Abstract
The mechanism underlying perinuclear calcium spiking induced during legume root endosymbioses is largely unknown. Lotus japonicus symbiosis-defective castor and pollux mutants are impaired in perinuclear calcium spiking. Homology modeling suggested that the related proteins CASTOR and POLLUX might be ion channels. Here, we show that CASTOR and POLLUX form two independent homocomplexes in planta. CASTOR reconstituted in planar lipid bilayers exhibited ion channel activity, and the channel characteristics were altered in a symbiosis-defective mutant carrying an amino acid replacement close to the selectivity filter. Permeability ratio determination and competition experiments reveled a weak preference of CASTOR for cations such as potassium over anions. POLLUX has an identical selectivity filter region and complemented a potassium transport–deficient yeast mutant, suggesting that POLLUX is also a potassium-permeable channel. Immunogold labeling localized the endogenous CASTOR protein to the nuclear envelope of Lotus root cells. Our data are consistent with a role of CASTOR and POLLUX in modulating the nuclear envelope membrane potential. They could either trigger the opening of calcium release channels or compensate the charge release during the calcium efflux as counter ion channels.
INTRODUCTION
Symbiosis between legumes and nitrogen-fixing rhizobia contributes significantly to the nitrogen supply of agricultural and natural ecosystems. A chemical communication between both partners accompanies the bacterial invasion of plant host cells and the development of a newly formed organ, the root nodule. In response to plant-released flavonoids, rhizobia produce lipo-chito-oligosaccharide signaling molecules, so-called Nod factors (Long, 1989; Lerouge et al., 1990). Early signal transduction in legumes, such as Lotus japonicus and Medicago truncatula, is associated with a succession of tightly orchestrated ion fluxes across different membrane systems of the host cell (Ehrhardt et al., 1996; Felle et al., 1999; Miwa et al., 2006). Nod factors alone are sufficient to trigger early plant responses, including cytosolic calcium (Ca2+) influx, cytosolic perinuclear Ca2+ oscillations, as well as transcription of symbiosis-related plant genes (Pichon et al., 1992; Ehrhardt et al., 1996; Felle et al., 1999). Following Nod factor application, a Ca2+ influx at the root hair tip can be observed within 1 min (Cardenas et al., 1999; Felle et al., 1999), whereas the perinuclear Ca2+ oscillations, known as Ca2+ spiking, occurs with a delay of 10 to 30 min (Ehrhardt et al., 1996). Due to the nucleoplasmic and perinuclear localization of the Ca2+ spiking, cisterns of the endoplasmic reticulum (ER) and the nuclear envelope are likely to be the corresponding Ca2+ stores (Oldroyd and Downie, 2006). In pea (Pisum sativum) root hair cells, perinuclear Ca2+ oscillations were induced by Nod factor–like molecules, N-acetylglucosamine tetramers, without induction of Ca2+ influx, suggesting that the plasma membrane Ca2+ influx and the perinuclear Ca2+ spiking are two distinct responses to Nod factors (Walker et al., 2000). Furthermore, the phenotype of nonnodulating mutants that are impaired in the perinuclear Ca2+ spiking but retain the Ca2+ influx demonstrates that the two calcium responses are genetically separable (Shaw and Long, 2003; Miwa et al., 2006).
In animal cells, Ca2+-permeable channels such as ryanodine receptors and inositol triphosphate receptors (InsP3-R) localized in the nuclear envelope or ER are involved in the generation of Ca2+ oscillation around the nucleus (Gerasimenko and Gerasimenko, 2004). These channels can be activated by secondary messengers such as nicotinic acid adenine dinucleotide phosphate, 1-(5-phospho-β-d-ribosyl) adenosine 5-phosphate cyclic anhydride, inositol-1,4,5-triphosphate (IP3), and Ca2+ (Gerasimenko and Gerasimenko, 2004). IP3 is liberated from phosphatidylinositol 4,5-bisphosphate by a phosphoinositide-dependent phospholipase C (PI-PLC). The mammalian PI-PLC is primarily activated via heterotrimeric G-proteins (Ross and Higashijima, 1994; Fukami, 2002). Mastoparan, an agonist of heterotrimeric G-proteins in animal cells, activates PI-PLC and induces Ca2+ fluxes (Ross and Higashijima, 1994). Although the precise mode of action of mastoparan in plant cells is unclear (Pingret et al., 1998; Miles et al., 2004), it triggers Ca2+ spiking (Pingret et al., 1998; Peiter et al., 2007; Sun et al., 2007) as well as nodulation gene expression in M. truncatula (Pingret et al., 1998). This suggests that similar transduction pathways lead to Ca2+ oscillations in plants and animals. However, ryanodine receptors or InsP3-R have not been identified in the completely sequenced plant genomes, suggesting that alternative yet unidentified channels are involved (Nagata et al., 2004). Despite the absence of clear homologs in plants, both nicotinic acid adenine dinucleotide phosphate and 1-(5-phospho-β-d-ribosyl) adenosine 5-phosphate cyclic anhydride elicit Ca2+ release from plant ER-derived vesicles (Navazio et al., 2000, 2001). Furthermore, an inhibitor of InsP3-activated channels, 2-APB, blocks the Nod factor–induced Ca2+ spiking (Engstrom et al., 2002). Collectively, these results suggest the presence of yet-to-be identified plant Ca2+ channels activated similarly to the Ca2+ channels responsible for Ca2+ oscillations in animal cells.
In Lotus japonicus, castor and pollux mutants are unable to establish bacterial and fungal symbioses and are impaired in the perinuclear Ca2+ spiking but retain the Ca2+ influx at the root hair tip (Miwa et al., 2006), indicating that both proteins are specifically involved in mediating perinuclear Ca2+ spiking. CASTOR, POLLUX, and proteins orthologous to POLLUX in M. truncatula (DOES NOT MAKE INFECTIONS1 [DMI1]) and SYM8 in pea are sequence-related and share a short stretch of predicted structural homology to the pore region of MthK, a calcium-activated potassium channel from Methanobacterium thermoautotrophicum (Ané et al., 2004; Imaizumi-Anraku et al., 2005; Oldroyd and Downie, 2006; Edwards et al., 2007). MthK is a highly selective potassium channel, and its selectivity is defined by a conserved motif, TVGYG, in the narrowest stretch of the pore known as the filter (Heginbotham et al., 1994; Jiang et al., 2002). However, the absence of this highly conserved potassium signature sequence and the low structural homology between both CASTOR and POLLUX and MthK left the functions of the plant proteins unclear and prompted us to study the biochemical and electrophysiological properties of CASTOR and POLLUX.
RESULTS
CASTOR and POLLUX Form Homocomplexes
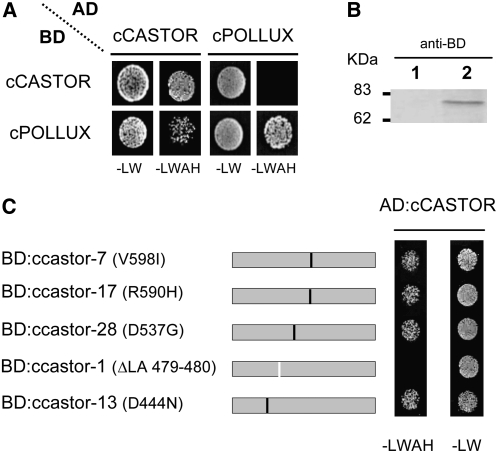
One common feature of ion channels is their organization in homotetrameric or heterotetrameric complexes of identical or similar subunits, respectively (Christie, 1995). We tested the potential of the C-terminal soluble domains of CASTOR (cCASTOR; H322 to E853) and POLLUX (cPOLLUX; H386 to D917) to form homomers or heteromers. In yeast two-hybrid assays, cCASTOR and cPOLLUX were each capable of self-interaction (Figure 1A). The interaction between POLLUX and CASTOR was only observed using cCASTOR fused to the GAL4 activator domain (AD) and cPOLLUX to the GAL4 binding domain (BD), but not when cCASTOR and cPOLLUX were fused to BD and AD, respectively (Figure 1A). Hence, the interaction between cCASTOR and cPOLLUX in yeast was only observed in one direction, an indication for a potentially false-positive interaction in this system (Bartel and Fields, 1997). To assess the specificity of the AD:cCASTOR interaction, we mutated cCASTOR according to a series of castor alleles carrying a point mutation or deletion (Imaizumi-Anraku et al., 2005). Five ccastor mutant versions, ccastor-1 (ΔLA 479-480), ccastor-7 (V598I), ccastor-13 (D444N), ccastor-17 (R590H), and ccastor-28 (D537G), were tested for cCASTOR interaction in yeast two-hybrid assays. AD:cCASTOR and BD:ccastor-1 expressed in yeast (Figure 1B) did not interact, indicating that interactions of AD:cCASTOR with either BD:cCASTOR or BD:cPOLLUX were specific (Figure 1C).
Figure 1.
Yeast Two-Hybrid Assay for Interactions of cCASTOR and cPOLLUX.
