Abstract
Vaccinia virus encodes a 90-kDa protein, conserved in all poxviruses, with DNA primase and nucleoside triphosphatase activities. DNA primase products, synthesized with a single stranded ϕX174 DNA template, were resolved as dinucleotides and long RNAs on denaturing polyacrylamide and agarose gels. Following phosphatase treatment, the dinucleotides GpC and ApC in a 4:1 ratio were identified by nearest neighbor analysis in which 32P was transferred from [α-32P]CTP to initiating purine nucleotides. Differences in the nucleotide binding sites for initiation and elongation were suggested by the absence of CpC and UpC dinucleotides as well as the inability of deoxynucleotides to mediate primer synthesis despite their incorporation into mixed RNA/DNA primers. Strong primase activity was detected with an oligo(dC) template. However, there was only weak activity with an oligo(dT) template and none with oligo(dA) or oligo(dG). The absence of stringent template specificity is consistent with a role for the enzyme in priming DNA synthesis at the replication fork.
Introduction
Poxvirus DNA replication occurs within specialized regions of the cytoplasm of infected cells by incompletely understood mechanisms. Self-priming, primer-dependent, and recombination models have all been proposed (Moss and De Silva, 2006). One model takes into account the hairpins at the ends of the linear, double-stranded poxvirus genome (Baroudy et al., 1982a) and resembles the rolling hairpin strand displacement mechanism proposed for parvoviruses (Baroudy et al., 1982b; Moyer and Graves, 1981). The evidence for nicking near the ends of the genome (Esteban and Holowczak, 1977b; Pogo et al., 1984; Pogo et al., 1981; Pogo, 1980) and the resolution of concatemeric forms of DNA by a virus-encoded by Holiday junction endonuclease (Garcia et al., 2000; Garcia and Moss, 2001) are compatible with a rolling hairpin mechanism. However, both linear minichromosomes with specific telomere ends (Du and Traktman, 1996) and closed circular DNA molecules without poxvirus-specific sequences replicate efficiently in infected cells (De Silva and Moss, 2005; DeLange and McFadden, 1986; Merchlinsky and Moss, 1988). In addition, there are reports of VACV DNA covalently linked to RNA (Olgiati et al., 1976) and of short nascent DNA segments resembling Okazaki fragments that could be chased into larger molecules in vivo (Esteban and Holowczak, 1977a). The recent finding of a DNA primase encoded by VACV (De Silva et al., 2007) provides a mechanism for the formation of RNA primers that could be used for initiation of DNA replication or lagging strand synthesis (Frick and Richardson, 2001).
The VACV DNA primase is encoded by the D5R gene (VACVWR110), which is conserved in all sequenced poxviruses (De Silva et al., 2007). D5 (the protein encoded by D5R) was shown to be required for DNA replication more than 20 years ago (Evans and Traktman, 1992; Roseman and Hruby, 1987; Seto et al., 1987). Analysis of the C-terminal region of D5 and homologs in other poxviruses as well as some distantly related large DNA viruses, revealed a domain characteristic of helicase superfamily III members of the AAA+ class of nucleoside triphosphatases (NTPases) (Gorbalenya and Koonin, 1989; Iyer et al., 2001). Furthermore, in vitro studies demonstrated DNA-independent NTPase activity (Evans et al., 1995) and mutations in the active site eliminated the ability of D5R to complement a temperature sensitive mutant (Boyle et al., 2007). The possibility of a second enzymatic activity in the N-terminal segment of D5 was suggested by a motif present in the archae-eukaryotic primase superfamily (Iyer et al., 2005). Recently, recombinant D5 was shown to have primase activity in vitro and mutation of the primase active site, which had no effect on NTPase activity, eliminated the ability of D5R to complement a conditional lethal mutant in vivo (De Silva et al., 2007). In the present study, we characterized the products of the D5 primase and provided information on the substrate and template specificities.
