Abstract
ASYMMETRIC LEAVES2 (AS2)/LATERAL ORGAN BOUNDARIES DOMAIN (LBD) family proteins are plant-specific nuclear proteins, and genes encoding several family members have been implicated in plant development. We investigated the function of two members of the Arabidopsis thaliana AS2/LBD family, AS2-LIKE19 (ASL19)/LBD30 and ASL20/LBD18, which encode homologous proteins. Both ASL19 and ASL20 were expressed in immature tracheary elements (TEs), and the expression was dependent on VASCULAR-RELATED NAC-DOMAIN PROTEIN6 (VND6) and VND7, which are transcription factors required for TE differentiation. Overexpression of ASL19 and ASL20 induced transdifferentiation of cells from nonvascular tissues into TE-like cells, similar to those induced upon VND6/7 overexpression. By contrast, aberrant TEs were formed when a cDNA encoding a fusion protein of ASL20 with an artificial repressor domain (ASL20-SRDX) was expressed from its native promoter. These results provide evidence that ASL proteins positively regulate TE differentiation. In transgenic plants overexpressing both ASL19 and ASL20, the xylem-deficient phenotype caused by the expression of dominant-negative versions of VND6/7 proteins was not rescued. However, ectopic expression of VND7 was detected in plants overexpressing ASL20. Moreover, VND genes and their downstream targets were downregulated in ASL20-SRDX plants. Therefore, ASL20 appears to be involved in a positive feedback loop for VND7 expression that regulates TE differentiation-related genes.
INTRODUCTION
Arabidopsis thaliana ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES DOMAIN6 (AS2/LBD6) and LATERAL ORGAN BOUNDARIES/AS2-LIKE4 (LOB/ASL4) genes were first discovered as genes encoding members of the AS2/LBD protein family (Iwakawa et al., 2002; Shuai et al., 2002). Whole genome sequence information revealed that there are 42 genes predicted to encode the AS2/LBD family proteins in Arabidopsis (Iwakawa et al., 2002; Shuai et al., 2002). Results obtained by forward and reverse genetics indicate that several members of the AS2/LBD protein family play important roles in regulating various aspects of plant development, such as determination of stem cell fate (Bortiri et al., 2006), development of the embryo sac (Evans, 2007), leaf development (Ori et al., 2000; Semiarti et al., 2001; Xu et al., 2003), and lateral root formation (Inukai et al., 2005; Liu et al., 2005; Okushima et al., 2007; Taramino et al., 2007). Based on its expression pattern, the LOB/ASL4 gene has been postulated to function in fate determination of a boundary between the initiating organ primordia and the stem cells from which they are generated (Shuai et al., 2002).
The AS2/LBD family of proteins are characterized by an N-terminal conserved domain with a CX2CX6CX3C motif and a Leu zipper–like sequence named the AS2/LOB domain. The AS2/LBD proteins likely execute their function in the nucleus, as the AS2/LBD–green fluorescent protein (GFP) fusion proteins localize to the nucleus (Iwakawa et al., 2002; Inukai et al., 2005; Naito et al., 2007). Consistent with this notion, the AS2/LOB domain of LOB protein was shown to bind to DNA in vitro (Husbands et al., 2007). The AS2 protein bound to sites in the promoter regions of the KNAT1 (for Knotted-like from Arabidopsis thaliana1) and KNAT2 genes, together with the AS1 protein (Guo et al., 2008), which also interacts with AS2 in vitro (Xu et al., 2003). KNAT1 is expressed in the peripheral zone of the shoot apical meristem (Lincoln et al., 1994). AS1 and AS2 are required for the maintenance of KNAT1 repression in developing leaves, once KNAT1 expression is repressed at the start of the formation of the leaf primordium (Ori et al., 2000; Semiarti et al., 2001; Lin et al., 2003). The AS1-AS2 complex binding sites in the KNAT1 promoter were found to be involved in KNAT1 repression in developing leaves (Guo et al., 2008). Thus, the AS2/LBD family of proteins seem to regulate gene expression by interacting with specific DNA sequences, although the molecular mechanisms of the AS2/LBD family of proteins are still poorly understood.
Here, we report that two members of the Arabidopsis AS2/LBD protein family are involved in the differentiation of tracheary elements (TEs), the basic units that constitute xylem vessels. Xylem vessels are specialized tissues for the transport of water, nutrients, and signaling molecules and also provide plants with mechanical stability. TE precursors are differentiated from procambial and cambial cells, which are stem-like cells able to differentiate into other vasculature elements as well. During TE differentiation, a series of coordinated cellular events occurs, including changes in cytoskeletal organization, patterned cell wall deposition, and autolysis (Fukuda, 2004; Turner et al., 2007). The resulting mature TEs are distinguishable by their characteristic secondary cell walls with annular, spiral, reticulate, or pitted wall thickenings. Several factors, such as auxin, cytokinin, brassinosteroid (BR), xylogen, and the CLE family of peptides, have been implicated in the regulation of TE differentiation (Szekeres et al., 1996; Yamamoto et al., 1997; Choe et al., 1999; Caño-Delgado et al., 2004; Fukuda, 2004; Motose et al., 2004; Ito et al., 2006; Mähönen et al., 2006).
A gene expression analysis of Arabidopsis suspension culture cells identified two NAC (for Petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2)–domain transcription factors that function as positive regulators of TE differentiation: VASCULAR-RELATED NAC-DOMAIN PROTEIN6 (VND6) and VND7 (Kubo et al., 2005). Although Kubo et al. (2005) proposed that VND6 and VND7 act as transcription switches in the formation of metaxylem and protoxylem vessels, respectively, both being required for TE differentiation, Yamaguchi et al. (2008) recently reported that VND7 regulates not only the formation of protoxylem vessels in roots but also the formation of metaxylem vessels in roots and protoxylem vessels in the aerial parts of seedlings. Therefore, elucidating the regulation of VND6 and VND7 expression and determining their downstream gene networks are clearly important issues for understanding the molecular mechanisms underlying xylogenesis.
In this study, we investigated the function of Arabidopsis ASL19/LBD30 and ASL20/LBD18 (hereafter referred to as ASL19 and ASL20, respectively), two homologous AS2/LBD family proteins. Borghi et al. (2007) reported that embryogenesis of jlo-1/asl19-1 mutants was arrested at the globular stage. However, asl19-2 (a novel allele identified in this study) mutants did not show the embryonic phenotype (see Supplemental Figures 1A to 1E online). Furthermore, the phenotypic traits observed in jlo-1 mutants were not suppressed when the recombinant ASL19 cDNA that was fused to the ASL19 promoter was introduced into the jlo-1 mutant (see Supplemental Figure 1F online). These results suggest that the embryonic lethality is not linked to the asl19 mutation. Our results showed that ASL19 and ASL20 were expressed in immature TEs. The expression was dependent on VND6 and VND7. Moreover, overexpression of ASL19 or ASL20 induced transdifferentiation of cells from nonvascular tissues into TE-like cells, similar to those formed upon VND6 and VND7 overexpression. These results suggest that ASL19 and ASL20 regulate TE differentiation downstream of VND6 and VND7. Further analyses showed that the generation of TE-like cells by ASL19/20 overexpression required VND6 and VND7 activities and that ASL20 overexpression induced ectopic expression of VND7. These results suggest that ASL19 and ASL20 are involved in a positive feedback loop that maintains or promotes VND7 expression. We discuss the relationship between ASL19/20 and VND6/7 during TE differentiation.