(A) Interaction assays were performed in the yeast strain AH109 cotransformed with the bait (BD) and prey (AD) vectors containing either cCASTOR (H322 to E853) or cPOLLUX (H386 to D917). The cotransformants and interacting partners were analyzed on synthetic dropout nutrient medium lacking Leu and Trp (−LW; nonselective condition) and lacking Leu, Trp, adenine, and His (−LWAH; selective condition), respectively.
(B) Expression analyses of BD:cCASTORΔLA 479-480 by immunoblot. Total extract from yeast transformed with empty vector (lane 1) and vector expressing BD:cCASTORΔLA 479-480 (lane 2).
(C) Yeast two-hybrid interaction analyses of ccastor mutant versions. All tested mutations originate from symbiosis-defective castor alleles: castor-1 (ΔLA 479-480), castor-7 (V598I), castor-13 (D444N), castor-17 (R590H) (Imaizumi-Anraku et al., 2005), and castor-28 (D537G) (Perry et al., 2003). Black lines, single amino acid changes; white line, deletion.
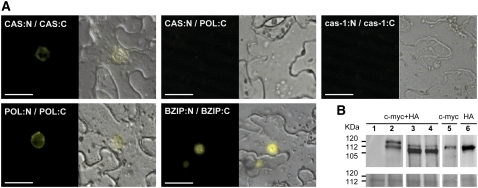
To address the question whether CASTOR and POLLUX form heterocomplexes or homocomplexes in planta, we used a bimolecular fluorescence complementation (BiFC) approach in tobacco (Nicotiana benthamiana) epidermal cells (Figure 2A). The full-length CASTOR and POLLUX were fused to either the N- or C-terminal half of the yellow fluorescent protein (YFP) gene. The expression of all fusion proteins was confirmed by immunoblots (Figure 2B). All BiFC CASTOR and POLLUX fusion constructs complemented the respective castor and pollux mutants, indicating that these fusions are functional (Table 1). In line with the yeast two-hybrid results, the full-length POLLUX and CASTOR were self-interacting. The CASTOR self-interaction was abolished by the castor-1 mutation (Figure 2A). The interaction between CASTOR and POLLUX, both tested in fusion with either the N- or C-terminal half of YFP, was never seen in tobacco epidermal cells (Figure 2A). Since the negative results for the heterocomplex formation were observed side by side with the positive results for the homocomplex, using the same constructs in the same agrobacterial suspensions and plants, we consider the lack of detectable heterocomplex formation in planta significant. Altogether, these observations suggest that, in planta, CASTOR and POLLUX form two distinct complexes for which the assembly involves the C-terminal region of the proteins.
Figure 2.
Formation of CASTOR and POLLUX Homocomplexes in Planta.
(A) BiFC analysis of CASTOR (CAS) and POLLUX (POL) interaction in transiently transformed tobacco leaf epidermal cells. CASTOR and POLLUX driven by the cauliflower mosaic virus 35S promoter were fused to the N-terminal half of YFP and a c-MYC tag (N) or to the C-terminal half of YFP and a HA tag (C). Self-interaction between CASTORΔLA 479-480 (cas-1) was used as a negative control, and the nucleus-localized transcription factor bZIP63 from Arabidopsis was used as a positive control. For each combination of constructs, 12 leaves on three plants were injected. After 2 d, 20 pieces of approximately 2 cm2 were cut out and the epidermal cell layer was peeled off and inspected. The whole experiment was repeated three times. Whereas the positive homocomplex formation was robust and reproducibly detected, an interaction between CASTOR and POLLUX was never observed. The experiment was visualized with an inverted epifluorescence microscope. Left panels, YFP fluorescence; right panels, bright-field and YFP fluorescence merged. Bars = 20 μm.
(B) Protein expression analysis by immunoblot. The protein extracts from leaves transiently transformed with empty vector (lane 1) or vectors expressing POLLUX (POL) or CASTOR (CAS), fused to either the N-terminal half of the yellow fluorescent c-MYC–tagged (N) or the C-terminal half of the yellow fluorescent HA-tagged (C) protein, were analyzed. Lane 2, POL:C and POL:N; lane 3, CAS:C and CAS:N; lane 4, cas-1:N and cas-1:C; lane 5, CAS:N and POL:C; lane 6, CAS:N and POL:C. Expected sizes: POL, 102 kD; CAS, 95 kD; N, 18 kD; C, 10 kD. Top panel, immunoblot; bottom panel, Coomassie blue–stained blot documenting protein loading.
Table 1.
Restoration of Root Nodule Symbioses in A. rhizogenes–Transformed Root Systems of L. japonicus Mutants pollux-2 and castor-12 with the Different POLLUX (POL) and CASTOR (CAS) Split YFP Versions, Respectively
| Plant Genotype | Transgene | Fraction of Nodulated Plants | Nodule/Nodulated Plants |
|---|---|---|---|
| Wild type | 35S:N | 32/49 | 4.6 |
| Wild type | 35S:C | 28/30 | 4.1 |
| pollux-2 | 35S:N | 0/89 | 0 |
| pollux-2 | 35S:POL:N | 30/57 | 4.2 |
| pollux-2 | 35S:POL:C | 43/72 | 3.8 |
| castor-12 | 35S:N | 0/64 | 0 |
| castor-12 | 35S:CAS:N | 59/85 | 3.4 |
| castor-12 | 35S:CAS:C | 54/78 | 5.1 |
The Endogenous CASTOR Protein Is Localized in the Nuclear Envelope
Using transgenic expression of green fluorescent protein (GFP) fusions, previous reports observed CASTOR, POLLUX, and DMI1 in different subcellular compartments (Imaizumi-Anraku et al., 2005; Riely et al., 2007). It has been reported previously that CASTOR:GFP and POLLUX:GFP, transiently transformed via particle bombardment and expressed under the control of the cauliflower mosaic virus 35S promoter (P35S), displayed a plastid localization pattern in onion (Allium cepa) epidermal and pea root cells (Imaizumi-Anraku et al., 2005). By contrast, M. truncatula root cells transformed by Agrobacterium rhizogenes with DMI1:GFP driven by a similar promoter resulted in fluorescence around the nucleus partially accumulating in patches of unknown identity (Riely et al., 2007). In tobacco leaf epidermal cells, the YFP fluorescence due to CASTOR or POLLUX self-interactions was clearly observed around the nucleus (Figure 2A). To further analyze the subcellular localization of CASTOR and POLLUX, we expressed GFP fusions driven either by P35S or double P35S (2xP35S) promoters. When expressed under the control of P35S, fluorescence of both constructs was observed around the nucleus, similar to what was observed with the BiFC constructs (see Supplemental Figures 1 and 2 online). However, when expressed under the control of 2xP35S, CASTOR:GFP and POLLUX:GFP were observed in plastids (see Supplemental Figure 3 online). These results demonstrate that entirely different localization patterns were detectable in transient assays.
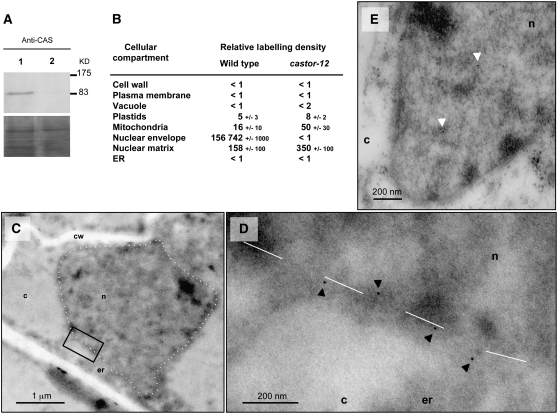
In order to assess the localization in L. japonicus root cells and to exclude possible artifacts associated with the transgenic expression of fusion proteins, we targeted the endogenous proteins using specific antibodies. Rabbit polyclonal antibodies were raised against hydrophilic peptides of CASTOR and POLLUX. In immunoblots, the purified CASTOR antibodies allowed a specific detection of CASTOR (Figure 3A), whereas the POLLUX antibody was unspecific (data not shown). The immunogold labeling with anti-CASTOR was performed on wild-type L. japonicus and on castor-12, a mutant carrying a single point mutation leading to a premature stop codon at position W93 (Imaizumi-Anraku et al., 2005). By electron microscopic analysis of immunogold-labeled sections, a strong and specific accumulation of gold particles was observed at the position of the nuclear envelope of the wild type. This immunodecoration was absent in samples of the castor-12 mutant, a result that demonstrates the specificity of the labeling (Figures 3B to 3E). Membrane structures were not well preserved due to the embedding method used. Therefore, we could not determine whether CASTOR resides in the inner or outer membrane of the nuclear envelope or in both. Only nonspecific background labeling was observed in plastids in both the wild type and castor-12 (Figure 3B). These data indicate a localization of the endogenous CASTOR protein in the nuclear envelope.
Figure 3.
Immunogold Localization of CASTOR in L. japonicus with Anti-CASTOR.
(A) Immunoblot detection with anti-CASTOR in total extract of transformed roots of L. japonicus mutant castor-12 (W93*). Lane 1, castor-12 complemented with P35S:CASTOR via A. rhizogenes hairy root transformation; lane 2, castor-12 transformed roots with empty vector as a negative control. Bottom panel, loading shown by Coomassie blue staining.
