Abstract
Light is a key stimulus for plant biological functions, several of which are controlled by light-activated kinases known as phototropins, a group of kinases that contain two light-sensing domains (LOV, Light-Oxygen-Voltage domains) and a C-terminal serine/threonine kinase domain. The second sensory domain, LOV2, plays a key role in regulating kinase enzymatic activity via the photochemical formation of a covalent adduct between a LOV2 cysteine residue and an internally-bound flavin mononucleotide (FMN) chromophore. Subsequent conformational changes in LOV2 lead to the unfolding of a peripheral Jα helix, and ultimately, phototropin kinase activation. To date, the mechanism coupling bond formation and helix dissociation has remained unclear. Previous studies found that a conserved glutamine residue (Q513 in the Avena sativa phototropin 1 LOV2 (AsLOV2) domain) switches its hydrogen-bonding pattern with FMN upon light stimulation. Located in the immediate vicinity of the FMN binding site, this Gln residue is provided by the Iβ strand that interacts with the Jα helix, suggesting a route for signal propagation from the core of the LOV domain to its peripheral Jα helix. To test whether Q513 plays a key role in tuning the photochemical and transduction properties of AsLOV2, we designed two point mutations, Q513L and Q513N, and monitored the effects on the chromophore and protein using a combination of UV-visible absorbance and circular dichroism spectroscopy, limited proteolysis, and solution NMR. The results show that these mutations significantly dampen the changes between the dark and lit state AsLOV2 structures, leaving the protein in a pseudo-dark state (Q513L) or a pseudo-lit state (Q513N) conformation. Further, both mutations changed the photochemical properties of this receptor, particularly the lifetime of the photoexcited signaling states. Together, these data establish that this residue plays a central role in both spectral tuning and signal propagation from the core of the LOV domain through the Iβ strand to the peripheral Jα helix.
Protein signaling cascades are central to organismal growth, adaptation, and communication; therefore, the regulation of these cascades is key to survival. PAS (Per-ARNT-Sim) domain-containing proteins are well characterized as vital members of many such regulatory paths, including adaptation to hypoxia (1), circadian rhythm-dependent gene transcription (2), and phototropism and chloroplast organization in plants (3). A specific subset of PAS proteins, the LOV (light-oxygen-voltage) domains (4), is capable of sensing blue light as an environmental signal and converting it into a biochemical signal in a wide variety of proteins.
LOV domains contain a series of highly conserved residues surrounding an internally bound flavin mononucleotide (FMN) or flavin adenine dinucleotide (FAD) chromophore (Fig. 1a, 1b) that converts blue light into protein structural changes. Spectroscopic studies on LOV-FMN and LOV-FAD complexes showed that blue light induces the formation of a covalent adduct between the isoalloxazine C4a position and a conserved cysteine residue within the LOV domain (Fig. 1c) (5, 6). The stability of this photoadduct is variable among LOV domains; most spontaneously relax back to the noncovalent dark state within several second to many hours (7, 8), but several systems appear to be effectively irreversible on a biological time scale in vitro (8, 9).
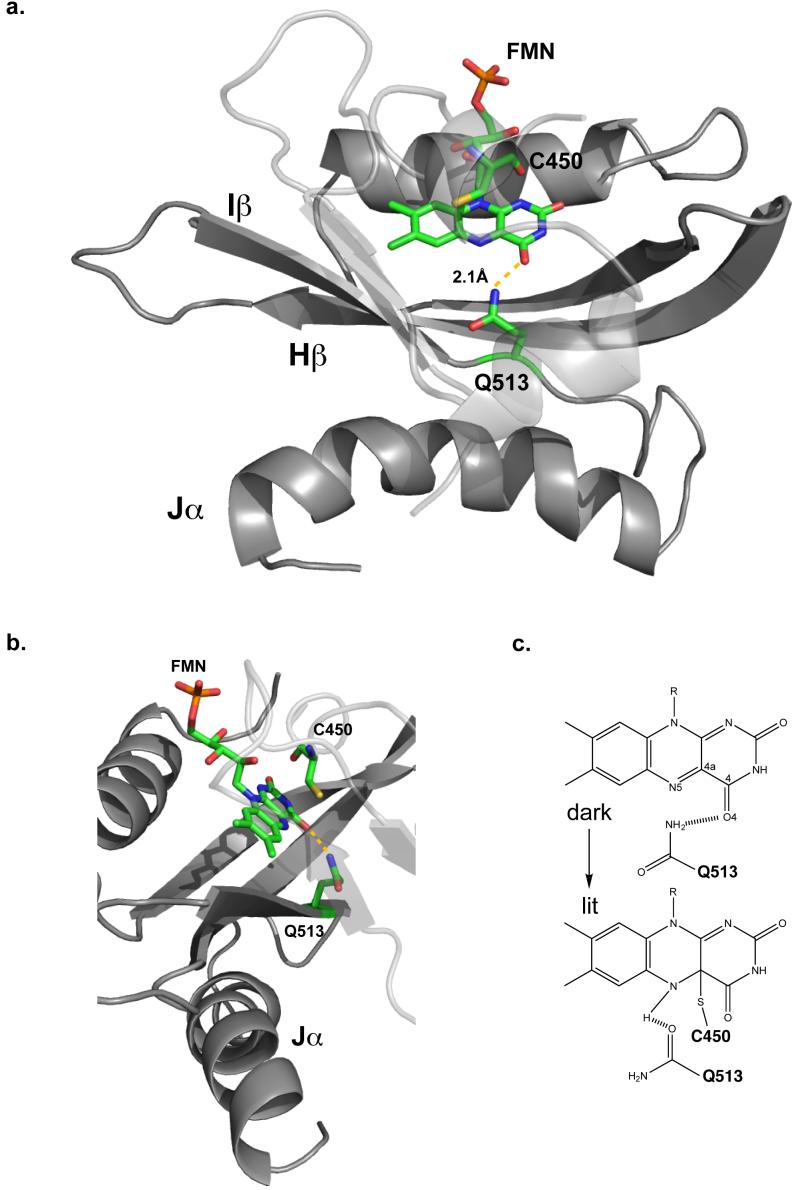
Figure 1. Proposed signal transduction pathway in the AsLOV2 domain.