RESULTS
Expression Profiles of ASL19 and ASL20
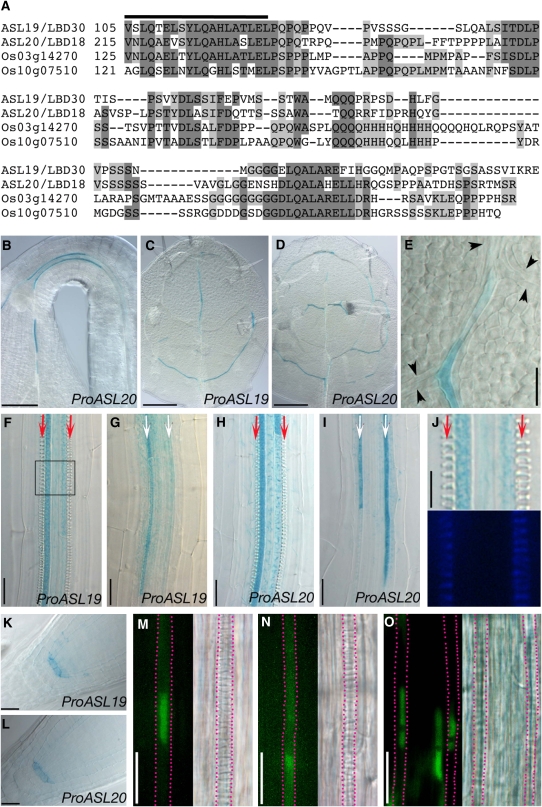
Although there is a high level of sequence conservation in the AS2/LOB domain, the C-terminal regions of the AS2/LBD proteins are remarkably variable. ASL19 and ASL20 appear to be exceptions, because their C-terminal regions are 46.1% identical in amino acid sequences (Figure 1A). The similarity in the primary structures of these two proteins suggested that their function might be conserved in Arabidopsis. In addition, a database homology search uncovered two rice (Oryza sativa) AS2/LBD proteins (encoded by Os03g14270 and Os10g07510) with C-terminal regions ∼28 to 33.3% identical to those of ASL19 and ASL20 (Figure 1A). Homologies in the C-terminal regions of Arabidopsis and rice ASL19/20 counterparts suggested a conserved role of these proteins in the development of both monocots and dicots.
Figure 1.
Similarity in Amino Acid Sequences of the C-Terminal Regions of Arabidopsis and Rice ASL Proteins, and Expression Patterns of ASL19 and ASL20.
(A) An alignment of amino acid sequences of Arabidopsis and rice ASL proteins. The thick line indicates the C-terminal ends of AS2/LOB domains.
(B) to (L) GUS staining in transgenic plants expressing ProASL19:EGFP-GUS ([C], [F], [G], [J], and [K]) and ProASL20:EGFP-GUS ([B], [D], [E], [H], [I], and [L]). Apical hook of an etiolated seedling (B), first or second leaf ([C] to [E]), maturation zones ([F], [H], and [J]; ∼1.5 to 2 mm from root tips) and differentiation zones ([G] and [I]; ∼0.4 to 0.8 mm from root tips) of roots and of root tips ([K] and [L]) of a seedling at 6 d after vernalization (DAV). (J) shows the boxed region in (F) at a higher magnification (top panel). Lignin deposited in protoxylem was visualized under UV illumination (bottom panel). Arrowheads in (E) indicate putative TE precursors. Note that putative TE precursors did not express EGFP-GUS. Arrows in (F) to (I) indicate cell files of protoxylem vessels with spiral wall thickenings (red) and undifferentiated TEs (white).
(M) to (O) Subcellular localization of the ASL20-EGFP protein. Fluorescence signals (left panels) due to ASL20-EGFP (M) and EGFP-GUS (N) driven by the ASL20 promoter and due to EGFP-NLS (O) driven by a 35S promoter and differential interference contrast images (right panels). Red dashed lines show immature TEs expressing EGFP fusion proteins.
Bars = 100 μm in (B) to (D), 20 μm in (E) to (I), (K), and (L), and 10 μm in (J) and (M) to (O).
To understand the functions of ASL19 and ASL20, we generated ProASL:EGFP (enhanced green fluorescent protein)–GUS (β-glucuronidase) reporter constructs using a 2-kb sequence upstream of their corresponding coding sequences (see Methods) and analyzed their expression profiles in transgenic plants. Histochemical GUS staining was detected in the vascular tissues of hypocotyls, leaves, roots, developing floral organs, and siliques of transgenic plants (Figures 1B to 1J; see Supplemental Figure 2 online). GUS staining patterns in vasculatures were not distinguishable between the reporter constructs for ASL19 and ASL20.
The Arabidopsis root has three or four files of metaxylem vessels surrounded by two files of protoxylem vessels in the vascular cylinder. Metaxylem vessels differentiate in the maturation zone of the root, whereas differentiation of protoxylem vessels occurs acropetally in the differentiation zone (Pyo et al., 2004). In the maturation zone of roots, strong GUS signals were detected in immature TEs of the outer two cell files of primary metaxylem vessels, although there was almost no staining in the central metaxylem vessels that differentiated later (Figures 1F and 1H). In the differentiation zone, expression of reporter genes was detected in immature TEs of the two cell files of protoxylem vessels but not in the cell files of metaxylem or other types of vascular cells (Figures 1G and 1I). The expression of the reporter genes was detected before the appearance of any obvious secondary cell wall thickening and lignin deposition (Figures 1F to 1J; compare with wall thickenings of mature TEs in Figure 2H or Figure 5I below), and GUS signals became stronger as the root developed. Similar patterns of EGFP-GUS expression in immature TEs were also observed in the expanding leaves (Figures 1C to 1E). In addition, GUS staining was detected around the quiescent centers of root tips (Figures 1K and 1L), although at a lower level than in immature TEs. These results indicate that ASL19 and ASL20 expression is closely associated with TE differentiation. The similar expression patterns of ASL19 and ASL20 are consistent with the hypothesis that these two genes have redundant functions.
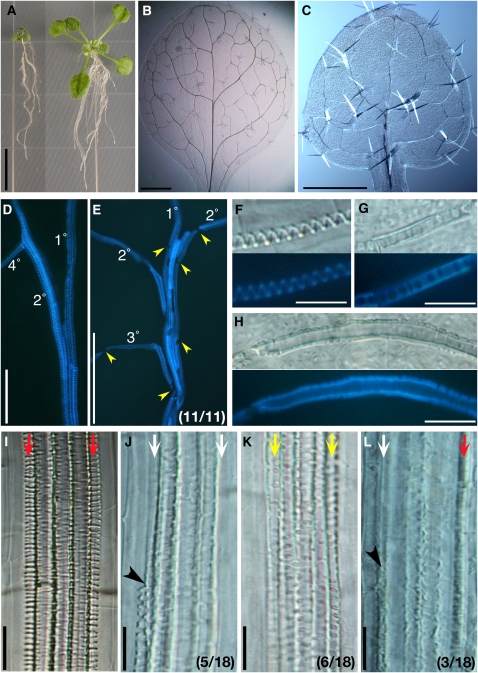
Figure 2.
