Abstract
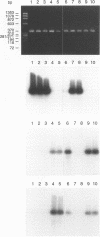
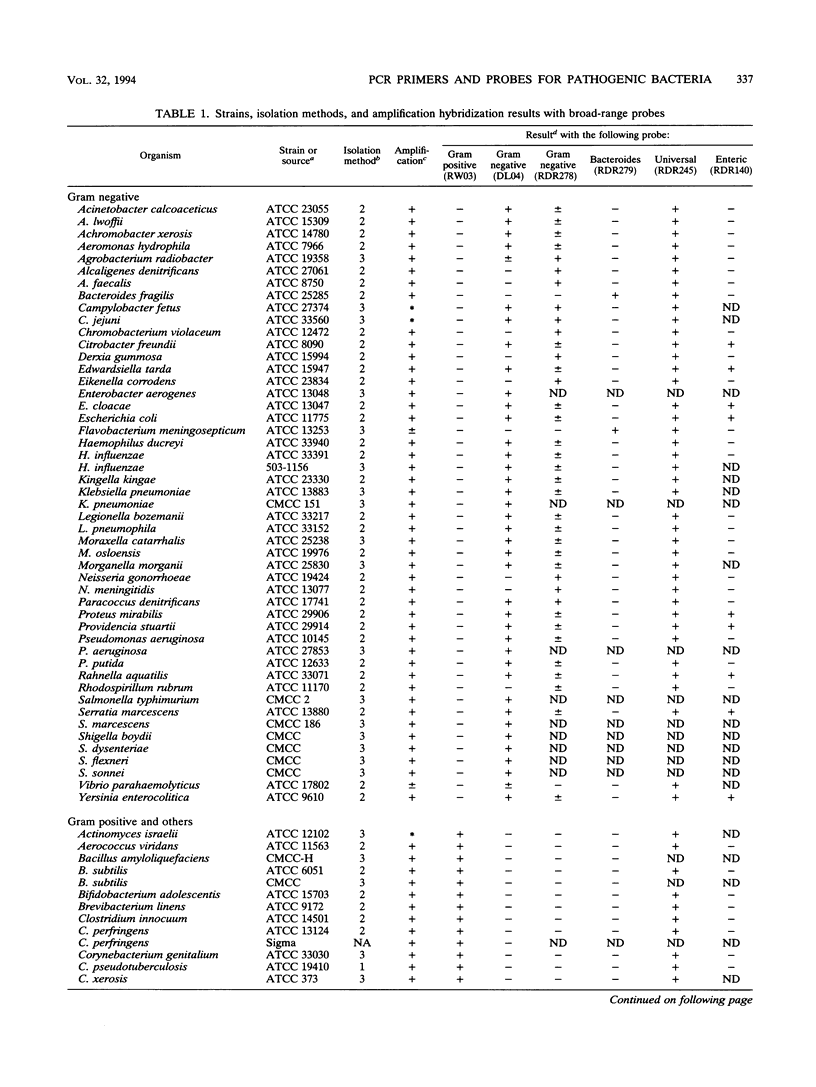
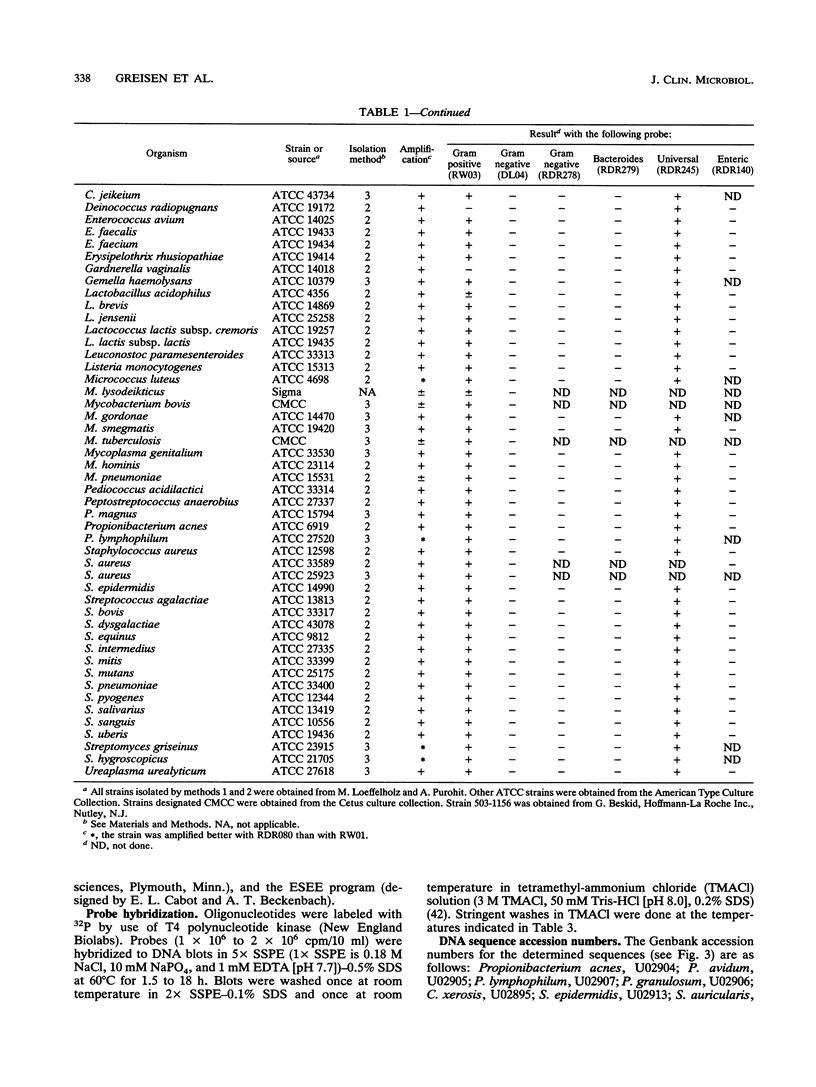
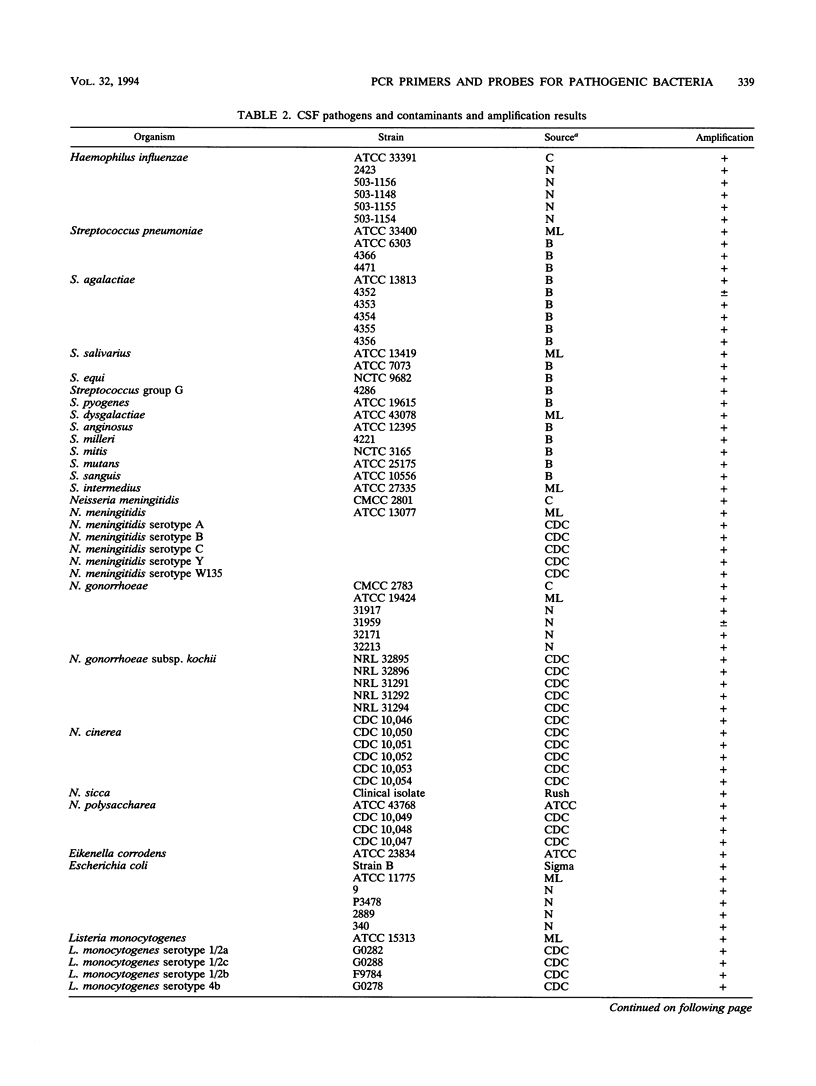
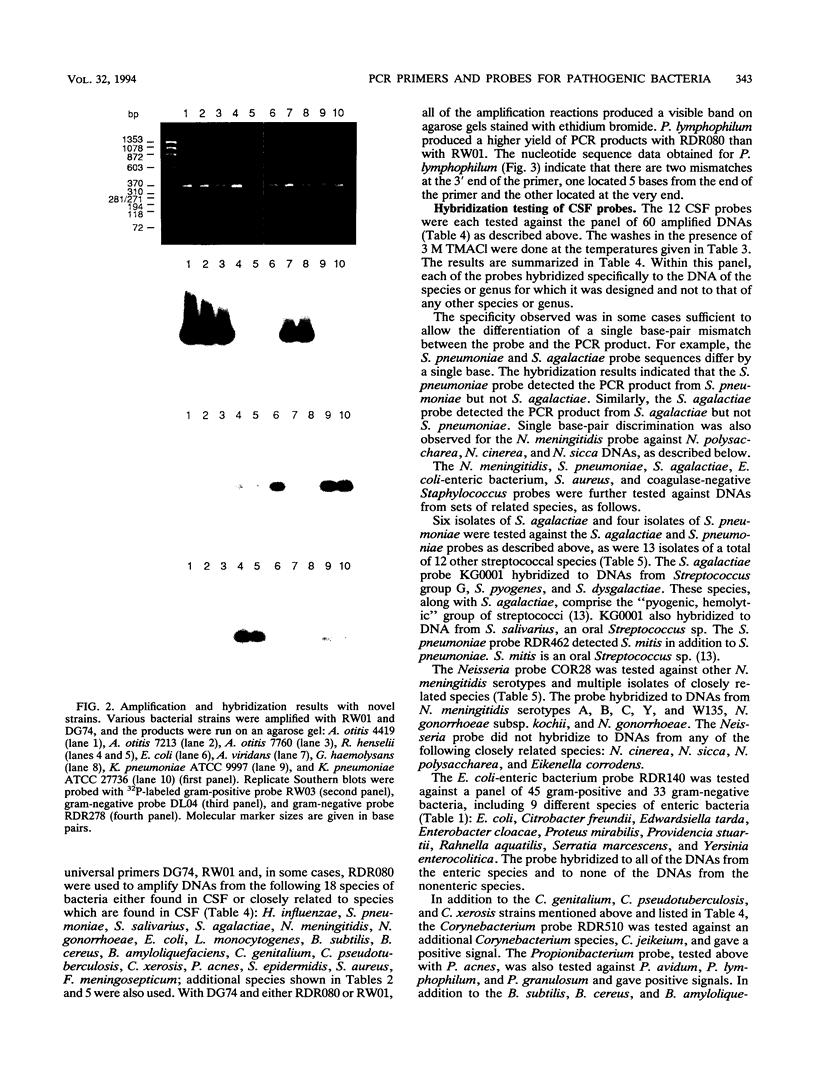
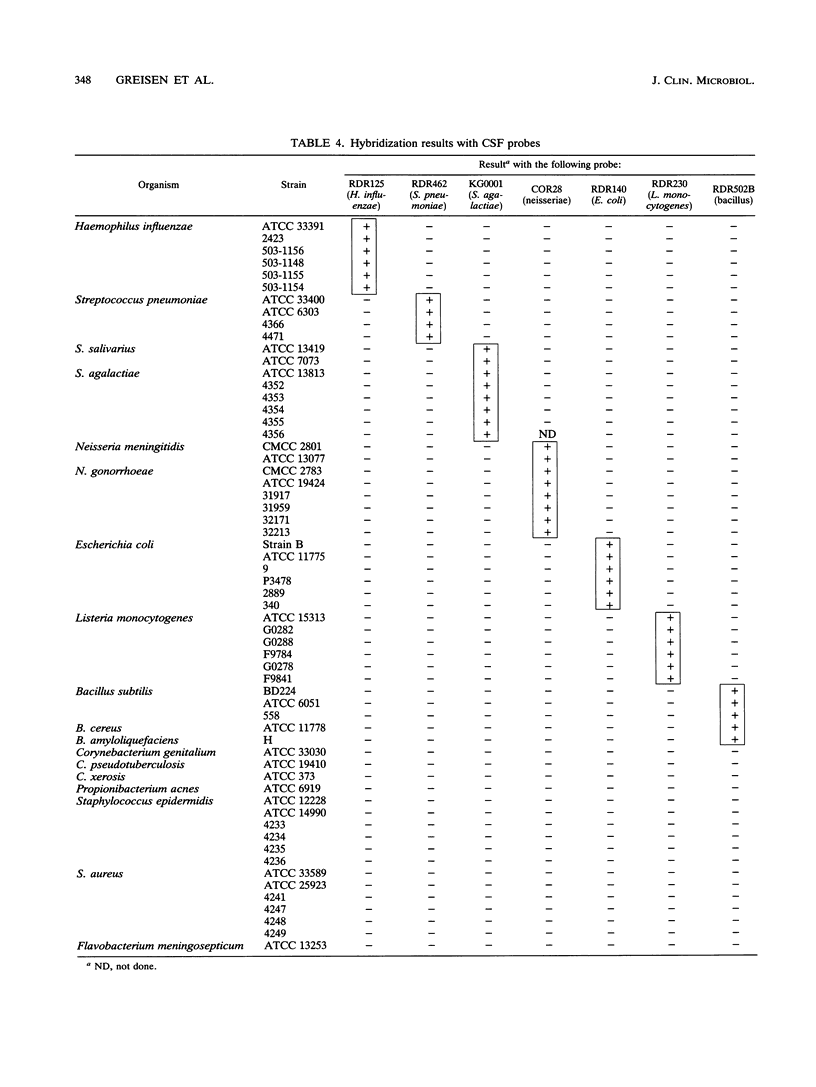
A set of broad-range PCR primers for the 16S rRNA gene in bacteria were tested, along with three series of oligonucleotide probes to detect the PCR product. The first series of probes is broad in range and consists of a universal bacterial probe, a gram-positive probe, a Bacteroides-Flavobacterium probe, and two probes for other gram-negative species. The second series was designed to detect PCR products from seven major bacterial species or groups frequently causing meningitis: Neisseria meningitidis, Haemophilus influenzae, Streptococcus pneumoniae, S. agalactiae, Escherichia coli and other enteric bacteria, Listeria monocytogenes, and Staphylococcus aureus. The third series was designed for the detection of DNA from species or genera commonly considered potential contaminants of clinical samples, including cerebrospinal fluid (CSF): Bacillus, Corynebacterium, Propionibacterium, and coagulase-negative Staphylococcus spp. The primers amplified DNA from all 124 different species of bacteria tested. Southern hybridization testing of the broad-range probes with washes