Abstract
Given their high alanine and glycine levels, plaque formation, α-helix to β-sheet interconversion and fusogenicity, FP (i.e., the N-terminal fusion peptide of HIV-1 gp41; 23 residues) and amyloids were proposed as belonging to the same protein superfamily. Here, we further test whether FP may exhibit ‘amyloid-like’ characteristics, by contrasting its structural and functional properties with those of Aβ(26–42), a 17-residue peptide from the C-terminus of the amyloid-beta protein responsible for Alzheimer’s. FTIR spectroscopy, electron microscopy, light scattering and predicted amyloid structure aggregation (PASTA) indicated that aqueous FP and Aβ(26–42) formed similar networked β-sheet fibrils, although the FP fibril interactions were weaker. FP and Aβ(26–42) both lysed and aggregated human erythrocytes, with the hemolysis-onsets correlated with the conversion of α-helix to β-sheet for each peptide in liposomes. Congo red (CR), a marker of amyloid plaques in situ, similarly inhibited either FP- or Aβ(26–42)-induced hemolysis, and surface plasmon resonance indicated that this may be due to direct CR-peptide binding. These findings suggest that membrane-bound β-sheets of FP may contribute to the cytopathicity of HIV in vivo through an amyloid-type mechanism, and support the classification of HIV-1 FP as an ‘amyloid homolog’ (or ‘amylog’).
Keywords: Fourier transform infrared, electron microscopy, light scattering, aggregation, hemolysis, prion, fibrils, Congo red, surface plasmon resonance, AIDS
1. Introduction
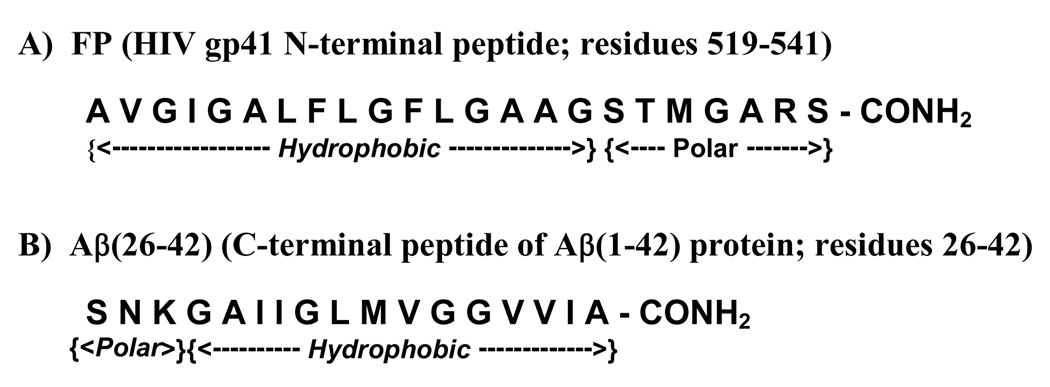
The amino-terminal ‘fusion peptide’ (FP; residues 519–541; Fig. 1) of glycoprotein 41,000 (gp41) is involved in the fusion processes underlying human immunodeficiency virus (HIV-1) infection of target cells [1,2]. Synthetic peptides based on the FP region induced leakage of lipid vesicles [3], in agreement with the hypothesis that FP is partially responsible for the cytolytic actions of HIV-1 virions. Consistent with this is the finding that the FPs of HIV or SIV lysed cultured cells [4–6] and erythrocytes [4,7–9]. FP addition to model liposomes also induced lipid mixing and aggregation [3]. With red cells, FP promotes not only rapid lipid mixing between cell membranes, but also multicell aggregates [7,10,11]. Although these results suggest that fusogenic actions of HIV-1 gp41 are mimicked by FPs, an understanding of how this N-terminal domain produces lysis and fusion has been complicated by its structural polymorphism. Residue-specific spectroscopy have shown that FP residues ~1–4 to 15–20 (i.e., gp41 residues 519–537; Fig. 1) are conformationally flexible, folding as either an α-helix in membrane mimics and membranes [3,12–16], or oligomers of extended β-sheet in aqueous and membrane lipid environments [3,14,17–20].
fig. 1.
Amino acid sequences of the native N-terminal peptide (FP) and the C-terminal domain of the amyloid-beta protein [Aβ(26–42)]. FP is based on the HIV-1 strain LAV1a and was numbered as described previously [7], while Aβ(26–42) is based on the C-terminal region (residues 26–42) from the human amyloid β-peptide (residues 1–42) [42].
Given the important roles of the FP domain in HIV-1 functions, it is of interest that the N-terminal gp41 peptide has been preliminarily assigned to the same class of proteins as amyloids [21–23]. Using sequence and hydrophobic analysis, Callebaut et al. [21] noted high sequence homologies and elevated alanine and glycine levels in viral fusion peptides such as HIV-1 FP and amyloid-derived peptides, including the conserved hydrophobic sequences of human prion protein (HPrP) responsible for peptide aggregation and neurotoxicity. A recent study of the corresponding hydrophobic sequence from bovine PrP (BPrP) also showed an interfacial hydrophobicity similar to HIV-1 FP, indicating comparable membrane partitioning for these peptides [23]. Additionally, suggestive of ‘amyloid-like’ properties for the N-terminal gp41 domain is that aqueous FPs produce either large spherical aggregates [14,24] or long filaments [14,23,25] that bear analogies to fibrils observed with the hydrophobic PrP peptides [23,26]. Moreover, FTIR spectroscopy of the HIV-1 FP [14,23] or the hydrophobic BPrP peptide [23] fibrils demonstrated high β-sheet levels. Another shared characteristic is polymorphism, as indicated by CD or FTIR spectroscopic studies showing that the FP (see above) and hydrophobic PrP peptides [23,26] may each adopt multiple conformations. The lytic and fusogenic actions reported for HIV-1 FP (see above) and the neurotoxic PrP sequences [23,26] may be due to these peptides converting between α-helix, β-sheet and random coils [21]. Nevertheless, HIV-1 FP may not act as a classic amyloid peptide, as solid-state 2D-NMR indicated that membrane-associated FP simultaneously folds as parallel and antiparallel β-sheets [27].
Because the assignment of HIV-1 FP as an ‘amyloid-like’ peptide remains preliminary, it would be worthwhile to further compare various structural and functional properties of this viral fusion peptide with those of peptides based on the Amyloid-Beta protein (i.e., Aβ). Aβ is a protein found in the extracellular deposits of Alzheimer plaques in the brain. The naturally-occurring Aβ proteins in Alzheimer plaques are approximately 39 to 43 residue peptides, with the 40-residue Aβ(1–40) and 42-residue Aβ(1–42) variants being the most abundant forms found in Alzheimer’s disease. Aβ(1–42) is an amphipathic protein, with a hydrophilic N-terminus region (1–28) and a hydrophobic C-terminal stalk (29–42) that anchors the protein in the membrane bilayer [28]. 2D-NMR [29–31] and CD [32] analyses of either the Aβ(1–40) or Aβ(1–42) proteins in membrane-mimics indicated high α-helix, with 2D-NMR showing α-helix between residues ~8–16 to 25 and ~28–31 to 35–38. The polymorphic nature of Aβ is emphasized by prior CD and FTIR findings that this protein may adopt β-sheet in either aqueous [33,34] or even membrane [32] environments. Recent solid-state 2D-NMR spectroscopy indicated that fibrillar Aβ(1–40) primarily folds with a parallel β-sheet, in register arrangement (PIRA) [35]. This is consistent with calculations using PASTA (i.e., prediction of amyloid structure aggregation) analysis, which systematically determines the relative energies of both parallel and antiparallel β-sheet arrangements for the various pairings of a protein, indicating that the most stable PIRA pairings for Aβ(1–40) are residues 12–20 and 31–40 [36,37]. Intermolecular hydrogen-bonding between Aβ molecules produces an ‘infinitely long' β-sheet with a fibril axis perpendicular to the extended Aβ chains. A second fibril axis, perpendicular to the plane of the β-sheet, is formed by successively layering these β-sheets on top of one another [28]. Also noteworthy are recent findings that full-length Aβ is neurotoxic [38], and also perturbs red blood cells [39,40].
Attention has focused lately on the critical roles that the hydrophobic C-terminal domain may play in the fibril formation and toxicity of the full-length Aβ protein. Based on separate approaches, which calculate minimum energies for various sequence pairings using the known 3D structures of proteins, Thompson et al. [41] and Trovato et al. [36,37] each predicted that the full-length Aβ protein would likely form PIRA alignments at the C-terminal region. It is of considerable interest, then, that synthetic fragments based on the Aβ sequence, which overlap the hydrophilic and hydrophobic domains at the C-terminus, mimic the fibrillar and cytotoxic actions of the full-length protein (see above). The C-terminal fragments Aβ(34–42) and Aβ(26–42) (Fig. 1) rapidly formed amyloid fibrils in aqueous buffers, and FTIR spectroscopy on each peptide fibril confirmed high β-sheet levels [42–44]. More recently, the overlapping Aβ(25–35) in either aqueous media [45] or aqueous media with liposomes [46] produced cross-beta fibrils comparable to those observed with the full-length Aβ protein [38]. C-terminal Aβ peptides have also been found to be similarly lytic and fusogenic with liposomes [47], and cytotoxic with neurons [6,48,49] and red cells [40,50]. Consequently, the use of C-terminal Aβ peptides provides a useful model system for probing key structural and functional aspects of the parent protein.
To further assess the ‘amyloid-like’ characteristics of HIV-1 FP, we here explore the properties of FP and Aβ(26–42) with a suite of assays, several of which have not been previously used. The Nomenclature Committee of the International Society of Amyloidosis has defined amyloids to include 25 human and 8 animal fibril proteins, which are all found as extracellular deposits in situ with amyloid characteristics such as cross-β structure, affinity for Congo red and cytotoxicity [51,52]. Neither FP nor Aβ(26–42) would fulfill this strict amyloid definition, because FP has not been observed as extracellular deposits in AIDS patients, while Alzheimer plaque contains full-length Aβ proteins. However, the Nomenclature Committee additionally defined ‘amyloid homologs’, a related group of proteins or peptides that forms fibril aggregates only in vitro, but which nevertheless exhibit ‘amyloid-like’ properties [51–53]. Because C-terminal Aβ peptides demonstrate typical ‘amyloid-like’ behaviors (see above), direct comparisons between FP and Aβ(26–42) may show whether FP like Aβ(26–42) is also an ‘amyloid homolog’ or ‘amylog’ [53]. Both peptides exhibit hydrophilic and hydrophobic regions, except that their positions have been reversed (Fig. 1). To make these comparisons, we here employ standard techniques such as FTIR spectroscopy, transmission electron microscopy (TEM) and hemolysis assays, and also newer ones such as dynamic light scattering (DLS), PASTA, and surface plasmon resonance (SPR) spectroscopy that have not been used previously with FP. FTIR, TEM, DLS and PASTA analysis indicate that aqueous FP and Aβ(26–42) each form β-sheet fibrils, although the FP fibril interactions are weaker due to its core β-sheet being less energetically favored. FP and Aβ(26–42) similarly lyse and aggregate red cells, with FTIR indicating that the hemolysis-onsets correlate well with the conformational shift of α-helix to β-sheet for each peptide in model liposomes. Congo red (CR), a dye-marker of amyloid plaques in vivo [51], inhibits either FP- or Aβ(26–42)-induced hemolysis at low doses, and SPR shows that this may be due to high-affinity CR-peptide interactions. Taken together, our results support the classification of HIV-1 FP as an amyloid homolog, and suggest that FP bound to membranes as β-sheets may contribute to the cytopathicity of HIV in vivo (e.g., depletion of CD4+-lymphocytes) through amyloid-type mechanisms.
2. Materials and methods
2.1. Materials
Peptide synthesis reagents, including FMOC amino acids and coupling solvents, were obtained from AnaSpec (San Jose, CA) and Pharmco Products (Brookfield, CT). Congo red was purchased from Sigma Chemical Co. (St. Louis, MO). The HBS-EP buffer was from Biacore (Uppsala, Sweden). All organic solvents used for sample synthesis were HPLC grade or better.
2.2. Solid-phase peptide synthesis, purification and characterization
The synthetic peptides used here (i.e., HIV-1 FP and Aβ(26–42); Fig. 1) were prepared with either an Applied Biosystems (Foster City, CA) 431A solid-phase peptide synthesizer or a Protein Technologies Symphony/Multiplex SPPS (Tucson, AZ) synthesizer using an FMOC strategy [4,10,54,55] (double-coupling 2 × 45 min). The peptides were cleaved using modified Reagent K (88.9% trifluoroacetic acid (TFA), 4.4% Triisopropyl silane (TIS), 2.2% thioanisol and 4.4% water), precipitated by addition of ice-cold diethyl ether and then collected by centrifugation. The peptides were purified with reverse-phase HPLC techniques [14]. The predicted molecular masses for each peptide were obtained by MALDI-TOF mass spectrometry using an ABI Voyager RP-RBT2 reflection time-of-flight mass spectrometer (Applied Biosystems, Foster City, CA).
2.3. Preparation of human red blood cells (RBCs)
Human RBCs were prepared using outdated units from the local blood bank [8,55]. Besides the blood type and Rh factor, each unit of blood is thoroughly tested for infectious diseases, including HIV-1 HIV-2, HTLV-1, syphilis, and hepatitis B and C. Red cells were washed three times in 0.85% NaCl, and packed cells from the last wash were suspended in isotonic phosphate-buffered saline (PBS) buffer, consisting of 120 mM NaCl-2.7 mM KCl-10 mM Na2HPO4 (pH 7.4).
2.4. Hemolysis of human RBCs
To determine peptide-induced lysis, 10 µl of packed, washed cells was added to 0.5 ml of the isotonic PBS buffer, and then small aliquots (≤ 30 µl) of concentrated FP and Aβ(26–42) (Fig. 1) in 0.1% acetic acid were added. After 30 min at 37°C (unless otherwise indicated), the suspension was centrifuged at approx. 5000 × g for 1.5 min and the supernatant analyzed spectrophotometrically for hemoglobin at 540 nm [4]. Percent lysis was calculated using a reference of the average of two tubes in which 10 µl of packed cells was incubated in 2% TX-100 in distilled water (0.5 ml) for 30 min at 37°C.