Ectopic Production of TE-Like Cells upon ASL Overexpression.
(A) to (C) Three-week-old seedlings of a wild-type plant (A), a Pro35S:ASL19 line (B), and a Pro35S:ASL20 line (C).
(D) to (J) TE-like cells generated in seedlings (10 DAV) of XVE:ASL19 ([D], [E], and [I]), XVE:ASL20 ([F] and [G]), and Pro35S:ASL20 (J) lines and wild-type control (H). Seedlings (3 DAV) of XVE:ASL19 and XVE:ASL20 lines were transferred onto plates with 10 μM β-estradiol and grown for 7 d.
(D) Basal margin of a cotyledon and its petiole. Fluorescence image of TE-like cells stained with fuchsin (top panel) and the corresponding Nomarski image (bottom panel).
(E) and (F) Cotyledons. Fluorescence images of TE-like cells visualized by UV illumination (left panels) and the corresponding Nomarski images (right panels).
(G) Living mesophyll cells observed under UV illumination. Fluorescence from chloroplasts (red; top and bottom panels) and merged with an image showing lignin (blue; top panel).
(H) to (J) Nomarski images of roots.
Bars = 1 cm in (A) to (C), 50 μm in (D) and (H) to (J), 0.2 mm in (E) and (F), and 20 μm in (G).
Figure 5.
Phenotypic Analysis of ProASL20:ASL20-SRDX Plants.
(A) ProASL20:ASL20-SRDX plants (12 DAV) with the dwarf phenotype (left) and with normal growth (right).
(B) to (H) First leaf of wild-type ([B], [D], and [F]) and dwarfed ProASL20:ASL20-SRDX ([C], [E], [G], and [H]) plants.
(B) and (C) Venation patterns.
(D) and (E) Xylem vessels of midveins, as visualized under UV illumination. Yellow arrowheads indicate gaps between TEs (E). The fraction of dwarfed plants showing the indicated feature is in parentheses (E).
(F) to (H) Magnified images of mature TEs. Nomarski images (top panels) and fluorescence images under UV illumination (bottom panels).
(I) to (L) Xylem vessels in roots of wild-type (I) and dwarfed ProASL20:ASL20-SRDX ([J] to [L]) plants. Arrows indicate cell files of protoxylem vessels with spiral wall thickenings (red), undifferentiated TEs (white), and TEs with pitted wall thickenings (yellow). Black arrowheads indicate discontinuities of protoxylem vessels. The fractions of plants showing the indicated features are in parentheses.
Bars = 1 cm in (A), 1 mm in (B), 0.5 mm in (C), 0.1 mm in (D) and (E), and 20 μm in (F) to (L).
Furthermore, we examined the subcellular localization of the ASL20-EGFP protein expressed from the native ASL20 promoter. We detected most fluorescence in the nucleus and a faint signal in the cytoplasm of differentiating TEs (Figure 1M). This result was similar to the subcellular localization of EGFP appended with a nuclear localization signal from SV40 (Figure 1O). Green fluorescence from ProASL20:EGFP-GUS plants was detected throughout the cytoplasm (Figure 1N).
Overexpression of ASL Genes
To investigate the functions of ASL19 and ASL20 proteins, we overexpressed their corresponding cDNAs from a cauliflower mosaic virus 35S promoter (Pro35S) or an estradiol-inducible system (XVE; for LexA-VP16-ER) (Zuo et al., 2000). Pro35S:ASL19 and Pro35S:ASL20 transgenic plants were dwarfed and produced short petioles and their leaves curled downward (Figures 2B and 2C). In addition, Pro35S:ASL19 plants appeared bushy because of the generation of ectopic shoots (in 58% [n = 85] of T1 plants) (Figure 2B) from the adaxial side of cotyledons. The root tips of Pro35S:ASL19 plants were shrunken, which inhibited root growth. Columella cells were also disorganized in Pro35S:ASL20 plants. The root tip phenotype was similar to that reported for jlo-D (Borghi et al., 2007).
Overexpression of ASL19 and ASL20 from either a 35S promoter or an XVE-inducible system did not influence the formation of xylem vessels. However, when transcription was induced by β-estradiol, multiple lines of XVE:ASL19 and XVE:ASL20 plants produced cells with wall thickenings, similar to those of protoxylem, in the mesophyll and epidermal tissues of cotyledons and in various tissues of the roots (Figures 2D to 2G and 2I). Similarly, Pro35S:ASL20 lines formed TE-like cells in the cortex and stele of the roots (Figure 2J). UV illumination and staining with fuchsin showed lignin deposition in the wall thickenings of TE-like cells in cotyledons and of mature TEs (Figures 2D to 2G). Fluorescence emitted from chloroplasts in living mesophyll cells was not detectable in TE-like cells produced in the mesophyll tissues of XVE:ASL20 plants (Figure 2G), suggesting that cell contents had been lost. TE-like cells in the roots were usually observed in the maturation region rather than in the apical region, which includes the differentiation zone. Generation of TE-like cells in plants overexpressing ASL19 was less frequent than in those overexpressing ASL20 (cf. Figures 2E and 2F). Control experiments showed that TE-like cells were not seen in plants overexpressing ASL17/LBD14 and ASL22/LBD31 (see Supplemental Figures 3C and 3D online), even though these plants were dwarfed and similar in morphology to plants overexpressing ASL19 and ASL20 (see Supplemental Figures 3A and 3B online). These results show that ASL19 and ASL20, but not ASL17 and ASL22, have the capacity to induce transdifferentiation of nonvascular cells into TE-like cells.
VND6 and VND7 Regulated the Expression of ASL Genes
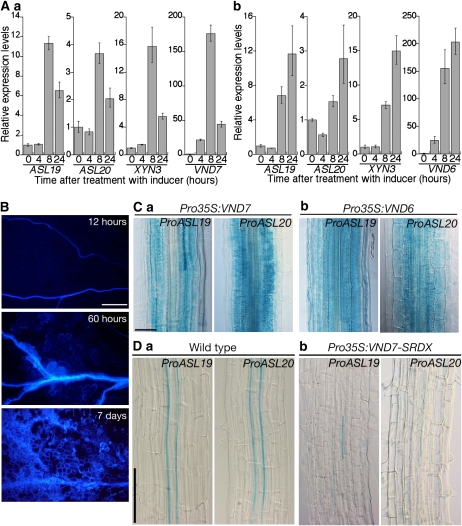
The overexpression of VND6 or VND7 produced TE-like cells from nonvascular tissues, similar to those formed when ASL19 or ASL20 was overexpressed (Kubo et al., 2005). We generated transgenic plants overexpressing VND6 and VND7 cDNAs using the inducible XVE system and performed real-time PCR to investigate the expression of ASL19 and ASL20 in these plants after treatment with β-estradiol. The levels of ASL19 and ASL20 mRNAs increased in both XVE:VND6 and XVE:VND7 plants within 8 h of β-estradiol treatment (Figure 3A). The expression of XYN3, encoding TE-specific xylanase (Sawa et al., 2005), was also induced. TE-like cells with deposited lignin appeared within 60 h of induction (Figure 3B). Induction of ASL19 and ASL20 expression preceded the appearance of visible TE-like cells. To investigate the effect of VND6 and VND7 on the expression pattern of ASL19 and ASL20, we introduced Pro35S:VND7 and Pro35S:VND6 into plants carrying ProASL19:EGFP-GUS and ProASL20:EGFP-GUS. Histochemical GUS staining assays showed ectopic expression of both promoter–reporter genes in the nonvascular tissues of plants overexpressing VND7 and VND6 (Figure 3C). These results indicate that VND6 and VND7 operate upstream of ASL19 and ASL20.
