Summary
In this paper, we carry out an in-depth theoretical investigation for inference with missing response and covariate data for general regression models. We assume that the missing data are Missing at Random (MAR) or Missing Completely at Random (MCAR) throughout. Previous theoretical investigations in the literature have focused only on missing covariates or missing responses, but not both. Here, we consider theoretical properties of the estimates under three different estimation settings: complete case analysis (CC), a complete response analysis (CR) that involves an analysis of those subjects with only completely observed responses, and the all case analysis (AC), which is an analysis based on all of the cases. Under each scenario, we derive general expressions for the likelihood and devise estimation schemes based on the EM algorithm. We carry out a theoretical investigation of the three estimation methods in the normal linear model and analytically characterize the loss of information for each method, as well as derive and compare the asymptotic variances for each method assuming the missing data are MAR or MCAR. In addition, a theoretical investigation of bias for the CC method is also carried out. A simulation study and real dataset are given to illustrate the methodology.
1 Introduction
Missing data arise in nearly every type of application in the statistical sciences. Over the past 30 years, there has been an enormous literature on likelihood-based methods of estimation and inference for a wide variety of missing data problems, including missing covariate data in linear models, generalized linear models, generalized linear mixed models, and survival models, as well as missing response data for models of longitudinal data. Since the literature is too vast to list here, we refer the reader to three review articles that discuss various methods for handling missing data: Little (1992), Horton and Laird (1999), and Ibrahim, Chen, Lipsitz, and Herring (2005). There has also been some literature for likelihood-based methods for establishing identifiability and asymptotic properties of estimators in missing covariate problems including Robins and Rotnitzky (1995), Lipsitz, Ibrahim, and Zhao (1999), Herring and Ibrahim (2001), and Chen, Ibrahim, and Shao (2004). There also has been some work on models for longitudinal data with nonignorable responses, including Baker and Laird (1988), Ibrahim, Chen, and Lipsitz (2001), and Tang, Little, and Raghunathan (2003). There has been some work done on maximum likelihood estimation in the presence of ignorable or nonignorable missing response and/or covariate data in longitudinal models, including Stubbendick and Ibrahim (2003, 2006) and Chen and Ibrahim (2006). However, there has been almost no literature examining theoretical properties of estimators in the presence of both MAR responses and covariates in regression models. This type of missing data problem presents many new challenges in estimation and theory that do not arise in missing covariate problems or missing response problems alone.
We refer to a regression problem with missing covariates and responses as a “missing (x, y) problem” throughout. An important issue in a missing (x, y) problem is the contribution to the information matrix of the cases with missing responses alone, the contributions of the cases with missing covariates alone, and the contributions of the cases with missing covariates and responses. In particular, we consider theoretical properties of the estimates under three different estimation settings: complete case analysis (CC), a complete response analysis (CR) that involves an analysis of those subjects with only completely observed responses, and the all case analysis (AC), which is an analysis based on all of the cases. Under each scenario, we derive general expressions for the likelihood and devise estimation schemes based on the EM algorithm. We compare the three estimation methods in the normal linear model and characterize the loss of information for each method as well as derive and compare the asymptotic variances for each method assuming the missing data are MAR. For the linear model, we show that AC analysis has more information than the CR and CC analyses in the sense that the Fisher information for the AC analysis has a greater determinant and trace compared to the Fisher information matrices for the CR and CC analyses, and the CR analysis yields a Fisher information with a greater determinant and trace compared to the Fisher information matrix for the CC analysis. Moreover, we show that the asymptotic variances of the estimates for the CC analyses are larger than the other two methods (CR or AC), and the asymptotic variances for some estimates in the AC analysis are larger than that of the corresponding estimates based on a CR analysis. We also carry out a theoretical investigation of bias for the CC method and analytically show that CC estimates under certain settings are biased.
The rest of this paper is organized as follows. In Section 2, we consider the basic data structure for a regression model with MAR response and/or covariate data. In Section 3, we consider the three analysis methods: CC, CR, and AC. For each method, we give the likelihood function corresponding to the method. Section 4 gets into the heart of the theory and properties of estimators for the three methods and several results are given characterizing the behavior of the Fisher information matrix and asymptotic variances for each method for the normal linear model with missing (x, y). Section 5 examines bias issues for MAR response and covariate data. Section 6 presents a simulation study and real dataset illustrating the theoretical results derived in Section 4. A brief discussion is given in Section 7. In the Appendix A, we devise the computational schemes based on the EM algorithm for obtaining the maximum likelihood estimates (MLE’s), and derive E and M-steps of the EM algorithm as well as the observed information matrix based on the observed data using Louis’s method for the missing (x, y) problem.
2 Model and Data Structure
2.1 Model
Suppose that {(xi, yi), i = 1, 2,…, n} are independent observations, where yi is the response variable, and xi = (xi1, … , xip)′ is a p × 1 random vector of covariates. We specify the joint distribution of (xi, yi) by specifying the conditional distribution of yi given xi, denoted [yi | xi], and the marginal distribution of xi, denoted [xi].
We let
denote the joint density for the marginal distribution [xi], where where α is the vector of model parameters for i = 1, 2, … , n. Assume that the distribution function for [yi | xi] is of the form
| (2.1) |
where β = (β1, β2,…, βp)′ denotes the p × 1 random vector of regression coefficients, and ζ is the column vector of nuisance parameters. In (2.1), we assume that the distribution [yi | xi] depends on xi and β only through x′β. If an intercept is included in the model, xi and β are modified accordingly.
The generalized linear model (GLM) is a special case of (2.1). In the GLM, the conditional density of [yi|xi] is given by
| (2.2) |
where θi = θ(ηi) is the canonical parameter, , and τ is a dispersion parameter. The functions b and c determine a particular family in the class, such as the binomial, normal, Poisson, etc. The functions ai(τ) are commonly of the form , where the wi’s are known weights. Thus, (2.1) reduces to the GLM with
and ζ = τ.
2.2 Missing Data Structures
We consider a general setting in which yi and some components of xi may be missing. Let Mi = {1 ≤ l ≤ p : xil is missing}, which denotes the set of indices for the ith missing covariates. We also let Ω = {1,2,…,p} denote the whole index space for xi. Table 1 gives the general data structure and characterizations of the various missing data patterns in the missing (x, y) problem.
Table 1.
The Data Structure with Various Missing Patterns
| Pattern (Block) | Response | Covariates |
|---|---|---|
| yi | xi | |
| B1 | observed | observed (Mi = θ) |
| B2 | Observed | partially or all missing (Mi ≠ θ or Mi = Ω) |
| B3 | missing | observed (Mi = θ) |
| B4 | missing | only partially missing (Mi ≠ θ and Mi ≠ Ω) |
| B5 | missing | completely missing (Mi = Ω) |
We denote each pattern above by Bj, j = 1,…, 5, and refer to Bj as the jth pattern or jth block. B1 denotes the portion of the data with both yi and xi completely observed. In B2, yi is observed while each xi is at least partially missing or completely missing. In B3, yi is missing and xi is completely observed, in B4, yi is missing but at least partial xi is observed, and both yi and xi are completely missing in B5.