Results
In vitro products of the VACV DNA primase
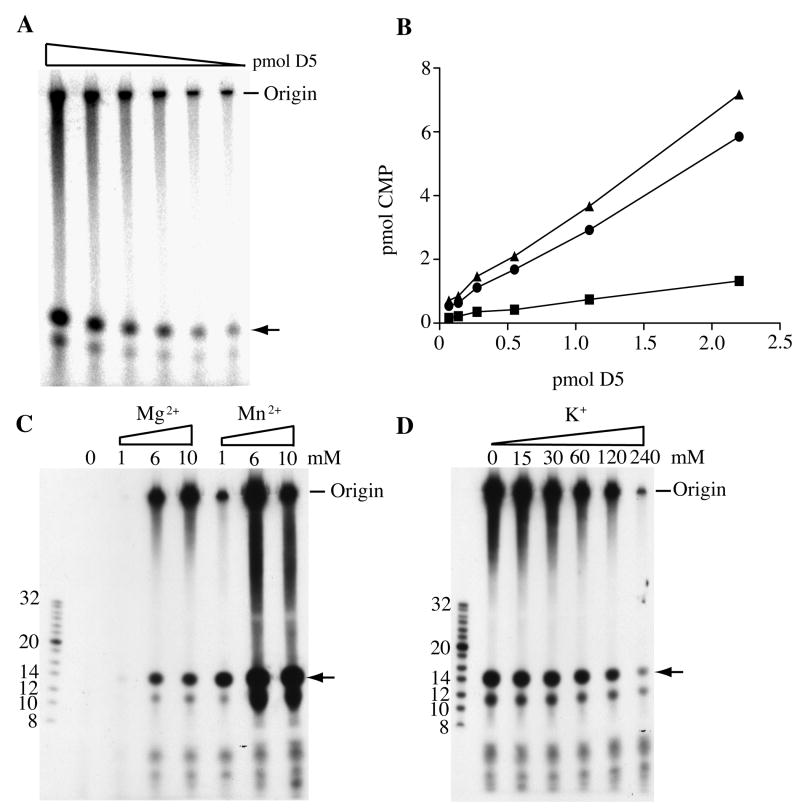
We previously demonstrated the DNA primase activity of purified recombinant D5 in a reaction containing single stranded ϕX174 or M13 phage DNA template, Mg2+ and the four rNTPs including either [α-32P]CTP or [α-32P]UTP (De Silva et al., 2007). Prior to analysis on denaturing 20% polyacrylamide gels, the products were treated with phosphatase, which reduces the background of unincorporated radioactive nucleotides and slows the migration of RNA of less than six nucleotides (Holstege et al., 1997). The RNase-sensitive products labeled with [α-32P]CTP consisted of a discrete band that migrated at the position of a phosphorylated 14-nucleotide marker and more slowly migrating products that extended to the top of the gel (Fig. 1A). Of the total radioactivity, approximately 20% migrated with the 14-nucleotide marker. Synthesis of these products was proportional to enzyme concentration and a specific activity of 3.6 nmol CMP incorporated in 30 min per nmol of enzyme with a ϕX174 DNA template was determined (Fig. 1B).
Fig.1.
Effects of enzyme concentration and divalent and monovalent cations on primase activity. (A) Primase reactions were carried out under standard conditions with a ϕX174 DNA template and 6 mM Mg2+. The amount of recombinant D5 primase varied from 0.07 to 2.2 pmol. Products were digested with calf intestinal alkaline phosphatase, denatured and resolved by electrophoresis on a 20% polyacrylamide/urea gel. A phosphoimager scan is shown. Origin indicates the position of the well. The arrow points to the position of the band migrating at the position of a phosphorylated 14-nucleotide marker. (B) The data in panel A were quantified by making a standard curve with known amounts of [α-32P]CTP. Symbols: ■, “14-nucleotide product”; ●, extended products; ▲, total. (C) Effect of Mg2+ and Mn2+ concentrations. Primase reactions were carried out under standard conditions with 1.1 pmol of primase except that the divalent cation concentrations were varied as shown. Positions of 32P-end labeled oligonucleotide markers are indicated on the left. (D) Effect of K+ concentration. Primase reactions were carried under standard conditions except that the monovalent cation concentraton was varied.
Before proceeding to further analyses of the primase products, we determined the effects of altering the divalent and monovalent cation concentrations, which can affect activity and processivity of polymerases. Our standard primase assay contains 6 mM Mg2+. In the experiment shown in Fig. 1C, Mg2+ and Mn2+ were varied from 0 to 10 mM, while total rNTPs were maintained at 1.5 mM. No primase activity was detected in the absence of added divalent cation. The effects of Mg2+ and Mn2+ were slightly different. Activity was greater with Mn2+ and peaked at 6 mM, whereas activity with Mg2+ increased up to 10 mM. The products appeared heterogenous at all divalent cation concentrations, although the proportion of the material migrating with the 14-nucleotide marker was about 5-fold higher with Mn2+ compared to Mg2+. Our standard primase assay contains 60 mM K+. However, we found that K+ decreased activity at concentrations above 15 mM (Fig. 1D). The greater activity with Mn2+ than Mg2+ and the inhibitory effect of monovalent cations are similar to the properties of eukaryotic DNA primases (Kirk and Kuchta, 1999).