Figure 3.
VND6- and VND7-Dependent Expression of ASL19 and ASL20.
(A) Real-time PCR was performed with total RNA isolated from seedlings (10 DVA) of XVE:VND7 (a) and XVE:VND6 (b) lines that were incubated with 10 μM β-estradiol for the indicated times. Results are presented as averages of normalized relative transcript levels ± sd of three replicates.
(B) TE-like cells in cotyledons of XVE:VND7 lines, as visualized under UV illumination. Seedlings (3 DAV) were treated with 10 μM β-estradiol for the indicated amounts of time.
(C) and (D) Expression of ProASL19:EGFP-GUS and ProASL20:EGFP-GUS in roots of Pro35S:VND7 (Ca), Pro35S:VND6 (Cb), and Pro35S:VND7-SRDX (Db) lines and in roots of control plants of the same age (Da).
Bars = 200 μm in (B), 50 μm in (C), and 100 μm in (D).
Since it has been reported that mutants of VND6 and VND7 do not exhibit significant phenotypes (Kubo et al., 2005), we constructed dominant-negative versions of VND6 and VND7 proteins by fusing with the SRDX artificial repression domain at their C termini (VND6-SRDX and VND7-SRDX) (Hiratsu et al., 2003; Kubo et al., 2005). We overexpressed cDNAs encoding these chimeric proteins using a 35S promoter in ProASL19:EGFP-GUS and ProASL20:EGFP-GUS lines. The expression of VND6-SRDX and VND7-SRDX in transgenic plants affected the formation of xylem vessels and produced discontinuous xylem vessels in the roots (Figure 3D; see Supplemental Figure 4A online) (Kubo et al., 2005). The expression of ProASL19:EGFP-GUS and ProASL20:EGFP-GUS was remarkably downregulated in these plants (Figure 3D; see Supplemental Figure 4A online). Weak and discontinuous GUS signals were observed in the cell files of protoxylem and metaxylem vessels. These results provide evidence that VND6 and VND7 are positive regulators of ASL19 and ASL20 expression and that their activities are required for ASL19/20 expression.
Since auxin and BR are required for the induction and progression of TE differentiation in an in vitro transdifferentiation system using isolated Zinnia mesophyll cells (Yamamoto et al., 1997; Fukuda, 2004), we also examined responses of ASL19 and ASL20 to these phytohormones. BR did not alter the expression of ASL19 and ASL20, even at 12 h after treatment. By contrast, consistent with the results of the microarray analysis by Okushima et al. (2005), levels of ASL20 mRNA, but not of ASL19 mRNA, increased within 30 min of naphthylacetic acid treatment (see Supplemental Figure 5A online), and this increase was dependent on AUXIN RESPONSE FACTOR7 (ARF7) (see Supplemental Figure 5B online). However, neither exogenous auxin nor the arf7 mutation altered the expression pattern of ProASL20:EGFP-GUS (see Supplemental Figure 5C online). Our results indicate that auxin enhances the expression of ASL20.
Overexpression of ASL20-Induced Ectopic Expression of VND7
Since ASL19 and ASL20 expression in immature TE was dependent on VND6 and VND7 activities, we examined whether ASL19 and ASL20 proteins could rescue a deficiency in VND7 activity. To this end, we introduced Pro35S:VND7-SRDX into XVE:ASL19 and XVE:ASL20 transgenic lines. In the absence of β-estradiol, these transgenic plants showed defects in the differentiation of protoxylem vessels around the differentiation zone of roots (Figures 4A and 4C). In the presence of β-estradiol, the induced expression of ASL19 and ASL20 was confirmed by RT-PCR (see Supplemental Figure 6 online). ASL19 and ASL20 overexpression only partially rescued the defects of xylem vessel formation (Figures 4B and 4D to 4F). Similar results were also obtained in experiments using Pro35S:VND6-SRDX (see Supplemental Figure 4B online). These results indicate that the activities of VND6 and VND7 are required for ASL19 and ASL20 proteins to fully induce TE differentiation.
Figure 4.
Overexpression of ASL20 Induced Ectopic Expression of VND7.
(A) to (F) Roots of Pro35S:VND7-SRDX lines with either XVE:ASL19 ([A] and [B]) or XVE:ASL20 ([C] and [D]). Seedlings (3 DAV) were transferred onto plates with (+) or without (−) the inducer and grown for 4 d. White lines indicate fragments of protoxylem ([B] and [D]). Magnified images of the boxed regions in (B) and (D) are shown in (E) and (F), respectively.
(G) to (N) GUS staining in cotyledons ([G] to [I] and [N]) and roots ([J] to [M]) of XVE:ASL20 lines with ProVND7:EGFP-GUS ([G] to [M]) and ProATHB-8:EGFP-GUS (N). Seedlings (3 DAV) were grown on plates with (+) or without (−) the inducer for 5 d. (I) shows a magnified image of a mesophyll cell with GUS staining transdifferentiating into TE-like cells. Maturation regions ([J] and [K]) and differentiation zones ([L] and [M]) of roots.
(O) Number of lignified (gray bars) and nonlignified (white bars) TEs with GUS signals in metaxylem vessels of roots. ProVND7:EGFP-GUS (ProVND7; n = 15), ProASL19:EGFP-GUS (ProASL19; n = 17), and ProASL20:EGFP-GUS (ProASL20; n = 16) lines (6 DAV). The error bars indicate sd.
Bars = 200 μm in (A) to (D) and (J) to (M), 50 μm in (E) and (F),100 μm in (G), (H), and (N), and 20 μm in (I).
Next, we introduced ProVND7:EGFP-GUS into XVE:ASL20 lines and performed a histochemical GUS staining assay. Note that the VND7 promoter region fully includes the promoter used by Kubo et al. (2005). In the absence of β-estradiol, we detected GUS signals in immature TEs of both metaxylem and protoxylem vessels (Figure 4L; see Supplemental Figure 7 online), which is consistent with the recent result of Yamaguchi et al. (2008). GUS signals were not detected in the mesophyll and epidermal tissues of cotyledons (Figure 4G). In the presence of β-estradiol, the expression of ProVND7:EGFP-GUS was detected in the mesophyll and epidermal cells of cotyledons (Figure 4H) and in the nonvascular cells in the maturation region of roots (Figure 4K). We observed transdifferentiation of the mesophyll cells with GUS signals into TE-like cells (Figure 4I). Although ectopic VND7 expression was not detected in the apical part of roots containing differentiating and immature TEs of protoxylem vessels, the intensity of GUS signals increased in immature TEs that were generated at developmentally normal positions (Figures 4L and 4M). Enhancement of EGFP-GUS expression in immature TEs was detected within 24 h of treatment. Ectopic expression of ProVND7:EGFP-GUS was detected from 3 d after inducer treatment.