Front (a) and side view (b) of AsLOV2 domain structure including Jα helix is shown in grey. FMN, Q513 and C450 residues are shown as stick figures with carbon (green), oxygen (red), nitrogen (blue), sulfur (yellow), and phosphorous (orange). (a) The hydrogen bond between Q513 side chain amide proton and FMN C4 carbonyl in the dark state are shown with a yellow dashed line. The side view (b) shows the relative orientations between C450, FMN, Q513, and the Jα helix. Bond formation between C450 and FMN leads to signal propagation through Q513 and ultimately to the dissociation of Jα helix from the Iβ strand. (c) Proposed side chain rotation and hydrogen bond switch by Q513 residue. Light-induced rotation of the Q513 side chain leads to breakage of a hydrogen bond between the Q513 amide and the C4 carbonyl of FMN and possibly formation of a new hydrogen bond between the Q513 carboxyl group and N5 of FMN.
The sensory role played by LOV domains is characterized in a variety of proteins, including transcription factors, ubiquitin ligases and kinases. Previous studies on phototropins, a group of plant photoreceptors that contain two LOV domains and a C-terminal serine/threonine kinase, demonstrated that they form the expected covalent cysteinyl adducts and exhibit a corresponding robust increase in autophosphorylation activity upon illumination (10). While the role of the N-terminal LOV (LOV1) domain remains poorly understood, light-induced changes in the C-terminal LOV (LOV2) domain structure are both necessary and sufficient for kinase activation (11). Despite our knowledge of LOV2 in the context of the full-length protein, the molecular mechanism by which the blue light signal is communicated to the kinase domain remains unclear. Harper et al. (12) proposed a mechanism for signal transduction in the Avena sativa phototropin 1 LOV2 domain (AsLOV2) that involves light-induced unfolding of a helix, termed Jα, that is C-terminal to the conserved LOV core domain. In the dark state, the Jα helix interacts with the β-sheet of the LOV domain, particularly the Gβ, Hβ and Iβ strands. Disruption of the Jα helix interaction with the Iβ strand by site-directed mutagenesis was sufficient to induce a pseudo-lit state structure of the LOV domain and constitutively activate kinase function in the absence of illumination (Fig. 1a, b) (13). Recent crystallographic data on AsLOV2 containing the Jα helix also support a role for Jα helix in signal transduction (14). While these studies clearly implicate the C-terminal Jα helix in communicating photodetection events to a downstream effector domain, it remains unclear how covalent adduct formation in the core leads to α-helical unfolding on the surface of the domain.
Insight into this question was provided by X-ray crystallography and molecular dynamics simulations that show a reorganization of the protein/FMN hydrogen-bonding network upon covalent adduct formation (15, 16). In the dark state, crystallography shows that the side chain amide of a conserved glutamine residue (Q1029) in the A. capillus-veneris neochrome1 LOV2 domain donates a hydrogen bond to the O4 atom of FMN (Fig. 1c) (17). Upon illumination, the glutamine side chain rotates, breaking this bond to O4 and instead allowing Q513 to accept a hydrogen bond from the newly-protonated N5 atom of FMN (15, 16). Additionally, FTIR studies demonstrated that mutation of Q1029 to leucine alters the electronic state of FMN and reduces the magnitude of light-induced protein structural changes (18). As a result, Nozaki et al. proposed that these changes are due to the absence of the glutamine carbonyl hydrogen bond to FMN in the lit state. Subsequent studies of the mutation at the corresponding glutamine in full-length Arabidopsis thaliana phototropin1 demonstrated attenuated autophosphorylation activity in the light versus wildtype protein (19). In addition, studies of the corresponding glutamine residue in the VIVID fungal photoreceptor also show a leucine mutant diminishes light-induced activity in vivo (20).
To better understand the role of this conserved glutamine residue in LOV domain signaling, we used a variety of biochemical and biophysical techniques to characterize how two mutations of this essential residue affect photochemistry and structural perturbations upon blue light illumination. Specifically, we made the corresponding glutamine to leucine (Q513L) mutation as well as a glutamine to asparagine (Q513N) mutation in the AsLOV2 domain. These mutations allowed us to probe how subtle perturbations of this side chain affect photochemistry and signal transmission. We observed significant changes in the electronic and structural properties of these mutants in comparison to wildtype AsLOV2 using a combination of UV-visible spectroscopy, limited proteolysis, circular dichroism, and NMR spectroscopy. While both mutant domains maintained photocycling capabilities and demonstrated light-induced structural changes, they appeared to dampen the degree of light-induced structural change, leaving one mutant (Q513L) in a pseudo-dark state while the other (Q513N) is more pseudo-lit state compared to wildtype AsLOV2. These data underscore the importance of hydrogen bond networks between FMN and the protein β-sheet in tuning properties of the chromophore and communicating light-induced structural changes throughout the domain.
MATERIALS AND METHODS
Cloning, Expression, and Purification of AsLOV2
Plasmid DNA encoding the AsLOV2 domain plus the Jα helix (residues 404-560 (12)) was used to generate Q513N and Q513L mutants. Mutagenesis was carried out according to the Quick Change II site-directed mutagenesis kit (Stratagene) following manufacturer's instructions and verified by DNA sequencing. Proteins were expressed in E. coli BL21(DE3) cells grown in M9 minimal medium supplemented with 15NH4Cl (1 g/L) at 37°C to an A600 of 0.6-0.8 and then induced with IPTG (0.12 g/L). After 16 hr induction at 20°C, cells were centrifuged and pellets resuspended in 50 mM Tris, 100 mM NaCl, pH 8 buffer. Cells were lysed using sonication and clarified with centrifugation at 10,000 g for 40 min. The soluble fraction was loaded onto a Ni+2-NTA column, allowing for rapid affinity purification of His-Gβ1 tagged (12) LOV fusions by eluting with 250 mM imidazole. After exchanging the LOV-containing fractions into 50 mM Tris, 100 mM NaCl pH 8.0 buffer, the His-Gβ1 tag was cleaved by adding 1 mg His6-TEV protease per 30 mg of fusion protein. Proteolysis reactions were allowed to proceed overnight at 4°C and stopped using a Ni+2-NTA column to remove the His-Gβ1 and His6-TEV protease. Post-cleavage, the resulting proteins contain only GEF (N-terminal) and G (C-terminal) residues as cloning artifacts.