VACV D5 forms dinucleotide and longer RNA products
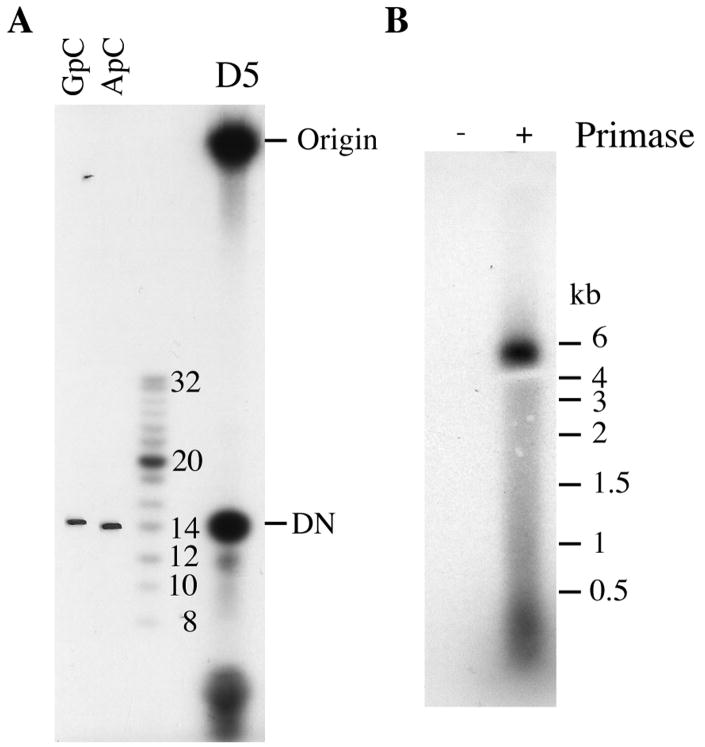
Phosphatase-treatment reduces the mobility of products less than 5 nucleotides long on high concentration polyacrylamide gels (Holstege et al., 1997). For these products, the mobility initially increases with size and is affected by base composition. Thus, the phosphatase-treated dinucleotide products of an archaeon primase migrated with a 14-nucleotide marker (Lao-Sirieix and Bell, 2004). Similarly, we found that corresponding mobility VACV DNA primase product(s) labeled with [α-32P]CTP and treated with phosphatase migrated as a band that overlapped with GpC and ApC on a denaturing 20% polyacrylamide gel (Fig. 2A). Although the product migrating as a dinucleotide was sensitive to RNase, the more rapidly migrating materials were insensitive (De Silva et al., 2007) and were not analyzed further.
Fig. 2.
Analysis of primase products. (A) The discrete primase product comigrates with dinucleotides GpC and ApC as well as with the phosphorylated 14-nucleotide marker. Primase assays were performed and the products analyzed alongside 32P-end labeled oligonucleotide markers. In addition, 2 μg of synthetic dinucleotides GpC and ApC were co-electrophoresed and detected by UV shadowing. Positions of GpC and ApC are marked on the left of the autoradiogram. The radioactive dinucleotide product is labeled DN. (B) Agarose gel electrophoresis. The products of reactions with primase (+) and without primase (-) were resolved on a formaldehyde/1% agarose gel. The positions of RNA size markers are on the right.
The labeled material near the top of the gel was previously shown to be sensitive to RNase and nuclease P1 but insensitive to DNase (De Silva et al., 2007). To investigate the possibility that the retarded electrophoresis was partly due to protein association that survived the formamide treatment, we treated the samples with proteinase K or proteinase K followed by phenol chloroform extraction prior to electrophoresis. However, these treatments had no effect suggesting that association with protein was not affecting the electrophoretic mobility of the RNA (data not shown). To determine the size range of the large RNA products, samples were resolved on a formaldehyde/1% agarose gel. Products that migrated with markers of up to nearly 6,000 nucleotides were found (Fig. 2B).
Sequence of the dinucleotide product
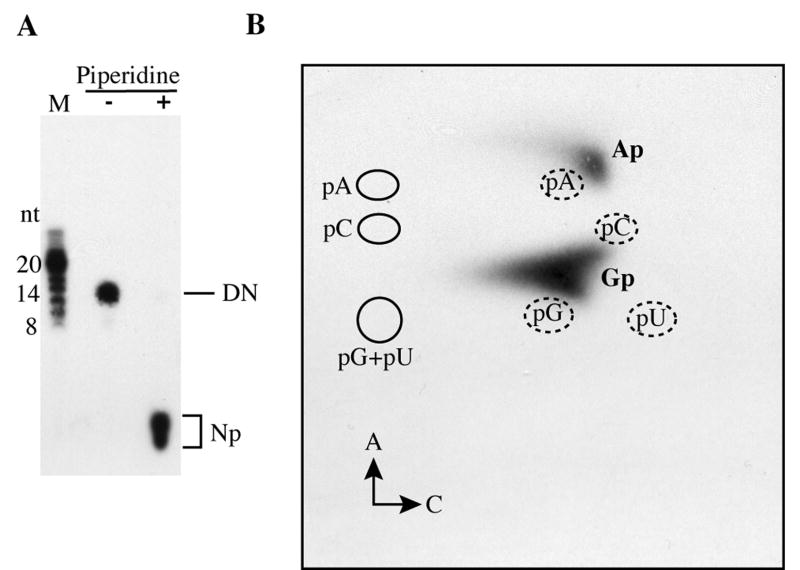
The primase products with the mobility of dinucleotides were examined by nearest neighbor analysis as follows. The primase reaction was carried out in the presence of [α-32P]CTP and unlabeled ATP, GTP and UTP, generating pppXp*C dinucleotide and longer products in which X could be any of the four bases and the starred phosphate derived from CTP was labeled. Following phosphatase treatment, the Xp*C dinucleoside monophosphate products were purified by electrophoresis and elution from a 20% polyacrylamide gel and hydrolyzed with piperidine (Fig. 3A). The latter step results in the formation of nucleoside-2’, 3’-phosphates with the resulting transfer of the 32P label to Xp*. The Xp* nucleotides were resolved by two-dimensional thin layer chromatography and located by autoradiography (Fig. 3B). The labeled nucleotides were identified as Gp and Ap by their position on the plates relative to unlabeled ribonucleoside-5’-phosphate markers detected by UV shadowing (Grosjean et al., 2007). The absence of Cp and Up indicated that the initiation of NpC dinucleotides had occurred exclusively with purine nucleoside triphosphates and also provided evidence that the product was indeed a dinucleotide. The ratio of Gp to Ap was approximately 4:1, indicating a preference for initiation with GTP when the second nucleotide was cytidine. Similar experiments have not been carried out to determine whether this preference holds when other second nucleotides are labeled.