To further confirm that ASL19 and ASL20 act downstream of VND7, we examined the metaxylem vessels of roots from plants transformed with promoter–GUS reporter constructs for VND7, ASL19, and ASL20 and counted the number of TEs with GUS signals. There was no significant difference in the number of lignified TEs with GUS signals among plants expressing these three different reporter constructs (Figure 4O). By contrast, ProVND7:EGFP-GUS was expressed in more nonlignified TEs than were the other two reporter genes (Figure 4O). This result indicates that, in differentiating TEs, VND7 expression occurs prior to that of ASL19 and ASL20.
We assumed that the patchy pattern of ProVND7:EGFP-GUS expression (Figure 4H) might be due to the generation of specialized cells with procambial cell–like identity. To address this issue, we examined the expression of ATHB-8, a molecular marker for preprocambial and procambial cells (Baima et al., 1995; Scarpella et al., 2004), in XVE:ASL20 plants using a ProATHB-8:EGFP-GUS reporter construct. Although GUS signals were detected in procambial cells along the vasculature of cotyledons, expression was not detected in the cells of other tissues, even when treated with β-estradiol (Figure 4N). This result suggests that the ectopic expression of VND7 is not due to the generation of procambial cell–like cells.
ASL Activity Is Critical for Appropriate TE Differentiation
To investigate loss-of-function phenotypes of ASL19 and ASL20, we obtained T-DNA and transposon insertion lines. The positions of T-DNA and transposon insertions are summarized in Supplemental Figure 1A online. RT-PCR analysis confirmed that these mutants are null, since ASL19 and ASL20 mRNAs were not detectable (see Supplemental Figure 1B online). Since the formation of xylem vessels was not significantly affected by asl19-2 and asl20-1 single mutations, we genetically created double mutants. However, double mutant plants did not exhibit any significant defect in xylem formation either.
We decided to use an alternative approach to investigate the loss-of-function phenotype. To this end, we constructed a cDNA encoding the ASL20 protein fused to the SRDX repressor domain at its C terminus (ASL20-SRDX). If the ASL20 protein acts on specific DNA sequences and positively regulates gene expression, the chimeric protein might downregulate gene expression downstream of ASL20. ASL20-SRDX cDNA was expressed from the native ASL20 promoter.
We found that 50.4% (n = 107) of T1 transgenic plants were markedly smaller than wild-type plants (Figure 5A). Although venation patterns of cotyledons and leaves were not significantly altered (Figures 5B and 5C), we found abnormalities in the xylem vessels of dwarfed transgenic plants (Figures 5E, 5G, 5H, and 5J to 5L). Xylem vessels of midveins in the leaves of wild-type plants were tightly associated with each other. Each TE in the same xylem vessel was connected to its neighboring TEs (Figure 5D). In dwarf plants, by contrast, we observed extra spaces between the xylem vessels in midveins and found gaps between mature TEs (Figure 5E). In addition, leaves of dwarf plants formed TEs with pitted wall thickenings (Figures 5G and 5H), instead of the typical spiral wall thickenings seen in leaves of wild-type plants (Figure 5F). The pitted pattern of wall thickenings was similar to that of metaxylem (Figure 5I) and was not observed in leaves of wild-type plants. In roots of dwarfed plants, we also discovered abnormalities, including discontinuities of protoxylem vessels (Figures 5J and 5L), formation of TEs with pitted wall thickenings in cell files of protoxylem vessels (Figure 5K), and TEs with obscure wall thickenings (Figure 5L). Pitted patterns on wall thickenings of metaxylem vessels in dwarf plants were larger than those in wild-type plants (Figures 5I and 5K). We also observed delayed differentiation of metaxylem vessels in roots (see Supplemental Figures 8A to 8A′′ online) and isolated fragments of xylem vessels in cotyledons and leaves of dwarf plants (see Supplemental Figures 8C and 8D online). These results indicate that the ASL20-SRDX protein affects the formation of patterned secondary cell wall thickenings and partially inhibits TE differentiation.
Effects of ASL20-SRDX on the Expression of Genes Related to TE Differentiation
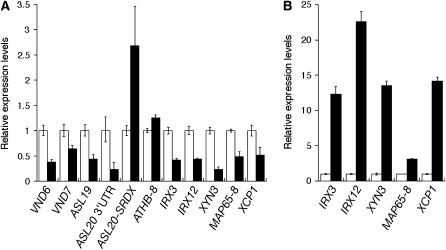
We analyzed the effects of the ASL20-SRDX protein on the expression of VND6 and VND7 genes. Because dwarfed ProASL20:ASL20-SRDX transgenic lines died before bolting on soil, we separately collected T1 plants that exhibited the dwarf phenotype and transgenic plants that grew normally and prepared total RNA from these samples. Real-time PCR revealed that both VND6 and VND7 mRNA levels were reduced in dwarfed plants (Figure 6A). Reduced levels of mRNAs for endogenous ASL19 and ASL20 were also detected (Figure 6A). However, no significant difference in ATHB-8 mRNA levels was detected between the two plant populations (Figure 6A). To further confirm ASL20-SRDX expression, we performed real-time PCR using a primer pair specific to the coding sequences for SRDX. We found that ASL20-SRDX mRNA accumulated to a higher level in this population than in plants with the wild-type phenotype (Figure 6A). Reduced levels of VND7 expression in dwarfed ProASL20:ASL20-SRDX plants are consistent with the ectopic expression of VND7 in plants overexpressing ASL20.
Figure 6.
Effects of ASL20-SRDX on Expression of Genes Related to TE Differentiation.
(A) Expression of TE differentiation-related genes in ProASL20:ASL20-SRDX plants. Real-time PCR was performed with total RNA prepared from ProASL20:ASL20-SRDX lines showing the dwarf phenotype (closed bars) and normal growth (open bars). To detect the expression of the endogenous ASL20 and that of ASL20-SRDX, primer pairs were used to amplify the 3′ untranslated region of ASL20 (ASL20 3′UTR) and the coding sequence of SRDX, respectively. Results are presented as averages of normalized relative transcript levels ± sd of three technical replicates.
(B) Induction of TE differentiation-related genes by VND7 overexpression. Real-time PCR was performed with total RNA prepared from XVE:VND7 lines that were (closed bars) or were not (open bars) treated with 10 μM β-estradiol for 8 h. Results are presented as averages of normalized relative transcript levels ± sd of three technical replicates.
We further analyzed the expression of several TE differentiation-related genes, including MAP65-8 (for MICROTUBULE-ASSOCIATED PROTEIN65 family protein), three secondary cell wall–related genes, IRREGULAR XYLEM3 (IRX3)/CesA7 (cellulose synthase) (Taylor et al., 1999), IRX12 (laccase) (Brown et al., 2005; Sawa et al., 2005), and XYN3 (xylanase) (Sawa et al., 2005), and a cell death–related gene, XCP1 (for XYLEM CYS PROTEASE) (Funk et al., 2002). A microarray analysis (Kubo et al., 2005) previously showed that the expression of these genes is induced during transdifferentiation of Arabidopsis suspension culture cells into TEs, along with the induction of ASL19, ASL20, and VND genes. Of the genes encoding the Arabidopsis MAP65 family of proteins, MAP65-8 was the only gene for which expression was upregulated in the Arabidopsis suspension culture cells after treatment to induce TE differentiation. We confirmed that the expression of these TE differentiation-related genes was induced by VND7 overexpression (Figure 6B). By contrast, reduced expression of MAP65-8, IRX3, IRX12, XYN3, and XCP1 was found in the population of dwarfed ProASL20:ASL20-SRDX lines (Figure 6A). These results show that the ASL20-SRDX protein downregulates the expression of genes related to TE differentiation, consistent with the downregulation of VND6/7 expression by the ASL20-SRDX protein.