Based on the data structure given in Table 1, we use yi if the ith response is observed and yi,mis if the ith response variable is missing. Also, we write where and xi,obs is the observed portion of xi.
3 Three Analysis Methods
In this section, we assume that the missing response and the missing covariates are missing at random (MAR). Under the MAR assumption, we only need to model [yi, xi]. We give the forms of the observed data log-likelihood functions under three analysis methods: complete case (CC) analysis, complete response (CR) analysis, and all case (AC) analysis.
3.1 Complete Case (CC) Analysis
Because standard techniques for regression models require full response and covariate information, one simple way to avoid the problem of missing data is to analyze only those subjects who are completely observed. This method is known as a complete case (CC) analysis.
Based on the data structure displayed in Table 1, the CC analysis uses the portion of data given in Block B1. Thus, the likelihood function under this method is given by
| (3.1) |
where θ = (β, ϕ, α), and the log-likelihood function is given by
| (3.2) |
3.2 Complete Response (CR) Analysis
The complete response cases (CR) analysis is to analyze only those subjects whose responses are completely observed. Thus, in the CR analysis, we only include the portion of data given in Blocks B1 and B2 of Table 1. The likelihood function under CR is given by
| (3.3) |
and the log-likelihood function is given by
| (3.4) |
3.3 All Case (AC) Analysis
The all cases (AC) analysis uses the whole data. The likelihood function is given by
| (3.5) |
Since
the likelihood function Lac(θ) reduces to
| (3.6) |
Thus, the portion of the data given in Block B5 of Table 1 does not make any contribution to the likelihood function even under the AC analysis under the MAR assumption.
Using (3.6), the log-likelihood function is given by
| (3.7) |
4 Theoretical Comparisons Between CC, CR, and AC for the Normal Linear Regression Model
In this section, we characterize the properties of the three analysis methods by examining the Fisher information matrix under each method and determining information loss (gain) for each method as well as comparing the asymptotic variances for each method. This comparison allows us to examine the efficiency of each method. To facilitate comparisons in this section, we assume that the missing data are MCAR, since closed-form analytic results for the Fisher information are available in the (x, y) missing problem only under MCAR.
4.1 Simple Linear Regression Model with Missing Responses and Covariates
We first consider a simple normal regression model with a single covariate and unit variances. In this case, we have
| (4.1) |
Write θ = (β′, α)′. Let nj = #(Bj) be the cardinality of Bj for j = 1, 2, 3 and n = n1 + n2 + n3. For the CC analysis, we have
and the Fisher information matrix is given by
| (4.2) |
For the CR analysis, we have
After some messy algebra, we obtain the Fisher information matrix given by
| (4.3) |
For the AC analysis, the log-likelihood function is given by
The corresponding Fisher information matrix is given by
| (4.4) |
Using (4.2), we have
| (4.5) |
Observe that
where
and . After some algebra, we have
Note that u′A−1u = n2/n1. Thus, we have
| (4.6) |
the trace of Icr is given by
| (4.7) |
and the inverse matrix of Icr is given by
| (4.8) |
Similarly, we can write
where . We have
| (4.9) |
the trace of Iac is given by
| (4.10) |
and the inverse matrix of Iac is given by
| (4.11) |
where .
Using (4.5) — (4.11), we are led to the following results.
Result 4.1
Based on either the determinant or the trace of Fisher information matrix, AC yields most gain in information over both CR and CC, and CR gains more information than CC. Specifically, we have
and
The inverse of the Fisher information gives the asymptotic variance and covariance matrix of the MLE’s under each analysis method. Now, let Var and Var (α̂.) denote the asymptotic variances under each of CC, CR, AC. Then, we have the following results.
Result 4.2
(i) CR leads to smaller asymptotic variances for all parameters than CC. Specifically, we have
(ii) AC improves the asymptotic variances for β0 and α over CR, but not for β1. Specifically, we have
where b* and are given in (4.11).
From Result 4.2, the additional information from Block B3 does improve the standard errors of and α̂. Surprisingly, the information from Block B3 does not help improve the standard error of .
4.2 Multiple Linear Regression Model with Missing Responses and Covariates
To further examine the theoretical relationship among these three analysis methods, we consider a multiple normal linear regression model with p ≥ 2. For illustrative purposes, it suffices to consider two missing covariates. Specifically, we assume that xi,p−1 and xi,p have at least one missing value and xi1, xi2, and xi,p−2 are observed in all cases as we have shown in Section 3.3 that the cases in Block 5 do not make any contribution to the log-likelihood function lac(θ). For notational convenience, we let zi1 = (1, xi1,…,xi,p−2)′. We further assume
where α1 = (α10, α11,…,α1,p−2)′, and
where α21 = (α20, α21,…, α2,p−2)′. We assume all variances are known. For ease of exploration, we choose .
In this setting, we need to consider the cases from Blocks 1 to 4.
For Blocks 2 and 4, we assume , where
for j = 2, 4, and B24 = {i : both xi,p−1 and xip are missing}. We further let njk = #(Bjk) be the cardinality of Bjk for j = 2, 4. Then we have .
Define , where j = 1, 22, 23, 24, 3, 42, 43, and . For the CC analysis, we have
and the Fisher information matrix is given by
where .
For the CR analysis, we have
where
and
After some messy algebra, we obtain the Fisher information matrix given by
where
.
For the AC analysis, the log-likelihood function is given by
The corresponding Fisher information matrix is given by
| (4.12) |
where .
For ease of exploration, we choose p=2, in other words, the completely observed covariates only include the intercept.
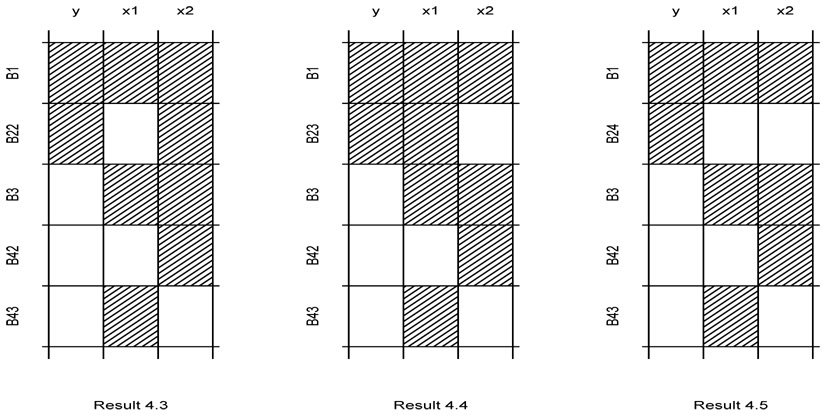
Result 4.3
(i) When n23 = n24 = 0, CR leads to smaller asymptotic variances for β1 and β2 than CC. Specifically, we have
where .
(ii) When n23 = n24 = 0, AC leads to smaller asymptotic variances for β1 and β2 than CR. Specifically, we have
where .
Result 4.4
(i) When n22 = n24 = 0, CR leads to smaller asymptotic variances for β1 and β2 than CC. Specifically, we have
(ii) When n22 = n24 = 0, AC improves the asymptotic variances for β1 over CR, but not for β1. Specifically, we have
where .
