [PubMed] [Google Scholar]
- Vactor DV, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- Winberg ML, Mitchell KJ, Goodman CS. Genetic analysis of the mechanisms controlling target selection: complementary and combinatorial functions of netrins, semaphorins, and IgCAMs. Cell. 1998;93:581–591. doi: 10.1016/s0092-8674(00)81187-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.