DISCUSSION
ASL19 and ASL20 Positively Regulate TE Differentiation
Here, we investigated the function of ASL19 and ASL20 genes, which encode members of the Arabidopsis AS2/LBD family of proteins. Three lines of evidence implicate these two ASL genes in the regulation of TE differentiation. (1) Promoter analysis of ASL19 and ASL20 showed that they are specifically expressed in immature TEs (Figure 1). (2) The overexpression of ASL19 and ASL20 induced transdifferentiation of cells from nonvascular tissues into TE-like cells (Figure 2). (3) Aberrant TEs were formed when ASL20-SRDX, a dominant-negative version of ASL20, was expressed (Figure 5). Taken together, these results provide evidence that ASL proteins promote TE differentiation. Furthermore, similar expression patterns and the generation of TE-like cells by overexpression (Figures 1 and 2) support the notion that ASL19 and ASL20 function redundantly to regulate TE differentiation. By contrast, differences in phenotypic traits between plants overexpressing ASL19 and ASL20 (for instance, formation of ectopic shoots in Pro35S:ASL19 plants and differences in efficiencies to induce TE-like cells; Figure 2) suggest that ASL19 and ASL20 have specific activities and actions as well.
Although double mutations in ASL19 and ASL20 did not affect the formation of xylem vessels, expression of ASL20-SRDX protein caused defects in TE differentiation (Figure 5; see Supplemental Figure 8 online). The partial inhibition of TE differentiation was an effect opposite to that seen when ASL19/20 were overexpressed (Figures 2 and 5). These results suggest that the ASL20-SRDX protein dominantly interfered with the function of endogenous ASL proteins. A similar inhibitory effect of the repressor domain SRDX was also reported for the ASL18 protein (Okushima et al., 2007). With the exception of as2, mutants of Arabidopsis AS2/LBD family genes reported so far have not displayed any obvious phenotypes (Shuai et al., 2002; Nakazawa et al., 2003; Chalfun-Junior et al., 2005; Okushima et al., 2007). Transgenic plants overexpressing ASL17, ASL19, ASL20, and ASL22 developed leaves with similar morphological alternations (Figures 2B and 2C; see Supplemental Figures 3A and 3B online), suggesting a high degree of functional redundancy among the AS2/LBD proteins. The effects of asl19 and asl20 mutations are most likely masked by functional redundancy. We used the native ASL20 promoter to express ASL20-SRDX in plants. Among the several AS2/LBD genes tested, only ASL19 and ASL20 showed the capacity to induce the formation of TE-like cells (Figure 2; see Supplemental Figures 3C and 3D online). Therefore, the phenotype caused by ASL20-SRDX protein is most likely due to the inhibition of ASL proteins that redundantly function with ASL20.
ASL Genes Mediate a Feedback Pathway Downstream of VND6 and VND7
VND6 and VND7 are key positive regulators of TE differentiation. The overexpression of these genes induces transdifferentiation of nonvascular cells into TE-like cells, similar to when ASL19 and ASL20 are overexpressed (Kubo et al., 2005). We demonstrated that VND6 and VND7 are positive regulators for ASL19 and ASL20 expression and are essential for ASL19/20 expression in immature TEs (Figure 3). Therefore, we proposed that ASL19 and ASL20 act downstream of VND6 and VND7 in the regulation of TE differentiation. However, the xylem-deficient phenotype caused by the overexpression of dominant-negative versions of VND proteins was only partially rescued by ASL19 and ASL20 overexpression (Figures 4A to 4F; see Supplemental Figure 4B online). This observation raised three possibilities: (1) other factors under the regulation of VND6 and VND7 are required for the ASL19 and ASL20 proteins to fully function; (2) generation of ectopic TE-like cells by the overexpression of ASL genes is mediated by the expression of VND6 and VND7; and (3) ASL19 and ASL20 are involved in only some events that occur during TE differentiation, such as biosynthesis of secondary cell wall compounds and their deposition. The third possibility appears unlikely, because we found loss of cell contents in TE-like cells (Figure 2G), suggesting the occurrence of autolysis, which is typical of the final step of TE differentiation; moreover, these TE-like cells displayed patterned formation of wall thickenings and lignin deposition. Ectopic expression of VND7 in plants overexpressing ASL20 (Figures 4H, 4I, and 4K) supported the second possibility, although the first possibility could not be excluded. Patchy patterns of VND7 expression were similar to the distribution of TE-like cells produced in the cotyledon in response to ASL20 overexpression (Figures 2F and 4H). Restriction of ectopic VND7 expression to the mature region of the root also corresponded to the root region where TE-like cells were generated due to ASL20 overexpression (Figures 2J, 4K, and 4M). Since ASL20 overexpression did not alter the expression pattern of ATHB-8 (Figure 4N), ASL20 probably affected VND7 expression and generated TE-like cells without acquiring procambial cell–like identity. Considering that VND6 and VND7 are required for the expression of ASL19 and ASL20 and that VND7 expression occurs prior to that of ASL19/20 genes in immature TEs, it is possible that ASL19/20 genes are involved in a positive-feedback loop for VND7 expression (Figure 7). This proposal is also consistent with the reduced levels of VND6 and VND7 mRNAs found in plants expressing ProASL20:ASL20-SRDX (Figure 6A).
Figure 7.
A Model Showing the Feedback Pathway Mediated by ASL19 and ASL20 to Maintain VND6/7 Expression during TE Differentiation.
SCW, secondary cell wall.
Kubo et al. (2005) reported that T-DNA insertion mutants and expression of RNA interference constructs for VND6 and VND7 did not yield any detectable defects in plant morphology, presumably due to gene function redundancy. However, the ASL20-SRDX protein interfered with the formation of TEs even if the expression of VND6 and VND7 was not completely repressed (Figures 5 and 6A). These results suggest that ASL20 regulates the expression of other genes involved in TE differentiation as well as VND6 and VND7. This notion is consistent with the partial suppression of the xylem-deficient phenotype of Pro35S:VND7-SRDX plants by ASL19 and ASL20 overexpression. Yamaguchi et al. (2008) reported that expression domains of VND genes, including VND6 and VND7, partially overlapped and that VND7 interacted with other VND proteins. ASL19 and ASL20 may also regulate the expression of VND genes other than VND6 and VND7.