(B) Relative density of gold particles in different cellular compartments of L. japonicus Gifu wild-type and mutant castor-12 roots after immunogold labeling with anti-CASTOR.
(C) to (E) Electron microscopic images of L. japonicus Gifu wild-type nucleus (C) and castor-12 nucleus (E). (D) shows an enlargement of the black box in (C). Dashed white line, delimitation of the nuclear envelopes. c, cytoplasm; cw, cell wall; er, endoplasmic reticulum; n, nucleus. Black arrowheads mark positive immunodetection of CASTOR, and white arrowheads mark background labeling in the nuclear matrix.
CASTOR Is an Ion Channel
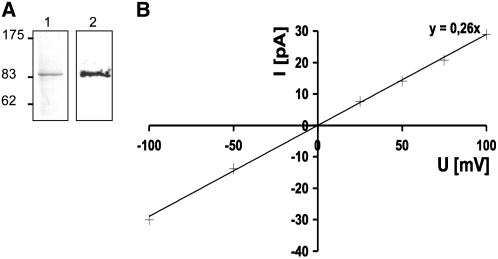
To determine whether CASTOR is an ion channel, we expressed CASTOR using an in vitro transcription–translation system (see Supplemental Figure 4 online). The protein was purified by selective solubilization, denatured, and subsequently reconstituted into liposomes by dialysis (Figure 4A). Proteoliposomes were then fused into planar lipid bilayers (Morera et al., 2007). The modeling of the CASTOR filter based on the structural homology with MthK predicted a decoration of the pore with carbonyl oxygen atoms from the peptide backbone, suggesting a selectivity for cations (Imaizumi-Anraku et al., 2005). To test this prediction experimentally, we checked the permeability of CASTOR for physiologically relevant cations over anions. In asymmetrical KCl or NaCl solutions of 250 and 20 mM, we recorded the reversal potential of CASTOR, which corresponds to the membrane potential at which the net flow of ions from one reservoir to the other is zero (Table 2; see Supplemental Figure 5 online). The calculation of the permeability ratio was based on the Goldman-Hodgkin-Katz current equation (Goldman, 1943; Hodgkin and Katz, 1949). The permeability ratios PK+/PCl− of 6.0, PNa+/PCl− of 1.6, and PCa2+/PCl− of 0.5 indicate a slight preference of CASTOR for potassium (Table 2). To test whether the preference for potassium was influenced by a conformational change of the filter due to the presence of only one cation in excess (Valiyaveetil et al., 2006), we performed competition assays between K+, Ca2+, and Na+ (Table 3). Solutions of K+ and Ca2+ or K+ and Na+ were applied to the cis or trans side of the planar lipid bilayer, respectively. The permeability ratios PK+/PNa+ and PK+/PCa2+(Table 2) indicated a moderate preference for K+ over Na+ and Ca2+.
Figure 4.
Potassium Conductivity of CASTOR.
(A) Purified CASTOR. Lane 1, Coomassie blue staining with 4 μg of purified CASTOR by selective solubilization; lane 2, immunoblot with anti-CASTOR using 2 μg of solubilized protein.
(B) Conductance of CASTOR at different voltages in the presence of potassium. The experiment was done with symmetrical 250 mM KCl buffer in each reservoir. The conductance was recorded at different voltages; crosses represent individual data points. The slope of the linear regression curve (y = 0.26x) indicated a conductance of 260 pS.
Table 2.
Reversal Potential (Erev) and Permeability Ratios (PX/PCl−) for CASTOR and castor-2
| CASTOR
|
castor-2
|
|||
|---|---|---|---|---|
| Ion (X) | Erev (mV) | PX/PCl− | Erev (mV) | PX/PCl− |
| K+ | 36 ± 2 (n = 40) | 6.0a | 28 ± 2 (n = 48) | 3.7a |
| Na+ | 11 ± 1 (n = 8) | 1.6a | 25 ± 3 (n = 13) | 3.2a |
| Ca2+ | −29 ± 2 (n = 13) | 0.5b | −32 ± 2 (n = 19) | 0.6b |
Permeability ratios calculated from reversal potentials using equation Erev = RT/zF × ln(PK+[K]o/PNa+[Na]i).
Permeability ratios calculated from reversal potentials using equation Erev= RT/F × ln(√((4PCa2+[Ca]o/PK+[K]i)+1/4) − 1/2), where RT/F = 25.26 mV and Z = 1. The junction potential correction was as follows: NaCl (250 mM/20 mM, agar bridge, 2 M KCl), Vm = Vcmd + 2.3 mV; KCl (250 mM/20 mM, agar bridge, 2 M KCl), Vm = Vcmd + 1.2 mV; CaCl2 (125 mM/10 mM, agar bridge, 2 M KCl), Vm = Vcmd + 3.1 mV.
Table 3.
Reversal Potential (Erev) and Permeability Ratios (PK+/PX) for CASTOR
Permeability ratios calculated from reversal potentials using equation Erev = RT/zF × ln(PK+[K]o/PNa+[Na]i).
Permeability ratios calculated from reversal potentials using equation Erev= RT/F × ln(√((4PCa2+[Ca]o/PK+[K]i)+1/4) − 1/2), where RT/F = 25.26 mV and Z = 1. The junction potential correction was as follows: KCl-NaCl, Vm = V − 1.1 mV; KCl-CaCl2, Vm = V − 2.3 mV.
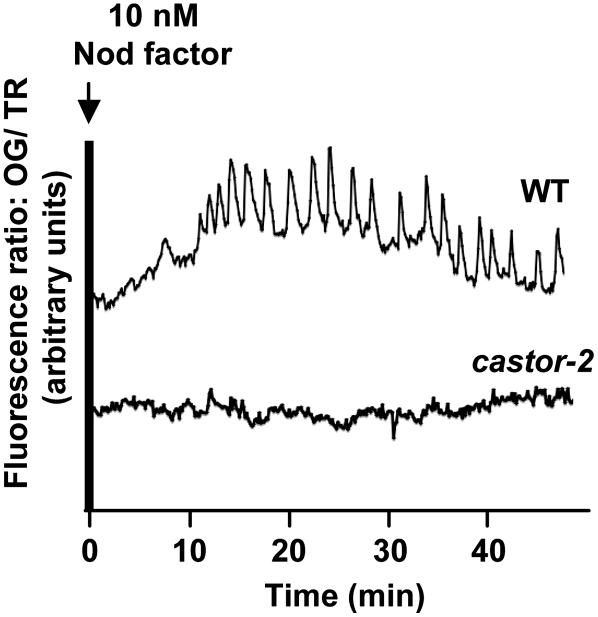
The castor-2 allele carries a single nucleotide replacement that leads to the substitution of Ala by Thr at position 264 (Imaizumi-Anraku et al., 2005). The castor-2 mutant is defective in Ca2+ spiking (Figure 5) and both root nodule and arbuscular mycorrhiza symbioses (Imaizumi-Anraku et al., 2005). The mutated Ala is positioned at the hinge of the selectivity filter, which could alter the selectivity of the channel (see Supplemental Figure 6 online). To test this hypothesis, we determined the permeability ratio of castor-2 for K+ and Na+ (Table 2). We observed that castor-2 was equally permeable to Na+ and K+ (Table 2). This result indicates that the A264T mutation alters the selectivity of the pore. The altered channel behavior of castor-2 is an important confirmation that the observed channel activities are indeed due to the reconstituted CASTOR protein.
Figure 5.
Calcium Spiking in Root Hairs of L. japonicus Ecotype Gifu Wild Type and the castor-2 Mutant.
Traces represent the fluorescence ratio between Oregon Green (OG; calcium-sensitive) and Texas Red (TR; reference) over time following application of 10 nM Nod factor to intact roots. Calcium spiking was observed in 22 of 28 wild-type root hair cells and in 0 of 23 castor-2 root hair cells.