Result 4.5
(i) When n22 = n23 = 0, CR leads to smaller asymptotic variances for β than CC. Specifically, we have
where .
(ii) When n22 = n23 = 0, AC leads to smaller asymptotic variances for β than CR. Specifically, we have
where .
Remark 1
The information in Block B43 does not improve the asymptotic variances of the estimates of β1 and β2 in all of the three situations considered here.
Remark 2
When n23 = n24 = 0 (Result 4.3), the differences of the asymptotic variances for β1 and β2 do not depend on β2 comparing CR to CC and AC to CR.
Remark 3
When n22 = n24 = 0 (Result 4.4), the differences of the asymptotic variances for β1 and β2 do not depend on β1 comparing CR to CC and AC to CR.
Remark 4
When n22 = n23 = 0 (Result 4.5), the ratios of the asymptotic variances improvement of β1 versus β2 equal to for CR versus CC and AC versus CR, i.e.
Remark 5
When n23 = n24 = 0, the differences of the asymptotic variances of β1 are monotone decreasing function of n1 for CR versus CC and AC versus CR. Other monotonic properties are listed in Table 2.
Table 2.
Monotonic Properties for CR versus CC and AC versus CR
| Missing Pattern | Parameter | n1 | n22 | n23 | n24 |
|---|---|---|---|---|---|
| NM | NM | ||||
| Result 4.3 | NM | NM | |||
| n23 = n24 = 0 | NM | NM | |||
| NM | NM | ||||
| ↘ | NM | ||||
| Result 4.4 | ↘ | ↗ | |||
| n22 = n24 = 0 | ↘ | ↗ | |||
| ↘ | ↗ | ||||
| Result 4.5 | ↘ | ↗ | |||
| n22 = n23 = 0 | ↘ | ↗ | |||
| ↘ | ↗ | ||||
5 Analysis of Bias
In this section, we examine bias in the CC situation when the missing data are MAR. For inference with only missing response data, Little and Rubin (2002, page 43.) note without proof that when the data are MAR, the CC estimates are not biased when the missing data mechanism depends only on the covariates and not the response. The estimates, however, are biased if the missing data mechanism depends on the response. We now examine this bias issue in the missing (x, y) problem, where missingness is MAR.
Based on the data structure given in Table 1, define missing data indicators as
and
for j = 1, 2,…,p.
Let f(ri|ϕ, yi, xi denote the distribution of ri, which may possibly depend on yi and xi, where ϕ is the vector of parameters in the ri model. Under the MAR assumption, following models for ri are possible:
| (5.1) |
| (5.2) |
where ○ denotes the direct product, or
| (5.3) |
Note that the model specified by (5.1) defines MCAR and some other versions of the MAR models can be considered as well.
Because standard techniques for regression models require full response and covariate information, one simple way to avoid the problem of missing data is to analyze only those subjects who are completely observed. This method is known as a complete case (CC) analysis.
Based on the data structure displayed in Table 1, the CC analysis uses the portion of data given in Block B1. Thus, the likelihood function under this method is given by
| (5.4) |
where θ = (β,α,ϕ), and the log-likelihood function is given by
| (5.5) |
Under CC, we will make conditional inference given ri = 1, where 1 = (1, 1,…,1)′. More specifically, we need to consider conditional distribution [yi, xi|ri = 1] in examining biasness of the MLE’s and in deriving the Fisher information matrix. We assume throughout that ϕ is distinct from β and α.
Under the model given by (5.1), we have
Thus, under MCAR, the conditional distribution of (yi, xi) given ri = 1 is the same as the unconditional distribution and hence, the MLE’s of β and α are unbiased or asymptotically consistent under certain usual regularity conditions.
Under MAR with the model given by (5.2) for ri, we have
| (5.6) |
From (5.6), it is easy to see that the MLE of β is unbiased or asymptotically consistent, but the MLE of α may not be in this case.
Under the MAR with the model given by (5.3) for ri, we obtain
| (5.7) |
In this case, the MLE’s for both β and α are likely to be biased.
To obtain the closed form analytical results for (5.6) and (5.7), we consider the simple normal regression model given by (4.1). For notational simplicity, we assume that both yi and xi are observed for i = 1, 2,…,m. Let yobs = (y1, y2,…, ym)′, , and robs = (1, 1,…,1)′. Then, the MLE of β is given by and the MLE of α is .
In (5.6), we assume . Then, (5.6) implies
and
| (5.8) |
Thus, we have
which is unbiased. However,
which may be biased. Also, an analytical derivation of the Fisher information matrix is not possible as the conditional distribution of xi given ri = 1 involves an analytically intractable integral.
Under MAR given by (5.3), we assume a logistic regression model for rix, i.e.,
From (5.7), we obtain
Thus, E[yi|β, α, ϕ, xi, ri = 1] ≠ β0 + β1xi. In this case, both and may be biased. Again, an analytical derivation of the Fisher information matrix is not possible.
6 Simulation Studies and a Real Data Example
In this section, we present two detailed simulation studies and a real data example, demonstrating the various properties of the CC, CR, and AC methodology for analyzing MCAR and MAR response and/or covariate data in linear regression and logistic regression. In particular, we study efficiency and bias in the estimates for the three methods for these two types of regression models.
6.1 Simulation Study I: Normal Linear Regression Model with MCAR Response and Covariate Data
We consider a multiple linear regression model with an intercept, a completely observed covariate and two missing covariates. 5000 replicates with n = 500 subjects are considered. The response model is yi ~ N(β0 + β1zi1 + β2xi1 + β3xi2, 1), where zi is simulated from Unif(0, 1), xi1 is simulated from N(α10 + α11zi1, 1), xi2 is simulated from N(α20 + α21zi1 + α22xi1, 1), and yi, xi1 and xi2 are missing for some subjects. In each simulation, the sizes for each missing pattern, n11, n2, n23, n24, n3, n42, n43 and n5, were varied in order to evaluate the various properties of the CC, CR, and AC methods. To better study the differences of the asymptotic variances of the estimates of the regression coefficients using the three methods, we calculated the variances in two ways: plugging in the true parameter values as well as plugging in the maximum likelihood estimates into the Fisher information matrix.
Table 3 gives the simulation results of the linear regression model with the variances evaluated at the true parameter values. We note here that the variance estimates decrease monotonically based on the three methods, CC, CR, and AC. In particular, Result 4.3 and Result 4.4 hold and Remark 1 – Remark 3 hold, but not Remark 4. We note that Remark 1 is only true for the regression coefficients of the missing covariates but not of the completely observed covariates. The information in Block B43 does improve the asymptotic variances of regression coefficients of the zi0 = 1 and zi1 when the AC method is used.
Table 3.