Protein-Flavin Stoichiometry Calculation
UV-visible absorbance spectra (from 250 nm to 550 nm) were recorded for all three freshly purified proteins following buffer exchange into 50 mM sodium phosphate, 100 mM NaCl, pH 6.0. During buffer exchange the flow through fraction was monitored by UV-visible spectroscopy for the presence of free FMN. Using the A280/A446 ratio for wildtype (2.60) as a reference for 1:1 protein/FMN stoichiometry (21), this same ratio was calculated for each of the mutant domains (2.62 for Q513N and 2.76 for Q513L), revealing an approximately 1:1 protein/FMN stoichiometry for both Q513N and Q513L, suggesting the mutations do not significantly affect flavin incorporation.
UV-visible Absorbance Spectroscopy and Photocycle Kinetics
All proteins were concentrated to <70 μM in buffer containing 50 mM sodium phosphate, 100 mM NaCl, pH 6.0. UV-visible absorbance spectra were measured on a Varian Cary Series 50 spectrophotometer from 250-550 nm. Dark state spectra were obtained on samples exposed only to red light for the past 24 hr, while lit state spectra were obtained immediately after exposing sample to illumination from a photographic flash. Kinetic experiments monitored the return of the A446 signal following illumination. Data points were fitted using a first order rate equation to obtained the time constant (τ).
Limited Proteolysis
Proteins were buffer exchanged to 50 mM sodium phosphate, 100 mM NaCl, pH 7.5 buffer. A 1:90 ratio (w/w) of chymotrypsin to protein was used in a single volume with subsequent samples collected from this larger quantity. Samples collected for each time point were stopped by the addition of SDS loading buffer containing 25% glycerol and visualized on 20% SDS-PAGE gel. Dark state experiments were done under dim red light while lit state experiments were performed under constant irradiation with 488 nm laser light at 50 mW power.
Circular Dichroism Spectroscopy
Proteins were buffer exchanged into buffer containing 50 mM sodium phosphate and 100 mM NaCl at pH 6.0. A total of 500 μl of 15 μM sample was used for each CD experiment. Dark state spectra were collected under dim red light; while lit state spectra were recorded following exposure to photographic flash every 10 s during the course of the experiment. CD data were collected using a wavelength range from 195 to 260 nm at 10°C with 1.5 nm bandwidth and 3 s averaging time. Final data were generated from an average of 3 repeats.
Nuclear Magnetic Resonance spectroscopy
Proteins were concentrated to 1 mM in pH 6.0 buffer containing 50 mM sodium phosphate, 50 μM FMN, and 100 mM NaCl, with 10% (v/v) D2O added to all samples prior to all NMR experiments. NMR experiments were performed on Varian Inova 500 and 600 MHz spectrometers at 25°C, using nmrPipe (22) for data processing and NMRview (23) for analysis. Lit state HSQC spectra were acquired with a 488 nm Coherent Sapphire laser. The output from this laser was focused into a 10 m long, 0.6 mm diameter quartz fiber optic. The other end of the fiber was placed into the bottom of a coaxial insert tube designed to hold external chemical shift standards inside a 5 mm NMR sample tube. This allowed the illuminated tip to be immersed in protein solution without contamination. Power level measurements were conducted prior to every experiment to establish the efficiency of coupling the laser output to the fiber optic, and all power levels reported here are those measured at the end of the fiber. Each 15N/1H HSQC spectrum was recorded by preceding each transient in the experiment with a 50 mW 200 ms laser pulse during the 1.06 sec delay between transients (12).
Sequence Alignment
A multiple sequence alignment of LOV domains was generated using CLUSTAL W (24) and sequences were displayed using ESPript.cgi Version 3.06 (25).
RESULTS
Effects of Q513 mutations on FMN spectral properties
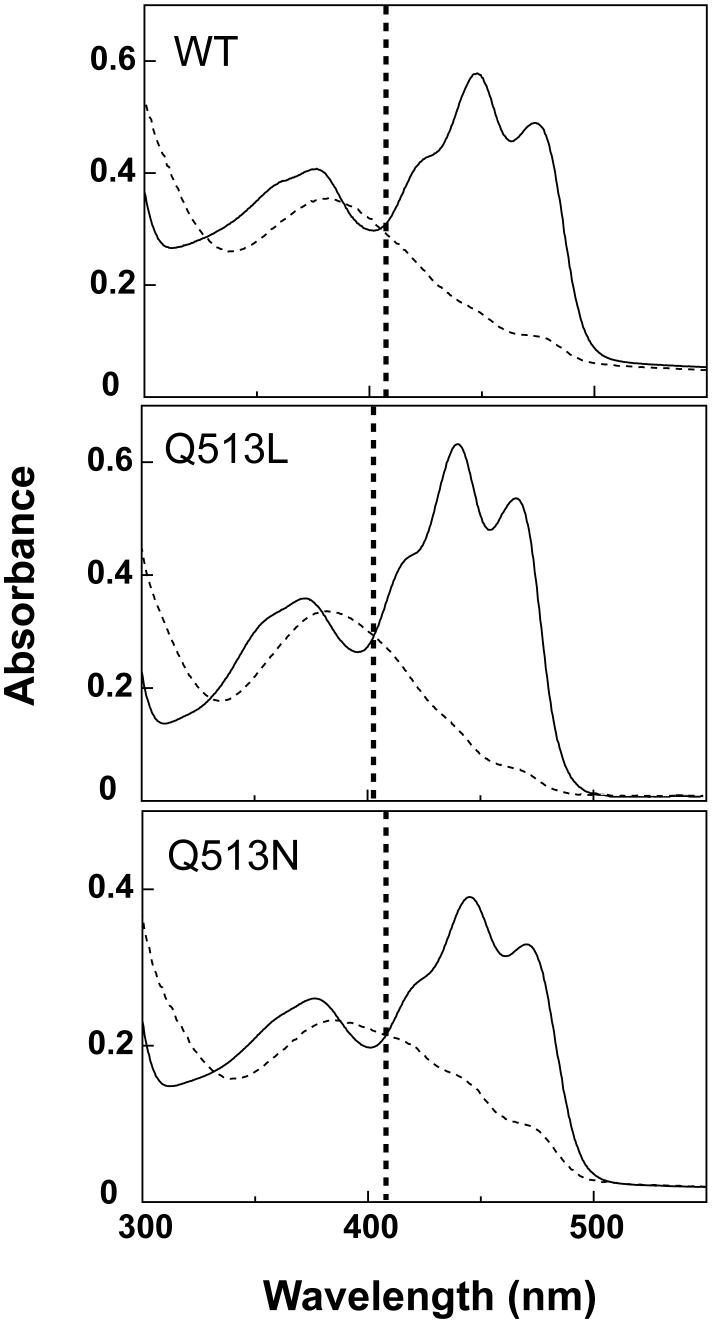