ASL20 Activity Is Required for Proper Development of Xylem Vessels
Plants expressing ASL20-SRDX protein from the ASL20 promoter were affected in several aspects of xylem vessel formation. We detected loose bundling of xylem vessels in the leaves of these plants, physical gaps between mature TEs in the same cell file, aberrant patterns of wall thickenings, and delayed formation and discontinuities of xylem vessels (Figures 5D to 5L). Formation of gaps between mature TEs and loose bundling of xylem vessels in the leaves were probably due to the reduced flexibility of pitted wall thickenings being unable to withstand the forces of stretching that occur during organ expansion. Arabidopsis mutants with discontinuities of xylem vessels are often dwarfed, similar to ProASL20:ASL20-SRDX plants (Turner et al., 2007). No alteration in the vascular patterns in the leaves or in the levels of ATHB-8 mRNA in transgenic plants indicates that ASL20-SRDX did not affect the formation of procambial cells (Figures 5C and 6A). Therefore, the delayed formation of wall thickenings and discontinuities of xylem vessels were due to a partial inhibition of TE differentiation by the ASL20-SRDX protein. Similar defects in TE differentiation were also observed in plants expressing dominant-negative versions of VND6 and VND7 proteins (Kubo et al., 2005).
Differentiation of TEs is a complex cellular process that is both temporally and spatially coordinated. This process includes cell expansion, rearrangement of cortical microtubules (MTs), secondary cell wall biosynthesis and deposition, and autolysis. Any changes in this series of events will result in the formation of inappropriate patterns of wall thickenings. Therefore, a positive feedback mechanism that could account for the expression of genes related to TE differentiation would be important to orchestrate this highly organized process. The sites for deposition of cellulose and other secondary cell wall compounds have been thought to be marked by bundles of cortical MTs, which are reorganized in developing TEs before and during the formation of secondary cell wall thickenings (Hogetsu, 1991; Gardiner et al., 2003; Oda et al., 2005). Interestingly, the formation of different types of wall thickenings by the overexpression of VND6 and VND7 suggests that they regulate the expression of genes involved in the reorganization of cortical MTs during TE differentiation. Furthermore, we showed that overexpression of the ASL20-SRDX protein resulted in the downregulation of MAP65-8 together with genes related to secondary cell wall formation and to autolysis in dwarf ProASL20:ASL20-SRDX transgenic lines (Figure 6A). MAP65 proteins are known to bundle MTs and localize to cortical MTs during TE differentiation (Mao et al., 2006). Expression of these TE differentiation–related genes was also induced by VND7 overexpression (Figure 6B). This result suggests that the ASL20-SRDX protein interferes with the expression of MAP65-8, IRX3, IRX12, XYN3, and XCP1 through the downregulation of VND6 and VND7, which leads to the formation of aberrant patterns of wall thickenings and the partial inhibition of TE differentiation. ASL19 and ASL20 may coordinate and stabilize gene expression status during the irreversible process of TE differentiation by maintaining or promoting VND6/VND7 expression (Figure 7).
The inhibitory effects of ASL20-SRDX on TE differentiation (Figures 5 and 6) suggest that ASL20 functions on specific DNA sequences to positively regulate gene expression. VND6 and VND7 are possible candidates for genes targeted by ASL19/20 proteins. However, the overexpression of VND6 and VND7 more efficiently generated TE-like cells than the overexpression of ASL19 and ASL20 (cf. Figures 2E and 2F with Figure 3B). Furthermore, ectopic expression of VND7 was detectable 3 d after the induction of ASL20 overexpression. By contrast, enhancement of VND7 expression in immature TEs was detected within 24 h of induction of ASL20 overexpression. These results suggest that additional factors are required for ASL proteins to induce transcription of VND6 and VND7. Identifying the components in protein complexes containing ASL19/20 is important to further elucidate the molecular functions of the ASL19 and ASL20 proteins.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Columbia) was used as the wild type. Plants were germinated on sterile Murashige and Skoog medium (0.8% agar) at 23°C under continuous light conditions after vernalization treatment (at 4°C in the dark for 2 d). Fourteen days after vernalization (DAV), plants grown on plates were transferred to soil and further grown in a greenhouse at 25°C with a photoperiod of 16 h of light and 8 h of darkness. Agrobacterium tumefaciens–mediated genetic transformation of Arabidopsis was done by the floral dip method (Clough and Bent, 1998).
Construction of Plasmids
cDNAs for ASL19, ASL20, VND6, and VND7 and promoter regions of ASL19 (2055 bp), ASL20 (2082 bp), VND7 (2000 bp), and ATHB-8 (2004 bp) were amplified from cDNA pools as well as from genomic DNA using PCR with specific primer pairs (see Supplemental Table 1 online). cDNA fragments for each gene and DNA fragments for the promoter regions of VND7 and ATHB-8 were digested at sites present in the linker sequences with restriction enzymes, and the resulting fragments were ligated into an entry vector, pENTR-1A (Invitrogen). Promoter regions of ASL19 and ASL20 were cloned into the pCR8/GW/TOPO vector (Invitrogen) and into the pGEM-T Easy vector (Promega), respectively. An AvaI-NotI fragment containing the ASL20 promoter in pGEM-T was inserted into pENTR-1A using SalI and NotI sites. A coding sequence for SRDX was translationally fused to the 3′ end of VND7 cDNA in pENTR-1A by PCR. The VND7 cDNA fragment of VND7-SRDX was replaced by VND6 and ASL20 cDNAs to generate VND6-SRDX and ASL20-SRDX. A translational fusion of ASL20 and EGFP was generated by the insertion of the EGFP coding sequence at the 3′ end of ASL20 cDNA from which the stop codon was removed in pENTR-1A. For ProASL20:ASL20-EGFP and ProASL20:ASL20-SRDX, fragments of ASL20-EGFP and ASL20-SRDX were inserted downstream of the ASL20 promoter in pENTR-1A, using EcoRI and XhoI sites. Promoter fragments and cDNAs in entry vectors were transferred into destination vectors pKGWFS7, pB2GW7 (Karimi et al., 2002), and pER8-DC by LR clonase (Invitrogen). ProASL20:ASL20-EGFP and ProASL20:ASL20-SRDX were transferred into pBA-DC-HA from which the cauliflower mosaic virus 35S promoter was removed.
Histochemical GUS Staining Assay
Transgenic plants were treated with 90% acetone for 20 min at 4°C, washed three times with 100 mM NaPO4 buffer (pH 7.0), and incubated with a staining solution [100 mM NaPO4 (pH 7.0), 10 mM EDTA, 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (LabScientific), 3 mM K4Fe(CN)6, 3 mM K3Fe(CN)6, and 0.1% Triton X-100] for 45 min to 2 h at 37°C. Samples were then incubated in acetic acid:ethanol (1:6) to fix the tissues and to remove chlorophyll.
Real-Time PCR
Total RNA was prepared with the RNeasy Plant Mini Kit (Qiagen) and the RNase-Free DNase Set (Qiagen). cDNA synthesis was performed using oligo(dT)20 primer (Invitrogen) and Ready-to-Go You Prime First-Strand Synthesis Beads (GE Healthcare). Real-time PCR was performed with the 7100 real-time PCR system and Power SYBER Green PCR Master Mix (AB) as described in the manufacturer's protocols. Sequences of primers used for real-time PCR are presented in Supplemental Table 1 online. Relative amounts of PCR products were calculated with the ΔΔCt method, with DNA fragments amplified with a primer pair specific to α-tubulin cDNA serving as an internal control.