2.5. Coulter counter sizing of human RBCs
Aggregation of red cells at fixed peptide concentrations was determined with Coulter counter sizing [7,8,10,11]. Aliquots (10 µl) of washed, packed erythrocytes were incubated in 0.5 ml of PBS with the indicated peptide at 37°C for 30 min. Peptides were added as described above for hemolysis assays. At the end of the incubation, an aliquot of the reaction mixture was diluted 1/500 in isotonic PBS in a 20-ml disposable blood vial (Baxter Diagnostics, Irvine, CA). Particle numbers in each of 6 size ranges were determined with a Coulter Counter-Model ZF-1 (Coulter Electronics, Hialeah, FL). The total particle volume (VT) was calculated as the summation over each of the 6 channels of the number (NC) of particles in each channel multiplied by the maximum particle volume (VMC) in each channel. Percent total volume in each channel (%TVC) is then defined as:
| (1) |
2.6. Congo red (CR)-inhibition of FP- and Aβ(26–42) induced hemolysis of human RBCs
Determinations of Congo red (CR) inhibition of FP- or Aβ(26–42)-induced erythrocyte lysis were performed with minor modifications of earlier protocols [7,8,54,56]. First, aliquots (10 µl) of the packed cells were added to 0.5 ml of the isotonic buffer, and then small aliquots (≤ 30 µl) of the concentrated CR (in 50% DMSO) were added. For testing CR inhibition of FP- or Aβ(26–42)-induced RBC lysis, small aliquots (≤ 30 µl) of the concentrated membrane-perturbing peptides (in 0.1% acetic acid) were added to the mixture within 30 sec after CR, and incubated for 30 min at 37°C. Hemolysis measurements were then performed as described above. Maximal hemolysis (100%) was defined as that obtained by incubating a high concentration (60 µM) of the membrane-perturbing peptide [i.e., FP or Aβ(26–42)] with 10 µl of packed erythrocytes in 0.5 ml of PBS for 30 min, without CR inhibitor. Control experiments indicated that the highest concentrations of carrier DMSO for CR inhibition or acetic acid for membrane-perturbing peptides did not increase hemolysis.
2.7. Congo red (CR) binding to FP and Aβ(26–42) by Surface Plasmon Resonance (Biacore)
Binding affinities of Congo red (CR) for FP and Aβ(26–42) were measured with a Biacore 3000 system (Biacore, Uppsala, Sweden) [57]. FP and Aβ(26–42) were each chemically linked to a CM5 sensor chip (BR-1000-14, research grade, containing a carboxymethylated dextran matrix covalently attached to a gold film) through N-terminal amine coupling. Solutions of CR in HBS-EP buffer (i.e., 10 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20) were then flowed over the chip-linked peptide at a flow rate of 50 µl/min to determine binding affinity at 37°C. Binding associated with control medium containing no CR was subtracted from final affinity curves, and mean “on” and “off” rate constants (kon and koff) and the dissociation equilibrium constant (KD = koff/kon) were calculated using BIAevaluation Software Version 4.1 based on curve fitting from measurements at 10 nM CR concentration. The sensor surface was regenerated using 10 mM HCl between each sample injection [58].
2.8. Dynamic light scattering (DLS)
DLS experiments were performed at 37°C with a DynaPro light scattering instrument (Wyatt Technologies, Santa Barbara CA) for various times (0–40 min), at 50% laser intensity with a wavelength of 830 nm. The scattering angle was 90°. Scattering measurements were obtained on solutions of 40 µM HIV-1 FP or Aβ(26–42) in phosphate-buffered saline (PBS) pH 7.4, prepared by introducing small aliquots of concentrated peptide stock solutions (5 mM) suspended in 0.1% acetic acid. Data was sampled over 2 min, and analyzed by comparing the raw scattered light, and through the use of an autocorrelation function and its ratio of decay employing Dynamics Version 6.0 software. Particle translational diffusion coefficients (DT) were determined from autocorrelated light intensity data and converted to hydrodynamic radii (RH) with the Stokes-Einstein equation. Histograms of percentage intensity vs. RH were calculated, and intensity weighted mean RH were calculated for each subpeak [59].
2.9. Prediction of amyloid structure aggregation (PASTA) analyses
FP and Aβ(26–42) were each subjected to PASTA (i.e., prediction of amyloid structure aggregation) analyses [36,37,60] to determine those peptide regions most likely to aggregate as β-sheet. The PASTA algorithm systematically determines the relative energies of the various pairing arrangements, by calculating a pair-wise energy function for residues facing one another within a β-sheet. Using a database of known 3D-native structures, PASTA determines two different propensity sets depending on the directionality (i.e., parallel or antiparallel β-sheets) of the neighboring strands. PASTA assigns relative energies to specific β-pairings of two sequence stretches of the same length, and assumes that lower relative energies promote aggregation by further stabilizing the cross-β core.
Theoretical PASTA predictions of FP and Aβ(26–42) aggregation were performed here by submitting the primary sequences of these peptides (Fig. 1) to the PASTA website (http://protein.cribi/unipd.it/pasta/) [36]. PASTA determines the relative energies for the most stable pairings (i.e., parallel or antiparallel β-sheets), and outputs them to the user as an HTML file (maximum pairings: 100). The assignment of the PASTA energies for the top-ranked pairings allows the prediction of whether the β-sheet domains of one peptide will be more stable than those of other peptides. The energies for these pairings may also be used to compute an aggregation profile of those regions within a peptide most likely to aggregate as β-sheets. Here, the normalized aggregation [i.e., h(k)] was determined as a function of amino acid position (k) of each peptide, calculated using Eq. 5 of Trovato et al. [37] and outputted from the PASTA website as a PDF file.
2.10. Attenuated Total Reflectance-Fourier transform infrared (ATR-FTIR) spectroscopy
Infrared spectra were recorded at 25°C using Bruker Vector 22 FTIR spectrometers (LA Biomedical Research Institute or California State Polytechnic University Pomona) equipped with deuterated triglycine sulfate (DTGS) detectors, averaged over 256 scans at a gain of 4 and a resolution of 2 cm−1 [3,10,14]. For FTIR spectra of FP or Aβ(26–42), peptide self-films were prepared by first filtering peptides suspended in 100% hexafluoroisopropanol (HFIP) with Whatman Nylon Syringe filters (0.2 µM pore size, 13 mm diameter; Fisher Scientific, Waltham, MA), and then air drying the filtrates onto 50 × 20 × 2 mm 45° attenuated total reflectance (ATR) crystals fitted for the Bruker spectrometers (Pike Technologies, Madison, WI). The dried peptide self-films were then overlaid with solutions containing 100% HFIP or deuterated 10 mM sodium phosphate buffer (pH 7.4) before spectral acquisition; control solvent samples were similarly prepared, but without peptide. Spectra of peptides in solvent were obtained by subtraction of the solvent spectrum from that of the peptide-solvent.
For measurements with FP or Aβ(26–42) in a lipid environment, large unilamellar liposomes (LUV) of POPC:POPG (1 mole:1 mole) lipids in 10 mM sodium phosphate buffer (pH 7.4) (500 nmoles lipid/ml) of approx. 100 nm diameter were prepared by freeze-thawing five times, followed by fivefold extrusion through polycarbonate filters using a LipoFast apparatus (Avestin, Ottawa, CA) [10,61]. Peptides were added to the liposomes from 100% HFIP, and allowed to incubate with the liposomes for 1 h. FTIR spectra were measured on the above unchromatographed peptide:liposome mixtures, and also on these peptide:lipid samples passed through a Sephadex G-50 column to remove non-liposome associated peptide [62]. Chromatographed and unchromatographed lipid-peptide samples were dried onto the ATR crystal. POPC:POPG lipid-peptide samples, and also control POPC:POPG lipid samples without peptide, were hydrated for 2 h with nitrogen-D2O vapor, before spectral measurement [3,10]. The peptide in POPC:POPG lipid spectrum was obtained by subtracting the POPC:POPG lipid with D2O hydration spectrum that of peptide in POPC:POPG with D2O hydration. Peptide/lipid (P/L) ratios were determined here using the earlier finding of Martin et al. [62] that the peptide concentration is proportional to the area (Samide) of the amide I band (1680-1600 cm−1), while the lipid concentration is proportional to the area (Sν(C=O)lipid) of the lipid ν(C=O) band (1770–1700 cm−1). Therefore, the peptide/lipid ratio is proportional to the following ratio: (Samide)/(Sν(C=O)lipid).
The amide I bands of FTIR spectra of FP or Aβ(26–42) were analyzed for various secondary structures [3,14]. The proportions of α-helix, β-turn, β-sheet and disordered conformations were determined by Fourier self-deconvolutions for band narrowing and area calculations of component peaks determined with curve-fitting software supplied by Galactic Software (GRAMS/32, version 5; Galactic Industries Corp., Salem, NH). The frequency limits for the different structures were as follows: α-helix (1662 to 1645 cm−1), β-sheet (1637 to 1613 and 1710 to 1682 cm−1), β-turns (1682 to 1662 cm−1) and disordered or random (1650 to 1637 cm−1) [63].
2.11 Transmission electron microscopy (TEM)
HIV-1 fusion peptide (FP) and the Aβ(26–42) peptide in 0.1% acetic acid were each diluted in PBS (pH 7.4) to a final concentration of 40 :M and incubated at room temperature directly on formvar-coated, carbon-stabilized 200-mesh copper grids (Ted Pella; Redding, CA) for 1 hr, 5 hrs, and 24 hrs [14]. At the end of incubation, the grids were stained with 1% aqueous uranyl acetate for 5 minutes. Samples were then gently washed five times with ultrapure water and allowed to dry. Control grids were treated in the same manner with the exception of peptide inclusion in PBS. To enhance staining of smaller structures, some samples were stained with 2% aqueous uranyl acetate for 10 minutes. Samples were examined in a Zeiss 10C transmission electron microscope at 80 kV. Images were collected on Kodak SO-163 film at 25,000X to 50,000X. The electron films were digitized at 1200 dpi giving rise to a final pixel size of 0.42 by 0.84 nm.
3. Results
3.1. FTIR spectroscopic analysis of FP and Aβ(26–42) in lipid-mimic and aqueous environments
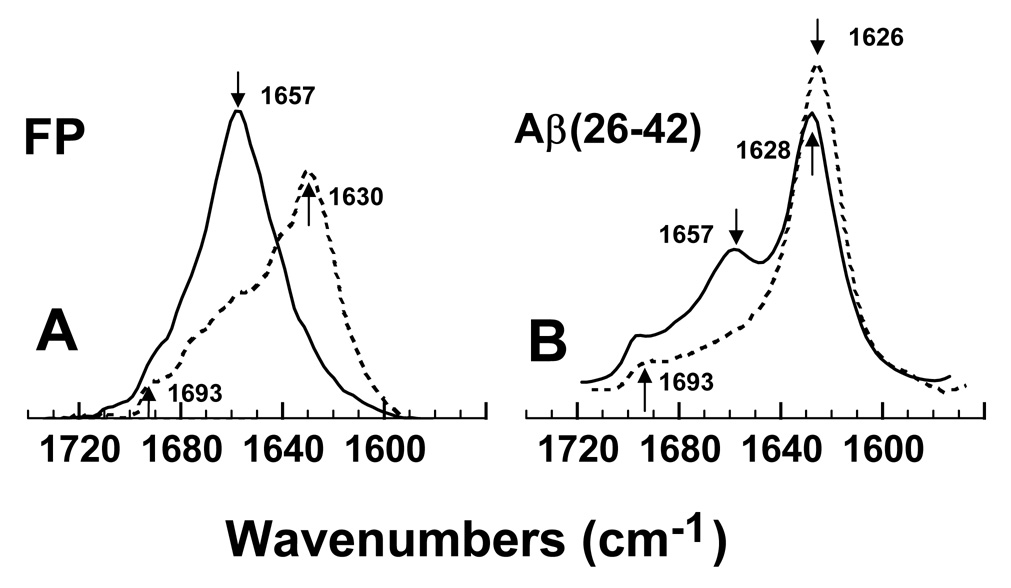
The secondary structures for FP and Aβ(26–42) in either the lipid-mimic hexafluoroisopropanol (HFIP) or phosphate buffer environments were investigated using conventional 12C-FTIR spectroscopy. Representative FTIR spectra of the amide I band for FP in the 100% HFIP and 10 mM sodium phosphate-buffer (pH 7.4) systems are shown in Fig. 2A. A principal band occurs at 1630 cm−1, and a minor high-field peak at ~1693 cm−1, for the phosphate-buffer spectrum (Fig. 2A). Consistent with these findings, earlier FTIR spectroscopic studies of N-terminal HIV-1 gp41 peptides (23-residues) in aqueous buffers each demonstrated principal peaks in the range ~1620–1628 cm−1, and less intense components at ~1689–1696 cm−1 [14,23,64]. It is likely that the spectral features in Fig. 2A reflect interstrand β-sheets for FP in the phosphate-buffer [14,63,65]. On the other hand, FTIR spectra of FP in the lipid-mimic HFIP solvent indicated a predominate band at 1657 cm−1 (Fig. 2A), and very similar spectra were previously reported for FP in HFIP, POPG liposomes and SDS micelles [3,14]. Because prior FTIR studies of proteins and peptides have assigned bands in the range of 1650–1659 cm−1 as α-helical [63,65], FP in the HFIP solvent likely assumes high α-helix levels. Fourier self-deconvolutions confirmed that FP is polymorphic, largely assuming α-helix (54.8%) in the lipid-mimic HFIP while principally converting to β-sheet (40.4%) in phosphate-buffer (Table 1). Because earlier circular dichroism (CD) spectra of FP in aqueous buffer indicated unacceptably low signal/noise (e.g., Fig. 4B of Ref. [56]) due to low peptide solubility and high light scattering, we here use ATR-FTIR spectroscopy that is not subject to these limitations.
fig. 2.
Fourier transform infrared (FTIR) spectra of the amide I bands of FP (A) and Aβ(26–42) (B) in either hexafluoroisopropanol (HFIP) or aqueous environments. (A) FTIR spectra of FP in 100% HFIP (solid line) or deuterated 10 mM sodium phosphate buffer (pH 7.4) (dashed line). The amide I band for FP in HFIP shows a dominant α-helical component centered at 1657 cm−1 (see arrow), while the corresponding band in phosphate buffer indicates a major 1630 cm−1 (arrow) peak denoting β-sheet. (B) FTIR spectra of Aβ(26–42) in 100% HFIP (solid line) or deuterated 10 mM sodium phosphate buffer (pH 7.4) (dashed line). The amide I band for Aβ(26–42) in HFIP shows predominant β-sheet (1628 cm−1 major peak, ~1693 cm−1 shoulder), and minor α-helix (1657 cm−1 secondary peak); the corresponding band in phosphate buffer indicates high β-sheet (1626 cm−1 major peak, ~1693 cm−1 shoulder). Peptide concentrations were 470 µM, and spectra were recorded at room temperature. The abscissa (left to right) is 1740–1560 cm−1, while the ordinate represents absorption (in arbitrary units).
Table 1.