Variance Comparison with True Parameters Plugged-in for Linear Regression
| (n1, n22, n23, n24, n3, n42, n43, n5) | Para | CC (VCC) | CR (VCC-VCR) | AC (VCR-VAC) |
|---|---|---|---|---|
| (50, 200, 0, 0, 100, 100, 50, 0) | β0 = 1.0 | 1.224 ×10−1 | 6.111 ×10−2 | 1.451 ×10−2 |
| β1 = 1.0 | 2.895 ×10−1 | 1.420 ×10−1 | 3.481 ×10−2 | |
| β2 = 2.0 | 1.000 ×10−1 | 6.419 ×10−2 | 1.117 ×10−3 | |
| β3 = 3.0 | 2.000 ×10−2 | 1.336 ×10−2 | 8.718 ×10−6 | |
| (50, 200, 0, 0, 100, 100, 50, 0) | β0 = 1.0 | 1.224 ×10−1 | 6.111 ×10−2 | 1.451 ×10−2 |
| β1 = 1.0 | 2.895 ×10−1 | 1.420 ×10−1 | 3.481 ×10−2 | |
| β2 = 2.0 | 1.000 ×10−1 | 6.419 ×10−2 | 1.117 ×10−3 | |
| β3 = 2.5 | 2.000 ×10−2 | 1.336 ×10−2 | 8.718 ×10−6 | |
| (50, 200, 0, 0, 100, 100, 20, 30) | β0 = 1.0 | 1.224 ×10−1 | 6.111 ×10−2 | 1.424 ×10−2 |
| β1 = 1.0 | 2.895 ×10−1 | 1.420 ×10−1 | 3.412 ×10−2 | |
| β2 = 2.0 | 1.000 ×10−1 | 6.419 ×10−2 | 1.117 ×10−3 | |
| β3 = 3.0 | 2.000 ×10−2 | 1.336 ×10−2 | 8.718 ×10−6 | |
| (50, 0, 200, 0, 100, 100, 50, 0) | β0 = 1.0 | 1.224 ×10−1 | 1.660 ×10−2 | 8.930 ×10−3 |
| β1 = 1.0 | 2.895 ×10−1 | 3.001 ×10−2 | 2.380 ×10−2 | |
| β2 = 2.0 | 1.000 ×10−1 | 3.509 ×10−2 | 1.749 ×10−3 | |
| β3 = 3.0 | 2.000 ×10−2 | 8.372 ×10−3 | 0 | |
| (50, 0, 200, 0, 100, 100, 50, 0) | β0 = 1.0 | 1.224 ×10−1 | 1.660 ×10−2 | 8.930 ×10−3 |
| β1 = 1.0 | 2.895 ×10−1 | 3.001 ×10−2 | 2.380 ×10−2 | |
| β2 = 1.5 | 1.000 ×10−1 | 3.509 ×10−2 | 1.749 ×10−3 | |
| β3 = 3.0 | 2.000 ×10−2 | 8.372 ×10−3 | 0 | |
| (50, 0, 200, 0, 100, 100, 20, 30) | β0 = 1.0 | 1.224 ×10−1 | 1.660 ×10−2 | 8.921 ×10−3 |
| β1 = 1.0 | 2.895 ×10−1 | 3.001 ×10−2 | 2.378 ×10−2 | |
| β2 = 2.0 | 1.000 ×10−1 | 3.509 ×10−2 | 1.749 ×10−3 | |
| β3 = 3.0 | 2.000 ×10−2 | 8.372 ×10−3 | 0 | |
| (50, 0, 0, 200, 100, 100, 50, 0) | β0 = 1.0 | 1.224 ×10−1 | 7.183 ×10−3 | 2.314 ×10−3 |
| β1 = 1.0 | 2.895 ×10−1 | 1.057 ×10−2 | 4.814 ×10−2 | |
| β2 = 2.0 | 1.000 ×10−1 | 1.439 ×10−2 | 8.327 ×10−3 | |
| β3 = 3.0 | 2.000 ×10−2 | 5.173 ×10−3 | 1.090 ×10−3 | |
| (50, 0, 0, 200, 100, 100, 50, 0) | β0 = 1.0 | 1.224 ×10−1 | 1.116 ×10−2 | 3.274 ×10−3 |
| β1 = 1.0 | 2.895 ×10−1 | 1.476 ×10−2 | 9.621 ×10−2 | |
| β2 = 2.0 | 1.000 ×10−1 | 2.191 ×10−2 | 1.112 ×10−3 | |
| β3 = 2.0 | 2.000 ×10−2 | 7.754 ×10−3 | 1.264 ×10−3 | |
| (50, 0, 0, 200, 100, 100, 20, 30) | β0 = 1.0 | 1.224 ×10−1 | 7.183 ×10−3 | 2.206 ×10−3 |
| β1 = 1.0 | 2.895 ×10−1 | 1.057 ×10−2 | 4.519 ×10−3 | |
| β2 = 2.0 | 1.000 ×10−1 | 1.439 ×10−2 | 8.327 ×10−3 | |
| β3 = 3.0 | 2.000 ×10−2 | 5.173 ×10−3 | 1.090 ×10−3 | |
Table 4 gives the simulation results of the linear regression model with the variances evaluated at the maximum likelihood estimates (MLE’s). The results show the gain in using the AC method over CR method, and using CR method over CC method. When when n22 = 200 and n23 = n24 = 0, the gain on the asymptotic variance of β3 is small compared to the AC to CR method and the difference of the empirical variances is slight.
Table 4.
Variance Comparison with MLE’s Plugged in for Linear Regression
| (n1, n22, n23n3, n42, n43, n5) | CC | CR | AC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Para | MLE | Sim:Vcc | Vcc | 95% CP | MLE | Sim:Vcc–Sim:Vcr | Vcc–Vcr | 95% CP | MLE | Sim:Vcr–Sim:Vac | Vcr–Vac | 95% CP | |
| (50, 200, 0, 0, 100, 100, 50, 0) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.2 | 0.994 | 6.922 ×10−2 | 6.214 ×10−2 | 97.4 | 0.992 | 1.448 ×10−2 | 1.490 ×10−2 | 96.8 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.2 | 0.995 | 1.500 ×10−1 | 1.442 ×10−1 | 97.4 | 0.994 | 4.252 ×10−2 | 3.566 ×10−2 | 97.5 | |
| β2 = 2.0 | 2.003 | 0.112 | 0.100 | 97.0 | 1.987 | 7.352 ×10−2 | 6.392 ×10−2 | 96.7 | 1.981 | 1.383 ×10−3 | 1.101 ×10−3 | 96.6 | |
| β3 = 3.0 | 3.000 | 0.022 | 0.020 | 97.1 | 3.006 | 1.483 ×10−2 | 1.332 ×10−2 | 97.7 | 3.009 | < 1.0 × 10−4 | < 1.0 × 10−4 | 98.0 | |
| (50, 200, 0, 0, 100, 100, 50, 0) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.2 | 0.994 | 6.922 ×10−2 | 6.214 ×10−2 | 97.4 | 0.992 | 1.448 ×10−2 | 1.490 ×10−2 | 96.8 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.2 | 0.995 | 1.500 ×10−1 | 1.442 ×10−1 | 97.4 | 0.994 | 4.252 ×10−2 | 3.566 ×10−2 | 97.5 | |
| β2 = 2.0 | 2.003 | 0.112 | 0.100 | 97.0 | 1.987 | 7.352 ×10−2 | 6.392 ×10−2 | 96.7 | 1.981 | 1.383 ×10−3 | 1.101 ×10−3 | 96.6 | |
| β3 = 3.0 | 2.500 | 0.022 | 0.020 | 97.1 | 2.506 | 1.483 ×10−2 | 1.332 ×10−2 | 97.7 | 2.509 | < 1.0 × 10−4 | < 1.0 × 10−4 | 98.0 | |