The electronic state of FMN within the LOV protein core is easily observed by UV-visible absorbance spectroscopy. Quantitative measurements of the flavin:protein stoichiometry indicated that both mutants retain FMN at approximately a 1:1 ratio with protein as previously described for wildtype AsLOV2(21) (Supporting Information, Supporting Fig. 2). As with wildtype, both the Q513L and Q513N mutants demonstrate typical LOV domain spectra with three characteristic absorbance peaks between 400 and 500 nm in the dark state and three isosbestic points (Fig. 2), with similar absorption coefficients to known flavoproteins at 446 nm (Table 1). Both mutants also display similar loss of this fine structure upon illumination with blue light, indicating the formation of covalent adduct. However, the Q513L absorbance profile is blue-shifted 9 nm in the dark state, as shown before (18, 19), indicating a change in the electronic environment surrounding the FMN. This contrasts with the Q513N absorbance spectra which do not significantly deviate from wildtype spectra, indicating comparatively little change in the electronic environment surrounding FMN in the dark state. Similarly, we observed that Q513L caused greater changes in the locations of the three isosbestic points compared to Q513N (Table 1). Overall, these spectroscopic data indicate that the electronic environment of the FMN exhibits a larger change in the Q513L mutant than the Q513N mutant, most likely due to loss of hydrogen bond contacts between FMN and the altered side chain.
Figure 2. UV-visible absorbance profiles of AsLOV2 wildtype protein and Q513 mutants.
Solid traces represent the dark state spectra and dashed traces represent the lit state spectra. The mutants all display the same characteristic dark state absorbance profile for typical LOV domains, with three distinctive maxima between 400 nm to 500 nm. These maxima diminish in the lit state in all three cases. The vertical dashed line is aligned with the largest wavelength isosbestic points of the LOV domains (406 nm for AsLOV2 and Q513N; 403 nm for Q513L).
Table 1. Comparisons of the kinetic time constants (τ) and the absorption coefficients (ε) at the isosbestic points for wildtype and Q513 mutants in the dark state.
The kinetics experiments are recorded at room temperature (22°C) for 200 s and the data points were fitted using a first order exponential rate equation to obtain the time constant (τ). The dark recovery time constant at A446 is measured by UV-visible absorbance spectroscopy while the dark recovery time constant at θ222 is measured via CD spectroscopy.
| Construct | τdark recovery[A446] (s) | τdark recovery[θ222] (s) | ε446 (M-1 cm-1) | Isosbestic points (nm) |
|---|---|---|---|---|
| WT | 68.3 | 72.4 | 11700 | 327, 388, and 406 |
| Q513N | 37.3 | 40.6 | 12400 | 330, 388, and 406 |
| Q513L | 1080 | >1000 | 10700 | 330, 380, and 403 |
Given the alterations in the environment surrounding the FMN cofactor, we sought to determine if these mutations would affect the photocycle of the LOV domain. We found the dark recovery time constant of the Q513L mutant followed by illumination is 1080 s, approximately 15-fold longer than wildtype (68 s, Table 1). In contrast, the Q513N mutation has a shorter recovery time constant (37 s, Table 1). These kinetic data indicate that the Q513L point mutation has a more significant effect on the relative energetics of AsLOV2 lit state and transition state it visits during the recovery process, complementing the steady state absorbance data which show a similarly larger effect of this mutation.
Structural effects of Q513 mutations by circular dichroism
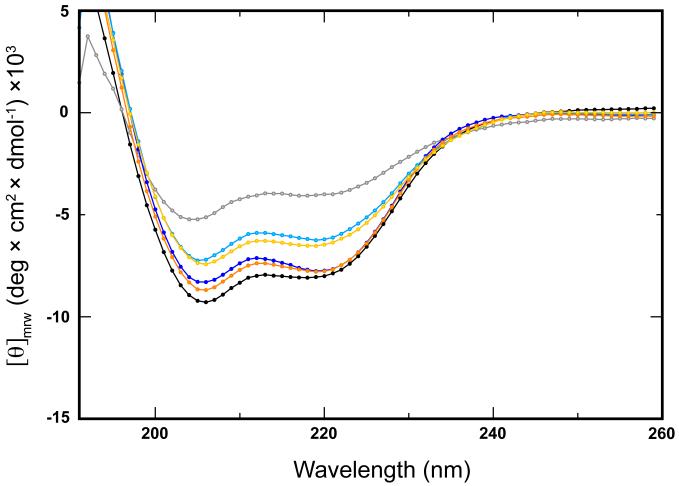
Prior solution NMR studies of wildtype AsLOV2 show that the C-terminal Jα helix dissociates from the core LOV domain and unfolds upon illumination, while the LOV domain itself remains intact and folded (12, 26). Circular dichroism reflects global secondary structure so the spectra presented represent the total mixed α/β fold of the LOV domains. The double minima at 208 nm and 222 nm and maximum at 195 nm are features of helical secondary structure (Fig. 3), which demonstrate a clear decrease in molar residue ellipticity in wildtype AsLOV2 following illumination with white light (27). In contrast, both the Q513L and Q513N mutants show reduced light-induced changes by CD (Fig. 3). This is consistent with the limited proteolysis and NMR results (vide infra) suggesting mutation of Q513 leads to some decoupling of covalent adduct formation from conformational changes.