Proportions of secondary structurea for FP and Aβ(26–42) in HFIP and phosphate buffer, as estimated from Fourier self-deconvolution of the FTIR spectrab of the peptide amide I band
| System | % Conformation | |||
|---|---|---|---|---|
| α-Helix | β-Sheet | β-Turn | Disordered | |
| FP | ||||
| HFIPc | 54.8 | 8.3 | 29.9 | 7.0 |
| Phosphate bufferd | 20.1 | 40.4 | 19.5 | 20.0 |
| Aβ(26–42) | ||||
| HFIPc | 22.6 | 49.4 | 14.1 | 13.9 |
| Phosphate bufferd | 15.3 | 66.1 | 2.3 | 16.3 |
Data are the means of four separate determinations and have an SE ± 5% or better.
FTIR spectra were deconvoluted as described in section 2.
Peptide [i.e., 470 µM FP or Aβ(26–42)] dried on to an ATR plate from 100% hexafluoroisopropanol (HFIP) and resolvated with 100% HFIP (Fig. 2).
Peptide [i.e, 470 µM FP or Aβ(26–42)] dried on to an ATR plate from 100% HFIP, then resolvated with Phosphate buffer (i.e., deuterated 10 mM sodium phosphate buffer, pH 7.4) (Fig. 2).
fig. 4.
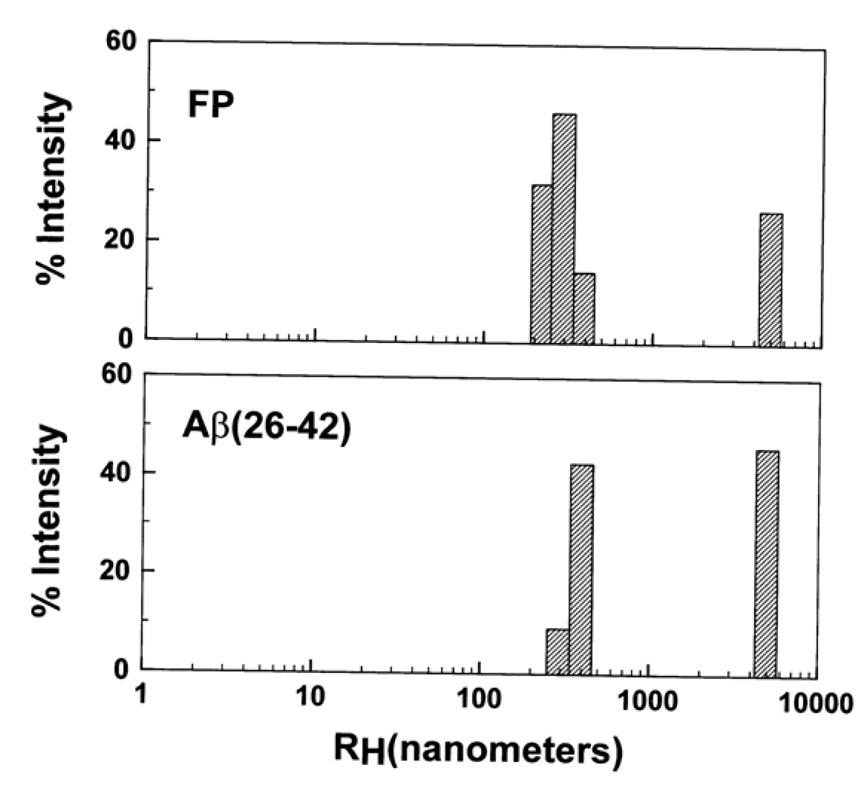
Dynamic light scattering (DLS) profile of HIV-1 FP (top) and Aβ(26–42) (bottom) in PBS, pH 7.4. Peptides were diluted with PBS to 40 µM from 5 mM in 0.1% acetic acid, and size readings were taken using a Dyna-Pro light-scattering instrument at 37°C every ten seconds for 2 min. Estimates of hydrodynamic radii (RH) were determined using the Dynamics Version 6.0 software (see section 2).
FTIR spectroscopy was also conducted to assess secondary structures of Aβ(26–42) in aqueous and HFIP environments. The FTIR spectrum of Aβ(26–42) in phosphate-buffer (pH 7.4) (Fig. 2B) indicates a major peak at 1626 cm−1, and a minor peak at 1693 cm−1, consistent with high amounts of extended β-sheet. Deconvolution of the aqueous Aβ(26–42) spectrum in Fig. 2B confirmed elevated levels of β-sheet (66.1%), with much smaller amounts of α-helix (15.3%) and other secondary structures (Table 1). Previous infrared spectroscopic results with either the full-length Aβ protein or overlapping C-terminal fragments are in agreement with these findings. For example, FTIR spectra of Aβ(1–40) in aqueous buffer demonstrated elevated β-sheet, as indicated by major (~1625 cm−1) and minor (~1685 cm−1) frequency bands [34]. In an early infrared study of C-terminal Aβ fragments dispersed in water-free KBr, predominate β-sheet conformations were identified from peaks at 1630 and 1695 cm−1 [66]. Moreover, aqueous fibrils of C-terminal Aβ peptides produced FTIR spectra with dominant maxima at ~1619–1630 cm−1 [42,43,46] and a high-field shoulder at ~1690 cm−1 [43], similar to the corresponding spectrum of the overlapping C-terminal Aβ(26–42) in phosphate buffer (Fig. 2B).
To assess whether multiple conformations also occur with this C-terminal Aβ peptide, FTIR spectroscopy was also performed with Aβ(26–42) in the lipid-mimic HFIP solvent. Fig. 2B shows the FTIR spectrum of Aβ(26–42) in 100% HFIP, indicating both high β-sheet (peaks at 1628 and 1693 cm−1) and low α-helix (secondary peak at 1657 cm−1). Spectral deconvolution verifies that Aβ(26–42) in HFIP largely retains a β-sheet conformation (49.4%), with small contributions due to α-helix, β-turn and disordered structures (Table 1). Consequently, Aβ(26–42) demonstrates only minor increases in α-helix (15.3 to 22.6%), and small decreases in β-sheet (66.1 to 49.4%), as the peptide is moved from aqueous to the lipid-mimic HFIP. Because previous studies of overlapping C-terminal Aβ peptides in membrane-mimics indicated either β-sheets [33,46] or α-helix [67,68], the limited polymorphism for Aβ(26–42) in the HFIP and aqueous environments (Fig. 2B) may reflect this peptide forming more stable β-sheet adducts than those of FP (see below).
3.2. TEM of FP and Aβ(26–42) in PBS buffer
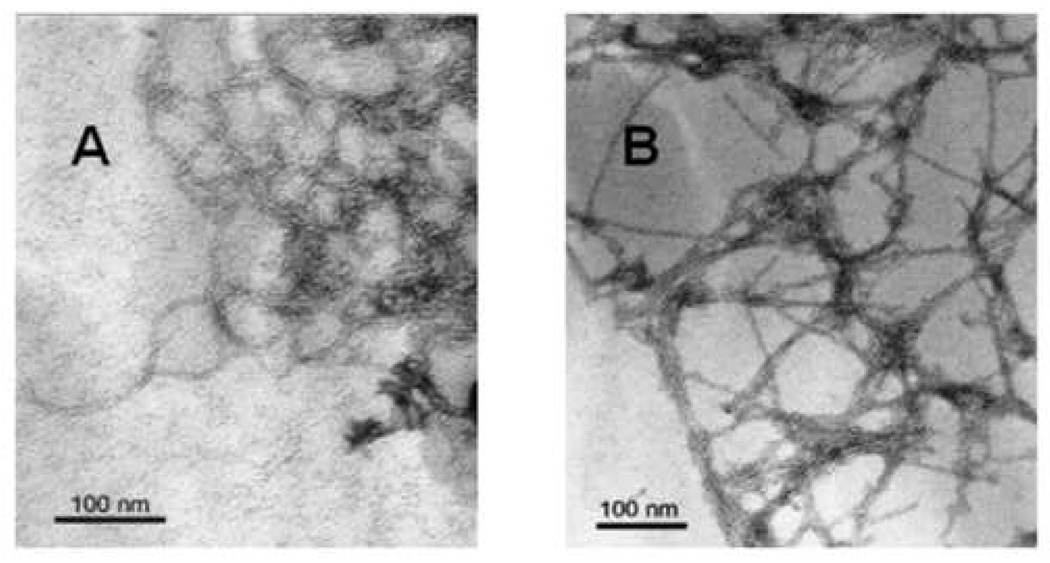
The peptide aggregates formed by FP and Aβ(26–42) in aqueous phosphate-buffered saline (PBS; pH 7.4) were examined with transmission electron microscopy (TEM). FP complexes are shown in Fig. 3A for peptide incubated on the formvar-coated grid in PBS at room temperature. For a 1-h incubation, the FP aggregates are a dense mat of networked fibrils or filaments. Individual fibrils are readily visualized with diameters of 7–8 nm. Although the length of these fibrils could not be determined precisely due to the extensive networking, discrete segments of loopy or curved filaments of 50–100 nm could be readily traced. At the nexus of the fibril branches are variably-sized aggregates or bundles (~50–75 nm in diameter), which themselves consist of smaller fibrils (Fig. 3A). Similar networked fibril aggregates (~75 nm in diameter), as well as spherical aggregates (~200 nm in diameter), were also identified in an earlier TEM study of FP incubated in PBS (pH 7.4) [14]. Comparable electron microscopic findings were made previously on HIV-1 FPs, in which large fibrillar bundles (~500–700 nm in diameter) were identified in aqueous buffers [23–25], assembled from long filaments ~5 nm in diameter [24].
fig. 3.
Transmission electron microscopy (TEM) of FP and Aβ(26–42) in phosphate-buffered saline (PBS), pH 7.4. Peptides were diluted with PBS to 40 µM from 5 mM in 0.1% acetic acid, and incubated at 37°C for 1 hr. (A) Image of uranyl acetate-stained grid in the presence of FP and PBS. (B) Image of uranyl acetate-stained grid in the presence of Aβ(26–42) and PBS. Scale bar represents 100 nm.
Aβ(26–42) aggregation was similarly studied here using TEM. For a 1-h incubation in PBS at room temperature, Aβ(26–42) forms a dense mat of networked fibrils (Fig. 3B). The individual fibrils that constitute this fibrillar network exhibited diameters of 7–8 nm, and straight fibrils of 100 nm in length (or longer) could be readily identified. As noted with electron micrographs of FP (Fig. 3A), however, the length of the Aβ(26–42) fibrils could not be accurately determined due to considerable networking. At the intersections of the fibril branches are aggregates (50–100 nm in diameter), some of them nearly spherical, and likely to be composed of smaller fibrillar bundles (Fig. 3B). Earlier electron microscopy of either Aβ(26–42) or overlapping C-terminal Aβ peptides in aqueous buffer showed fibril formation [42,43,46,66] similar to that observed for Aβ(26–42) (Fig. 3B). Incubation of Aβ(1–40) in aqueous media also demonstrated fibrils (8 nm in diameter), although these are typically ~500 nm or more in length [34], and thus distinct from the shorter Aβ(26–42) fibrils in Fig. 3B.
It is worthwhile to compare the respective fibrils formed upon incubation of FP and Aβ(26–42) in aqueous buffer. Electron microscopy of either FP (Fig. 3A) or Aβ(26–42) (Fig. 3B) indicated networks composed of constituent fibrils (~7–8 nm), with dense aggregates (~50–100 in diameter) at the branch intersections. The major difference between the FP and Aβ(26–42) networks lies in the connecting fibrils, which are loopy (curved) fibrils for FP (Fig. 3A) but are much straighter for Aβ(26–42) (Fig. 3B). This morphological discrepancy may be due to the differences observed in the FTIR spectra of FP and Aβ(26–42) in aqueous buffer. As noted in Fig. 2 and Table 1, deconvolution of the FTIR spectra of aqueous FP indicated lower β-sheet content, and higher levels of α-helix and β-turns, in comparison to that determined for aqueous Aβ(26–42). Alternatively, the shift in the absorption maximum for aqueous Aβ(26–42) in comparison to that of aqueous FP (i.e., 1626 vs. 1630 cm−1, respectively; Fig. 2) may be a consequence of the β-sheet structure of Aβ(26–42) being somewhat more extended and aggregated than that of FP [64,69]. Thus, stronger β-sheet interactions may be responsible for aqueous Aβ(26–42) exhibiting straighter fibrils than those observed for aqueous FP (Fig. 3). Such an interpretation would be consistent with the findings from a joint electron microscopy and FTIR study that limited heat-treated insulin (i.e., in aqueous buffer for 3 hr at 70°C) forms curved fibrils with high α-helix content and relatively low β-sheet [70]. Additional heat-treatment of insulin (i.e., in aqueous buffer for 48 h at 70°C) converts these ‘young’ fibrils into ‘mature’ ones more characteristic of amyloid fibrils; these mature fibrils are straighter on electron microscopy, and exhibit FTIR spectra with an enhanced β-sheet component shifted to lower frequencies [69–71].
3.3. DLS of FP and Aβ(26–42) in PBS buffer
A further assessment of the respective aggregation of HIV-1 FP and Aβ(26–42) in aqueous environments (PBS) was made using dynamic light scattering (DLS). Light scattering has been used to quantitate the aggregation state of full-length Aβ proteins [59,72]. In our present studies, FP or Aβ(26–42) was initially stored in 0.1% acetic acid to minimize peptide aggregation [17]. Figure 4 shows the hydrodynamic radii (RH) for FP and Aβ(26–42) at 2 min, indicating similar aggregation profiles for both peptides with peaks centered at 300 and 5000 nm. The FP and Aβ(26–42) aggregates detected in DLS may be related to those in electron microscopy (Fig. 3A and B) by noting that, for non-interacting rod diameters of 8 nm, our 300 and 5000 nm peaks approximately correspond to lengths of 900 to 50000 nm. As pointed out earlier [72], these lengths would be smaller if rod-rod interactions were occurring, while the actual lengths would be larger if the rods were flexible. Even with these caveats, however, our DLS results are consistent with the large fibrillar networks observed in TEM of HIV-1 FP and Aβ(26–42) in PBS (Fig. 3A and B). It should be emphasized that the aggregate sizes for either FP or Aβ(26–42) from DLS are larger than the much smaller fibrils reported for the full-length Aβ(1–40) protein. In earlier studies [72,73], light scattering of Aβ(1–40) indicated RH values from 10 to 50 nm, with corresponding lengths of noninteracting rods of 30–500 nm for these ‘protofibrils’ that are much smaller than our fibrils. However, the Aβ(1–40) samples were subjected to centrifugation and size-exclusion chromatography to exclude large aggregates that might obscure the detection of small oligomers [72,73].