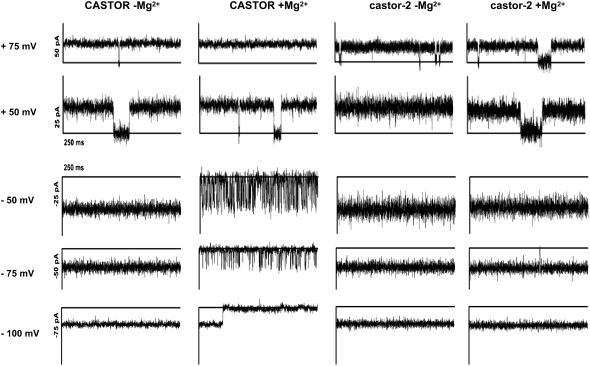
Voltage-Dependent Magnesium Blockage of CASTOR Is Absent in the castor-2 Mutant
In response to stimuli, ion channels can switch between closed- and open-state conformations, thereby acting as regulators of ion fluxes in cells. To explore the gating behavior of CASTOR, we applied different stimuli in order to influence its opening and closing. Voltage gating was evaluated by modulating the membrane potential from 100 to −100 mV in a solution of 250 mM KCl on both sides of the lipid bilayer. The current–voltage relation reveals that the channel was active over the large voltage range from −100 to 100 mV without any rectification of the current (Figure 4B). In the absence of magnesium, the current–voltage relationship of CASTOR was linear over this voltage range with a conductance of 260 pS (Figure 4B), which is of the same range as other well-characterized channels such as MthK (Jiang et al., 2002). We further tested the influence of divalent cations such as magnesium on the gating behavior of CASTOR. At positive voltage, random closing steps of CASTOR and castor-2 were recorded, attesting to the presence of the channels (Figure 6). At positive voltages, no differences were observed between CASTOR and castor-2 in the presence or absence of 3 mM MgCl2 on the cis side. However, at negative voltages of −50 and −75 mV, and specifically in the presence of MgCl2, an increased closing/opening frequency of CASTOR was observed (see Supplemental Figure 7 online), while no changes were observed for castor-2 (Figure 6). At −100 mV, CASTOR closed completely, but this closure was reversible by increasing the voltage (data not shown). By contrast, castor-2 did not show this magnesium-dependent closing behavior and remained open throughout the voltage range. Only random opening and closing steps were observed. The differences in gating observed between the mutant and the wild type channels confirms again that the observed currents are indeed a function of the reconstituted plant channels. Moreover the behavior of castor-2 indicates that the A264T mutation modifies the pore conformation in such a way that Mg2+-dependent blockage is no longer occurring.
Figure 6.
Voltage-Dependent Magnesium Blockage of CASTOR.
CASTOR and castor-2 channel currents were recorded at the voltages indicated at left in the presence and absence of 3 mM MgCl2. In the presence of MgCl2, high-frequency opening/closing steps of CASTOR were observed at negative voltages of −50 and −75 mV. At −100 mV, CASTOR closed completely, while castor-2 remained open.
Because InsP3 and Ca2+ are discussed as likely modulators of Ca2+ spiking, we analyzed the sensitivity of CASTOR gating to calcium and InsP3 at a holding potential from 100 to −100 mV in a recording solution containing 250 mM KCl. However, neither InsP3 nor Ca2+ influenced CASTOR gating at the concentrations tested (see Supplemental Figure 8 online).
POLLUX Complementation of a Yeast Potassium Import and Export Mutant
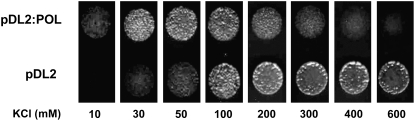
CASTOR and POLLUX share identical filter sequences, suggesting similar ion selectivity. Because the expression of POLLUX in the cell-free system was unsuccessful (see Supplemental Figure 4 online), we tested if POLLUX could complement a yeast mutant, MAB 2d, in which the main potassium importers and exporters are deleted (trk1Δ trk2Δ ena1-4Δ nha1Δ) (Rodriguez-Navarro, 2000; Kolacna et al., 2005; Maresova and Sychrova, 2005). The deletion of both the potassium efflux and influx systems enables the MAB 2d strain to grow on acidic minimal medium supplied with otherwise toxic potassium concentrations of 600 mM (Kolacna et al., 2005). A second aspect of the potassium uptake–deficient phenotype is the inability of the mutant to grow at potassium concentrations from 10 to 50 mM (Kolacna et al., 2005). The expression of POLLUX under the regulation of the Saccharomyces cerevisiae alcohol dehydrogenase promoter in the strain MAB 2d impaired the growth of the mutant at potassium concentrations between 300 and 600 mM KCl and promoted the growth on medium containing 10 to 50 mM KCl (Figure 7). These results demonstrate that in MAB 2d, the expression of POLLUX mediates an influx of potassium. Considering the sequence identity within the selectivity filter and strong overall similarity with the CASTOR ion channel, we conclude that POLLUX is also an ion channel with permeability for potassium.
Figure 7.
Complementation of MAB 2d.
Growth of MAB 2d cells transformed with pDL2:POLLUX (POL) under the control of the S. cerevisiae alcohol dehydrogenase 1 promoter or the empty vector (pDL2) at the indicated KCl concentrations on minimal medium, pH 4. Twenty microliters of a suspension of transformed yeast cells in water (OD600 = 0.06) was dropped on each medium. The colony growth was recorded after incubation for 48 h at 30°C.
P35S:POLLUX Can Suppress the castor-2 Nodulation Phenotype
Together, the electrophysiological and yeast complementation experiments provide evidence that CASTOR and POLLUX are ion channels. Both channels are constitutively expressed in L. japonicus roots, flowers, and leaves (Imaizumi-Anraku et al., 2005). To determine the spatial expression pattern of CASTOR and POLLUX in L. japonicus roots, we used the β-glucuronidase (GUS) reporter gene fused with the 5′ upstream region of CASTOR and POLLUX. In A. rhizogenes–transformed roots expressing the GUS reporter gene driven by either CASTOR or POLLUX promoters, the patterns of GUS activity were similar in all roots. Staining was strongest at the growing tips (see Supplemental Figure 9 online), and no differences in the expression of either construct were observed before and after inoculation with the endosymbiotic bacterium Mesorhizobium loti.
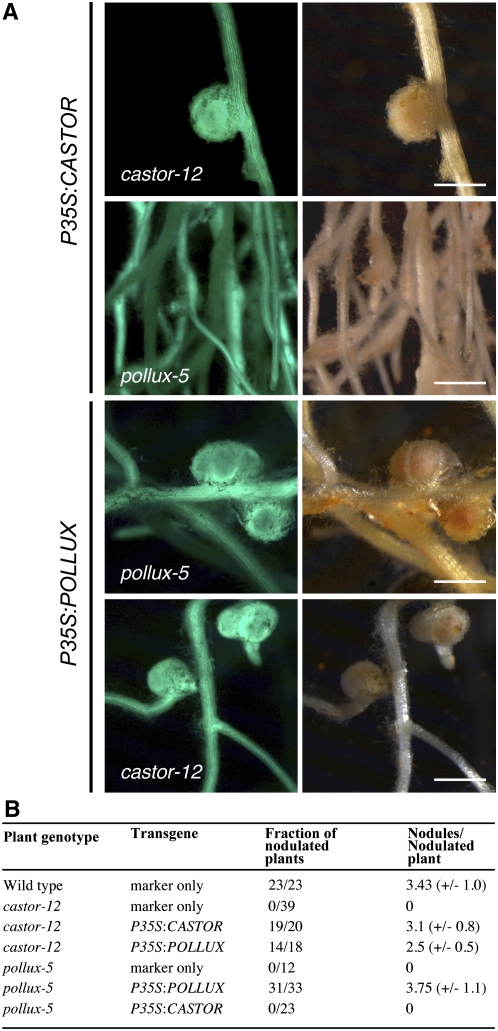
The similar expression and localization patterns and identical ion selectivity filter sequences of CASTOR and POLLUX suggest that they have similar functions. However, mutations in either castor or pollux alone lead to symbiosis deficiency. It is possible, therefore, that expression levels are limiting and that a certain minimum amount of CASTOR and/or POLLUX is required for nodulation. In order to test this, we expressed CASTOR and POLLUX under the control of a single P35S via A. rhizogenes transformation in castor-12 and pollux-5 hairy roots. The pollux-5 allele contains a nonsense mutation, W321*, leading to a truncated protein with three transmembrane domains, while castor-12 (W93*) does not retain any (Imaizumi-Anraku et al., 2005). The P35S:CASTOR construct complemented the nodulation deficiency in castor-12 but not in pollux-5 (Figure 8). However, the P35S:POLLUX construct restored the ability to nodulate in both pollux-5 and castor-12 (Figure 8). This result demonstrates that POLLUX can compensate for the loss of CASTOR when expressed under the control of P35S, which suggests that CASTOR and POLLUX have similar functions.
Figure 8.
Restoration of Root Nodule Symbiosis in A. rhizogenes–Transformed Root Systems of L. japonicus Mutants.
(A) Images of transgenic hairy roots 3 weeks after inoculation with M. loti R7A. The T-DNA carried GFP as a marker gene. Left panel, GFP fluorescence; right panel, bright field. Bars = 1 mm.
(B) Nodulation of transgenic roots. The fraction of nodulated plants and the number of nodules per nodulated plant were determined 3 weeks after inoculation with the M. loti R7A strain. sd values are indicated in parentheses.
DISCUSSION
Nod factor perception triggers perinuclear Ca2+ oscillations in legume root epidermal cells (Ehrhardt et al., 1996). CASTOR and POLLUX are required for Nod factor–induced Ca2+ spiking (Miwa et al., 2006). In this work, we provided evidence that CASTOR and POLLUX are ion channels. Furthermore, we showed that the endogenous CASTOR protein is localized in the nuclear envelope. These results provide important clues about the function of CASTOR and POLLUX in the Ca2+-spiking machinery.