Figure 3. Structural effects of Q513 mutations in the dark and lit states monitored by circular dichroism.
The room-temperature far UV region (190 nm to 250 nm) CD spectra of the wildtype dark (black) and lit (grey) states, Q513L dark (orange) and lit (yellow) states, and the Q513N dark (dark blue) and lit (light blue) states are overlaid here for comparison. The differences between the dark and lit states are more pronounced in the wildtype compared to the Q513 mutants.
Despite the reduced amplitude of light-induced changes in the CD signals of Q513L and Q513N, we were still able to monitor the kinetics of dark state recovery via changes in the secondary structure during this process. As observed with UV-visible absorbance spectroscopy, both mutants undergo a normal photocycle with complete recovery to the dark state following illumination. Also consistent with the UV-visible absorbance results, Q513L shows significantly slower recovery kinetics while Q513N is slightly accelerated (Table 1). We observed similar recovery kinetics regardless of whether we monitored the change via protein (CD) or chromophore (UV-visible absorbance), suggesting that these processes have a common rate-limiting step (7, 27).
Structural effects of Q513 mutations by limited proteolysis
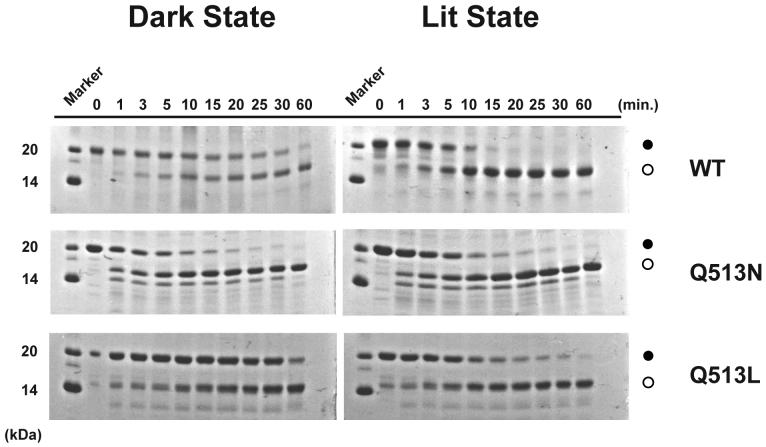
To further document how the Q513 mutations affected the overall stability of the LOV domain, we used limited proteolysis. Wildtype AsLOV2 becomes more susceptible to proteolytic cleavage by chymotrypsin upon illumination, specifically at Met530 located in the middle of the Jα helix (12). This is reflected in the light-induced acceleration in the appearance of a lower molecular weight species in SDS-PAGE (Fig. 4). Notably, neither mutant domain demonstrates as dramatic an increase in proteolysis after covalent adduct formation. The Q513L mutant is less susceptible to proteolysis from chymotrypsin in the lit state than wildtype AsLOV2, while the dark state demonstrates resistance similar to wildtype. In contrast, the Q513N mutant displays the opposite effect, with a protease-susceptible dark state and a lit state that is as easily proteolyzed as wildtype. The primary species formed upon cleavage of both mutants is consistent with that formed by similar treatment of wildtype AsLOV2, suggesting that Met530 is the likely cleavage site. In addition, chymotrypsin treatment of Q513N produces a lower molecular weight species that is formed very quickly upon addition of protease to the lit state. This additional band suggests that Q513N may adopt another domain conformation or have increased dynamics that allow protease accessibility to an otherwise inaccessible residue.
Figure 4. Structural effects of Q513 mutations in the dark and lit states monitored by limited proteolysis.
Limited proteolysis by chymotrypsin digestion was performed at room temperature for the wildtype protein and Q513 mutants in both the dark (left panel) and lit states (right panel). Time points ranging from 0 to 60 min are shown above the gels. The molecular weight marker indicating 14 kDa and 20 kDa are also shown on the left side of the gels. Black filled circles represent the full-length undigested domain and open circles represent the largest stable digested product.
Structural effects of Q513 mutations characterized using NMR spectroscopy
The low-resolution structural information provided by CD spectroscopy and limited proteolysis clearly show that both Q513L and Q513N mutants have fewer conformational changes upon illumination compared to wildtype AsLOV2. To examine this in further detail, we used two-dimensional 15N-1H HSQC spectra to monitor the environments of the pairs of J-coupled 15N-1H nuclei within the domain in both the dark and lit state. The 15N-1H HSQC spectrum of wildtype AsLOV2 in the dark state shows well-dispersed peaks consistent with our previous NMR results (Supporting Fig. 1a) (12). Upon illumination, we observe a general loss of amide proton chemical shift dispersion and the appearance of several new intense peaks in the center of the spectrum, indicative of increased dynamics in the LOV domain and dissociation of the Jα helix.