3.4. Prediction of amyloid structure aggregation (PASTA) analyses of FP and Aβ(26–42)
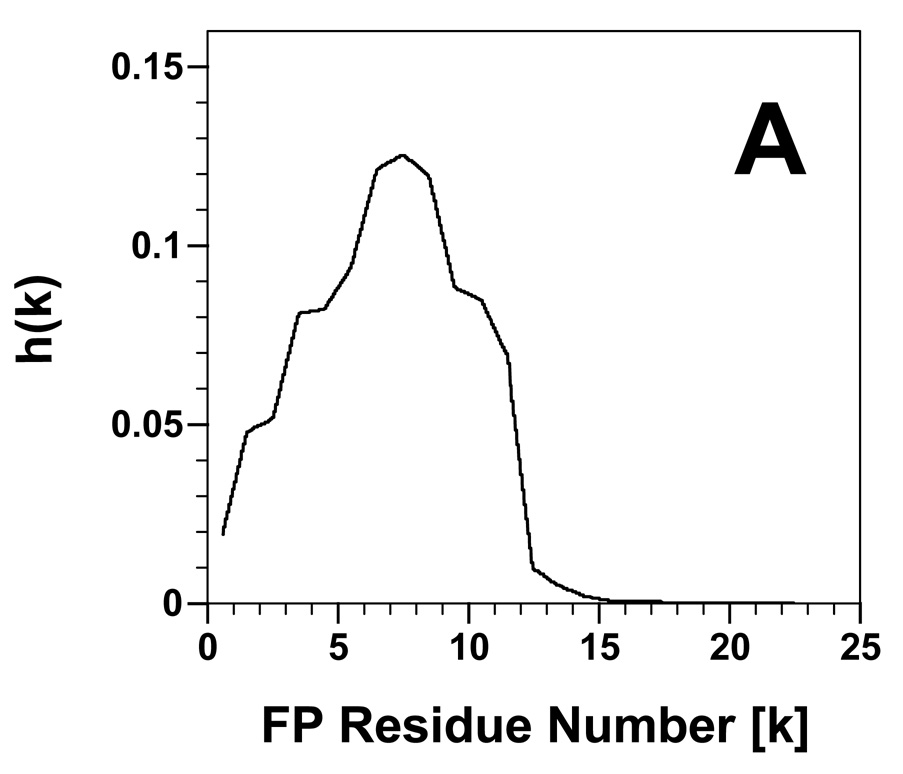
To further investigate the networked β-sheet fibrils identified above, energy calculations using PASTA analysis [36,37] were performed on both FP and Aβ(26–42). PASTA indicated that FP will most likely form a parallel β-sheet, in register arrangement (PIRA) for residues 2–11 with a relative energy of −3.47. Using as a benchmark a database of 179 peptides derived from the literature [36], a PASTA energy threshold of −3.47 indicates that the probability that FP is amyloid-like is ~72% (true positive rate). An aggregation profile was constructed for FP (Fig. 5A) (see section 2), and also showed that the sequence 2–11 would likely form β-sheets. Experimental confirmation of β-sheet for FP residues ~1–16 has been earlier obtained in residue-specific spectroscopy of N-terminal gp41 peptides in PBS buffer [14], model liposomes [17,19,20], and RBC ghost membranes or liposomes at high peptide loading [3].
fig. 5.
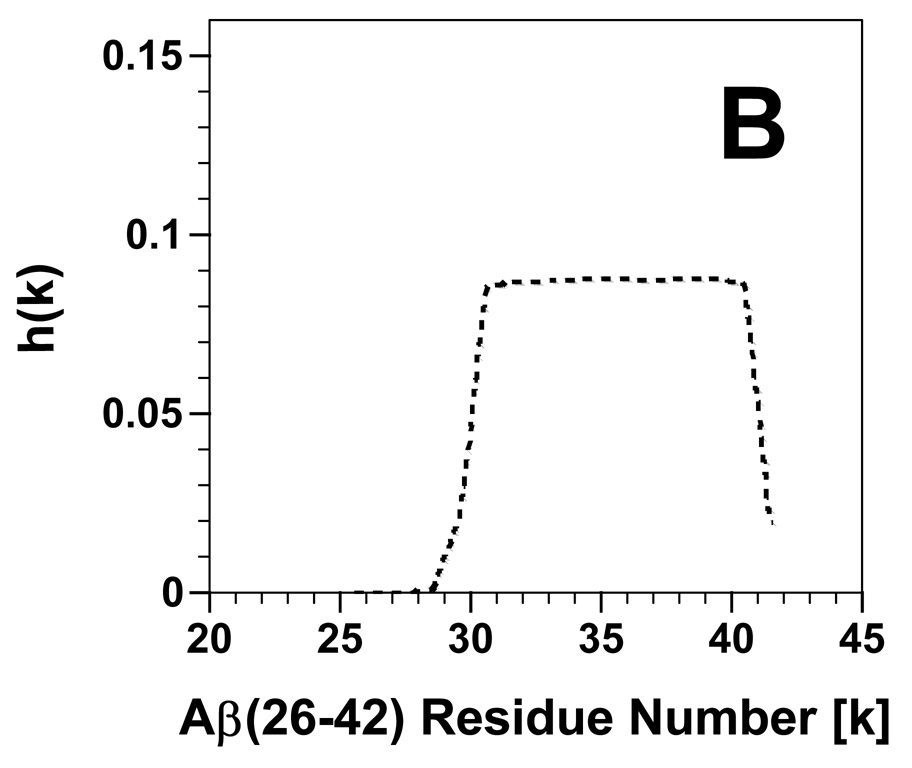
Propensity for β-sheet aggregation [i.e., h(k)] plots determined for FP (A) and Aβ(26–42) (B) from PASTA analyses [36,37]. The h(k) values are plotted for each peptide residue (k), and normalized so that the summation of all h(k)s for a peptide equals 1.0. The sequence and numbering for FP and Aβ(26–42) are in Fig. 1.
PASTA calculations were next conducted on Aβ(26–42), and also indicated that the fibrils identified above for this C-terminal Aβ peptide are probably due to parallel, in register (PIRA) β-sheets. The most stable PIRA pairing was for sequence 31–41, and showed a very low PASTA energy of −8.08 with a probability of ~100% (true positive rate) that this sequence is amyloid-like. The aggregation profile for Aβ(26–42) in Fig. 5B also demonstrates that residues ~31–41 will form β-sheets. These findings agree with those of the original PASTA analysis of the C-terminal region of the Aβ protein [36,37], and are verified by earlier residue-specific spectroscopic results on either the full-length Aβ protein or C-terminal Aβ peptides. For example, solid-state NMR spectroscopy of Aβ(1–40) fibrils reported that the C-terminal residues 30–40 form PIRA arrangements [35], while 13C-FTIR spectroscopy of Aβ(34–42) in self films indicated that the sequence 36–41 folds as β-sheet [44]. It is of particular interest that much lower PASTA energies of interactions are observed for the top-ranked Aβ(26–42) pairings than the corresponding FP ones. A stronger cross-β structure might explain the FTIR absorption maxima occurring at lower frequencies for Aβ(26–42) than for FP in aqueous media (Fig. 2A and B), and also account for the aqueous Aβ(26–42) fibrils being straighter than those for FP on TEM (Fig. 3A and B).
3.5. Lytic and aggregating actions of FP and Aβ(26–42) on human erythrocytes
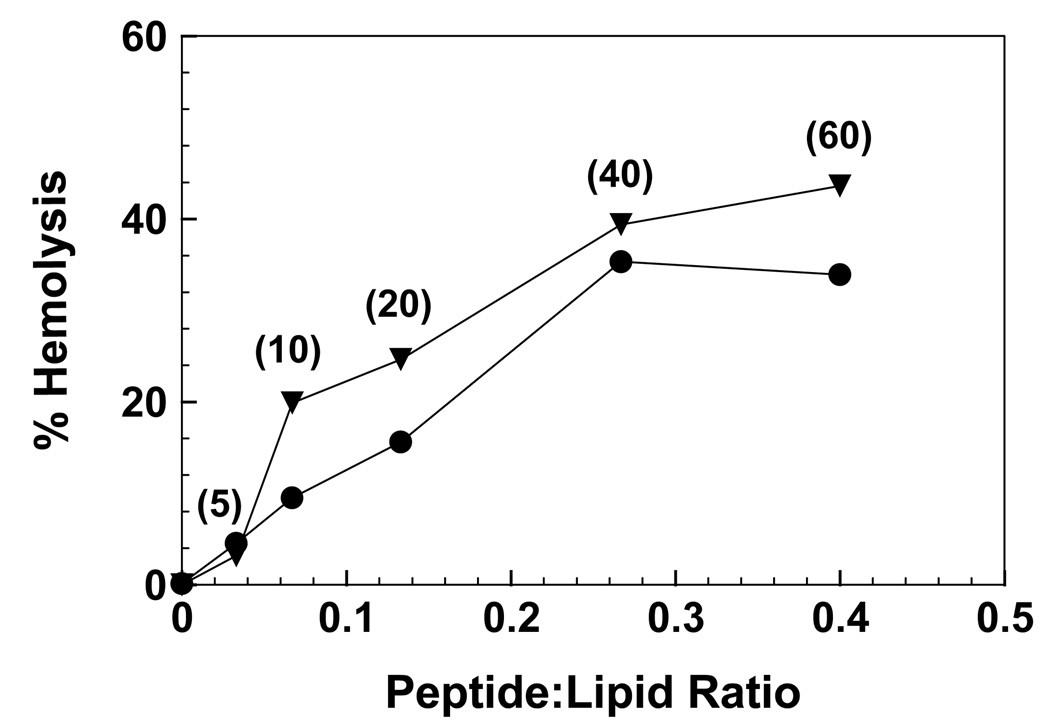
The respective hemolytic actions of the synthetic peptides FP (gp41 residues 519–541) and Aβ(26–42) (Fig. 1) were studied with human RBCs suspended in isotonic buffer at 37°C for 30 min. Fig. 6 shows that FP induces approximately 35% hemolysis, for peptide concentrations between 40 and 60 µM. Prior batches of synthetic FP produced comparable hemolysis with human red cells [4,7,8,10,11,54]. Lytic activity may be a general property of HIV FP, as the fusion peptides from both HIV-1 gp41 and HIV-2/SIV gp32 have been reported to be hemolytic with human red cells [9]. Fig. 6 further shows that Aβ(26–42) produces a similar dose-response profile to that of FP, with a slightly higher maximum hemolysis of ~40%. Comparable to that shown here with Aβ(26–42), high hemolysis was observed for incubations of the overlapping C-terminal Aβ(25–35) peptide with human red blood cells [40,50].
fig. 6.
Effects of the HIV-1 FP (●) and Aβ(26–42) (▼) peptides on the isotonic hemolysis of erythrocytes (see Fig. 1 for sequences). Red cells (1 × 1011 cells/liter) were incubated with peptide for 30 min at 37°C. Percent hemolysis is determined as described in Section 2; 100% hemolysis is defined by incubating 10 µl of packed cells in 2% Triton X-100 in distilled water (0.5 ml) for 30 min at room temperature. Peptide:lipid ratios were calculated assuming quantitative uptake of each peptide by the red cells [4,10]. Numbers in parentheses next to the FP and Aβ(26–42) curves indicate the peptide concentrations (µM) used in the incubations. Results are reported as the mean of duplicate measurements, and are representative of three independent measurements. The estimated SD for each value is ±5–8%.
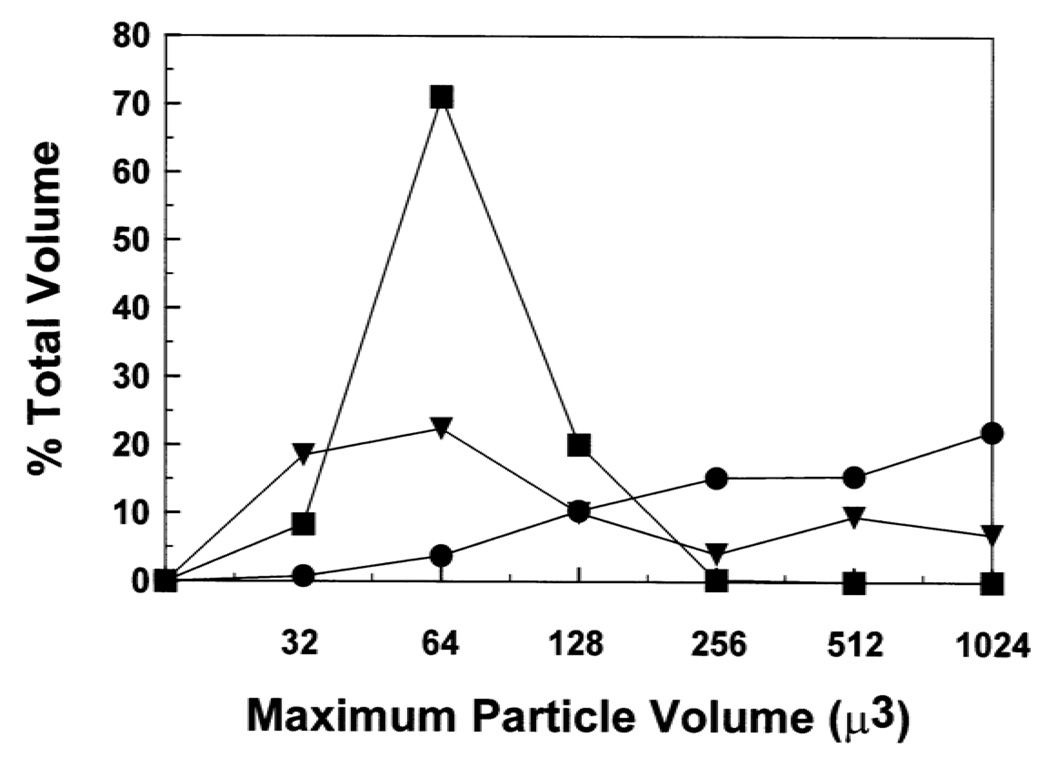
Because FP and Aβ(26–42) each induce similar hemolytic activities, these peptides were next tested for aggregation of human red cells. Coulter counter studies were performed on washed, packed red cells incubated in isotonic PBS with 60 µM doses of FP or Aβ(26–42). Counts were made for particles in channels ranging in maximum particle volume (µ3) from 32 to 1024. Plots of the percentage of total volume, %TV, versus the maximum particle volume are shown in Fig. 7, where %TV is the percent of red cell particle volume in a given channel relative to the total volume of the particles in all channels (see Section 2). Control experiments with erythrocytes incubated in PBS alone showed a well-defined maximum centered at the 64 µ3 channel, denoting single cells. For incubations with 60 µM FP, however, there was a large decline in the 64 µ3 channel representing single cells, and an increase in the occupancy of larger channels (i.e., ≥ 256 µ3) due to aggregated cells (Fig. 7). These results are consistent with previous Coulter counter and electron microscopic analyses showing that FP promotes the formation of red cell multimers [7,8,10,11], and also agree with earlier findings that N-terminal gp41 peptides aggregate model liposomes [64]. Figure 7 additionally shows that 60 µM Aβ(26–42) reduces the number of single cells and increases the proportion of multimeric cells, although the aggregation induced by Aβ(26–42) is less than that of FP. The aggregation activity of Aβ(26–42) with red cells reported here is in accord with prior experiments indicating that the full-length Aβ protein mediated lipid vesicle association with cells [74], and also that overlapping C-terminal Aβ peptides promoted aggregation of model liposomes [46,47,75].
fig. 7.