Our electrophysiological measurements on individual CASTOR channels reconstituted in planar lipid bilayers demonstrated that CASTOR has ion channel activity. This is important confirmation of the previous functional prediction, which was solely based on sequence analysis. With a selectivity ratio of ∼6:1 for PK/PCl, CASTOR displays a relatively weak preference for cations over anions. In competition assays confronting K+ with Na+ or K+ with Ca2+, permeability ratios of K+ over Na+ and Ca2+ were only 2 and 1.4, respectively (Table 3), indicating a relatively low preference for potassium. Importantly, these data revealed a calcium permeability of CASTOR, and it is at least formally possible that CASTOR is directly responsible for the calcium release during spiking. However, although weak, CASTOR consistently showed a preference for potassium, which, in combination with the relative cellular concentrations of potassium and calcium, makes potassium the more likely substrate in planta. The complementation of the potassium growth phenotype of the yeast strain MAB 2d provides evidence that POLLUX is a potassium-permeable. However, POLLUX is likely to be permeable also for additional ions, because the identical selectivity filter sequences of CASTOR and POLLUX make similar if not identical ion selectivities of the two channels very likely. In planta, the restoration of nodulation of a castor null allele by P35S:POLLUX demonstrates that POLLUX is able to compensate for the absence of CASTOR when overexpressed. Altogether, these data support a similar biochemical function of CASTOR and POLLUX. However, additional experiments will be required to determine whether subtle differences in subcellular localization, ion permeability, or regulation of channel activity are responsible for the inability of CASTOR to compensate for the lack of POLLUX.
To explore possible opening and closing mechanisms of CASTOR, we tested magnesium, which is known to be required for gating of some cation channels. At negative voltage and in the presence of Mg2+, a blockage of CASTOR conductance was observed. This behavior is reminiscent of voltage-dependent divalent cation blockage, a phenomenon that has been studied in detail in potassium and cation channels (Armstrong et al., 1982; Stern et al., 1987; Lu and MacKinnon, 1994; Zhang et al., 2006). Different mechanisms have been observed by which divalent cations block ion channels. A ring of negatively charged amino acids in the conduction pathway formed by the inner helices is involved in this electrostatic mechanism in a subset of potassium channels (Zhang et al., 2006). By contrast, in cation channels, the external site of the selectivity filter can bind divalent cations that serve to block monovalent cation conduction (Armstrong et al., 1982; Stern et al., 1987). The absence of blocking in castor-2 revealed that the A264T mutation modifies the structure of the pore and filter in such a way that Mg2+ blockage can no longer occur. A structural modification of the pore of castor-2 is further supported by the equal selectivity to K+ and Na+. Our data suggest that the phenomenon of voltage-dependent Mg2+ blockage also occurs in CASTOR. The absence of this blocking mechanism in castor-2 potentially leads to ion fluxes that are inappropriate to support Ca2+ spiking. Magnesium is constitutively present in millimolar concentrations within the cytoplasm and nucleoplasm (Allen and Sanders, 1996). The Mg2+ blockage mechanism is likely to be involved in keeping the channels closed in a nonstimulated ground state.
CASTOR and POLLUX each has a large C-terminal domain hypothetically involved in regulating the channels' conductance. It is possible, therefore, that allosteric effectors may regulate the opening of the channels. We tested candidate effectors proposed to be involved in the signaling pathway leading from Nod factor perception to Ca2+ spiking. As in M. truncatula, pharmacological studies implicated phospholipase D and PLC upstream of Ca2+ spiking (Engstrom et al., 2002; den Hartog et al., 2003), and phospholipid-derived signaling compounds such as IP3 may be directly or indirectly involved in triggering the Ca2+ oscillation. Our experiment did not reveal a direct influence of IP3 or Ca2+ on CASTOR gating, suggesting that if CASTOR is a ligand-activated channel, other compounds are involved in its gating mechanism.
Because Ca2+ spiking occurs in both the nucleoplasm and the nucleus-associated cytoplasm, the ER and nuclear envelope complex have been proposed to serve as the corresponding Ca2+ stores (Oldroyd and Downie, 2006). The immunogold labeling of endogenous CASTOR in L. japonicus clearly demonstrates the localization of CASTOR in the nuclear envelope. Although immunolocalization data for POLLUX are not available due to the lack of a specific antibody, accumulating evidence makes a localization of POLLUX and the Medicago ortholog DMI1 in the nuclear envelope very likely. Fluorescence from a complementing DMI1:GFP fusion was observed around the nucleus of Medicago root cells (Riely et al., 2007). We observed perinuclear localization of CASTOR and POLLUX in the BiFC experiment using constructs that complemented corresponding Lotus mutants. Moreover, the suppression of the castor-12 nodulation phenotype by P35S:POLLUX suggests a similar localization for both proteins. WoLF PSORT (Horton et al., 2007) analyses predicted three and one nuclear localization signals in POLLUX and CASTOR, respectively. However, the difference between CASTOR and POLLUX in numbers and sequences of predicted nuclear localization signals may reflect subtle differences in subcellular localization. The inability of P35S:CASTOR to restore nodulation of the pollux-5 mutant could be explained by a possible distinct targeting. In a hypothetical scenario, CASTOR resides in the outer nuclear envelope while POLLUX is required in both the inner and the outer nuclear membranes. However, higher resolution localization data for CASTOR and POLLUX will be required to test this hypothesis.
Previously, the localization of CASTOR and POLLUX, predicted by TargetP to carry a chloroplast transit peptide, has been reported in plastids of onion and pea root cells (Imaizumi-Anraku et al., 2005). In this study, we made contradictory observations when expressing CASTOR and POLLUX GFP fusion constructs in tobacco epidermal cells. Using the same Agrobacterium-based transformation system of the same cell type, we could observe either plastid localization or localization in the nuclear envelope. Although there was a correlation between the promoter and the resulting localization (nucleus when expressed under the control of P35S, plastid when driven by 2xP35S), it is difficult to rationalize these observations, since both promoters result in strong overexpression. However, these results and the predicted localization signals (both proteins carry nuclear localization signals in addition to chloroplast transit peptides) suggest that CASTOR and POLLUX have the potential to be targeted to either the plastid or the nucleus. Such exotic protein targeting is not without precedent. A protein initially targeted to the ER was found to be subsequently delivered to the plastid (Villarejo et al., 2005), but it is unclear from our study whether CASTOR and POLLUX are localized to the plastid after being targeted to the secretory pathway or delivered directly to the plastid when highly expressed in heterologous cells. Whether or not the endogenous CASTOR and POLLUX proteins also experience such differential localization under specific conditions or in specific cell types remains to be established.
Collectively, our results demonstrate that CASTOR and POLLUX are ion channels permeable to cations such as potassium. The mutant phenotype and nuclear localization of CASTOR and POLLUX suggest that they may be part of the Ca2+-spiking machinery. Considering that potassium is the cation with the highest concentration within the cell, together with the permeability of CASTOR and POLLUX for potassium, we conclude that potassium is likely to be the major ion going through these cation channels in vivo. In analogy to the role of potassium transporters in calcium signature formation in animal systems, there are at least two possible mechanisms by which CASTOR and POLLUX could contribute to Ca2+ spiking, and these are not mutually exclusive. The high conductance of CASTOR is similar to that of the potassium channels acting as counter-ion channels balancing the calcium charge released from the sarcoplasmic reticulum (Wang and Best, 1994). CASTOR and POLLUX may act as counter-ion channels facilitating an influx of potassium, which compensates for the rapid release of positive charge from the calcium store during each spike. This direction of ion flow into the nuclear envelope cavity would predict an orientation of the pore in such a way that the C-terminal portion is exposed to the outside of the nuclear envelope membranes. Since potassium contributes significantly to endomembrane potentials, an opening of CASTOR and POLLUX is likely to influence the potential of the membrane they reside in. In mammals, the membrane potential influences the activation of calcium-induced calcium-release channels (Stehno-Bittel et al., 1995). Therefore, the activation of CASTOR and POLLUX could affect the nuclear membrane potential, leading to an activation of Ca2+ channels on the same membrane system. In either scenario, the calcium store would subsequently be replenished by the activity of calcium ATPases. In previous studies, cyclopiazonic acid, an inhibitor of type IIa calcium-ATPase, inhibited Nod factor–induced Ca2+ spiking (Engstrom et al., 2002), suggesting their involvement in the generation of Ca2+ spiking.
Here, we show that nodulation (and, by inference, Ca2+ spiking) can be restored in a castor mutant by overexpressing POLLUX. These data suggest that quantitative gene expression levels are critical in determining the channel mutant phenotypes. Mastoparan was observed to induce Ca2+ spiking in M. truncatula wild-type as well as in dmi1 mutant root hairs (Sun et al., 2007). Taken at face value, this result made DMI1 (and, by analogy, POLLUX) an unlikely component of the Ca2+-spiking machinery. However, mastoparan treatment appears to be a much stronger stimulus than Nod factor treatment, because it leads to a massive activation of calcium release even at ectopic sites away from the nucleus (Peiter et al., 2007). By interrogation of public sequence databases, we identified an EST sequence likely representing the CASTOR ortholog of M. truncatula, Mt CASTOR (GenBank accession number CX525932). It is possible, therefore, that the mastoparan-induced Ca2+ spiking in the absence of DMI1 works via an overactivated Mt CASTOR sufficient to compensate for the loss of DMI1. Furthermore, the dominant negative dmi1-2 allele encoding a stop codon after the fourth transmembrane domain was found to suppress the mastoparan-induced Ca2+ spiking (Peiter et al., 2007). The remaining N-terminal portion in DMI1-2 retains the entire pore region, and its dominant effect could potentially result from the induction of inappropriate ion fluxes. In order to determine the relevance of the twin channels in mastoparan-induced Ca2+ spiking, a double mutant, dmi1/castor, will have to be tested.