Fig. 3.
Nearest neighbor analysis. Primase reactions were carried out in the presence of [α-32P]CTP and 1 mM Mn2+. (A) Samples were resolved on a denaturing 20% polyacrylamide gel (not shown) and the dinucleotide band was excised, eluted into nuclease free water and digested with piperidine. Chemically treated or untreated products were resolved on a denaturing 20% polyacrylamide gel. Np and DN represent the positions of mononucleotides and dinucleotides, respectively. (B) Chemically treated samples and 20 μg of a mixture of pA, pC, pG and pU were spotted at the bottom left edge of cellulose plates (intersection of arrows A and C). After two-dimensional chromatography (solvent A, 1st dimension; solvent C, 2nd dimension), plates were exposed to a 254 nm UV light source to detect nucleotide markers and to X-ray film to detect radioactive products. The migrations of the 5’ nucleotide markers are indicated by full circles in the first dimension and dashed circles in the second. Positions of Ap and Gp radioactive products relative to markers are indicated. Note that 2’, 3’ nucleotides move slightly faster than 5’ nucleotides in the first and second dimensions (Grosjean et al., 2007).
VACV primase catalyzed the incorporation deoxyribonucleotides into primers
We tested whether the VACV primase can use deoxyribonucleotides as a substrate. Reactions contained either [α-32P]CTP or [α-32P]dCTP in the presence of rNTPs or dNTPs or both. Incorporation of [α-32P]CTP into dinucleotide or longer products was insignificant when rNTPs were replaced with dNTPs, suggesting that neither dATP or dGTP can initiate oligonucleotide synthesis (Fig. 4). In addition dNTPs reduced the incorporation of [α-32P]CTP in the presence rNTPs. Nevertheless, [α-32P]dCTP was incorporated into a discrete band that co-migrated with a 24-nucleotide marker and longer products in the presence of rNTPs but not dNTPs (Fig. 4). Based on the pattern, we presume that the “24-nucleotide” product is GpdC and ApdC, which have slower migrations than GpC and ApC on 20% polyacrylamide gels (Lao-Sirieix and Bell, 2004). Both the presumed dinucleotide and longer RNA products were sensitive to nuclease P1, whereas the more rapidly migrating material was not (data not shown). Also, dNTPs reduced the incorporation of [α-32P]dCTP in the presence of rNTPs (Fig. 4). The dNTPs appeared to have a greater effect on reducing the amount of long polynucleotides than of dinucleotides.
Fig. 4.
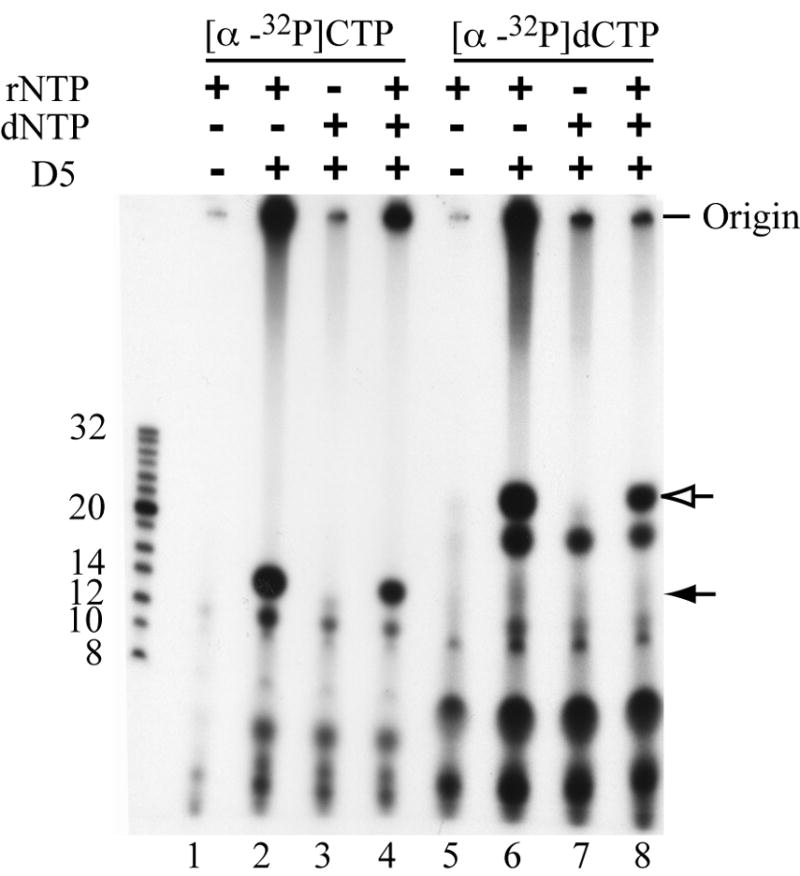
Incorporation of ribo- and deoxyribonucleotides. Primase assays were performed using [α-32P]CTP (lanes 1-4) or [α-32P]dCTP (lanes 5-8) supplemented with combinations of 1.5 mM rNTPs and/or 1.5 mM dNTPs. Positions of 32P-end labeled oligonucleotide markers are indicated on the left. Origin indicates the top of the well; filled arrow represents dinucleotide product formed with [α-32P]CTP; open arrow represents the presumed dinucleotide product formed with [α-32P]dCTP.