containing 3 M tetramethylammonium chloride indicated that this set of probes correctly identified all but two of the 102 bacterial species tested, the exceptions being Deinococcus radiopugnans and Gardnerella vaginalis. The gram-negative and gram-positive probes hybridized to isolates of two newly characterized bacteria, Alloiococcus otitis and Rochalimaea henselii, as predicted by Gram stain characteristics. The CSF pathogen and contaminant probe sequences were compared with available sequence information and with sequencing data for 32 different species. Testing of the CSF pathogen and contaminant probes against DNA from over 60 different strains indicated that, with the exception of the coagulase-negative Staphylococcus probes, these probes provided the correct identification of bacterial species known to be found in CSF.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguirre M., Collins M. D. Phylogenetic analysis of Alloiococcus otitis gen. nov., sp. nov., an organism from human middle ear fluid. Int J Syst Bacteriol. 1992 Jan;42(1):79–83. doi: 10.1099/00207713-42-1-79. [DOI] [PubMed] [Google Scholar]
- Bentley R. W., Leigh J. A., Collins M. D. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int J Syst Bacteriol. 1991 Oct;41(4):487–494. doi: 10.1099/00207713-41-4-487. [DOI] [PubMed] [Google Scholar]
- Bingen E., Lambert-Zechovsky N., Mariani-Kurkdjian P., Doit C., Aujard Y., Fournerie F., Mathieu H. Bacterial counts in cerebrospinal fluid of children with meningitis. Eur J Clin Microbiol Infect Dis. 1990 Apr;9(4):278–281. doi: 10.1007/BF01968060. [DOI] [PubMed] [Google Scholar]
- Bowman B. H., Palumbi S. R. Rapid production of single-stranded sequencing template from amplified DNA using magnetic beads. Methods Enzymol. 1993;224:399–406. doi: 10.1016/0076-6879(93)24030-x. [DOI] [PubMed] [Google Scholar]
- Böttger E. C. Rapid determination of bacterial ribosomal RNA sequences by direct sequencing of enzymatically amplified DNA. FEMS Microbiol Lett. 1989 Nov;53(1-2):171–176. doi: 10.1016/0378-1097(89)90386-8. [DOI] [PubMed] [Google Scholar]
- Collins M. D., Wallbanks S., Lane D. J., Shah J., Nietupski R., Smida J., Dorsch M., Stackebrandt E. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991 Apr;41(2):240–246. doi: 10.1099/00207713-41-2-240. [DOI] [PubMed] [Google Scholar]
- Dauga C., Grimont F., Grimont P. A. Nucleotide sequences of 16S rRNA from ten Serratia species. Res Microbiol. 1990 Nov-Dec;141(9):1139–1149. doi: 10.1016/0923-2508(90)90087-7. [DOI] [PubMed] [Google Scholar]