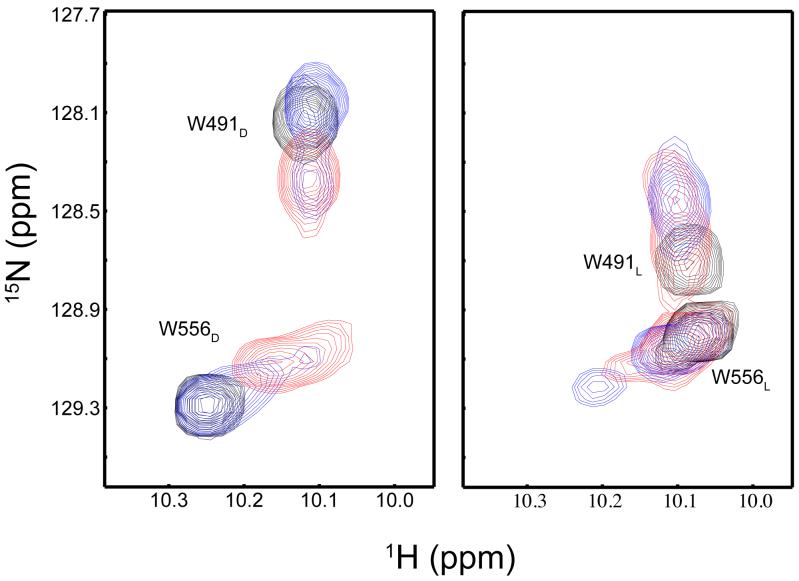
Specific analysis of two tryptophan side chain indole (Hε1-Nε1) crosspeaks highlights the light-induced structural changes surrounding residues W491 and W556, located near the LOV-Jα helix hinge region and C-terminal to the Jα helix respectively (12). In the dark state of wildtype AsLOV2, these indole crosspeaks are clearly separated (Fig. 5a). Upon illumination, they collapse towards a central position that is near the average location for protein tryptophans in general (Fig. 5b) (28), also consistent with the Jα helix unfolding and the tryptophans adopting less distinctive chemical environments after the covalent adduct is formed. Comparison with this same region in the Q513L spectrum again shows two clearly resolved crosspeaks that overlay with the wildtype dark state (Fig. 5a). After light induction, there is a small chemical shift perturbation of the Q513L crosspeaks (Fig. 5b), indicating that a significant majority of the Q513L population still remains closer to the dark state conformation after light induction. Conversely, the Q513N mutant displays very different NMR spectra in the tryptophan indole region from Q513L or wildtype. Prior to light irradiation, the tryptophan signals are closer to their counterparts in the wildtype lit state (Fig. 5a). Additionally, the crosspeaks do not collapse toward each other to the same extent as wildtype upon illumination (Fig. 5b), suggesting that the Q513N mutant adopts a pseudo-lit structure that resembles the wildtype lit state and hence undergoes relatively few light-induced structural changes. While we discuss Q513L and Q513N as pseudo-dark and -lit structures here, we suggest that both are significantly more dynamic than wildtype given the increased linebroadening present in both spectra (Fig. 5). Further, signs of peak doubling can be observed in several of these spectra, suggestive of slow (τ~ms or longer) interconversion between states. Overall, these data support and extend both the proteolysis and CD data, showing that both Q513L and Q513N undergo more limited structural changes with illumination and appear to be poised more towards either the dark- or lit-state structure of the wildtype.
Figure 5. 15N/1H HSQC spectra of the tryptophan Hε1-Nε1 region.
The overlaid spectra of the dark (a) and lit (b) states of the wildtype AsLOV2 (black), Q513L (blue), and Q513N (red) are shown. The tryptophan indole assignments are indicated in both the dark and lit state panels with subscript D or L respectively. In the lit state, there is a significant shift in this region in wildtype protein (b).
An analysis of the full 15N/1H HSQC spectra of wildtype, Q513L and Q513N (Supporting Fig. 1) supports the assignment of Q513L as a pseudo-dark state structure and Q513N as pseudo-lit state. The dark state spectrum of Q513L is quite similar to wildtype, consistent with limited proteolysis. In contrast, the Q513N dark state shows significant chemical shift changes and/or linebroadening, and is reminiscent of the spectra of the wildtype lit state. These data clearly indicate that the Q513N point mutation causes a greater structural perturbation of the wildtype structure than Q513L. As with the wildtype protein, illumination causes significant spectral changes for both the Q513L and Q513N mutants. Unfortunately, these perturbations cannot be unambiguously interpreted to provide independent confirmation of the reduced conformational changes in the two Q513 mutants as reported above by proteolysis and CD. This is due to the fact that chemical shift changes originate from two interrelated sources: bona fide protein conformational changes and the significant alteration in the electronic structure of the FMN isoalloxazine ring upon adduct formation. Given that the adduct forms successfully in all three AsLOV2 variants tested here, we expect significant chemical shift changes in these proteins regardless of their ability to couple this photochemical event with protein conformational changes. Despite this caveat, these NMR spectra support the assignment of Q513L and Q513N domains adopting pseudo-dark and pseudo-lit state structures in the dark.
DISCUSSION
While the connection between light-induced covalent adduct formation and protein conformational changes in LOV domains is well established (7, 12-14, 18, 20, 29), the mechanism through which this occurs remains unclear. A highly conserved glutamine residue (Q513 in AsLOV2) in the core of the LOV domain was previously suggested to be crucial for this signaling process (18, 20). While FTIR studies show that illumination breaks a hydrogen bond between this residue and the FMN O4 position upon adduct formation (18), formation of a proposed new hydrogen bond between Q513 and the FMN N5 position has been more difficult to demonstrate. Some crystal structures show that the side chain of this glutamine rotates with illumination, consistent with formation of this new hydrogen bond (15, 16, 20, 29), while other structures argue against it (14). In light of this ambiguity, we targeted our point mutations to test the importance of the Q513/FMN interaction for intradomain signal communication. Ground state structures, light-induced structural changes and dark state recovery rates are all altered by mutations at this position, demonstrating an important role for Q513 in AsLOV2 signaling.
Structural effects of Q513L and Q513N point mutations
Residue Q513 is located on the Iβ strand, on the opposite side as the Jα-helix binding surface, thus suggesting a direct path from the internally bound FMN to the Jα-helix on the surface (Fig. 1a). As such, it seemed reasonable that changing hydrogen-bonding patterns between Q513 and FMN would alter the structure of the anchoring Iβ strand in such a way as to interfere with this pathway. The Q513L mutation was designed to disrupt all hydrogen bonding with FMN while only slightly increasing the volume of a glutamine residue. The UV-visible absorbance profile of Q513L is blue-shifted in the dark state, consistent with the loss of this hydrogen bond to the O4 carbonyl oxygen of FMN. Without this hydrogen bonding capability, we anticipated that the Q513 side chain would not rotate in an organized fashion upon covalent adduct formation. With loss of this rotation, the Iβ strand structure and dynamics would likely remain unchanged with illumination. Our data bear out these predictions, as the Q513L mutant demonstrated similar structural properties to the wildtype dark state and also had reduced light-induced conformational changes. Previous findings of the analogous Q1029L mutation in neo1 LOV2 (18, 30) support this view.