Homooligomer template specificity of VACV primase
Defined length homooligomers were tested for their ability to serve as templates. Primase assays were performed using synthetic 18-mers of dA, dC, dG, and dT with complementary rNTPs. Each reaction was divided into two aliquots and one was treated with nuclease P1 before gel electrophoresis. Oligo(dC) was by far the best template resulting in a range of nuclease P1-sensitive products that migrated between the phosphorylated markers of 16 and 87 nucleotides (Fig. 5). Products longer than the template could arise from primase slippage, primer reannealing, or terminal nucleotidyl transferase activity. Oligo(dT) was a much poorer template than oligo(dC) and no nuclease P1 sensitive material was obtained with either oligo(dG) or oligo(dA) (Fig. 5). There was, however, some undefined nuclease-resistant material at the origin in all cases.
Fig. 5.
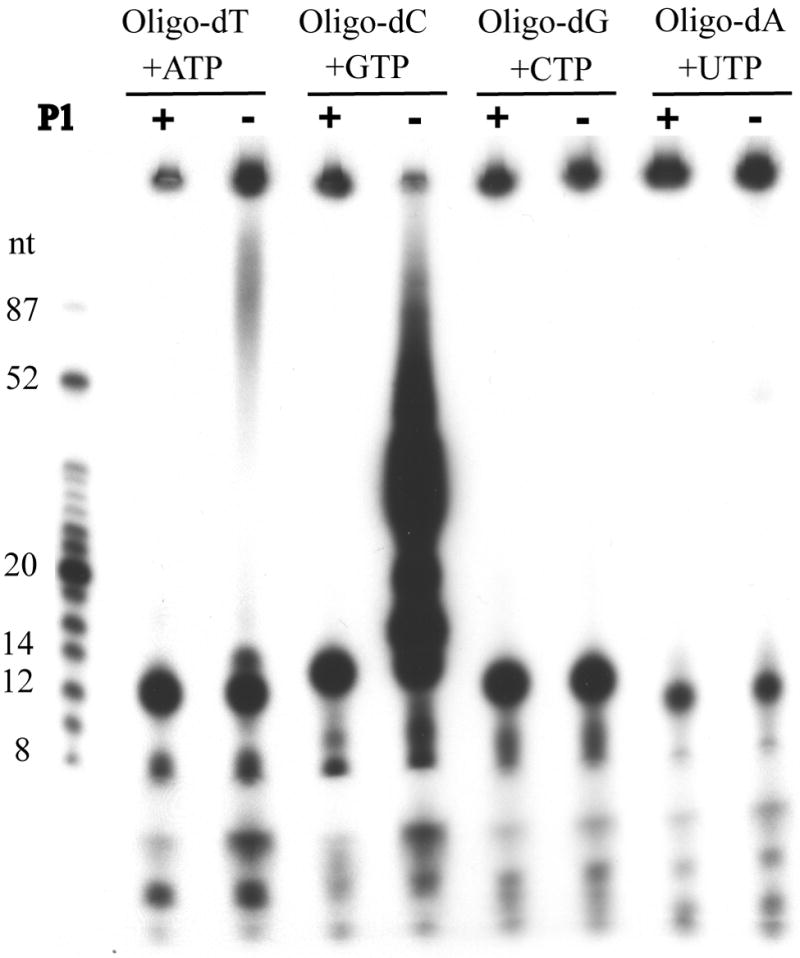
Template recognition of homooligomer templates. Primase reactions were carried out using 100 ng of dT, dC, dG, and dA 18mer homooligomers in the presence of the complementary [α-32P]NTP and 1 mM Mn2+. Products were treated with calf intestinal alkaline phosphatase and then with (+) or without (-) nuclease P1. An autoradiograph of a 20% polyacrylamide gel is shown with the positions of 32P-end labeled oligonucleotide markers on the left.