Coulter counter sizing of erythrocytes treated with HIV-1 FP and Aβ(26–42). Erythrocytes were tested for aggregation by incubating 10 µl of packed red blood cells, diluted to 0.5 ml in PS buffer, with the following agents at 37°C for 30 min: 60 µM FP (●); 60 µM Aβ(26–42) (▼); control PBS (■). At the end of each incubation, an aliquot of the reaction mixture was diluted 1/500 in isotonic phosphate-buffered saline. The number of particles in each size range (i.e., maximum particle volume µ3,x axis) shown was then measured with a Coulter counter. Percent (total volume) for the particles in each channel (y-axis) is calculated as in Section 2. Results are reported as the mean of duplicate determinations, and are representative of three independent experiments.
3.6. FTIR spectroscopic analysis of FP and Aβ(26–42) in model liposomes
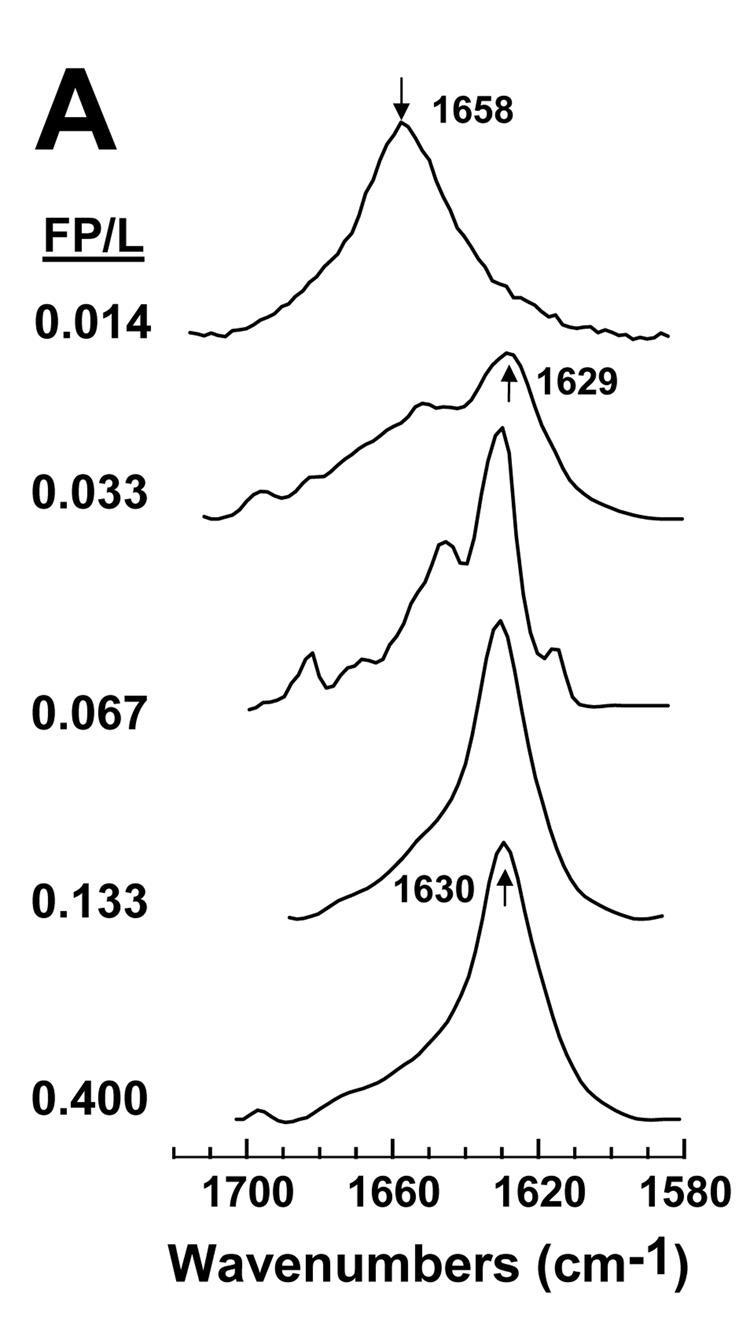
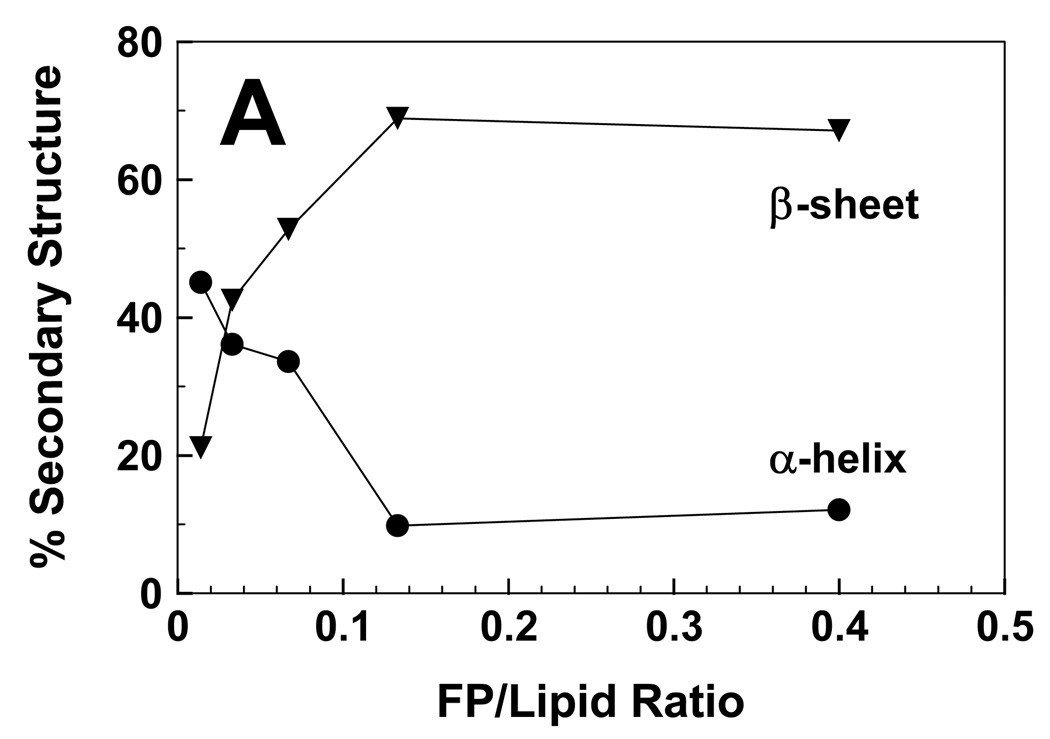
It is also worthwhile to characterize the secondary structures of FP and Aβ(26–42) in environments in LUV of POPC:POPG (1:1). Because the zwitterionic POPC and the anionic POPG are comparable to lipids found in surface membranes, this mixture should be a more accurate mimic of the lipid bilayer than HFIP solutions. With POPC:POPG (1:1) at a low FP/L of 0.014, the FTIR spectrum of the amide I band indicated a dominant helical component centered at 1658 cm−1 (Fig. 8A). Importantly, nearly identical FTIR spectra were previously reported at these low FP/L in either POPC:POPG (1:1) [10] or erythrocyte ghost liposomes [3]. Subsequent deconvolution of the FP in POPC:POPG (P/L = 0.014) spectrum (Fig. 8A) confirmed a high proportion of α-helix and much lower levels of β-sheet (Fig. 9A) for this low P/L, which is non-lytic in hemolysis assays (Fig. 6). FTIR spectra are also shown for FP in POPC:POPG liposomes recorded over peptide concentrations (Fig. 8A) that match those used in hemolysis assays (Fig. 6), and spectral deconvolutions reveal the relative proportions of α-helix and β-sheet as a function of P/L (Fig. 9A). As the FP/L increases from low (0.014) to high (0.400) high ratios, Fig. 8A and Fig. 9A demonstrate that a second spectral component arises centered at 1629–1630 cm−1 due to β-sheet, while concurrently the α-helical component at 1658 cm−1 becomes diminished. The predominate β-sheet component obtained here for FTIR spectra of POPC:POPG liposomes at high loading (i.e., FP/L of 0.133 or 0.400) (Fig. 8A) is similar to those observed for human RBC ghost liposomes at high loading (FP/L of 0.100) [3], again indicating that POPC:POPG liposomes are useful for assessing peptide conformations in biomembranes. The most likely explanation of the spectral results in Fig. 8A and Fig. 9A is that raising the peptide level converts lipid-associated FP from α-helix at low loading to β-sheet at high loading. It is also interesting that the FP/L ratios that induce hemolysis (Fig. 6) are closely correlated with the onset of significant β-sheet (Fig. 8A and Fig. 9A), raising the possibility that the lytic FP species is membrane-bound β-sheet.
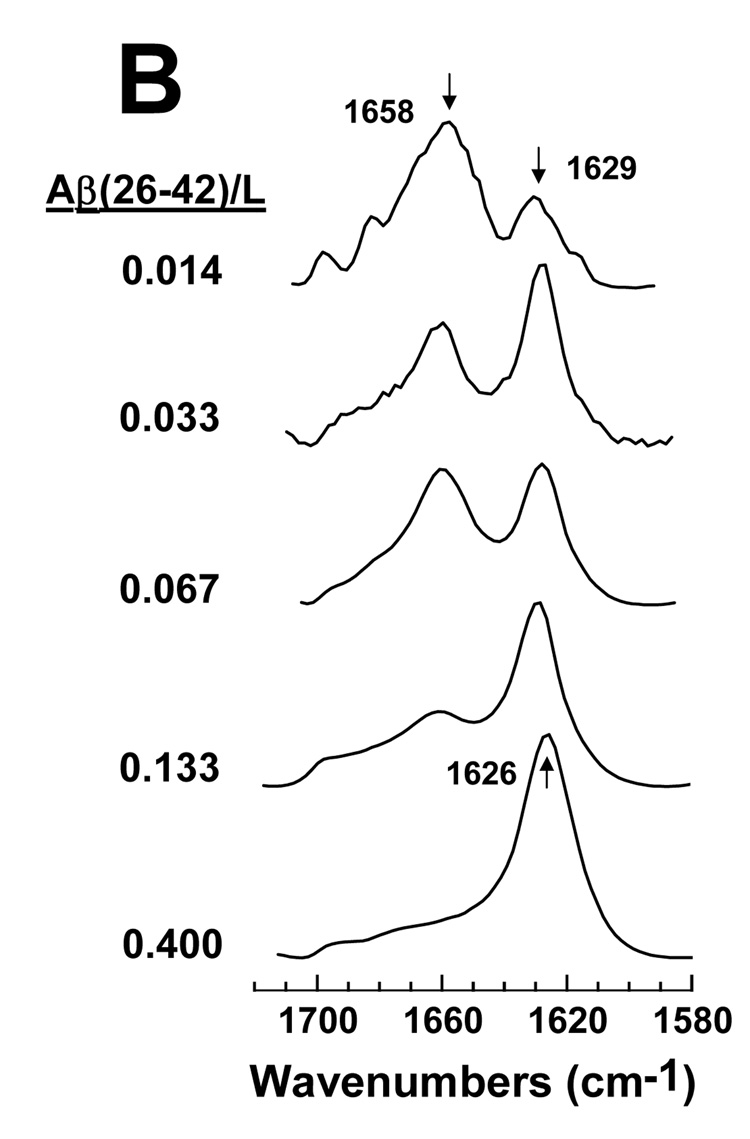
fig. 8.
FTIR spectra of FP (A) and Aβ(26–42) (B) in POPC:POPG liposomes at the indicated peptide/lipid (FP/L or Aβ(26–42)/L) ratios. FP and the Aβ(26–42) in a stock solution of 100% HFIP were added to LUV of POPC:POPG (1:1) lipids in 10 mM sodium phosphate buffer (pH 7.4). Peptide:liposomes were chromatographed to remove non-lipid-associated peptide, as described in Section 2. Spectra were recorded on chromatographed peptide:liposomes that were dried on the ATR and hydrated with D2O for 2 h. The arrows at 1658 cm−1 in A and B indicate the α-helix component, while the arrows at 1629 and 1630 cm−1 in A and 1629 and 1626 cm−1 denote the β-sheet component.
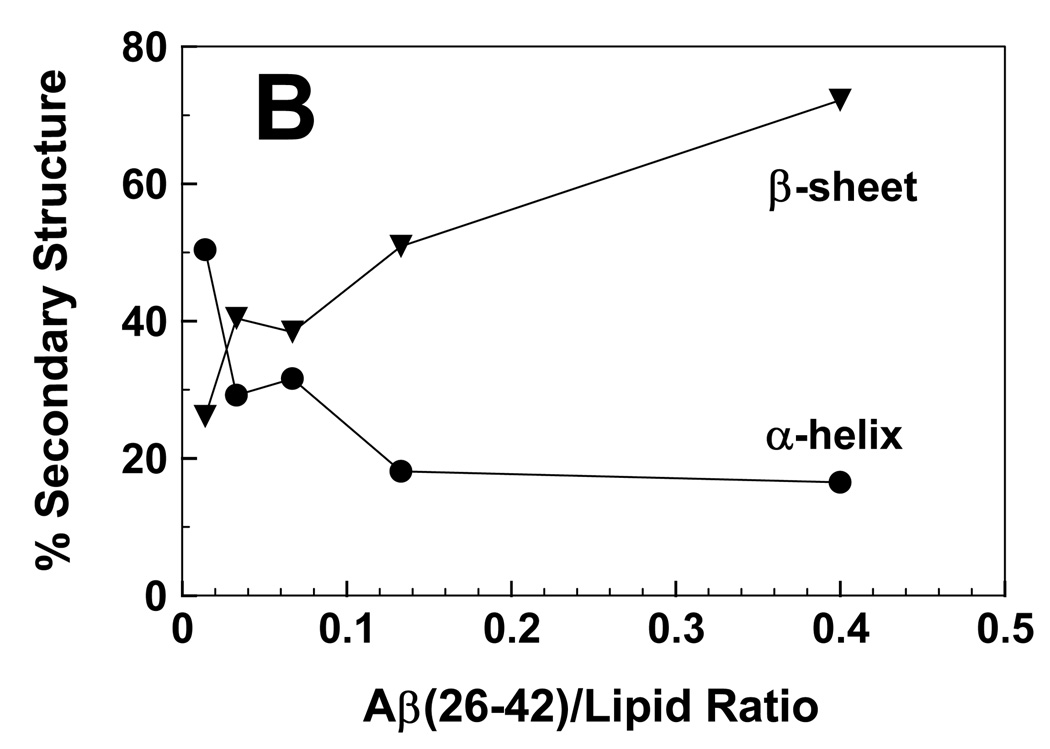
fig. 9.