- Dewhirst F. E., Paster B. J., Olsen I., Fraser G. J. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J Bacteriol. 1992 Mar;174(6):2002–2013. doi: 10.1128/jb.174.6.2002-2013.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faden H., Dryja D. Recovery of a unique bacterial organism in human middle ear fluid and its possible role in chronic otitis media. J Clin Microbiol. 1989 Nov;27(11):2488–2491. doi: 10.1128/jcm.27.11.2488-2491.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel G., Giron J. A., Valmassoi J., Schoolnik G. K. Multi-gene amplification: simultaneous detection of three virulence genes in diarrhoeal stool. Mol Microbiol. 1989 Dec;3(12):1729–1734. doi: 10.1111/j.1365-2958.1989.tb00158.x. [DOI] [PubMed] [Google Scholar]
- Knapp J. S. Historical perspectives and identification of Neisseria and related species. Clin Microbiol Rev. 1988 Oct;1(4):415–431. doi: 10.1128/cmr.1.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scolea L. J., Jr, Dryja D. Quantitation of bacteria in cerebrospinal fluid and blood of children with meningitis and its diagnostic significance. J Clin Microbiol. 1984 Feb;19(2):187–190. doi: 10.1128/jcm.19.2.187-190.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffelholz M. J., Lewinski C. A., Silver S. R., Purohit A. P., Herman S. A., Buonagurio D. A., Dragon E. A. Detection of Chlamydia trachomatis in endocervical specimens by polymerase chain reaction. J Clin Microbiol. 