Discussion
DNA primases catalyze the synthesis of RNA in at least four discrete steps: template binding, NTP binding, phosphodiester bond formation, and chain extension (Frick and Richardson, 2001). Most primases do not exhibit template specificity, although they may have sequence preferences, as they must initiate RNA synthesis frequently during lagging strand synthesis. In this context, the VACV DNA primase was able to use single stranded phage DNAs as well as oligo(dC) and to a lesser extent oligo(dT) as templates. Extensive studies with bacteriophage T7 and other primases indicate the presence of separate NTP binding sites for initiation and elongation. Primer synthesis is initiated by the formation of a dinucleotide with release of pyrophosphate. The product is then transferred to the initiation site allowing the incorporation of a third nucleotide in the empty elongation site. This step is repeated as the primer elongates. Purified VACV primase synthesized dinucleotides as well as longer RNAs in the presence of ϕX174 single stranded DNA template and either Mg2+ or Mn2+. Although there was greater incorporation of labeled CMP into the extended products compared to the dinucleotide, there was several hundred times more dinucleotide on a molar basis. The rate of primer synthesis and ratio of dinucleotide to extended products remained constant with time, suggesting that primer initiation is the rate-limiting step and that elongation is processive. Formation of dinucleotides by primases and other RNA polymerases is common and is thought to represent abortive synthesis (Frick and Richardson, 2001).
We previously determined that ATP or GTP was required for VACV primer synthesis and that omission of GTP reduced activity more than omission of ATP (De Silva et al., 2007). An explanation for this was found upon nearest neighbor sequencing of the dinucleotide products labeled with [α-32P]CTP, which revealed four times more G than A and neither C nor U in the first position. Although we do not know whether a similar ratio of G to A would be found by nearest neighbor analysis with other labeled ribonucleoside triphosphates, preferential initiation with G can also explain the finding that the template activity of poly(dC) was much higher than oligo(dT) and that oligo(dG) and oligo(dA) were inactive. The VACV primase resembles other primases in regard to initiation with a purine (Frick and Richardson, 2001). The VACV primase was unable to initiate oligonucleotide synthesis in the presence of dNTPs, though dNTPs could be incorporated in the presence of rNTPs. Thus, it seems likely that the VACV primase discriminates between rNTPs and dNTPs in the initiation site but to a lesser degree in the elongation site, similar to that of bacterial and eukaryotic DNA primases (Conaway and Lehman, 1982; Rowen and Kornberg, 1978) but different from archael DNA primases, which appear to have additional functions including gap filling and 3’-terminal nucleotidyl transferase activities (Bocquier et al., 2001; Lao-Sirieix and Bell, 2004; Le Breton et al., 2007). Although DNA primases typically form short RNAs (Frick and Richardson, 2001), the VACV DNA primase synthesized RNAs that were several kb long, like archael (Bocquier et al., 2001; Lao-Sirieix and Bell, 2004) and some viral (Mikhailov and Rohrmann, 2002) primases.
In an infected cell, D5 primase and NTPase activities function in conjunction with other viral proteins. It is known that the A20 protein interacts with both D5 and the D4 uracil DNA glycosylase (McCraith et al., 2000) and that A20 and D4 form a processivity factor for the DNA polymerase (Stanitsa et al., 2006). These proteins presumably form a complex that allows DNA synthesis at the replication fork. It will be interesting to attempt to reconstitute this complex in vitro and couple primer synthesis with DNA replication.
Materials and methods
Chemicals
Adenylyl (3ʹ→5ʹ) cytidine [ApC], guanylyl (3ʹ→5ʹ) cytidine [GpC], nuclease P1, isobutyric acid, and concentrated ammonia were purchased from Sigma-Aldrich (St. Louis, MO). Sequa gel solutions for making denaturing polyacrylamide gels were obtained from National Diagnostics Inc. (Atlanta, GA). Ribonucleotide and deoxynucleotide triphosphates were from Roche Diagnostics Corp. (Indianapolis, IN). The 18mer oligo(dA), oligo(dC), oligo(dG), and oligo(dT) were purchased from US Biologicals (Swampscott, MA). Fluor coated plates were from Ambion Inc. (Austin, TX). Isopropanol and hydrochloric acid were from, Mallinckrodt Baker, Inc. (Phillipsburg, NJ). Proteinase K and phenol/chloroform were from Invitrogen (Carlsbad, CA).
Primase assays
Primase reactions were carried out as previously described (De Silva et al., 2007). Unless stated otherwise, reaction mixtures (5 μl) contained 25 mM Tris acetate (pH 7.5), 60 mM K acetate, 6 mM Mg acetate, 10 mM DTT, 100 μg/ml BSA, 1 mM ATP, 250 μM UTP, 250 μM GTP, 25 μM CTP, 0.66 μM [α-32P]CTP (3,000 Ci/mmol) (Perkin–Elmer Life Sciences, Shelton, CT), 1 μg (0.57 pmol) ϕX174 phage DNA (New England Biolabs, Beverly, MA), 100 ng (1.1 pmol) recombinant D5 with a 10-histidine tag (De Silva et al., 2007). Reactions were incubated at 37°C for 30 min, then stopped by heating at 80°C for 10 min. Products were treated with 4 units of calf intestinal alkaline phosphatase (New England Biolabs, Beverly, MA) at 37°C for 1 h and denatured by adding an equal volume of formamide stop solution (95% deionized formamide, 20 mM EDTA, plus bromophenol blue and xylene cyanol). Reaction products were heated at 90°C for 2 min and analyzed on a denaturing 20% polyacrylamide, 7.5 M urea gel followed by autoradiography or analysis on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Oligonucleotide size markers ((GE Healthcare, Piscataway, NJ) or chemically synthesized oligonucleotides were 32P-end-labeled using polynucleotide kinase (New England Biolabs, Beverly, MA) and [γ-32P]ATP [3,000 Ci/mmol] (Perkin–Elmer Life Sciences, Shelton, CT).