Proportions of α-helix (●) and β-sheet (▼) for FP (A) and Aβ(26–42) (B) in POPC:POPG (1:1) liposomes, at the indicated peptide/lipid ratios. The % secondary structure conformations were determined from the FTIR spectra of FP (Fig. 8A) or Aβ(26–42) (Fig. 8B) in lipid, as described in section 2.
FTIR spectra were also recorded with Aβ(26–42) added to POPC:POPG liposomes at the same P/L ratios used to study hemolysis in Fig. 6. For Aβ(26–42) in POPC:POPG (1:1) at a low P/L ratio of 0.014, the FTIR spectrum in Fig. 8B indicated primarily α-helix at 1658 cm−1, and lower levels of β-sheet at 1629 cm−1. Spectral deconvolution of the Aβ(26–42) spectrum (P/L = 0.014) confirmed that α-helix was the primary conformation at low loading, with smaller contributions due to β-sheet (Fig. 9B). Similar to that noted for FP in Fig. 8A and Fig. 9A, increasing the Aβ(26–42) concentration substantially elevated the proportion of the β-sheet component, such that the β-sheet became the dominant conformation above P/L ratios of 0.014 (Fig. 8B and Fig. 9B). The Aβ(26–42)/lipid ratios that induce hemolysis (Fig. 6) are correlated with the development of substantial β-sheet (Fig. 8B and Fig. 9B), suggesting that Aβ(26–42) like FP lyses RBCs by binding to membranes in a β-sheet conformation.
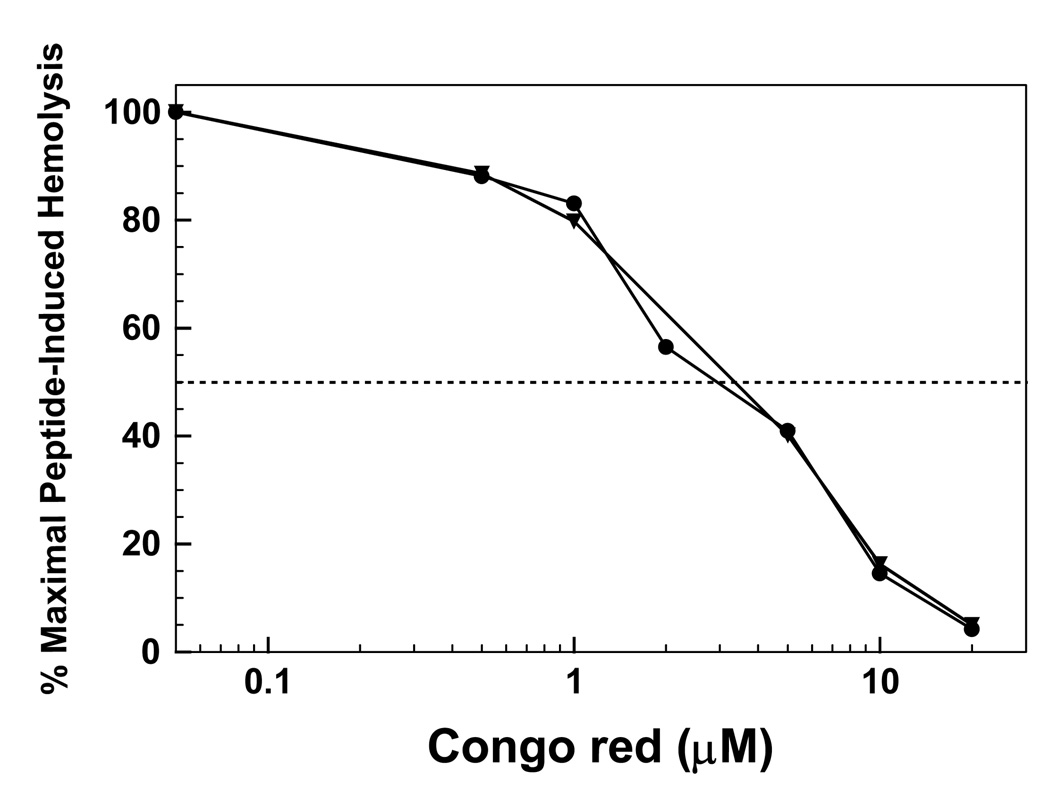
3.7. Congo red (CR) inhibition of FP- and Aβ(26–42)-induced hemolysis
Because HIV-1 FP and Aβ (26–42) induce similar lytic activities with human erythrocytes, FP and Aβ(26–42) may each be further assessed for amyloid-like properties by testing the cross-inhibition of their lytic activities by Congo red (CR). The actions of CR are noteworthy, not only because this dye has been widely used as a marker of amyloid deposits in situ [51,52,76], but also because it reduces the cytotoxicity of various amyloids, including Aβ proteins and peptides [38,40,77]. The concentration (60 µM) of membrane-perturbing FP and Aβ(26–42) was chosen from hemolytic curves that gave substantial release of hemoglobin (Fig. 6). For red cells preincubated with a range of CR doses, inhibition plots were constructed as the percent maximal hemolysis versus the concentration of CR (Fig. 10). Here, the percent maximal hemolysis for each membrane-perturbing peptide [i.e., 60 µM FP or Aβ(26–42)] is defined as 100, without inhibitor (Fig. 6). The relative potency of CR in reducing the membrane actions of FP and Aβ(26–42) may then be determined from ID50s, calculated as those doses that produce 50% inhibition for 60 µM FP or Aβ(26–42). For comparisons with archival inhibition of HIV-1 FP- or Aβ peptide-induced toxicity, Congo red IR100 values were also calculated as the ratios of the CR concentration to that of the membrane-perturbing peptide that produce 100% inhibition.
fig. 10.
Congo red (CR) inhibition of hemolysis induced by FP (●) or Aβ(26–42) (▼). Erythrocytes (1 × 1011 cells/liter) were incubated with 60 µM FP or Aβ(26–42), and the indicated concentrations CR for 30 min at 37°C. Percent hemolysis was determined as described in Section 2, and is reported as the mean of duplicate determinations. One hundred percent maximal hemolysis for the inhibition curves is defined as that obtained by incubating 60 µM FP or 60 µM Aβ(26–42) with 10 µl of packed erythrocytes in 0.5 ml of PBS, without inhibitor. The dashed horizontal line represents 50% of the maximal hemolysis, observed without CR.
The inhibition profiles of CR on FP- and Aβ(26–42)-induced hemolysis are shown in Fig. 10. CR is an effective inhibitor of 60 µM Aβ(26–42)-induced hemolysis, yielding an ID50 of 4.2 µM; at a higher dose of 20 µM, CR completely blocked Aβ(26–42)-induced hemolysis, with an IR100 of 0.33. These results are in good agreement with previous studies of the human RBC lysis caused by either the overlapping Aβ(25–35) peptide or full-length Aβ(1–42), in which pretreatment with CR achieved IR100 values of ~0.2 [40]. It is of particular interest that CR also lowered FP-induced hemolysis with ID50 (4.2 µM) and IR100 (0.33) values identical to those seen with Aβ(26–42) (Fig. 10). Our finding of very similar CR-inhibition patterns for both FP and Aβ(26–42), combined with prior results showing that CR likewise blocks the toxic activity of classic amyloid proteins found as plaques in situ [38,40,77], further supports the assignment of both FP and Aβ(26–42) as ‘amyloid homologs’.
3.8. Surface plasmon resonance (SPR) detection of Congo red (CR) binding to FP and Aβ(26–42)
Given the high activity for Congo red in reducing either FP- or Aβ(26–42)-induced hemolysis, it is important to determine whether this inhibition may be due to the direct binding of CR to these peptides. Here, we use surface plasmon resonance (SPR) spectroscopy to assess CR binding to either FP or Aβ(26–42). SPR is a surface-sensitive methodology, in which the ligand is chemically-linked to a sensor surface, and the solute is then flowed past the ‘chipped’ molecule. The binding of the solute to the immobilized ligand produces an evanescent response, which is proportional to the bound mass. SPR experiments have recently detected high-affinity binding of CR to amyloid proteins (e.g., PrP [58,78] and Aβ(1–42) [79] ) or amyloid homologs (e.g., the C-terminal Aβ(10–35) peptide [80] ). In the present studies, FP and Aβ(26–42) are each chemically-linked to chips at their respective N-terminal amine groups, while the solute CR in buffer is flowed past the immobilized peptides. Molecular binding affinities (associations) between the soluble CR and chip-linked films of FP or Aβ(26–42) were measured at 37°C using a Biacore apparatus [57]. Results in Table 2 showed that either FP or Aβ(26–42) had substantial binding (association) affinity for CR based on high uptake rate constants (kon) and low dissociation rate constants (koff). The respective equilibrium dissociation constants (KD = koff/kon) for either FP or Aβ(26–42) binding to CR were similarly low in Table 2, indicating that CR had high affinity for both peptides (Table 2).
Table 2.
Mean association and dissociation kinetic rate constants (kon, koff) and equilibrium dissociation constants (KD), calculated from surface plasmon resonance (SPR) measurements for aqueous Congo red (CR) flowing past chip-linked FP and Aβ(26–42) monolayersa
| Chip-linked peptide | kon (1/Ms) | koff (1/s) | KD (nM) |
|---|---|---|---|
| FP | 62.9 × 105 | 18.7 × 10−3 | 2.97 |
| Aβ(26–42) | 19.2 × 105 | 4.0 × 10−3 | 2.10 |
Congo red in HBS-EP buffer was flowed past monolayers of FP or Aβ(26–42), linked via their respective N-terminal amides to CM5 sensor chips in a Biacore 3000 system (see section 2). Mean kinetic rate constants (kon, koff) and equilibrium dissociation constants (KD = koff/kon) were determined from curve fitting analyses of SPR results at 10 nM CR.
4. Discussion
The present structural and functional comparisons of HIV-1 FP and a C-terminal Aβ(26–42) peptide further support the hypothesis that the N-terminal peptide of HIV-1 gp41 exhibits ‘amyloid-like’ characteristics. Although FP and Aβ(26–42) do not fulfill the currently-accepted definition of ‘amyloid’ because neither is found in extracellular plaque in situ, these peptides may nevertheless belong to a related class of ‘amyloid homologs’ that was recently described by the International Society of Amyloidosis [51–53]. As noted in the Introduction and Fig. 1, FP and Aβ(26–42) each have elevated alanine and glycine levels, which is a typical of amyloid-related peptides [21]. Another shared property of amyloid-like peptides is structural polymorphism [21], and here we show that FP and Aβ(26–42) exhibit high α-helix at low peptide loading in liposomes (Fig. 8 and Fig. 9), while converting to β-sheet for peptides in either liposomes at high loading (Fig. 8 and Fig. 9) or aqueous buffer (Fig. 2; Table 1). Results from FTIR, TEM and DLS experiments show that these peptides readily form extensive cross-β networks in vitro (Fig. 2–Fig. 4) that mimic those formed by actual amyloid proteins in vivo. FP and Aβ(26–42) similarly lysed and aggregated human RBCs (Fig. 6 and Fig. 7), sharing the toxic and fusogenic properties that are hallmarks of amyloid proteins [21]. Congo red potently inhibited the hemolytic actions of FP and Aβ(26–42) (Fig. 10), probably due to the dye binding to these peptides with high-affinity (Table 2). Congo red has been widely used as a marker of amyloid deposits in vivo [51,52,76], and also blocks the cytotoxic activities of classically-defined amyloid proteins [38,40,77]. Taken together, our results support the classification of both FP and Aβ(26–42) as ‘amyloid-like’ or ‘amyloid homologs’ [51].
Despite the findings here indicating that both FP and Aβ(26–42) form networked β-fibrils in aqueous buffer, results from energy predictions and FTIR and TEM experiments also show that the FP fibrils are more easily deformed due to weaker core β-sheet interactions. PASTA calculations indicated that FP and Aβ(26–42) will each form parallel β-sheet, in register arrangements (PIRA) (Fig. 5A and B), but that the cross-β structure will be considerably stronger for the Aβ(26–42) peptide (see section 3.4). Weaker β-sheet interactions in the FP fibrils may account for the FTIR maxima of aqueous FP occurring at higher frequencies than those observed for aqueous Aβ(26–42) (i.e., 1630 vs. 1626 cm−1; Fig. 2). As noted in section 3.2, the FP fibrils are more loopy (curved) than those of Aβ(26–42) in TEM (Fig. 3), probably due to the β-sheet of FP being less extended and aggregated than that of Aβ(26–42). Interestingly, a recent survey indicated that native β-sheet proteins typically yield FTIR spectra with maxima at 1630 cm−1 and higher frequencies, while the corresponding spectra of amyloid fibrils exhibit maxima at 1630 cm−1 and lower [69]. With this FTIR classification scheme, we would assign Aβ(26–42) as an amyloid, while FP might exhibit characteristics between those of amyloid and native β-sheet proteins. PASTA analysis also here categorizes FP as an amyloid with somewhat less confidence than Aβ(26–42) (i.e., true positive rates of ~72 and ~100%, respectively), due to the prediction of weaker cross-β interactions in FP fibrils (see section 3.4).