| (50, 200, 0, 0, 100, 100, 50, 0) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.2 | 0.994 | 6.922 ×10−2 | 6.214 ×10−2 | 97.4 | 0.992 | 1.405 ×10−2 | 1.461 ×10−2 | 96.8 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.2 | 0.995 | 1.500 ×10−1 | 1.442 ×10−1 | 97.4 | 0.994 | 4.117 ×10−2 | 3.495 ×10−2 | 97.4 | |
| β2 = 2.0 | 2.003 | 0.112 | 0.100 | 97.0 | 1.987 | 7.352 ×10−2 | 6.392 ×10−2 | 96.7 | 1.981 | 1.383 ×10−3 | 1.100 ×10−3 | 96.6 | |
| β3 = 3.0 | 3.000 | 0.022 | 0.020 | 97.1 | 3.006 | 1.483 ×10−2 | 1.332 ×10−2 | 97.7 | 3.009 | < 1.0 × 10−4 | < 1.0 × 10−4 | 98.0 | |
| (50, 0, 200, 0, 100, 100, 50, 0) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.3 | 0.987 | 1.939 ×10−2 | 1.686 ×10−2 | 97.4 | 0.991 | 8.583 ×10−3 | 9.371 ×10−3 | 97.2 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.3 | 1.010 | 4.199 ×10−2 | 3.087 ×10−2 | 97.7 | 1.008 | 2.295 ×10−2 | 2.459 ×10−2 | 97.9 | |
| β2 = 2.0 | 2.003 | 0.112 | 0.100 | 97.0 | 2.003 | 4.255 ×10−2 | 3.509 ×10−2 | 97.3 | 2.005 | 3.047 ×10−3 | 1.784 ×10−3 | 97.4 | |
| β3 = 3.0 | 3.000 | 0.022 | 0.020 | 97.1 | 3.000 | 1.017 ×10−2 | 8.376 ×10−3 | 97.1 | 2.999 | < 1.0 × 10−4 | < 1.0 × 10−4 | 97.0 | |
| (50, 0, 200, 0, 100, 100, 50, 0) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.3 | 0.987 | 1.939 ×10−2 | 1.686 ×10−2 | 97.4 | 0.991 | 8.583 ×10−3 | 9.371 ×10−3 | 97.2 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.3 | 1.010 | 4.199 ×10−2 | 3.087 ×10−2 | 97.7 | 1.008 | 2.295 ×10−2 | 2.459 ×10−2 | 97.9 | |
| β2 = 1.5 | 1.503 | 0.112 | 0.100 | 97.0 | 1.503 | 4.255 ×10−2 | 3.509 ×10−2 | 97.3 | 1.503 | 3.047 ×10−3 | 1.784 ×10−3 | 97.4 | |
| β3 = 3.0 | 3.000 | 0.022 | 0.020 | 97.1 | 3.000 | 1.017 ×10−2 | 8.376 ×10−3 | 97.1 | 2.999 | < 1.0 × 10−4 | < 1.0 × 10−4 | 97.0 | |
| (50, 0, 200, 0, 100, 100, 20, 30) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.3 | 0.987 | 1.939 ×10−2 | 1.686 ×10−2 | 97.4 | 0.991 | 8.625 ×10−3 | 9.341 ×10−3 | 97.2 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.3 | 1.010 | 4.199 ×10−2 | 3.087 ×10−2 | 97.7 | 1.008 | 2.305 ×10−2 | 2.456 ×10−2 | 97.9 | |
| β2 = 2.0 | 2.003 | 0.112 | 0.100 | 97.0 | 2.003 | 4.255 ×10−2 | 3.509 ×10−2 | 97.3 | 2.005 | 3.046 ×10−3 | 1.784 ×10−3 | 97.4 | |
| β3 = 3.0 | 3.000 | 0.022 | 0.020 | 97.1 | 3.000 | 1.017 ×10−2 | 8.376 ×10−3 | 97.1 | 2.999 | < 1.0 × 10−4 | < 1.0 × 10−4 | 97.0 | |
| (50, 0, 200, 0, 100, 100, 50, 0) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.2 | 0.983 | 2.849 ×10−3 | 6.160 ×10−3 | 96.9 | 0.987 | 3.209 ×10−3 | 3.779 ×10−3 | 96.9 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.2 | 1.005 | 5.615 ×10−3 | 8.373 ×10−3 | 97.4 | 1.008 | 8.295 ×10−3 | 8.444 ×10−3 | 97.5 | |
| β2 = 2.0 | 2.003 | 0.112 | 0.100 | 97.0 | 1.998 | 2.241 ×10−4 | 1.448 ×10−2 | 95.9 | 2.001 | 1.159 ×10−4 | 8.268 ×10−3 | 95.1 | |
| β3 = 3.0 | 3.000 | 0.022 | 0.020 | 97.1 | 3.003 | 1.849 ×10−4 | 5.171 ×10−3 | 94.9 | 3.001 | < 1.0 × 10−4 | 1.088 ×10−3 | 94.0 | |
| (50, 0, 200, 0, 100, 100, 50, 0) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.2 | 0.983 | 5.208 ×10−3 | 1.032 ×10−2 | 96.6 | 0.988 | 5.290 ×10−3 | 4.636 ×10−3 | 96.8 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.2 | 1.004 | 9.803 ×10−3 | 1.305 ×10−2 | 97.4 | 1.007 | 1.362 ×10−2 | 1.395 ×10−2 | 97.5 | |
| β2 = 2.0 | 2.003 | 0.112 | 0.100 | 97.0 | 1.996 | 5.067 ×10−4 | 2.205 ×10−2 | 95.3 | 2.000 | 2.442 ×10−4 | 1.101 ×10−2 | 93.6 | |
| β3 = 2.0 | 2.000 | 0.022 | 0.020 | 97.1 | 2.004 | 2.720 ×10−4 | 7.745 ×10−3 | 93.1 | 2.001 | 1.257 ×10−4 | 1.264 ×10−3 | 91.9 | |
| (50, 0, 200, 0, 100, 100, 50, 0) | β0 = 1.0 | 0.988 | 0.130 | 0.124 | 97.2 | 0.983 | 2.849 ×10−3 | 6.160 ×10−3 | 97.4 | 0.987 | 2.939 ×10−3 | 3.632 ×10−3 | 96.9 |
| β1 = 1.0 | 1.011 | 0.305 | 0.292 | 97.2 | 1.005 | 5.615 ×10−3 | 8.373 ×10−3 | 97.7 | 1.008 | 7.312 ×10−3 | 8.101 ×10−3 | 97.6 | |
| β2 = 2.0 | 2.003 | 0.112 | 0.100 | 97.0 | 1.998 | 2.241 ×10−4 | 1.448 ×10−2 | 97.3 | 2.001 | 9.505 ×10−5 | 8.266 ×10−3 | 95.1 | |
| β3 = 3.0 | 3.000 | 0.022 | 0.020 | 97.1 | 3.003 | 1.849 ×10−4 | 5.171 ×10−3 | 97.1 | 3.001 | < 1.0 × 10−4 | 1.088 ×10−3 | 93.9 | |
Sim:Vcc, Sim:Vcr and Sim:Vac are the simulated variances of CC, CR and AC methods. Vcc, Vcr and Vac are the variances of CC, CR and AC methods using the formula derived in Section 4 with MLE’s plug-in. 95% CP is the 95% coverage probability.