In comparison, the Q513N mutant also had reduced amplitude of light-induced structural changes but appeared to adopt a pseudo-lit state structure in the dark. The Q513N mutation was designed to maintain hydrogen-bonding contacts, and the similarity of the Q513N and wildtype UV-visible absorbance spectra is consistent with hydrogen bonds being maintained between this residue and the FMN O4 and N5 atoms (if we assume this interaction occurs in the lit state). To maintain these bonds, the Iβ strand may be distorted to allow for FMN interaction with the shorter asparagine side chain. This type of stress may somehow be similar to the type of movement or changes that normally induce Jα release, giving rise to a pseudo-lit state type structure. Given that Q513N is already in this pseudo-lit state in the dark, we suggest that illumination and cysteinyl/C4a adduct formation cannot induce further conformational changes, consistent with our results.
Dark state recovery kinetic effects of Q513L and Q513N point mutations
While the Q513L and Q513N mutants were created to study their roles in the structural changes accompanying signal transduction, we found that both also affected dark state recovery rates. Other work has identified several solution parameters that perturb these rates, including the presence of basic compounds (e.g. imidazole) (31), alkaline pH (32, 33) and ionic strength (32, 33); however, the most relevant factors for our results are related to the conformations of the lit state and the transition state between the dark and lit structures. Chemical transition state theory establishes that the energetics of these two states influence the kinetics of the return rate. In AsLOV2, the difference between these two states is approximately ΔG‡~14.5 kcal mol-1 based on the temperature dependence of dark state recovery (7). In parallel, the spontaneous relaxation of the lit state establishes that it is energetically less favorable than the dark state, leading to the suggestion that the lit state is somehow conformationally strained (34), which is experimentally supported by a light-dependent increase in 2H exchange rates (7). As such, changes that lower ΔG‡ by either destabilizing the lit state or stabilizing the transition state are predicted to accelerate dark state recovery, and vice-versa. This is supported by the accelerated recovery rates of an AsLOV2 I427V point mutant, which removes a methyl group that is predicted to stabilize the lit state Cys-C4a adduct (35).
Our data on the recovery rates of Q513L and Q513N are consistent with this model. Q513L exhibits a fifteen-fold slowing of the dark state return rate, as well as data indicating that this protein undergoes much smaller light-induced structural changes than the wildtype domain. These data are consistent with stabilization of the lit state and a corresponding increase in ΔG‡, possibly by relieving tension that would otherwise be maintained by the Q513:FMN hydrogen bonds in the lit state. In agreement with this, an analogous Q1029L point mutation in A. capillus-veneris neo1 LOV2 slowed dark state recovery significantly (7-fold, (30)), as does a F1010L mutation at an adjacent position on the neighboring Hβ strand (10-fold, (36)). In contrast, we find that the Q513N point mutant accelerates dark state recovery two-fold. We suggest that this protein retains dark state hydrogen bonding between the Q513 amide and the O4 carbonyl oxygen based on visible absorbance spectroscopy, and may retain similar interactions with the flavin cofactor in the light. The maintenance of these interactions despite the loss of a methylene group would likely lead to a more destabilized lit state, consistent with the rate acceleration we observe. While a detailed understanding of this process remains to be established, we suggest that these and other rate-perturbing mutations are providing useful evidence for the features of AsLOV2 that establish the lifetime of the signaling state.
Models of light-induced movement in Q513
The combination of molecular dynamics simulations and x-ray crystallography has led to models for light-induced Q513 movement with somewhat opposing conclusions. Simulations of AsLOV2 minus the Jα helix identified breakage of the dark state hydrogen bond between Q513 and FMN and further demonstrated light-induced hydrogen bond formation at the FMN N5 position (16). In addition, these simulations also suggested a second conformation in which Q513 interacted with neighboring residues on the Iβ strand, thereby increasing dynamics in this region. Such an alteration at the LOV-Jα interface may contribute towards Jα release and signal transduction. In contrast, recent crystal structures of AsLOV2 containing the Jα (14) demonstrate neither any rotation of the Q513 side chain upon illumination nor a bent conformation. Consistently, crystal structures that fail to demonstrate side chain rotation also fail to demonstrate the previously described loss of hydrogen bonding to FMN O4. These inconsistencies among computational models, solution studies, and crystallographic structural methods suggest the role of Q513 side chain rotation in signal transduction merits further investigation.
Role of Q513 in full-length LOV-containing proteins
Studies on a single isolated domain, such as those discussed here, provide interesting results from which we can postulate the role of Q513 in signal transduction, but the behavior in full-length proteins requires further investigation. Limited proteolysis, circular dichroism and NMR data all demonstrate fewer light/dark conformational changes for the mutant domains. Specifically, Q513L appears locked into a dark state-like conformation and Q513N retains lit state-like characteristics regardless of FMN electronic state. In vitro biochemical experiments with full-length phototropin containing a mutation analogous to Q513L support our findings, showing that this mutation attenuates light-activated autophosphorylation activity (19). These data are consistent with the Q513L mutant AsLOV2 domain maintaining a dark state-like, inactive conformation, as we have found. This residue also plays a central role in the FAD-bound LOV domain photoreceptor, VIVID (20). A comparison of dark and lit state crystal structures of this protein shows a network of light-induced rearrangements in hydrogen bond contacts between the protein and FAD. In the wildtype protein, these lead to a series of side chain reorientations that ultimately alter the protein surface. Introduction of a leucine mutation at the equivalent glutamine position in VIVID (Q182 in VIVID) disrupts these changes, as shown by differential elution times in size exclusion chromatography. While this work further extends the results of the domain studies to full-length proteins, it also suggests that this conserved glutamine is important for signal communication in non-phototropin related LOV domains.