1992 Nov;30(11):2847–2851. doi: 10.1128/jcm.30.11.2847-2851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucey D., Dolan M. J., Moss C. W., Garcia M., Hollis D. G., Wegner S., Morgan G., Almeida R., Leong D., Greisen K. S. Relapsing illness due to Rochalimaea henselae in immunocompetent hosts: implication for therapy and new epidemiological associations. Clin Infect Dis. 1992 Mar;14(3):683–688. doi: 10.1093/clinids/14.3.683. [DOI] [PubMed] [Google Scholar]
- Mahbubani M. H., Bej A. K., Miller R., Haff L., DiCesare J., Atlas R. M. Detection of Legionella with polymerase chain reaction and gene probe methods. Mol Cell Probes. 1990 Jun;4(3):175–187. doi: 10.1016/0890-8508(90)90051-z. [DOI] [PubMed] [Google Scholar]
- Mazloum H., Totten P. A., Brooks G. F., Dawson C. R., Falkow S., James J. F., Knapp J. S., Koomey J. M., Lammel C. J., Peters D. An unusual Neisseria isolated from conjunctival cultures in rural Egypt. J Infect Dis. 1986 Aug;154(2):212–224. doi: 10.1093/infdis/154.2.212. [DOI] [PubMed] [Google Scholar]
- Mitchell L. G., Merril C. R. Affinity generation of single-stranded DNA for dideoxy sequencing following the polymerase chain reaction. Anal Biochem. 1989 May 1;178(2):239–242. doi: 10.1016/0003-2697(89)90631-3. [DOI] [PubMed] [Google Scholar]