6.2 Simulation Study II: Logistic Regression Model with MAR Response and Covariate Data
A simulation with 1000 replicates was conducted to numerically compare the CC, CR and AC methods in a logistic regression model. The estimates using the full data (FD) before missing are also provided as a benchmark of other methods. In each simulation, we generated 500 binary samples from a logistic regression model logit(P(yi = 1)) = β0 + β1zi1 + β2xi1, where zi1 was simulated from Unif(0, 1) and xi1 was simulated from a Bernoulli distribution with the success probability modeled as logit(P(xi1 = 1)) = α0 + α1zi1. The covariate zi1 is completely observed for all subjects, and xi1 and the response yi are missing at random (MAR) for some subjects. The missing mechanisms for yi and xi1 are logit(P(riy = 1)) = ϕ20 + ϕ21zi1 and logit(P(rix = 1)) = ϕ10 + ϕ11zi1 + ϕ12riyyi, where riy = 1 or rix = 1 if yi or xi1 is observed, 0 otherwise. On average, 31.4% samples have completely observed covariate and response, 34.8% have missing covariate but observed response, 15.0% have observed covariate but missing response, and 18.8% have missing covariate and missing response.
Table 5 gives the simulation results of the logistic regression model. The AC method provides estimates with higher precision (smaller standard error) and lower mean square error (MSE) than the CR method for all the parameters. Both the CR and AC methods are uniformly better than the CC method in terms of bias, simulated standard error and MSE.
Table 5.
Simulation for Logistic Regression Model
| Method | β0 = −1.8 | β1 = 1.0 | β2 = 2.0 | α0 = −0.5 | α1 = 2.0 | |
|---|---|---|---|---|---|---|
| CC | Bias | 0.224 | 0.483 | 0.059 | 0.104 | 0.273 |
| SE | 0.450 | 0.723 | 0.408 | 0.351 | 0.672 | |
| SSE | 0.466 | 0.720 | 0.419 | 0.351 | 0.680 | |
| CP% | 99.5 | 99.6 | 97.8 | 98.8 | 98.4 | |
| MSE | 0.484 | 1.268 | 0.355 | 0.257 | 1.000 | |
| CR | Bias | −0.036 | −0.008 | 0.059 | −0.017 | 0.056 |
| SE | 0.378 | 0.544 | 0.437 | 0.372 | 0.687 | |
| SSE | 0.408 | 0.527 | 0.419 | 0.339 | 0.650 | |
| CP% | 97.9 | 98.7 | 97.9 | 98.7 | 97.7 | |
| MSE | 0.333 | 0.555 | 0.355 | 0.230 | 0.847 | |
| AC | Bias | −0.035 | −0.007 | 0.055 | −0.019 | 0.051 |
| SE | 0.377 | 0.528 | 0.417 | 0.262 | 0.539 | |
| SSE | 0.402 | 0.516 | 0.415 | 0.250 | 0.533 | |
| CP% | 97.7 | 98.3 | 97.6 | 98.5 | 97.7 | |
| MSE | 0.324 | 0.533 | 0.347 | 0.126 | 0.571 | |
| FD | Bias | −0.020 | 0.006 | 0.023 | −0.011 | 0.016 |
| SE | 0.245 | 0.366 | 0.223 | 0.186 | 0.344 | |
| SSE | 0.247 | 0.364 | 0.225 | 0.181 | 0.337 | |
| CP% | 97.4 | 97.5 | 97.4 | 97.4 | 98.0 | |
| MSE | 0.123 | 0.264 | 0.102 | 0.065 | 0.227 | |
, SE is the mean of the standard error calculated by Louis’s formula, SSE is the simulated standard error, CP is the coverage probability, MSE = Bias2 + SSE2 is the mean square error.
6.3 Analysis of Small Cell Lung Cancer Data
We consider a real dataset to compare the three analysis methods in terms of bias and efficiency. We consider a lung cancer dataset from a recent phase III clinical trial (Socinski et al., 2002) of non-small-cell lung cancer (NSCLC), which is the leading cause of cancer-related mortality. In the year 2001, among approximately 170,000 patients newly diagnosed, more than 90% died from NSCLC and approximately 35% of all new cases were stage IIIB/IV (malignant pleural effusion) the disease. A randomized, two-armed, multi-center trial was initiated in 1998 with the aim to determine the optimal duration of chemotherapy by comparing four cycles of therapy versus continuous therapy in advanced NSCLC. Patients were randomized to two treatment arms: four cycles of carboplatin at an area under the curve of 6 and paclitaxel 200 mg/m2 every 21 days (arm A), or continuous treatment with carboplatin/paclitaxel until progression (arm B). At progression, all patients on both arms received second-line weekly paclitaxel at 80 mg/m2. One of the primary endpoints was quality of life (QOL). There were n = 230 patients in this dataset. The response variable considered in this analysis is the quality of life (QOL) factg score. The covariates included in the model were treatment (trt, 0=arm A, 1=arm B), gender (0=female, 1=male), Histology (hist, 0=Non-Squamous, 1=Squamous), age at entry in years, highest grade toxicity (recorded by cycle) (apex, 0 if highest grade toxicity =0 and 1 if highest grade toxicity > 0), and recovery status (recov, 0 if recovered and 1 otherwise). For these six covariates, apex and recov had missing information and trt, gender, hist, and age were completely observed for all cases. In this population, 63% of the patients were male, and the age at entry ranged from 32 to 82 with a mean of 62. The missing data fractions were 28% in apex, 54% in recov, and 35% in factg. There was a total missing data fraction of 74% on apex, recov, and factg.
We use a linear regression to model the response variable, factg, as
We consider two models for the missing covariates recov and apex as follows.
Model M1
Model M2
Table 6 shows the results for the CC, CR and AC methods discussed in Section 4. We assume that the missing data are MAR so that a missing data mechanism need not be considered in the estimation scheme for β. As shown in the table, the overall conclusions are the same for the CR and AC methods, as these two methods yield similar p-values for the various regression coefficients. significance level. However, the CR and AC methods yield more significant p-values than the CC analysis, especially for the age effect. Table 6 also shows that the estimates of β and the standard errors of the estimated regression coefficients are quite similar for model M1 and model M2, indicating robustness of estimates to the the choice of covariate distribution. The EM algorithm was implemented for computing all maximum likelihood estimates. The convergence criterion for the EM algorithm was that the squared distance between the kth and (k + 10)th iterations was less than 10−7. The EM algorithm required 25 iterations to converge under both model M1 and model M2.
Table 6.