CASTOR and POLLUX are the founder members of a novel cation channel family. Sequence-related genes to CASTOR and POLLUX are widespread across higher plants (Ané et al., 2004; Imaizumi-Anraku et al., 2005; Gutjahr et al., 2008). The presence of members in Arabidopsis thaliana, an asymbiotic species (Imaizumi-Anraku et al., 2005), suggests additional roles of this class of potassium-permeable channels. Ca2+ oscillations are involved in the regulation of different biological processes such as pollen tube growth and guard cell turgor (McAinsh et al., 1995; Franklin-Tong et al., 1996). Interestingly, guard cells can integrate information from multiple stimuli, such as abscisic acid, external K+, or Ca2+, to mount appropriate Ca2+ signatures that lead to stomatal closure (McAinsh et al., 1995; Leckie et al., 1998). It will be interesting to determine whether the Arabidopsis channels related to CASTOR and POLLUX are involved in the generation of Ca2+ oscillations in guard cells.
METHODS
Yeast-Two-Hybrid Interaction Assays
The following primer combinations were used to amplify cCASTOR, cPOLLUX, and the mutated versions of cCASTOR (all primer sequences are shown in Supplemental Table 1 online): cCASTOR (P5/P2) and cPOLLUX (P6/P4) to generate Gateway-compatible inserts. The mutations were generated by site-directed mutagenesis as described (Horton et al., 1989), using the primer pairs (forward and reverse) P7/P8 for ccastor-1, P9/P10 for ccastor-7, P11/P12 for ccastor-13, P13/P14 for ccastor-17, and P15/P16 for ccastor-28. The PCR fragments were cloned into the Gateway entry vector pENTR/D-TOPO (Invitrogen). The original pBD-GAL4 Cam bait and pAD-GAL4 prey vectors (Clontech) were converted into appropriate Gateway-compatible destination pBD-GAL4-GW and pAD-GAL4-GW vectors following the instructions of the manufacturer and subsequently used as destination vectors to generate prey and bait cCASTOR, cPOLLUX, ccastor-1, ccastor-7, ccastor-13, ccastor-17, and ccastor-28 plasmids. Interaction analyses were performed in the yeast strain AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2∷GAL1UAS-GAL1 TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, ura3∷MEL1UAS-MEL1TATA-lacZ) according to the user manual (Yeast Protocols Handbook, PT3024-1; Clontech). AH109 was transformed using the lithium acetate method (Gietz and Woods, 2001). Transformants were screened on SD medium containing 0.67% yeast nitrogen base, 2% glucose, and lacking appropriate nutrients. The crude yeast protein extraction was performed as described in the user manual (DUALmembrane kit 3, P01001; Dualsystems Biotech). The protein expression levels were monitored by immunoblotting using mouse monoclonal anti-BD (Clontech) at a dilution of 1:1000.
BiFC Analyses, CASTOR and POLLUX Localization in Tobacco, Protein Extraction, and Immunoblot Analysis
castor-1 full-length cDNA was generated by site-directed mutagenesis according to Horton et al. (1989) using primers P1, P7, P8, and P2 (see Supplemental Table 1 online). castor-1, CASTOR, and POLLUX coding sequences were cloned into the Gateway-compatible destination vectors pSPYNE 35S-GW and pSPYCE 35S-GW (Walter et al., 2004), pK7FWG2 (P35S) (Karimi et al., 2002), and pAMPAT-MCS (2xP35S) (GenBank accession number AY436765) to generate CASTOR:GFP and POLLUX:GFP fusion proteins. To generate the plastid-localized GFP control, the transit peptide of spinach (Spinacia oleracea) ferredoxin NADP(H) oxidoreductase (Lohse et al., 2005) was amplified with forward and reverse primers P19 and P20 from clone pBIN:FNR (kindly provided by T. Fester), subcloned into the Gateway entry vector pENTR/D-TOPO (Invitrogen), and subsequently cloned into the pK7FWG2 vector (Karimi et al., 2002). BiFC and the GFP fusion protein localization were performed in 5-week-old tobacco (Nicotiana benthamiana) plants following Agrobacterium tumefaciens GV3101pMP90RK (Koncz and Schell, 1986) or Agl1 strain–mediated transient transformation, respectively, as described (Lazo et al., 1991; Walter et al., 2004). Total crude protein from 2 cm2 of transformed leaves was extracted as described (Walter et al., 2004). Fifty-microgram protein samples per well were resolved by 6% SDS-PAGE. Mouse monoclonal anti-c-MYC (Roche), anti-hemagglutinin (HA; Roche), or anti-GFP (Roche) at a dilution of 1:5000 was used for detection.
Complementation of L. japonicus Mutant Root Systems via Agrobacterium rhizogenes Transformation
The Agrobacterium rhizogenes strain AR1193 (Stougaard et al., 1987) was used to transform Lotus japonicus (ecotype Gifu) wild type and mutants. Seeds were sterilized as described previously (Hansen et al., 1989) and incubated at 4°C overnight, followed by germination on B5 medium (Gamborg et al., 1968) in a growth chamber (3 d of dark, 3 d of short light, 70% humidity, 24°C). The emerging roots were cut, and hypocotyls were dipped in AR1193 carrying the relevant plasmids (OD600 = 1). Transformed plants were grown on B5 medium (containing 40 μg/mL kanamycin for plants transformed with the BiFC construct) in a growth chamber (2 d of dark, 3 d of short light) and then transferred to B5 medium containing cefotaxime (300 mg/mL). Shoots with developed hairy roots were dipped into a dilution of a Mesorhizobium loti strain R7A culture (OD600 = 0.01) and transplanted into sterile pots containing 300 mL of expanded clay particles (Seramis) and 100 mL of Fahraeus medium (Fahraeus, 1957). Roots were scored for nodulation 3 weeks after inoculation.
Localization of CASTOR
CASTOR and POLLUX peptide antibodies were raised in rabbits against the peptides H2N-SSPSQYGRRFHTNSNT-CONH2 and H2N-CRRGSLPKDFVYPKSP-CONH2 for CASTOR and H2N-TTRKRRPSSVKPPST-CONH2 and H2N-GFFPRIPDAPKYPEK-CONH2 for POLLUX. One-week-old seedlings were used for immunogold electron microscopy. Pieces of roots were fixed immediately after collection with 2.5% formaldehyde in 75 mM sodium cacodylate and 2 mM MgCl2, pH 7.0, for 2 h at room temperature. The tissue was washed four times at room temperature with 1% Gly and 2.5% formaldehyde in 75 mM sodium cacodylate and 2 mM MgCl2, pH 7.0. After two washing steps in distilled water, the tissue pieces were dehydrated with a graded series of acetone (20 to 100%). Tissue samples were then infiltrated and embedded in Spurr's low-viscosity resin. After polymerization, sections with a thickness of 70 to 90 nm were cut with a diamond knife and mounted on collodion-coated nickel grids. Ultrathin sections were incubated first in 50 mM Gly in PBS buffer (136 mM NaCl, 2 mM KCl, 4 mM Na2HPO4, and 1 mM KH2PO4, pH 7.4) for 20 min and then in blocking buffer (0.5% BSA, 0.01% Tween 20, and 0.05% gelatin, in PBS buffer) for 1 h. Sections were incubated with purified anti-CASTOR serum (Eurogentec) in blocking buffer (diluted 1:20 to 1:100) overnight at 4°C. After six washing steps with blocking buffer, anti-rabbit IgG conjugated with 5-nm gold (Sigma-Aldrich) was added at a dilution of 1:20 (with blocking buffer) and incubated for 90 min at room temperature. After three washings with blocking buffer, 1% Gly in PBS buffer, and distilled water, sections were used either unstained or poststained with aqueous lead citrate (100 mM, pH 13.0). Micrographs were taken with a Zeiss EM 912 electron microscope equipped with an integrated OMEGA energy filter operated in the zero loss mode. Gold particles were counted on 15 micrographs (magnification ×40,000) as described (Lucocq, 1994). For the nuclear envelope, the covered area was a 5-mm-wide strip around the nucleus. The surface of each organelle was calculated and normalized as a percentage of the cell. The density of gold particles was calculated by dividing the number of gold particles in each compartment by their normalized surface (in percentage).