Formaldehyde agarose gel electrophoresis of primase products
The primase reaction was performed as described above except the reaction was stopped by the addition of RNA loading buffer (5X buffer: 0.25% (w/v) bromophenol blue, 40 mM EDTA (pH 8.0), 0.88 M formaldehyde, 20% [v/v] glycerol, 31% [v/v] formamide, 40% [v/v] 10X gel buffer (0.2 M MOPS, 50 mM sodium acetate, 10 mM EDTA [pH 7.0]). The samples were resolved on a 1% agarose gel made up in 1X gel buffer with 18% (v/v) 37% formaldehyde. The gel was run at 5 V/cm for 3 h, rinsed in diethylpyrocarbonate-treated water, incubated in ten gel volumes of 50 mM NaOH, 1.5 M NaCl for 30 min and neutralized in 0.5 M Tris–HCl (pH 7.4), 1.5 M NaCl for 20 min before transfer to 10X SSC (1X SSC is 0.15 M NaCl, 0.015 M trisodium citrate pH 7.0). After 45 min incubation, the RNAs were transferred to a nylon membrane by downward capillary action. RNA products were detected by autoradiography. A 0.5-10 kb RNA ladder (Invitrogen, Carlsbad, CA) was used as a marker.
Nearest Neighbor analysis
The analysis was carried out as previously described (Grosjean et al., 2007) with some modifications. Primase assays were carried out with 1 mM Mn2+ instead of Mg2+. Products were treated with Antarctic phosphatase (New England Biolabs, Beverly, MA) instead of calf alkaline phosphatase and heat inactivated for 10 min at 65°C. Products were resolved on a 20% denaturing polyacrylamide gel and visualized by autoradiography. Bands were cut out from gel and incubated with an equal volume of nuclease free water overnight at 4°C on a rotating platform. Eluted samples were then digested with 15% piperidine (Sigma-Aldrich, St. Louis, MO) for 90 min at 99°C. Approximately 20 μg of a mix of commercially available mononucleotides (AMP, GMP, UMP, CMP) (Sigma-Aldrich, St. Louis, MO) was spotted 2 cm from the bottom left edges of plastic-coated cellulose MN 300 plates (Macherey-Nagel, Bethlehem, PA). Piperidine-treated samples (approximately 250 cts/min) were co-spotted and allowed to dry. Samples were resolved in the first dimension using solvent A containing isobutyric acid/concentrate ammonia/water (66/1/33[v/v/v]) for approximately 4 h. After allowing the plate to dry, it was oriented in a perpendicular direction to the first dimension and resolved in solvent C containing isopropanol/concentrated HCl/water (68/18/14[v/v/v]) for 7 h, then allowed to dry. Plates were exposed to UV light with a filter at 254 nm to determine positions of the nucleotide marker. Radiolabeled nucleotides were visualized by exposure of plates to X-ray film. The identities of the radioactive spots were determined by position relative to markers and published chromatographic profiles (Grosjean et al., 2007). For quantitation of radioactive bands, data were acquired on a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and quantified with ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Acknowledgments
The work was supported by the Division of Intramural Research, National Institute of allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baroudy BM, Venkatesan S, Moss B. Incompletely base-paired flipflop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982a;28:315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- Baroudy BM, Venkatesan S, Moss B. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp Quant Biol. 1982b;47:723–729. doi: 10.1101/sqb.1983.047.01.083. [DOI] [PubMed] [Google Scholar]
- Bocquier AA, Liu L, Cann IK, Komori K, Kohda D, Ishino Y. Archaeal primase: bridging the gap between RNA and DNA polymerases. Curr Biol. 2001;11:452–456. doi: 10.1016/s0960-9822(01)00119-1. [DOI] [PubMed] [Google Scholar]
- Boyle KA, Arps L, Traktman P. Biochemical and genetic analysis of the vaccinia virus D5 protein: Multimerization-dependent ATPase activity is required to support viral DNA replication. J Virol. 2007;81:844–859. doi: 10.1128/JVI.02217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Lehman IR. Synthesis by the DNA primase of Drosophila melanogaster of a primer with a unique chain length. Proc Natl Acad Sci USA. 1982;79:4585–4588. doi: 10.1073/pnas.79.15.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Lewis W, Berglund P, Koonin EV, Moss B. Poxvirus DNA primase. Proc Natl Acad Sci USA. 2007;104:18724–18729. doi: 10.1073/pnas.0709276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Moss B. Origin-independent plasmid replication occurs in vaccinia virus cytoplasmic factories and requires all five known poxvirus replication factors. Virol J. 2005;2:23. doi: 10.1186/1743-422X-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange AM, McFadden G. Sequence-nonspecific replication of transfected plasmid DNA in poxvirus-infected cells. Proc Natl Acad Sci USA. 1986;83:614–618. doi: 10.1073/pnas.83.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Traktman P. Vaccinia virus DNA replication: Two hundred base pairs of telomeric sequence confer optimal replication efficiency on minichromsome templates. Proc Natl Acad Sci USA. 1996;93:9693–9698. doi: 10.1073/pnas.93.18.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M, Holowczak JA. Replication of vaccinia DNA in mouse L cells. I. In vivo DNA synthesis. Virology. 1977a;78:57–75. doi: 10.1016/0042-6822(77)90078-2. [DOI] [PubMed] [Google Scholar]