Lung Cancer Data Analysis
| Model M1 | Model M2 | ||||||
|---|---|---|---|---|---|---|---|
| Method | Effect | Estimate | SE | P-value | Estimate | SE | P-value |
| CC | Intercept | 79.008 | 3.536 | < 0.001 | 79.008 | 3.536 | < 0.001 |
| trt | −2.366 | 3.222 | 0.463 | −2.366 | 3.222 | 0.463 | |
| gender | −3.033 | 3.431 | 0.377 | −3.033 | 3.431 | 0.377 | |
| hist | 3.011 | 3.694 | 0.415 | 3.011 | 3.694 | 0.415 | |
| age | 3.049 | 1.587 | 0.055 | 3.049 | 1.587 | 0.055 | |
| apex | 4.825 | 5.004 | 0.335 | 4.825 | 5.004 | 0.335 | |
| recov | −0.485 | 3.421 | 0.887 | −0.485 | 3.421 | 0.887 | |
| 147.896 | 27.230 | < 0.001 | 147.896 | 27.230 | < 0.001 | ||
| CR | Intercept | 81.565 | 2.912 | < 0.001 | 81.573 | 2.910 | < 0.001 |
| trt | 0.743 | 2.543 | 0.770 | 0.730 | 2.540 | 0.774 | |
| gender | −6.048 | 2.580 | 0.019 | −6.034 | 2.576 | 0.019 | |
| hist | 2.003 | 3.076 | 0.515 | 2.007 | 3.076 | 0.514 | |
| age | 3.640 | 1.244 | 0.003 | 3.641 | 1.244 | 0.003 | |
| apex | 3.473 | 6.088 | 0.568 | 3.433 | 6.078 | 0.572 | |
| recov | −4.676 | 4.319 | 0.279 | −4.700 | 4.317 | 0.276 | |
| 210.197 | 25.511 | < 0.001 | 210.183 | 25.525 | < 0.001 | ||
| AC | Intercept | 81.533 | 2.768 | < 0.001 | 81.535 | 2.762 | < 0.001 |
| trt | 0.905 | 2.556 | 0.723 | 0.905 | 2.550 | 0.723 | |
| gender | −5.778 | 2.516 | 0.021 | −5.777 | 2.508 | 0.021 | |
| hist | 2.067 | 3.064 | 0.500 | 2.074 | 3.057 | 0.497 | |
| age | 3.527 | 1.234 | 0.004 | 3.532 | 1.232 | 0.004 | |
| apex | 3.227 | 6.168 | 0.601 | 3.199 | 6.154 | 0.603 | |
| recov | −5.326 | 4.362 | 0.222 | −5.343 | 4.348 | 0.219 | |
| 208.812 | 25.667 | < 0.001 | 208.436 | 25.573 | < 0.001 | ||
Although we have assumed that data are MAR and a missing data mechanism need not be modeled, it is of some interest if we could determine the best fitting MAR missing data mechanism, so that we could at least (though somewhat ad-hoc) determine whether the missing data are MAR or MCAR. Towards this goal, we posited several different MAR and MCAR missing data mechanisms, and using the complete cases to fit these models as logistic regression in SAS. We then computed the log-likelihood statistic to determine the best fitting missing data mechanism. We considered 5 missing data mechanism, two of them are MCAR, and the other three are MAR. Let ri,factg, ri,apex, and ri,recov denote the missing data indicators for factg, apex, and recov, respectively. To determine the final log-likelihood statistic value, we added the three contributions from the three parts of the binary regression models for ri,factg, ri,apex, and ri,recov. Two MCAR models are [ri,factg][ri,apex][ri,recov] (MCAR1) and [ri,factg|ri,apex, ri,recov][ri,apex][ri,recov|ri,apex] (MCAR2), where, for example, [ri,factg] denotes a logistic regression model with intercept only and [ri,factg|ri,apex, ri,recov] is a logistic regression model with intercept and covariates ri,apex and ri,recov. Let xi,obs = (trti, genderi, histi, agei). Three MAR models include [ri,factg|xi,obs, apexiri,apex, recoviri,recov] [ri,apex|xi,obs][ri,recov|xi,obs, apexiri,apex] (MAR1), [ri,factg|xi,obs, ri,apex, ri,recov][ri,apex|xi,obs] [ri,recov|xi,obs, ri,apex] (MAR2), and [ri,factg|trti, genderi, ri,apex, ri,recov][ri,apex|trti, genderi] [ri,recov|trti, genderi, ri,apex] (MAR3). For the lung cancer data, the log-likelihood statistics, −2 log(likelihood), are 889.4, 873.5, 868.5, 856.6, and 860.5 under models MCAR1, MCAR2, MAR1, MAR2, and MAR3, respectively. We see from these results that the best fitting model is MAR2 as the missing data mechanism suggesting that the missing data are missing at random.
7 Discussion
We have given several results regarding bias and efficiency of estimates in missing (x, y) regression problems, and have shown that the AC analysis provides the most efficient estimates and the least biased estimates in the MAR setting. The results derived in Section 4, Section 5, and Section 6 are new and important and shed light on the bias and efficiency of estimates in regression problems with MCAR or MAR responses and/or covariates. In Section 4.1 and 4.2, the variances are assumed to be known. This assumption can be relaxed. The asymptotic variance and covariance matrix of the MLE’s under each analysis method for the simple linear regression model with unknown variances is derived in Appendix B. With the known variances, as shown in Result 4.2, the AC analysis does not improve the efficiency of the MLE for β1 over the CR analysis. When the variances are unknown, it is interesting to see, from Appendix B, that the AC analysis improves the efficiency of the MLE for ²1 over both the CC and CR analsyese. Thus, the AC analysis becomes even more important in this case. However, the derivation of the asymptotic variance and covariance matrix of the MLE’s under each analysis method for the multiple linear regression model with unknown variances becomes very lengthy and hence, detailed derivations are omitted for brevity.
Finally, we mention that we have assumed throughout that jointly, (xi, yi) are iid. This is by far the most common approach in regression settings with missing covariate and/or response data. We note here, however, that since inference typically focuses on the parameters of [yi|xi], the yi’s conditional on the xi’s are not iid, but rather only independent. This development is still quite general since is covers settings such as the linear model and generalized linear models with MAR covariate and/or response data. Future work involves examination of the proposed methods for dependent responses, including dynamic linear models, models for longitudinal data, and generalized linear mixed models. Such theoretical investigations are currently being examined. The initial investigation taken here is the first of its kind, and should lead to fruitful results for other types of models.
Figure 1. Missing Patterns.
Note: Blank block stands for missing, marked block stands for observed.
Acknowledgements
The authors wish to thank the Editor, the Associate Editor and two referees for helpful comments and suggestions which have improved the paper. Dr. Ibrahim’s and Dr. Chen’s research was partially supported by National Institute of Health (NIH) grant numbers GM 70335 and CA 74015.
Appendix A
Computational Development
We describe the model fitting and computational procedures for each of the analysis methods. For CC, the MLE of θ can be obtained by standard statistical software such as SAS. Here, we consider only for AC as the computation of the MLE’s under CR is similar to and even easier than AC.
We first consider the case where all xi,mis’s are categorical. In this case, we use the EM algorithm via the method of weights proposed by Ibrahim (1990). Let θ(t) = (β(t), ϕ(t), α(t)) denote the value of θ at the tth iteration of EM algorithm.