A large-scale sequence alignment of LOV domain sequences suggests that Q513 is highly, but not absolutely, conserved (Supporting Fig. 2). In particular, we see that several proteins contain naturally occurring leucine substitutions at this critical site. While most of the LOV domains with leucine substitutions have not been studied to the extent of determining structural information or dark state recovery time constants, a small cohort of A. thaliana proteins are of particular interest. These three proteins, FKF1, LKP2 and ZTL have extremely stable cysteinyl-flavin adducts: FKF1 demonstrates a dark-state recovery half-life of 62.5 hr (9), while the other two have been described as effectively irreversible (2). Intriguingly, LKP2 contains a leucine at the position equivalent to Q513 in AsLOV2, suggesting it may play a role in extending the dark state recovery of this protein. While this is an enticing hypothesis, neither FKF1 nor ZTL have a leucine at this position, indicating there must be other factors influencing dark state recovery rates. Mutational studies have determined several other positions that contribute to dark state recovery kinetics (5, 35-38). Of these, a phenylalanine to leucine mutation at the position equivalent to AsLOV2 F494 led to a 10-fold increase in half-life of the excited state (36). FKF1, ZTL and LKP2 all contain this naturally-occurring leucine substitution, which occurs on the Hβ strand immediately adjacent to Q513, positing an important role for this residue in tuning photocycle kinetics. These data, combined with the studies on Q513 presented here, indicate that several residues of the chromophore-binding pocket of LOV domains collectively play roles in critical aspects of signaling, including signal transmission and regulation of signaling state lifetimes. A combination of further biochemical and biophysical measurements are needed to characterize the detailed basis of this control, perhaps allowing artificial control of these features (26).
Supplementary Material
Acknowledgements
We thank Charles Dann III and members of the Gardner lab for helpful discussions.
This work was supported by grants from the NIH (R01 GM081875) and Robert A. Welch Foundation (I-1424) to K.H.G. A.I.N. was supported by an NIH Predoctoral Training Grant in Molecular Biophysics (T32 GM008297).
Abbreviations
- PAS
Period-ARNT-Single Minded (Per-ARNT-Sim)
- LOV
Light-Oxygen-Voltage
- FMN
flavin mononucleotide
- FAD
flavin adenine dinucleotide
- AsLOV2
Avena sativa phototropin 1 LOV2 domain
References
- 1.Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 2.Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 3.Liscum E, Briggs WR. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 5.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 6.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 7.Harper SM, Neil LC, Day IJ, Hore PJ, Gardner KH. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. J Am Chem Soc. 2004;126:3390–3391. doi: 10.1021/ja038224f. [DOI] [PubMed] [Google Scholar]
- 8.He Q, Liu Y. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zikihara K, Iwata T, Matsuoka D, Kandori H, Todo T, Tokutomi S. Photoreaction cycle of the light, oxygen, and voltage domain in FKF1 determined by low-temperature absorption spectroscopy. Biochemistry. 2006;45:10828–10837. doi: 10.1021/bi0607857. [DOI] [PubMed] [Google Scholar]
- 10.Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 11.Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32:205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- 12.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 13.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 14.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 15.Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophys J. 2006;91:3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci U S A. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nozaki D, Iwata T, Ishikawa T, Todo T, Tokutomi S, Kandori H. Role of Gln1029 in the photoactivation processes of the LOV2 domain in adiantum phytochrome3. Biochemistry. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 19.Jones MA, Feeney KA, Kelly SM, Christie JM. Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J Biol Chem. 2007;282:6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 20.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci U S A. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:227–339. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 26.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-Jalpha photoswitch. Nat Chem Biol. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corchnoy SB, Swartz TE, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J Biol Chem. 2003;278:724–731. doi: 10.1074/jbc.M209119200. [DOI] [PubMed] [Google Scholar]
- 28.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fedorov R, Schlichting I, Hartmann E, Domratcheva T, Fuhrmann M, Hegemann P. Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys J. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwata T, Nozaki D, Tokutomi S, Kandori H. Comparative investigation of the LOV1 and LOV2 domains in Adiantum phytochrome3. Biochemistry. 2005;44:7427–7434. doi: 10.1021/bi047281y. [DOI] [PubMed] [Google Scholar]
- 31.Alexandre MT, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JT. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry. 2007;46:3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 32.Kottke T, Heberle J, Hehn D, Dick B, Hegemann P. Phot-LOV1: photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys J. 2003;84:1192–1201. doi: 10.1016/S0006-3495(03)74933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H, Kottke T, Hegemann P, Dick B. The phot LOV2 domain and its interaction with LOV1. Biophys J. 2005;89:402–412. doi: 10.1529/biophysj.104.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Losi A, Kottke T, Hegemann P. Recording of blue light-induced energy and volume changes within the wild-type and mutated phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys J. 2004;86:1051–1060. doi: 10.1016/S0006-3495(04)74180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christie JM, Corchnoy SB, Swartz TE, Hokenson M, Han IS, Briggs WR, Bogomolni RA. Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry. 2007;46:9310–9319. doi: 10.1021/bi700852w. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto A, Iwata T, Tokutomi S, Kandori H. Role of Phe1010 in light-induced structural changes of the neo1-LOV2 domain of Adiantum. Biochemistry. 2008;47:922–928. doi: 10.1021/bi701851v. [DOI] [PubMed] [Google Scholar]
- 37.Kottke T, Dick B, Fedorov R, Schlichting I, Deutzmann R, Hegemann P. Irreversible photoreduction of flavin in a mutated Phot-LOV1 domain. Biochemistry. 2003;42:9854–9862. doi: 10.1021/bi034863r. [DOI] [PubMed] [Google Scholar]
- 38.Song SH, Dick B, Penzkofer A, Hegemann P. Photo-reduction of flavin mononucleotide to semiquinone form in LOV domain mutants of blue-light receptor phot from Chlamydomonas reinhardtii. J Photochem Photobiol B. 2007;87:37–48. doi: 10.1016/j.jphotobiol.2006.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.