Close correlations were noted here between the onset of hemolysis induced by either FP or Aβ(26–42) (Fig. 6) and the conversion of α-helix to β-sheet for each peptide in liposomes (Fig. 8 and Fig. 9). Although both FP and the C-terminal Aβ peptides have been extensively investigated, there are no generally accepted models for how these peptides exert their respective fusogenic and lytic activities on cells and liposomes. Earlier work on HIV-1 fusion peptides (for review, see [3]) or Aβ proteins and peptides [32,47,75,81,82] indicated possible roles for the deep insertion of peptides into lipid bilayers, oblique intercalation of α-helical peptides into the membrane bilayer, membrane aggregation of peptides, formation of membrane-associated peptide in β-conformations, peptide-mediated membrane fluidization, peptide induction of non-bilayer lipid, and/or creation of peptide-membrane pores. In the present studies, the FP/L or Aβ(26–42)/L ratios that induce hemolysis (Fig. 6) are correlated with the development of substantial β-sheet for these peptides in liposomes (Fig. 8 and Fig. 9), suggesting that each peptide lyses RBCs by binding to membranes in a perturbing β-sheet conformation. Earlier residue-specific 13C-labeled FTIR studies of RBC ghost membranes or liposomes [3] are in agreement with the above assignment of membrane-bound, β-sheet FP as the hemolytic species. For example, FTIR spectra of 13C-labeled FP in ghost liposomes indicated that FP residues Gly-5 through Ala-15 were α-helix at a low, non-lytic FP/L ratio of 0.014, but instead folded as β-sheet at a higher, lytic P/L of 0.10 [3]. These FP β-sheet adducts may require a highly-conserved, stereospecific structure to produce membrane perturbations. Prior studies indicated that several synthetic FP analogs (i.e., FP2V/E with Val → Glu at residue 2; FP11F/Y with Phe → Tyr at residue 11; FPCLP1 with FP truncated at residue 7) were non-hemolytic, despite each analog converting from α-helix at a low P/L (= 0.013) to β-sheet at a high P/L (= 0.100) when bound to liposomes [10]. Interestingly, our results are largely supportive of a mechanism proposed earlier by Sackett and Shai [19], based on 13CFTIR spectroscopy and model building, which posited that the N-terminal gp41 domain would mediate its membrane actions by first forming trimeric FP sequences folded with parallel β-sheet, in register alignments (PIRA) for residues ~1–16. An analogous model was also forwarded to explain the fusogenic properties of the full-length Aβ protein and Aβ peptides [19], and our results indicating that membrane-bound, β-sheet Aβ(26–42) is hemolytic (Fig. 6 and Fig. 9) are consistent with this hypothesis.
The observation that HIV-1 FP perturbs membranes through ‘amyloid-like’ mechanisms may aid our understanding for how this N-terminal gp41 domain participates in both the HIV life-cycle and AIDS pathogenesis. Considerable evidence has now confirmed the involvement of the FP region in HIV-1 mediated cytolytic and fusogenic processes. For example, site-directed mutagenesis indicated defective gp41 fusion activity for various modifications in the N-terminal domain [10]. Moreover, synthetic peptides based on the FP region promoted leakage of lipid vesicles [3], and also lysed erythrocytes [4,7–9] and cultured cells, including CD4+-lymphocytes [4–6]. Here, we showed that the N-terminal gp41 peptide may induce RBC lysis at high FP/L ratios (e.g., ≥ 0.10) (Fig. 6) through the formation of membrane-bound β-sheet FP, as was detected adding FP to either erythrocyte ghost lipids [3] or ghost lipid mimics (Fig. 8A and Fig. 9A). Given that the lipid composition of RBC membranes is similar to that of lymphocyte plasma membranes [83,84], it is likely that the synthetic FP-induced lysis of CD4+-lymphocytes noted in the above in vitro studies may also be due to amyloidogenic perturbations involving surface membrane-bound, β-sheet FP adducts. This raises the possibility that the in vivo depletion of CD4+-lymphocytes that is a hallmark of AIDS patients may be partially due to amyloid-like actions on lymphocyte plasma membranes triggered by the N-terminal gp41 domain of HIV-1 virions. To the extent that FP shares amyloid properties, it is also tempting to speculate that anti-HIV agents that block the membrane perturbations of FP may also reduce the cytotoxic actions of amyloids and amyloid homologs.
Acknowledgments
This study was supported by NIH SCORE Grants GM 53933 (P. Mobley, S. Eskandari). The ABI 431A peptide synthesizer was obtained with NIH Small Equipment Grant GM 50483 (A. Waring, L. Gordon), the Protein Technologies Symphony®/Multiplex SPPS synthesizer was acquired with a NIH National Center for Research Resources Shared Instrumentation Grant 1 S10 RR14867-01A1 (M. R. Yeaman, A. Waring) and the REI Bruker Vector 22™ FTIR spectrometer was funded by a grant from the Harbor-UCLA REI Common Use and Replacement Equipment Program (A. Waring).
Abbreviations
- HIV-1
Human Immunodeficiency Virus, Type 1
- HIV-2
Human Immunodeficiency Virus, Type 2
- gp41
glycoprotein 41,000 of HIV-1
- FP
fusion peptide (23-residues) at the N-terminus of HIV-1 gp41 (LAV1a strain)
- SIV
simian immunodeficiency virus
- gp32
glycoprotein 32,000 of HIV-2/SIV
- Aβ
amyloid-beta protein
- Aβ(1–40) or Aβ(1–42)
amyloid-beta protein residues 1–40 or 1–42
- Aβ(26–42)
C-terminal peptide 26–42 of amyloid-beta protein
- DLS
dynamic light scattering
- TEM
transmission electron microscopy
- PIRA
parallel β-sheet, in register arrangement
- PASTA
prediction of amyloid structure aggregation
- PBS
phosphate buffered saline; Phosphate buffer, 10 mM sodium phosphate buffer, pH 7.4
- HBS-EP buffer
10 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20
- DMSO
dimethyl sulfoxide
- HFIP
hexafluoroisopropanol
- SDS
sodium dodecyl sulfate
- TFE
trifluoroethanol
- RBC
red blood cells
- POPG
1-palmitoyl-2-oleoyl phosphatidylglycerol
- LUV
large unilamellar vesicles
- POPC
1-palmitoyl-2-oleoylphosphatidylcholine
- POPG
1-palmitoyl-2-oleoylphosphatidylglycerol
- P/L
peptide/lipid molar ratio
- CD
circular dichroism
- FTIR
Fourier transform infrared
- ATR
attenuated-total-reflectance
- 2D-NMR
two dimensional Nuclear Magnetic Resonance
- IR50
inhibitory ratio (IR) of the agent concentration to that the membrane-active agent that induces 50% inhibition
- IR100
inhibitory ratio (IR) of the agent concentration to that the membrane-active agent that induces 100% inhibition
- ID50
inhibitor concentration that yields 50% inhibition for a stated dose of membrane-active agent
- PrP
prion protein
- HPrP
human prion protein
- BPrP
bovine prion protein
- MD
molecular dynamics
- SPR
surface plasmon resonance
- CR
Congo red
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson MR, Nowak MA, Shaw GM, Saag MS. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 2.Jiang S, Zhao Q, Debnath AK. Peptide and non-peptide HIV fusion inhibitors. Curr. Pharm. Des. 2002;8:563–580. doi: 10.2174/1381612024607180. [DOI] [PubMed] [Google Scholar]
- 3.Gordon LM, Mobley PW, Pilpa R, Sherman MA, Waring AJ. Conformational mapping of the N-terminal peptide of HIV-1 gp41 in membrane environments using 13C-enhanced Fourier transform infrared spectroscopy. Biochim. Biophys. Acta. 2002;1559:96–120. doi: 10.1016/s0005-2736(01)00443-6. [DOI] [PubMed] [Google Scholar]
- 4.Mobley PW, Curtain CC, Kirkpatrick A, Rostamkhani M, Waring AJ, Gordon LM. The amino-terminal peptide of HIV-1 glycoprotein 41 lyses human erythrocytes and CD4+ lymphocytes. Biochim. Biophys. Acta. 1992;1139:251–256. doi: 10.1016/0925-4439(92)90098-8. [DOI] [PubMed] [Google Scholar]
- 5.Dimitrov AS, Xiao X, Dimitrov DS, Blumenthal R. Early intermediates in HIV-1 envelope glycoprotein-mediated fusion triggered by CD4 and co-receptor complexes. J. Biol. Chem. 2001;276:30335–30341. doi: 10.1074/jbc.M103788200. [DOI] [PubMed] [Google Scholar]
- 6.Pillot T, Drouet B, Queille S, Labeur C, Vandekerckhove J, Rosseneu M, Pincon-Raymond M, Chambaz J. The nonfibrillar amyloid beta-peptide induces apoptotic neuronal cell death: involvement of its C-terminal fusogenic domain. J. Neurochem. 1999;73:1626–1634. doi: 10.1046/j.1471-4159.1999.0731626.x. [DOI] [PubMed] [Google Scholar]
- 7.Mobley PW, Pilpa R, Brown C, Waring AJ, Gordon LM. Membrane-perturbing domains of HIV Type 1 glycoprotein 41. AIDS Res. Hum. Retroviruses. 2001;17:311–327. doi: 10.1089/08892220150503681. [DOI] [PubMed] [Google Scholar]
- 8.Mobley PW, Barry JA, Waring AJ, Sherman MA, Gordon LM. Membrane perturbing actions of HIV Type 1 glycoprotein 41 domains are inhibited by helical C-peptides. AIDS Res. Hum. Retroviruses. 2007;23:224–242. doi: 10.1089/aid.2006.0046. [DOI] [PubMed] [Google Scholar]
- 9.Munch J, Standker L, Andermann K, Schulz A, Schindler M, Chinnadurai R, Pohlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing W, Jiang S, Forssmann W-G, Kirchhoff F. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129:263–275. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 10.Mobley PW, Waring AJ, Sherman MA, Gordon LM. Membrane interactions of the synthetic N-terminal peptide of HIV-1 gp41 and its structural analogs. Biochim. Biophys. Acta. 1999;1418:1–18. doi: 10.1016/s0005-2736(99)00014-0. [DOI] [PubMed] [Google Scholar]
- 11.Mobley PW, Lee H-F, Curtain CC, Kirkpatrick A, Waring AJ, Gordon LM. The amino-terminal peptide of HIV-1 glycoprotein 41 fuses human erythrocytes. Biochim. Biophys. Acta. 1995;1271:304–315. doi: 10.1016/0925-4439(95)00048-9. [DOI] [PubMed] [Google Scholar]
- 12.Chang D-K, Cheng S-F, Chien W-J. The amino-terminal fusion domain peptide of Human Immunodeficiency Virus Type 1 gp41 inserts into the sodium dodecyl sulfate micelle primarily as a helix with a conserved glycine at the micelle-water interface. J. Virol. 1997;71:6593–6602. doi: 10.1128/jvi.71.9.6593-6602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris KF, Gao X, Wong TC. The interactions of HIV gp41 fusion peptides with zwitterionic membrane mimics determined by NMR spectroscopy. Biochim. Biophys. Acta. 2004;1667:67–81. doi: 10.1016/j.bbamem.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Gordon LM, Mobley PW, Lee W, Eskandari S, Kaznessis YN, Sherman MA, Waring AJ. Conformational mapping of the N-terminal peptide of HIV-1 gp41 in lipid detergent and aqueous environments using 13C-enhanced Fourier transform infrared spectroscopy. Protein Sci. 2004;13:1012–1030. doi: 10.1110/ps.03407704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and dynamics of micelle-associated Human Immunodeficiency Virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Tamm LK. Structure and plasticity of the Human Immunodeficiency Virus gp41 fusion domain in lipid micelles and bilayers. Biophys. J. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Gabrys CM, Weliky DP. Solid-state nuclear magnetic resonance evidence for an extended β strand conformation of the membrane-bound HIV-1 fusion peptide. Biochemistry. 2001;40:8126–8137. doi: 10.1021/bi0100283. [DOI] [PubMed] [Google Scholar]
- 18.Kanyalkar M, Srivastava S, Saran A, Coutinho E. Conformational study of fragments of envelope proteins (gp120: 254–274 and gp41: 519–541) of HIV-1 by NMR and MD simulations. J. Pept. Sci. 2004;10:363–380. doi: 10.1002/psc.530. [DOI] [PubMed] [Google Scholar]
- 19.Sackett K, Shai Y. The HIV fusion peptide adopts intermolecular parallel beta-sheet structure in membranes when stabilized by the adjacent N-terminal heptad repeat: A 13C FTIR study. J. Mol. Biol. 2005;350:790–805. doi: 10.1016/j.jmb.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 20.Qiang W, Yang J, Weliky DP. Solid-state nuclear magnetic resonance measurements of HIV fusion peptide to lipid distances reveal the intimate contact of beta strand peptide with membranes and the proximity of the Ala-14-Gly-16 region with lipid headgroups. Biochemistry. 2007;46:4997–5008. doi: 10.1021/bi6024808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callebaut I, Tasso A, Brasseur R, Burny A, Portetelle D, Mornon JP. Common prevalence of alanine and glycine in mobile reactive centre loops of serpins and viral fusion peptides: Do prions possess a fusion peptide? J. Comput. Aided Mol. Des. 1994;8:175–191. doi: 10.1007/BF00119866. [DOI] [PubMed] [Google Scholar]
- 22.Tamm LK, Han X. Viral fusion peptides: A tool set to disrupt and connect biological membranes. Biosci. Rep. 2000;20:501–518. doi: 10.1023/a:1010406920417. [DOI] [PubMed] [Google Scholar]
- 23.Saez-Cirion A, Nieva JL, Gallaher WR. The hydrophobic internal region of bovine prion protein shares structural and functional properties with HIV type 1 fusion peptide. AIDS Res. Hum. Retroviruses. 2003;19:969–978. doi: 10.1089/088922203322588323. [DOI] [PubMed] [Google Scholar]
- 24.Slepushkin VA, Andreev SM, Sidorova MV, Melikyan GB, Grigoriev VB, Chumakov VM, Grinfeldt AE, Manukyan RA, Karamov EV. Investigation of human immunodeficiency virus fusion peptides. Analysis of interrelations between their structure and function. AIDS Res. Hum. Retroviruses. 1992;8:9–18. doi: 10.1089/aid.1992.8.9. [DOI] [PubMed] [Google Scholar]
- 25.Pereira FB, Goni FM, Nieva JL. Permeabilization and fusion of uncharged lipid vesicles induced by the HIV-1 fusion peptide adopting an extended conformation: Dose and sequence effects. Biophys. J. 1997;73:1977–1986. doi: 10.1016/S0006-3495(97)78228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillot T, Lins L, Goethals M, Vanloo B, Baert J, Vandekerckhove J, Rosseneu M, Brasseur R. The 118–135 peptide of the human prion protein forms amyloid fibrils and induces liposome fusion. J. Mol. Biol. 1997;274:381–393. doi: 10.1006/jmbi.1997.1382. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Weliky DP. Solid-state Nuclear Magnetic Resonance evidence for parallel and antiparallel strand arrangements in the membrane-associated HIV-1 fusion peptide. Biochemistry. 2003;42:11879–11890. doi: 10.1021/bi0348157. [DOI] [PubMed] [Google Scholar]