The E-step at the (t + 1)st iteration can be written as
| (A.1) |
where ,
| (A.2) |
and
| (A.3) |
The inner sum extends over all of the possible values of the missing components of the covariate vector, with j indexing the distinct covariate patterns for subject i.
The weights, wij,(t), are the conditional probabilities corresponding to [xi,mis|xi,obs, yi θ] or [xi,mis|xi,obs, α] and are given by
or
The M-step at the (t + 1)st iteration proceeds as follows. We first compute
and
When we use the saturated model for xi,mis, we have α = (α(j)), where α(j) denotes the probability of the jth missing pattern. In this case, we update
where when xi is completely observed, wij,(t) = 1 if xi(j) = xi and wij,(t) = 0 if xi(j) ≠ xi.
Let θ̂ denote the estimate of θ at EM convergence. We use Louis’s method (Louis, 1982) to compute the estimated observed information matrix of θ based on the observed data. Write the matrix of second derivatives of Q(θ|θ(t)) as
where γ = (β′, ϕ′)′,
and
Write the gradient vector of Qi(θ|θ(t)) for the ith observation as
In addition, write the complete data score vector Si(θ|xi, yi) as
Then, the estimated observed information matrix of θ̂ is given by
| (A.4) |
where the weights, wij,(t), are computed at EM convergence. Thus, the estimate of the asymptotic covariance matrix of θ̂ is [I (θ̂)]−1.
When missing covariates are continuous or mixed Continuous and categorical, a Monte Carlo EM (MCEM) algorithm is required. The implementation of the MCEM is similar to the EM algorithm for the categorical missing covariates, and is developed in detail in Ibrahim, Lipsitz and Chen (1999), and Ibrahim, Chen, and Lipsitz (1999). Specifically, we replace the weight average in (A.2), (A.3) and (A.4) by a Monte Carlo average. For example, in the E-Step, for missing covariates in the Block B2, we take an MCMC sample of size from
Then, we compute
. We then take m(t+1) = m(t) + Δm, where Δm > 0. In such a way, the MCEM algorithm requires much less computational time, as a large m(t) is not needed in early iterations of the algorithm.
Appendix B
Simple Linear Regression Model with Unknown Variances
We consider a simple normal regression model with a single covariate and unknown variances here. In this case, we have
Write θ = (β′, σ2, α, τ2)′. Let nj = #(Bj) be the cardinality of Bj for j = 1, 2, 3 and n = n1 + n2 + n3. For the CC analysis, we have
and the Fisher information matrix is given by
For the CR analysis, we have
where . After some messy algebra, we obtain the Fisher information matrix given by
For the AC analysis, the log-likelihood function is given by
The corresponding Fisher information matrix is given by
Based on either the determinant or the trace of Fisher information matrix, AC yields most gain in information over both CR and CC, and CR gains more information than CC. Specifically, we have
and
CR leads to smaller asymptotic variances for all parameters than CC. Specifically, we have
where .
In addition, AC improves the asymptotic variances over CR. Specifically, we have
where .
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker SG, Laird NM. Regression Analysis for Categorical Variables with Outcome Subject to Nonignorable Nonresponse. Journal of the American Statistical Association. 1988;83:62–69. [Google Scholar]
- Chen Q, Ibrahim JG. Semiparametric Models for Missing Covariate and Response Data in Regression Models. Biometrics. 2006;62:177–184. doi: 10.1111/j.1541-0420.2005.00438.x. [DOI] [PubMed] [Google Scholar]
- Chen M-H, Ibrahim JG, Shao Q-M. Propriety of the Posterior Distribution and Existence of the Maximum Likelihood Estimator for Regression Models with Covariates Missing at Random. Journal of the American Statistical Association. 2004;99:421–438. [Google Scholar]
- Herring AH, Ibrahim JG. Likelihood-based Methods for Missing Covariates in the Cox Proportional Hazards Model. Journal of the American Statistical Association. 2001;96:292–302. [Google Scholar]
- Horton NJ, Laird NM. Maximum Likelihood Analysis of Generalized Linear Models with Missing Covariates. Statistical Methods in Medical Research. 1999;8:37–50. doi: 10.1177/096228029900800104. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG. Incomplete data in generalized linear models. Journal of the American Statistical Association. 1990;85:765–769. [Google Scholar]
- Ibrahim JG, Chen M-H, Lipsitz SR. Monte Carlo EM for Missing Covariates in Parametric Regression Models. Biometrics. 1999;55:591–596. doi: 10.1111/j.0006-341x.1999.00591.x. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG, Chen M-H, Lipsitz SR. Missing Responses in Generalized Linear Mixed Models When the Missing Data Mechanism Is Nonignorable. Biometrika. 2001;88:551–564. [Google Scholar]
- Ibrahim JG, Chen M-H, Lipsitz SR, Herring AH. Missing Data Methods for Generalized Linear Models: A Comparative Review. Journal of the American Statistical Association. 2005;100:332–346. [Google Scholar]
- Ibrahim JG, Lipsitz SR, Chen M-H. Missing Covariates in Generalized Linear Models When the Missing Data Mechanism is Nonignorable. Journal of the Royal Statistical Society, Series B. 1999;61:173–190. [Google Scholar]
- Lipsitz SR, Ibrahim JG, Zhao LP. A New Weighted Estimating Equation for Missing Covariate Data with Properties Similar to Maximum Likelihood. Journal of the American Statistical Association. 1999;94:1147–1160. [Google Scholar]
- Little RJA. Regression with Missing X’s: A Review. Journal of the American Statistical Association. 1992;87:1227–1237. [Google Scholar]
- Louis TA. Finding the Observed Information Matrix When Using the EM Algorithm. Journal of the Royal Statistical Society, Series B. 1982;44:226–233. [Google Scholar]
- Robins JM, Rotnitzky A. Semiparametric Efficiency in Multivariate Regression Models with Missing Data. Journal of the American Statistical Association. 1995;90:122–129. [Google Scholar]
- Socinski MA, Schell MJ, Peterman A, Bakri K, Yates S, Gitten R, Unger P, Lee J, Lee Ji, Tynan M, Moore M, Kies M. Phase III Trial Comparing Defined Duration of Therapy Versus Continuous Therapy Followed by Second-LineTherapy in Advanced-Stage IIIB/IV Non-Small-Cell Lung Cancer. Journal of Clinical Oncology. 2002;20:1335–1343. doi: 10.1200/JCO.2002.20.5.1335. [DOI] [PubMed] [Google Scholar]
- Stubbendick AL, Ibrahim JG. Maximum Likelihood Methods for Nonignorable Responses and Covariates in Random Effects Models. Biometrics. 2003;59:1140–1150. doi: 10.1111/j.0006-341x.2003.00131.x. [DOI] [PubMed] [Google Scholar]
- Stubbendick AL, Ibrahim JG. Likelihood-based Inference with Nonignorable Missing Responses and Covariates in Models for Discrete Longitudinal Data. Statistica Sinica. 2006;16:1143–1167. [Google Scholar]
- Tang G, Little RJA, Raghunathan TE. Analysis of Multivariate Missing Data with Nonignorable Nonresponse. Biometrika. 2003;90:747–764. [Google Scholar]