- Esteban M, Holowczak JA. Replication of vaccinia DNA in mouse L cells. III. Intracellular forms of viral DNA. Virology. 1977b;82:308–322. doi: 10.1016/0042-6822(77)90006-x. [DOI] [PubMed] [Google Scholar]
- Evans E, Klemperer N, Ghosh R, Traktman P. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleotide triphosphatase. J Virol. 1995;69:5353–5361. doi: 10.1128/jvi.69.9.5353-5361.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Traktman P. Characterization of vaccinia virus DNA replication mutants with lesions in the D5 gene. Chromosoma. 1992;102:S72–S82. doi: 10.1007/BF02451789. [DOI] [PubMed] [Google Scholar]
- Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- Garcia AD, Aravind L, Koonin EV, Moss B. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc Natl Acad Sci USA. 2000;97:8926–8931. doi: 10.1073/pnas.150238697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Moss B. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J Virol. 2001;75:6460–6471. doi: 10.1128/JVI.75.14.6460-6471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Viral proteins containing the purine NTP-binding sequence pattern. Nucl Acids Res. 1989;17:8413–8440. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Fiedler U, Timmers HT. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk BW, Kuchta RD. Human DNA primase: anion inhibition, manganese stimulation, and their effects on in vitro start-site selection. Biochemistry. 1999;38:10126–10134. doi: 10.1021/bi990351u. [DOI] [PubMed] [Google Scholar]
- Lao-Sirieix SH, Bell SD. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3’-terminal nucleotidyl transferase activities. J Mol Biol. 2004;344:1251–1263. doi: 10.1016/j.jmb.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Le Breton M, Henneke G, Norais C, Flament D, Myllykallio H, Querellou J, Raffin JP. The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and repair? J Mol Biol. 2007;374:1172–1185. doi: 10.1016/j.jmb.2007.10.015. [DOI] [PubMed] [Google Scholar]
- McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchlinsky M, Moss B. Sequence-independent replication and sequence-specific resolution of plasmids containing the vaccinia virus concatemer junction: Requirements for early and late trans-acting factors. In: Kelly T, Stillman B, editors. Cancer Cells 6/Eukaryotic DNA Replication. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. pp. 87–93. [Google Scholar]
- Mikhailov VS, Rohrmann GF. Baculovirus replication factor LEF-1 is a DNA primase. J Virol. 2002;76:2287–2297. doi: 10.1128/jvi.76.5.2287-2297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B, De Silva F. Poxvirus DNA replication and human disease. In: DePamphilis ML, editor. DNA Replication & Human Disease. Cold Spring Harbor Laboratory Press; Cold Spring Hrbor: 2006. pp. 707–727. [Google Scholar]
- Moyer RW, Graves RL. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981;27:391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Olgiati DD, Pogo BG, Dales S. Evidence for RNA linked to nascent DNA in HeLa cells. J Cell Biol. 1976;68:557–566. doi: 10.1083/jcb.68.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogo BG, Berkowitz EM, Dales S. Investigation of vaccinia virus DNA replication employing a conditional lethal mutant defective in DNA. Virology. 1984;132:436–444. doi: 10.1016/0042-6822(84)90048-5. [DOI] [PubMed] [Google Scholar]
- Pogo BGT, O’Shea M, Freimuth P. Initiation and termination of vaccinia virus DNA replication. Virology. 1981;108:241–248. doi: 10.1016/0042-6822(81)90543-2. [DOI] [PubMed] [Google Scholar]
- Pogo BT. Changes in parental vaccinia virus DNA after viral penetration into cells. Virology. 1980;101:520–524. doi: 10.1016/0042-6822(80)90466-3. [DOI] [PubMed] [Google Scholar]
- Roseman NA, Hruby DE. Nucleotide sequence and transcript organization of a region of the vaccinia virus genome which encodes a constitutively expresssed gene required for DNA replication. J Virol. 1987;61:1398–1406. doi: 10.1128/jvi.61.5.1398-1406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen L, Kornberg A. A ribo-deoxyribonucleotide primer synthesized by primase. J Biol Chem. 1978;253:770–774. [PubMed] [Google Scholar]
- Seto J, Celenza LM, Condit RC, Niles EG. Genetic map of the vaccinia virus HindIII D fragment. Virology. 1987;160:110–119. doi: 10.1016/0042-6822(87)90051-1. [DOI] [PubMed] [Google Scholar]
- Stanitsa ES, Arps L, Traktman P. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J Biol Chem. 2006;281:3439–3451. doi: 10.1074/jbc.M511239200. [DOI] [PubMed] [Google Scholar]