- 28.Serpell LC. Alzheimer's amyloid fibrils: structure and assembly. Biochim. Biophys. Acta. 2000;1502:16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 29.Sticht H, Bayer P, Willbold D, Dames S, Hilbich C, Beyreuther K, Frank RW, Rosch P. Structure of amyloid A4-(1–40)-peptide of Alzheimer's disease. Eur. J. Biochem. 1995;233:293–298. doi: 10.1111/j.1432-1033.1995.293_1.x. [DOI] [PubMed] [Google Scholar]
- 30.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Solution structure of amyloid beta-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 31.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D'Ursi AM, Temussi PA, Picone D. Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment. Similiarity with a viral fusion domain. Eur. J. Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 32.de Planque MR, Raussens V, Contera SA, Rijkers DT, Liskamp RM, Ruysschaert J-M, Ryan JF, Separovic F, Watts A. Beta-sheet structured beta-amyloid(1–40) perturbs phosphatidylcholine model membranes. J. Mol. Biol. 2007;368:982–997. doi: 10.1016/j.jmb.2007.02.063. [DOI] [PubMed] [Google Scholar]
- 33.Barrow CJ, Zagorski MG. Solution structures of beta peptide and its constituent fragments: Relation to amyloid deposition. Science. 1991;253:179–182. doi: 10.1126/science.1853202. [DOI] [PubMed] [Google Scholar]
- 34.Fraser PE, Nguyen JT, Inouye H, Surewicz WK, Selkoe DJ, Podlisny MB, Kirschner DA. Fibril formation by primate, rodent and Dutch-hemorrhagic analogues of Alzheimer amyloid beta-protein. Biochemistry. 1992;31:10716–10723. doi: 10.1021/bi00159a011. [DOI] [PubMed] [Google Scholar]
- 35.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer's beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trovato A, Seno F, Tosatto SCE. The PASTA server for protein aggregation prediction. Protein Eng. Des. Sel. 2007;20:521–523. doi: 10.1093/protein/gzm042. [DOI] [PubMed] [Google Scholar]
- 37.Trovato A, Chiti F, Maritan A, Seno F. Insight into the structure of amyloid fibrils from the analysis of globular proteins. PloS Comput. Biol. 2006;2:e170. doi: 10.1371/journal.pcbi.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenzo A, Yankner BA. Beta-amyloid neurotoxicity requires fibril formation and is inhibited by Congo red. Proc. Natl. Acad. Sci. USA. 1994;91:12243–12247. doi: 10.1073/pnas.91.25.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayakumar R, Kusiak JW, Chrest FJ, Demehin AA, Murali J, Wersto RP, Nagababu E, Ravi L, Rifkind JM. Red cell perturbations by amyloid beta-protein. Biochim. Biophys. Acta. 2003;1622:20–28. doi: 10.1016/s0304-4165(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 40.Mattson MP, Begley JG, Mark RJ, Furukawa K. Abeta25–35 induces rapid lysis of red blood cells: contrast with Aβ1-42 and examination of underlying mechanisms. Brain Res. 1997;771:147–153. doi: 10.1016/s0006-8993(97)00824-x. [DOI] [PubMed] [Google Scholar]
- 41.Thompson MJ, Sievers SA, Karanicolas J, Ivanova MI, Baker D, Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl. Acad. Sci. USA. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halverson K, Fraser PE, Kirschner DA, Lansbury PT., Jr Molecular determinants of amyloid deposition in Alzheimer's disease: Conformational studies of synthetic β-protein fragments. Biochemistry. 1990;29:2639–2644. doi: 10.1021/bi00463a003. [DOI] [PubMed] [Google Scholar]
- 43.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer's disease. Biochemistry. 1993;32:4693–4967. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 44.Halverson KJ, Sucholeiki I, Ashburn TT, Lansbury PT., Jr Location of beta-sheet-forming sequences in amyloid proteins by FTIR. J. Am. Chem. Soc. 1991;113:6701–6703. [Google Scholar]
- 45.Hughes E, Burke RM, Doig AJ. Inhibition of toxicity in the beta-amyloid peptide fragment beta-(25–35) using N-methylated derivatives: a general strategy to prevent amyloid formation. J. Biol. Chem. 2000;275:25109–25115. doi: 10.1074/jbc.M003554200. [DOI] [PubMed] [Google Scholar]
- 46.Del Mar Martinez-Senac M, Villalain J, Gomez-Fernandez JC. Structure of the Alzheimer beta-amyloid peptide (25–35) and its interaction with negatively charged phospholipid vesicles. Eur. J. Biochem. 1999;265:744–753. doi: 10.1046/j.1432-1327.1999.00775.x. [DOI] [PubMed] [Google Scholar]
- 47.Pillot T, Goethals M, Vanloo B, Talussot C, Brasseur R, Vandekerckhove J, Rosseneu M, Lins L. Fusogenic properties of the C-terminal domain of the Alzheimer β-amyloid peptide. J. Biol. Chem. 1996;271:28757–28765. doi: 10.1074/jbc.271.46.28757. [DOI] [PubMed] [Google Scholar]
- 48.Yankner BA, Duffy LK, Kirschner DA. Neurotrophic and neurotoxic effects of Amyloid Beta protein: reversal by tachykinin neuropeptides. Science. 1990;250:279–282. doi: 10.1126/science.2218531. [DOI] [PubMed] [Google Scholar]
- 49.Demeester N, Baier G, Enzinger C, Goethals M, Vandekerckhove J, Rosseneu M, Labeur C. Apoptosis induced in neuronal cells by C-terminal amyloid beta-fragments is correlated with their aggregation properties in phospholipid membranes. Mol. Membr. Biol. 2000;17:219–228. doi: 10.1080/09687680010015838. [DOI] [PubMed] [Google Scholar]
- 50.Galeazzi L, Galeazzi R, Valli MB, Corder EH, Giunta S. Albumin protects human red blood cells against Abeta25–32-induced lysis more effectively than ApoE. NeuroReport. 2002;13:2149–2154. doi: 10.1097/00001756-200211150-00032. [DOI] [PubMed] [Google Scholar]
- 51.Westermark P, Benson MD, Buxbaum JN, Cohen AS, Frangione B, Ikeda S, Masters CL, Merlini G, Saraiva MJ, Sipe JD. Amyloid: Toward terminology clarification. Report from the Nomenclature Committee of the International Society of Amyloidosis. Amyloid. 2005;12:1–4. doi: 10.1080/13506120500032196. [DOI] [PubMed] [Google Scholar]
- 52.Westermark P. Aspects of human amyloid forms and their fibril polypeptides. FEBS J. 2005;272:5942–5949. doi: 10.1111/j.1742-4658.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- 53.Buxbaum JN. Diseases of protein conformation: what do in vitro experiments tells us about in vivo diseases? TIBS. 2003;28:585–592. doi: 10.1016/j.tibs.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 54.Gordon LM, Waring AJ, Curtain CC, Kirkpatrick A, Leung C, Faull K, Mobley PW. Antivirals that target the amino-terminal domain of HIV-1 gp41. AIDS Res. Hum. Retroviruses. 1995;11:677–686. doi: 10.1089/aid.1995.11.677. [DOI] [PubMed] [Google Scholar]
- 55.Gordon LM, Curtain CC, Zhong YC, Kirkpatrick A, Mobley PW, Waring AJ. The amino terminal peptide of HIV-1 glycoprotein 41 interacts with human erythrocyte membranes: peptide conformation, orientation and aggregation. Biochim. Biophys. Acta. 1992;1139:257–274. doi: 10.1016/0925-4439(92)90099-9. [DOI] [PubMed] [Google Scholar]
- 56.Gordon LM, Curtain CC, McCloyn V, Kirkpatrick A, Mobley PW, Waring AJ. The amino-terminal peptide of HIV-1 gp41 interacts with human serum albumin. AIDS Res. Hum. Retroviruses. 1993;9:1145–1156. doi: 10.1089/aid.1993.9.1145. [DOI] [PubMed] [Google Scholar]
- 57.Walther FJ, Waring AJ, Hernandez-Juviel J, Gordon LM, Schwan AL, Jung C-L, Chang Y, Wang Z, Notter RH. Dynamic surface activity of a fully synthetic phospholipase-resistant lipid/peptide lung surfactant. PLoS ONE. 2007;2:e1039. doi: 10.1371/journal.pone.0001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawatake S, Nishimura Y, Sakaguchi S, Iwaki T, Doh-ura K. Surface plasmon resonance analysis for the screening of anti-prion compounds. Biol. Pharm. Bull. 2006;29:927–932. doi: 10.1248/bpb.29.927. [DOI] [PubMed] [Google Scholar]
- 59.Nichols MR, Moss MA, Reed DK, Cratic-McDaniel S, Hoh JH, Rosenberry TL. Amyloid-beta protofibrils differ from amyloid-beta aggregates induced in dilute hexafluoroisopropanol in stability and morphology. J. Biol. Chem. 2005;280:2471–2480. doi: 10.1074/jbc.M410553200. [DOI] [PubMed] [Google Scholar]
- 60.Trovato A, Maritan A, Seno F. Aggregation of natively folded proteins: a theoretical approach. J. Phys. Condens. Matter. 2007;19:285221. [Google Scholar]
- 61.Waring AJ, Harwig SSL, Lehrer RI. Structure and activity of protegrin-1 in model lipid membranes. Protein Pept. Lett. 1996;3:177–184. [Google Scholar]
- 62.Martin I, Defrise-Quertain F, Decroly E, Vandenbranden M, Brasseur R, Ruysschaert J-M. Orientation and structure of the NH2-terminal HIV-1 gp41 peptide in fused and aggregated liposomes. Biochim. Biophys. Acta. 1993;1145:124–133. doi: 10.1016/0005-2736(93)90389-h. [DOI] [PubMed] [Google Scholar]
- 63.Byler DM, Susi H. Examination of the secondary structure of protein by deconvolved FTIR spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 64.Agirre A, Flach C, Goni FM, Mendelsohn R, Valpuesta JM, Wu F, Nieva JL. Interactions of the HIV-1 fusion peptide with large unilamellar vesicles and monolayers. A cryo-TEM and spectroscopic study. Biochim. Biophys. Acta. 2000;1467:153–164. doi: 10.1016/s0005-2736(00)00214-5. [DOI] [PubMed] [Google Scholar]
- 65.Surewicz WK, Mantsch HH. New insight into protein secondary structure from resolution-enhanced infrared spectra. Biochim. Biophys. Acta. 1988;952:115–130. doi: 10.1016/0167-4838(88)90107-0. [DOI] [PubMed] [Google Scholar]
- 66.Hilbich C, Kisters-Woike B, Reed J, Masters CL, Beyreuther K. Aggregation and secondary structure of synthetic Amyloid BetaA4 peptides of Alzheimer's disease. J. Mol. Biol. 1991;218:149–163. doi: 10.1016/0022-2836(91)90881-6. [DOI] [PubMed] [Google Scholar]
- 67.Kohno T, Kobayashi K, Maeda T, Sato K, Takashima A. Three-dimensional structures of the amyloid βpeptide (25–35) in membrane-mimicking environment. Biochemistry. 1996;35:16094–16104. doi: 10.1021/bi961598j. [DOI] [PubMed] [Google Scholar]
- 68.D'Ursi AM, Armenante MR, Guerrini R, Salvadori S, Sorrentino G, Picone D. Solution structure of amyloid beta-peptide (25–35) in different media. J. Med. Chem. 2004;47:4231–4238. doi: 10.1021/jm040773o. [DOI] [PubMed] [Google Scholar]
- 69.Zandomeneghi G, Krebs MRH, McCammon MG, Fandrich M. FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nettleton EJ, Tito P, Sunde M, Bouchard M, Dobson CM, Robinson CV. Characterization of the oligomeric states of insulin in self-assembly and amyloid fibril formation by mass spectrometry. Biophys. J. 2000;79:1053–1065. doi: 10.1016/S0006-3495(00)76359-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouchard M, Zurdo J, Nettleton EJ, Dobson CM, Robinson CV. Formation of insulin amyloid fibrils followed by FTIR simultaneous with CD and electron microscopy. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB. Amyloid beta-protein fibrillogenesis. Detection of a protofibrillar intermediate. J. Biol. Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 73.Walsh DM, Hartley DM, Kusumoto Y, Fezoui Y, Condron MM, Lomakin A, Benedek GB, Selkoe DJ, Teplow DB. Amyloid beta-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J. Biol. Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 74.Wojtowicz WM, Farzan M, Joyal JL, Carter K, Babcock GJ, Israel DI, Sodroski J, Mirzabekov T. Stimulation of enveloped virus infection by beta-amyloid fibrils. J. Biol. Chem. 2002;277:35019–35024. doi: 10.1074/jbc.M203518200. [DOI] [PubMed] [Google Scholar]
- 75.Mingeot-Leclercq MP, Lins L, Bensliman M, Van Bambeke F, Van Der Smissen P, Peuvot J, Schanck A, Brasseur R. Membrane destabilization induced by Beta-amyloid peptide 29–42: Importance of the amino terminus. Chem. Phys. Lipids. 2002;120:57–74. doi: 10.1016/s0009-3084(02)00108-1. [DOI] [PubMed] [Google Scholar]
- 76.Westermark GT, Johnson KH, Westermark P. Staining methods for identification of amyloid in tissue. Meth. Enzymol. 1999;309:3–25. doi: 10.1016/s0076-6879(99)09003-5. [DOI] [PubMed] [Google Scholar]
- 77.Caughey B, Ernst D, Race RE. Congo red inhibition of scrapie agent replication. J. Virol. 1993;67:6270–6272. doi: 10.1128/jvi.67.10.6270-6272.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Touil F, Pratt S, Mutter R, Chen B. Screening a library of potential prion therapeutics against cellular prion proteins and insights into their mode of biological activities by surface plasmon resonance. J. Pharm. Biomed. Anal. 2006;40:822–832. doi: 10.1016/j.jpba.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 79.Maezawa I, Hong H-S, Liu R, Wu C-Y, Cheng RH, Kung M-P, Kung HF, Lam KS, Oddo S, Laferla FM, Jin L-W. Congo red and thioflavin-T analogs detect Abeta oligomers. J. Neurochem. 2008;104:457–468. doi: 10.1111/j.1471-4159.2007.04972.x. [DOI] [PubMed] [Google Scholar]
- 80.Cairo CW, Strzelec A, Murphy RM, Kiessling LL. Affinity-based inhibition of beta-amyloid toxicity. Biochemistry. 2002;41:8620–8629. doi: 10.1021/bi0156254. [DOI] [PubMed] [Google Scholar]
- 81.Avdulov NA, Chochina SV, Igbavboa U, O'Hare EO, Schroeder F, Cleary JP, Wood WG. Amyloid β-peptides increase annular and bulk fluidity and induce lipid peroxidation in brain synaptic plasma membranes. J. Neurochem. 1997;68:2086–2091. doi: 10.1046/j.1471-4159.1997.68052086.x. [DOI] [PubMed] [Google Scholar]
- 82.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Amyloid pores from pathogenic mutations. Nature. 2002;418:291–291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 83.Aloia RC, Jensen FC, Curtain CC, Mobley PW, Gordon LM. Lipid composition and fluidity of the human immunodeficiency virus type-1. Proc. Natl. Acad. Sci. USA. 1988;85:900–904. doi: 10.1073/pnas.85.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]