Microscopic Analysis
Plant samples were fixed with acetic acid:ethanol (1:6) for 1 h or overnight. The fixed samples were serially treated with 70% ethanol, 35% ethanol, and 10% ethanol for 30 min. Staining of TE-like cells with fuchsin was performed according to Mähönen et al. (2006). Samples were mounted on a clearing solution (8 g of chloral hydrate, 1 mL of glycerol, and 2 mL of water) before observation. Images of leaf venation patterns were captured using a stereoscopic microscope (SMZ-U; Nikon) equipped with a digital camera (COOLPIX5400; Nikon). Fluorescence and Nomarski images were captured using Axioskop (Carl Zeiss) equipped with a HBO100 microscope illuminator (Carl Zeiss) and a cooled CCD camera system (Diagnostic Instruments).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative databases under the following accession numbers: ATHB-8 (At4g32880), ASL19 (At4g00220), ASL20 (Atg45420), IRX3 (At5g17427), IRX12 (At2g38030), MAP65-8 (At1g27920), VND6 (At5g62380), VND7 (At1g71930), XCP1 (At4g35350), and XYN3 (At4g08160).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mutations in ASL19 and ASL20.
Supplemental Figure 2. Expression of ProASL20:EGFP-GUS and ProASL19:EGFP-GUS in Floral Organs and Siliques.
Supplemental Figure 3. Overexpression of ASL17 and ASL22 cDNAs.
Supplemental Figure 4. Overexpression of VND6-SRDX.
Supplemental Figure 5. Enhancement of ASL20 Expression by Auxin.
Supplemental Figure 6. Expression of ASL19 and ASL20 in XVE-ASL19 Pro35S:VND7-SRDX Plants and XVE-ASL20 Pro35S:VND7-SRDX Plants.
Supplemental Figure 7. GUS Staining in Transgenic Plants Expressing ProVND7:EGFP-GUS.
Supplemental Figure 8. Delayed Formation and Discontinuities of Xylem Vessels.
Supplemental Table 1. Sequences of Primers.
Supplementary Material
Acknowledgments
We thank the Cold Spring Harbor Laboratory and the ABRC for providing seeds. We thank Yoshihisa Ueno (Nagoya University) and Rossana Henriques (The Rockefeller University) for critical reading of the manuscript and discussion. T.S. was supported by a fellowship from the Human Frontier Science Program Organization (Grant LT00194/2005-L/5). This work was supported by National Institutes of Health Grant GM-44640 to N.-H.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author (www.plantcell.org) is: Nam-Hai Chua (chua@mail.rockefeller.edu).
Online version contains Web-only data.
References
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121 4171–4182. [DOI] [PubMed] [Google Scholar]
- Borghi, L., Bureau, M., and Simon, R. (2007). Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri, E., Chuck, G., Vollbrecht, E., Rocheford, T., Martienssen, R., and Hake, S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.M., Zeef, L.A., Ellis, J., Goodacre, R., and Turner, S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-Garcia, S., Cheng, J.C., Nam, K.H., Li, J., and Chory, J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131 5341–5351. [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior, A., Franken, J., Mes, J.J., Marsch-Martinez, N., Pereira, A., and Angenent, G.C. (2005). ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal–distal patterning in Arabidopsis petals. Plant Mol. Biol. 57 559–575. [DOI] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Gregory, B.D., Ross, A.S., Yuan, H., Noguchi, T., Fujioka, S., Takatsuto, S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Evans, M.M. (2007). The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19 46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5 379–391. [DOI] [PubMed] [Google Scholar]
- Funk, V., Kositsup, B., Zhao, C., and Beers, E.P. (2002). The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128 84–94. [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J.C., Taylor, N.G., and Turner, S.R. (2003). Control of cellulose synthase complex localization in developing xylem. Plant Cell 15 1740–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M., Thomas, J., Collins, G., and Timmermansa, M.C.P. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Hogetsu, T. (1991). Mechanism for formation of the secondary wall thickening in tracheary elements—Microtubules and microfibrils of tracheary elements of Pisum sativum L and Commelina communis L and the effects of amiprophosmethyl. Planta 185 190–200. [DOI] [PubMed] [Google Scholar]
- Husbands, A., Bell, E.M., Shuai, B., Smith, H.M., and Springer, P.S. (2007). LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K., Umemura, I., Hasegawa, Y., Ashikari, M., Kitano, H., and Matsuoka, M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, Y., Nakanomyo, I., Motose, H., Iwamoto, K., Sawa, S., Dohmae, N., and Fukuda, H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313 842–845. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.C., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 12 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Wang, S., Yu, X., Yu, J., He, X., Zhang, S., Shou, H., and Wu, P. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43 47–56. [DOI] [PubMed] [Google Scholar]
- Mähönen, A.P., Bishopp, A., Higuchi, M., Nieminen, K.M., Kinoshita, K., Törmäkangas, K., Ikeda, Y., Oka, A., Kakimoto, T., and Helariutta, Y. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311 94–98. [DOI] [PubMed] [Google Scholar]
- Mao, G., Buschmann, H., Doonan, J.H., and Lloyd, C.W. (2006). The role of MAP65-1 in microtubule bundling during Zinnia tracheary element formation. J. Cell Sci. 119 753–758. [DOI] [PubMed] [Google Scholar]
- Motose, H., Sugiyama, M., and Fukuda, H. (2004). A proteoglycan mediates inductive interaction during plant vascular development. Nature 429 873–878. [DOI] [PubMed] [Google Scholar]
- Naito, T., Yamashino, T., Kiba, T., Koizumi, N., Kojima, M., Sakakibara, H., and Mizuno, T. (2007). A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71 1269–1278. [DOI] [PubMed] [Google Scholar]
- Nakazawa, M., Ichikawa, T., Ishikawa, A., Kobayashi, H., Tsuhara, Y., Kawashima, M., Suzuki, K., Muto, S., and Matsui, M. (2003). Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 34 741–750. [DOI] [PubMed] [Google Scholar]
- Oda, Y., Mimura, T., and Hasezawa, S. (2005). Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 137 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Pyo, H., Demura, T., and Fukuda, H. (2004). Spatial and temporal tracing of vessel differentiation in young Arabidopsis seedlings by the expression of an immature tracheary element-specific promoter. Plant Cell Physiol. 45 1529–1536. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Demura, T., Horiguchi, G., Kubo, M., and Fukuda, H. (2005). The ATE genes are responsible for repression of transdifferentiation into xylem cells in Arabidopsis. Plant Physiol. 137 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella, E., Francis, P., and Berleth, T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131 3445–3455. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The LATERAL ORGANBOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85 171–182. [DOI] [PubMed] [Google Scholar]
- Taramino, G., Sauer, M., Stauffer, J.L., Jr., Multani, D., Niu, X., Sakai, H., and Hochholdinger, F. (2007). The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 50 649–659. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S., Gallois, P., and Brown, D. (2007). Tracheary element differentiation. Annu. Rev. Plant Biol. 58 407–433. [DOI] [PubMed] [Google Scholar]
- Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., Xu, Y., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 4097–4107. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, M., Kubo, M., Fukuda, H., and Demura, T. (2008). VASCULAR-RELATED NAC-DOMAIN7 is involved in differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55 652–664. [DOI] [PubMed] [Google Scholar]
- Yamamoto, R., Demura, T., and Fukuda, H. (1997). Brassinosteroids induce entry into the final stage of tracheary element differentiation in cultured Zinnia cells. Plant Cell Physiol. 38 980–983. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.-H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.