In Vitro Expression and Purification of CASTOR
CASTOR, castor-2, and POLLUX coding sequences were subcloned into the pDEST17 vector (Invitrogen). castor-2 full-length sequence was generated by site-directed mutagenesis according to Horton et al. (1989) using primers P1, P17, P18, and P2 (see Supplemental Table 1 online). To test the expression of CASTOR and POLLUX in a cell-free system, a rapid expression screen was performed with the RTS 100 Escherichia coli HY kit (Roche) according to the manufacturer's instructions. To scale up the protein amount, CASTOR and castor-2 were expressed using the RTS 500 ProteoMaster E. coli HY Kit (Roche) according to the manufacturer's instructions. The solubilization and purification of proteins were performed by selective solubilization. The pellets of the cell-free reaction containing CASTOR or castor-2 were washed twice in 1 mL of washing buffer (100 mM NaH2PO4 and 10 mM Tris, pH 7.2) and centrifuged for 5 min at 5000g. To remove E. coli proteins, the pellet was suspended in E. coli protein solubilization buffer (100 mM NaH2PO4, 2% MEGA-9, and 10 mM Tris, pH 7.2) for 45 min at 40°C under shaking and centrifuged for 30 min at 50,000g. This step was repeated twice. Protein was solubilized in 100 mM NaH2PO4, 0.05% SDS, and 10 mM Tris, pH 7.2, for 2 h at room temperature with shaking. Insoluble proteins were precipitated by centrifugation for 30 min at 50,000g. The production of soluble CASTOR and castor-2 was checked by SDS-PAGE (Coomassie Brilliant Blue staining) and immunoblot analysis using rabbit polyclonal anti-CASTOR (Eurogentec) used at a dilution of 1:200.
Reconstitution of CASTOR in Proteoliposomes and Electrophysiological Measurements
Purified CASTOR proteins were dialyzed against 100 mM NaH2PO4, 10 mM Tris, and 8 M urea, pH 7.2, for 2 h at room temperature. To remove any remaining SDS, the dialyzed proteins were incubated with Serdolit PAD I beads (Serva) for 3 h. The protein concentrations were determined by Bradford assays (Bio-Rad). The reconstitution of proteoliposomes and electrophysiological measurements were performed according to Ertel et al. (2005) with the following modification: after formation of a stable lipid bilayer, the solution of the cis chamber was exchanged with 250 mM KCl and 10 mM MOPS/Tris, pH 7.0. The recording of the reversal potential was done in a 10 mM MOPS/Tris, pH 7.0, solution containing in the cis/trans chambers 250/20 mM KCl, 250/20 mM NaCl, or 125/10 mM CaCl2. The conductance and current properties were determined in equal solutions of 250 mM KCl and 10 mM MOPS/Tris, pH 7.0. The background noise of the bilayer was around 8 pS.
Homology Modeling
Modeling of CASTOR and castor-1 pores based on the x-ray structure of MthK was done using the protein-modeling program DeepView Swiss-PdbViewer (http://www.expasy.org/spdbv/).
Microinjection and Calcium Imaging
Calcium imaging was performed on a Leica inverted microscope (DMI6000B) equipped with a 40× dry objective (numerical aperture 0.6). Seedlings of L. japonicus were grown in a phytochamber at 24°C in the dark for 2 d and bathed in BNM medium (Ehrhardt et al., 1996) for microinjection. Injection needles were made from capillary tubes (GB150F-8P; Science Product) with a puller (P-97; Sutter Instruments) and manipulated with a micromanipulator (PS-7000C; Scientifica). The calcium indicator dye, Oregon Green 488 BAPTA-1 dextran MW10,000 (Invitrogen), and the reference dye, Texas Red dextran MW10,000 (Invitrogen), were injected by iontophoresis using a bridge amplifier equipped with a timer module (BRAMP-01R, TMR-01; NPI Electronic). The injected root hairs were treated with 10 nM Nod factor prepared as described (Shibata et al., 2005), and the fluorescence images were acquired every 5 s with filter system GFP and TX2 (Leica). The acquired images were analyzed using the Leica LAS-AF software package, and ratios were calculated using Microsoft Excel.
Yeast Complementation and Immunoblot Analyses
CASTOR and POLLUX coding sequences were amplified with the primers P25/P26 and P27/P28, respectively. The PCR fragments were subcloned into the yeast expression vector pDL2xN (Dualsystems Biotech) at the SfiI restriction site as described (DUALmembrane kit 3, P01001; Dualsystems Biotech). The yeast strain MAB 2d was transformed using the LiAc method (Gietz and Woods, 2001). The crude yeast protein extraction was performed as described in the user manual (DUALmembrane kit 3, P01001; Dualsystems Biotech). The protein expression levels were monitored by immunoblotting using mouse monoclonal anti-HA (Roche) at a dilution of 1:5000.
GUS Assay
Promoter regions of CASTOR (2309 bp) and POLLUX (2843 bp) were amplified using primers P21/P22 and P23/P24, respectively, by PCR and cloned upstream of the GUS gene of pKGWFS7 (Karimi et al., 2002). The A. rhizogenes transformation of L. japonicus (ecotype Gifu) wild type was performed according to the description above. In order to perform the GUS staining, transformed seedlings were incubated for 40 min at 37°C with 2 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 50 mM NaPO4, pH 7, 1 mM EDTA, and 0.1% Triton X-100 and subsequently washed in 70% ethanol.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Mt CASTOR (CX525932), CASTOR (BAD89019), POLLUX (BAD89022), FNR (X07981).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Localization of CASTOR:GFP and POLLUX:GFP in the Nucleus upon Expression in Tobacco Leaf Epidermal Cells.
Supplemental Figure 2. Detection of P35S:CASTOR:GFP and P35S:POLLUX:GFP Fusion Proteins in Transiently Transformed Tobacco Leaves by Immunoblot.
Supplemental Figure 3. Localization of CASTOR:GFP and POLLUX:GFP in Plastids upon Expression in Tobacco Leaf Epidermal Cells.
Supplemental Figure 4. Cell-Free Expression of CASTOR, castor-2, and POLLUX.
Supplemental Figure 5. Reversal Potentials of CASTOR.
Supplemental Figure 6. Modelization of the CASTOR Pore and the castor-2 Mutant Pore.
Supplemental Figure 7. Magnesium-Dependent Opening and Closing Behavior of CASTOR.
Supplemental Figure 8. Effect of IP3 or Calcium on CASTOR Gating.
Supplemental Figure 9. Expression Patterns of CASTOR and POLLUX Promoter:GUS Fusions in L. japonicus Roots.
Supplemental Table 1. Primer Sequences.
Supplementary Material
Acknowledgments
We thank Hana Sychrova for providing the MAB 2d strain, Jörg Kudla for providing the vectors pSPYCE-GW and pSPYNE-GW as well as the constructs bZIP63-YFPC and bZIP63-YFPN, and Andreas Brachmann and Esben B. Madsen for critically reading the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft (TR1-SFB “Endosymbiosis”), by the European Union (Marie Curie Research Training Network “INTEGRAL”), by the Volkswagen Foundation (to R.B.), and by a postdoctoral fellowship of the Japan Society for the Promotion of Science (to N.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Martin Parniske (parniske@lmu.de).
Online version contains Web-only data.
References
- Allen, G.J., and Sanders, D. (1996). Control of ionic currents in guard cell vacuoles by cytosolic and luminal calcium. Plant J. 10 1055–1069. [DOI] [PubMed] [Google Scholar]
- Ané, J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. [DOI] [PubMed] [Google Scholar]
- Armstrong, C.M., Swenson, R.P., Jr., and Taylor, S.R. (1982). Block of squid axon K channels by internally and externally applied barium ions. J. Gen. Physiol. 80 663–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, P.L., and Fields, S. (1997). The Yeast Two-Hybrid System. (Oxford, UK: Oxford University Press).