- Olive D. M. Detection of enterotoxigenic Escherichia coli after polymerase chain reaction amplification with a thermostable DNA polymerase. J Clin Microbiol. 1989 Feb;27(2):261–265. doi: 10.1128/jcm.27.2.261-265.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R., Kvach J. T., Mounts P. Isolation and restriction endonuclease analysis of mycobacterial DNA. J Gen Microbiol. 1986 Feb;132(2):541–551. doi: 10.1099/00221287-132-2-541. [DOI] [PubMed] [Google Scholar]
- Pitcher D. G. Rapid identification of cell wall components as a guide to the classification of aerobic coryneform bacteria from human skin. J Med Microbiol. 1977 Nov;10(4):439–445. doi: 10.1099/00222615-10-4-439. [DOI] [PubMed] [Google Scholar]
- Rosa P. A., Schwan T. G. A specific and sensitive assay for the Lyme disease spirochete Borrelia burgdorferi using the polymerase chain reaction. J Infect Dis. 1989 Dec;160(6):1018–1029. doi: 10.1093/infdis/160.6.1018. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Walsh P. S., Levenson C. H., Erlich H. A. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Madej R., Persing D. H. The polymerase chain reaction: clinical applications. Adv Clin Chem. 1992;29:161–196. doi: 10.1016/s0065-2423(08)60224-3. [DOI] [PubMed] [Google Scholar]
- Wilson K. H., Blitchington R. B., Greene R. C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990 Sep;28(9):1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]