- Cardenas, L., Feijo, J.A., Kunkel, J.G., Sanchez, F., Holdaway-Clarke, T., Hepler, P.K., and Quinto, C. (1999). Rhizobium nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. Plant J. 19 347–352. [DOI] [PubMed] [Google Scholar]
- Christie, M.J. (1995). Molecular and functional diversity of K+ channels. Clin. Exp. Pharmacol. Physiol. 22 944–951. [DOI] [PubMed] [Google Scholar]
- den Hartog, M., Verhoef, N., and Munnik, T. (2003). Nod factor and elicitors activate different phospholipid signaling pathways in suspension-cultured alfalfa cells. Plant Physiol. 132 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, A., Heckmann, A.B., Yousafzai, F., Duc, G., and Downie, J.A. (2007). Structural implications of mutations in the pea SYM8 symbiosis gene, the DMI1 ortholog, encoding a predicted ion channel. Mol. Plant Microbe Interact. 20 1183–1191. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- Engstrom, E.M., Ehrhardt, D.W., Mitra, R.M., and Long, S.R. (2002). Pharmacological analysis of nod factor-induced calcium spiking in Medicago truncatula. Evidence for the requirement of type IIA calcium pumps and phosphoinositide signaling. Plant Physiol. 128 1390–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertel, F., Mirus, O., Bredemeier, R., Moslavac, S., Becker, T., and Schleiff, E. (2005). The evolutionarily related beta-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J. Biol. Chem. 280 28281–28289. [DOI] [PubMed] [Google Scholar]
- Fahraeus, G. (1957). The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16 374–381. [DOI] [PubMed] [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1999). Elevation of the cytosolic free [Ca2+] is indispensable for the transduction of the Nod factor signal in alfalfa. Plant Physiol. 121 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong, V.E., Drobak, B.K., Allan, A.C., Watkins, P., and Trewavas, A.J. (1996). Growth of pollen tubes of Papaver rhoeas is regulated by a slow-moving calcium wave propagated by inositol 1,4,5-trisphosphate. Plant Cell 8 1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami, K. (2002). Structure, regulation, and function of phospholipase C isozymes. J. Biochem. 131 293–299. [DOI] [PubMed] [Google Scholar]
- Gamborg, O.L., Miller, R.A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50 151–158. [DOI] [PubMed] [Google Scholar]
- Gerasimenko, O., and Gerasimenko, J. (2004). New aspects of nuclear calcium signalling. J. Cell Sci. 117 3087–3094. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2001). Genetic transformation of yeast. Biotechniques 30 816–820, 822–826, 828. [DOI] [PubMed] [Google Scholar]
- Goldman, D.E. (1943). Potential, impedance, and rectification in membranes. J. Gen. Physiol. 27 37–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr, C., Banba, M., Croset, V., An, K., Miyao, A., An, G., Hirochika, H., Imaizumi-Anraku, H., and Paszkowski, U. (2008). Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. http://dx.doi.org/10.1105/tpc.108.062414. [DOI] [PMC free article] [PubMed]
- Hansen, J., Jorgensen, J.-E., Stougaard, J., and Marker, K. (1989). Hairy roots—A shortcut to transgenic root nodules. Plant Cell Rep. 8 12–15. [DOI] [PubMed] [Google Scholar]
- Heginbotham, L., Lu, Z., Abramson, T., and MacKinnon, R. (1994). Mutations in the K+ channel signature sequence. Biophys. J. 66 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, A.L., and Katz, B. (1949). The effect of sodium ions on the electrical activity of the giant axon of the squid. J. Physiol. 108 37–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, P., Park, K.J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C.J., and Nakai, K. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35 W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, R.M., Hunt, H.D., Ho, S.N., Pullen, J.K., and Pease, L.R. (1989). Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 77 61–68. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku, H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B.T., and MacKinnon, R. (2002). Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417 515–522. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kolacna, L., Zimmermannova, O., Hasenbrink, G., Schwarzer, S., Ludwig, J., Lichtenberg-Frate, H., and Sychrova, H. (2005). New phenotypes of functional expression of the mKir2.1 channel in potassium efflux-deficient Saccharomyces cerevisiae strains. Yeast 22 1315–1323. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Lazo, G.R., Stein, P.A., and Ludwig, R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (New York) 9 963–967. [DOI] [PubMed] [Google Scholar]
- Leckie, C.P., McAinsh, M.R., Allen, G.J., Sanders, D., and Hetherington, A.M. (1998). Abscisic acid-induced stomatal closure mediated by cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95 15837–15842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge, P., Roche, P., Faucher, C., Maillet, F., Truchet, G., Prome, J.C., and Denarie, J. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344 781–784. [DOI] [PubMed] [Google Scholar]
- Lohse, S., Schliemann, W., Ammer, C., Kopka, J., Strack, D., and Fester, T. (2005). Organization and metabolism of plastids and mitochondria in arbuscular mycorrhizal roots of Medicago truncatula. Plant Physiol. 139 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, S.R. (1989). Rhizobium-legume nodulation: Life together in the underground. Cell 56 203–214. [DOI] [PubMed] [Google Scholar]
- Lu, Z., and MacKinnon, R. (1994). Electrostatic tuning of Mg2+ affinity in an inward-rectifier K+ channel. Nature 371 243–246. [DOI] [PubMed] [Google Scholar]
- Lucocq, J. (1994). Quantitation of gold labelling and antigens in immunolabelled ultrathin sections. J. Anat. 184 1–13. [PMC free article] [PubMed] [Google Scholar]
- Maresova, L., and Sychrova, H. (2005). Physiological characterization of Saccharomyces cerevisiae kha1 deletion mutants. Mol. Microbiol. 55 588–600. [DOI] [PubMed] [Google Scholar]
- McAinsh, M.R., Webb, A., Taylor, J.E., and Hetherington, A.M. (1995). Stimulus-induced oscillations in guard cell cytosolic free calcium. Plant Cell 7 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, G.P., Samuel, M.A., Jones, A.M., and Ellis, B.E. (2004). Mastoparan rapidly activates plant MAP kinase signaling independent of heterotrimeric G proteins. Plant Physiol. 134 1332–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, H., Sun, J., Oldroyd, G.E., and Downie, J.A. (2006). Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 19 914–923. [DOI] [PubMed] [Google Scholar]
- Morera, F.J., Vargas, G., Gonzalez, C., Rosenmann, E., and Latorre, R. (2007). Ion-channel reconstitution. Methods Mol. Biol. 400 571–585. [DOI] [PubMed] [Google Scholar]
- Nagata, T., Iizumi, S., Satoh, K., Ooka, H., Kawai, J., Carninci, P., Hayashizaki, Y., Otomo, Y., Murakami, K., Matsubara, K., and Kikuchi, S. (2004). Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Mol. Biol. Evol. 21 1855–1870. [DOI] [PubMed] [Google Scholar]
- Navazio, L., Bewell, M.A., Siddiqua, A., Dickinson, G.D., Galione, A., and Sanders, D. (2000). Calcium release from the endoplasmic reticulum of higher plants elicited by the NADP metabolite nicotinic acid adenine dinucleotide phosphate. Proc. Natl. Acad. Sci. USA 97 8693–8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navazio, L., Mariani, P., and Sanders, D. (2001). Mobilization of Ca2+ by cyclic ADP-ribose from the endoplasmic reticulum of cauliflower florets. Plant Physiol. 125 2129–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Downie, J.A. (2006). Nuclear calcium changes at the core of symbiosis signalling. Curr. Opin. Plant Biol. 9 351–357. [DOI] [PubMed] [Google Scholar]
- Peiter, E., et al. (2007). The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol. 145 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J.A., Wang, T.L., Welham, T.J., Gardner, S., Pike, J.M., Yoshida, S., and Parniske, M. (2003). A TILLING reverse genetics tool and a Web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 131 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon, M., Journet, E.P., Dedieu, A., de Billy, F., Truchet, G., and Barker, D.G. (1992). Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingret, J.L., Journet, E.P., and Barker, D.G. (1998). Rhizobium nod factor signaling. Evidence for a G protein-mediated transduction mechanism. Plant Cell 10 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely, B.K., Lougnon, G., Ane, J.M., and Cook, D.R. (2007). The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J. 49 208–216. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro, A. (2000). Potassium transport in fungi and plants. Biochim. Biophys. Acta 1469 1–30. [DOI] [PubMed] [Google Scholar]
- Ross, E.M., and Higashijima, T. (1994). Regulation of G-protein activation by mastoparans and other cationic peptides. Methods Enzymol. 237 26–37. [DOI] [PubMed] [Google Scholar]
- Shaw, S.L., and Long, S.R. (2003). Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol. 131 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, S., Mitsui, H., and Kouchi, H. (2005). Acetylation of a fucosyl residue at the reducing end of Mesorhizobium loti nod factors is not essential for nodulation of Lotus japonicus. Plant Cell Physiol. 46 1016–1020. [DOI] [PubMed] [Google Scholar]
- Stehno-Bittel, L., Lückhoff, A., and Clapham, D.E. (1995). Calcium release from the nucleus by InsP3 receptor channels. Neuron 14 163–167. [DOI] [PubMed] [Google Scholar]
- Stern, J.H., Knutsson, H., and MacLeish, P.R. (1987). Divalent cations directly affect the conductance of excised patches of rod photoreceptor membrane. Science 236 1674–1678. [DOI] [PubMed] [Google Scholar]
- Stougaard, J., Abildsten, D., and Marcker, K.A. (1987). The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol. Gen. Genet. 207 251–255. [Google Scholar]
- Sun, J., Miwa, H., Downie, J.A., and Oldroyd, G.E. (2007). Mastoparan activates calcium spiking analogous to Nod factor-induced responses in Medicago truncatula root hair cells. Plant Physiol. 144 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil, F.I., Leonetti, M., Muir, T.W., and Mackinnon, R. (2006). Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science 314 1004–1007. [DOI] [PubMed] [Google Scholar]
- Villarejo, A., et al. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7 1224–1231. [DOI] [PubMed] [Google Scholar]
- Walker, S.A., Viprey, V., and Downie, J.A. (2000). Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by nod factors and chitin oligomers. Proc. Natl. Acad. Sci. USA 97 13413–13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, J., and Best, P.M. (1994). Characterization of the potassium channel from frog skeletal muscle sarcoplasmic reticulum membrane. J. Physiol. 477 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Niu, X., Brelidze, T.I., and Magleby, K.L. (2006). Ring of negative charge in BK channels facilitates block by intracellular Mg2+ and polyamines through electrostatics. J. Gen. Physiol. 128 185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.