Abstract
The microtubule-associated protein, MAP65, is a member of a family of divergent microtubule-associated proteins from different organisms generally involved in maintaining the integrity of the central spindle in mitosis. The dicotyledon Arabidopsis thaliana and the monocotyledon rice (Oryza sativa) genomes contain 9 and 11 MAP65 genes, respectively. In this work, we show that the majority of these proteins fall into five phylogenetic clades, with the greatest variation between clades being in the C-terminal random coil domain. At least one Arabidopsis and one rice isotype is within each clade, indicating a functional specification for the C terminus. In At MAP65-1, the C-terminal domain is a microtubule binding region (MTB2) harboring the phosphorylation sites that control its activity. The At MAP65 isotypes show differential localization to microtubule arrays and promote microtubule polymerization with variable efficiency in a MTB2-dependent manner. In vivo studies demonstrate that the dynamics of the association and dissociation of different MAP65 isotypes with microtubules can vary up to 10-fold and that this correlates with their ability to promote microtubule polymerization. Our data demonstrate that the C-terminal variable region, MTB2, determines the dynamic properties of individual isotypes and suggest that slower turnover is conditional for more efficient microtubule polymerization.
INTRODUCTION
Microtubules form networks of filaments and are organized into structurally distinct arrays, three of which are peculiar to plant cells: the interphase array of cortical microtubules located within the subplasmalemma layer of cytoplasm, the preprophase band, and the cytokinetic phragmoplast (Goddard et al., 1994). The interphase cortical array governs the direction of cell expansion and aligns cellulose microfibrils of the cell wall. Mutations in interphase microtubule organization can cause twisting of the cell files (reviewed in Ishida et al., 2007). The preprophase band is necessary to determine the division plane, and abolishing this structure causes disorganized cell divisions, resulting in aberrant tissue organization (Traas et al., 1995). The phragmoplast creates the cell plate that divides the daughter cells, and disruption of this network leads to defective cytokinesis (Yasuhara et al., 1993).
Microtubules are composed of tubulin, which is a heterodimeric protein of α and β subunits. Tubulin is capable of spontaneous polymerization dependent upon optimal environmental conditions. During polymerization, tubulin molecules are positioned within a microtubule so that α-tubulin faces the slow-growing minus end and β-tubulin faces the fast-growing plus end (Bayley et al., 1994; Wade and Hyman, 1997). With this polarity, microtubules can treadmill and also undergo rapid elongation or shortening excursions, known as dynamic instability. The importance of microtubule dynamics in plant cell growth and development has been demonstrated in several drug studies. For example, increasing microtubule elongation and stabilization using taxol or promoting microtubule shortening using propyzamide disturbs normal plant cell shape and plant growth (Yasuhara et al., 1993; Baskin et al., 1994; Anthony and Hussey, 1999). This indicates that microtubule dynamics must be tightly regulated in vivo, and this is controlled by a set of heterogeneous microtubule-associated proteins (MAPs).
De novo formation of new microtubule arrays or transformation of an existing array requires a set of minus-end binding proteins that promote the initiation of new microtubules (reviewed in Pastuglia and Bouchez, 2007) and a set of proteins that bind plus ends and determine microtubule lifetime via the control of elongation or shortening (reviewed in Hamada, 2007; Sedbrook and Kaloriti, 2008). In addition, existing microtubules need to be stabilized or targeted to a particular site within the cell by proteins that bind at sites along the microtubule surface (e.g., phospholipase D [Gardiner et al., 2001] can link cortical microtubules to plasmalemma). MAPs that bind along the length of microtubules can cause microtubules to bundle, which is important for the structural organization of all microtubule arrays (Hamada, 2007). The structure of these bundles is determined by the distance between the microtubules: 8- to 10-nm cross-bridges generated by Microtubule Organization1/Gemini1 (Yasuhara et al., 2002), whereas Wave Dampened2 appears to glue microtubules together (Perrin et al., 2007).
The microtubule-bundling protein MAP65 was originally isolated biochemically from tobacco (Nicotiana tabacum) tissue culture cells (Jiang and Sonobe, 1993). Characterizing the corresponding MAP65 cDNA, the localization of MAP65 protein, and the biochemical properties of recombinant MAP65 (Smertenko et al., 2000, 2004; Hussey et al., 2002) identified it as a functional homolog of members of a family of divergent proteins that includes Saccharomyces cerevisiae Ase1p (Pellman et al., 1995) and Homo sapiens PRC1 (Jiang et al., 1998). These proteins are involved in the maintenance of the spindle midzone, a structure that is formed between daughter chromatids during anaphase B and is essential for the completion of mitosis. MAP65-like proteins have been suggested to maintain the integrity of the midzone by cross-linking and stabilizing interdigitating antiparallel microtubules. Carrot (Daucus carota) MAP65 (Chan et al., 1999) and Arabidopsis thaliana MAP65-1 (Smertenko et al., 2004) bundle microtubules via 25-nm cross-bridges. MAP65-1 has a weak effect on microtubule polymerization (Smertenko et al., 2004; Mao et al., 2005b), and fluorescence recovery after photobleaching (FRAP) analysis of green fluorescent protein (GFP):MAP65-1 reveals that it has a very high turnover on microtubules (Chang et al., 2005). Polymerization rates and the frequency of catastrophes of microtubules decorated with MAP65-1:GFP in suspension culture cells were similar to those in the control; however, depolymerization rates were reduced (Van Damme et al., 2004b). Taken together, these data suggest that bundling of microtubules by At MAP65-1 in vivo has a weak effect on their dynamics, making MAP65-1 ideally suited for the spatial organization of highly dynamic plant microtubule arrays (Chang et al., 2005).
Arabidopsis MAP65-1 has two microtubule binding regions located in the C-terminal half of the protein (Smertenko et al., 2004). The sequence of one of these regions is highly conserved among all members of the Ase1p/PRC1/MAP65 family (Schuyler et al., 2003), while the second region at the extreme C terminus is not conserved and is composed of a random coil domain and contains a series of phosphorylation sites for several different kinases, including mitogen-activated protein kinase and cyclin-dependent kinase (Sasabe et al., 2006; Smertenko et al., 2006). Phosphorylation weakens the binding of MAP65-1 to microtubules. Although both microtubule binding regions can bind microtubules independently, successful microtubule bundling requires dimerization, which occurs within the N-terminal half of the protein (Smertenko et al., 2004).
Unlike animal and fungal genomes, which contain one or two MAP65 homologs, the Arabidopsis genome contains a family of nine MAP65 genes (Hussey et al., 2002) sharing between 25 and 78% amino acid sequence identity. Diversity in primary sequence underlies the variability in MAP-65 isotype activity. For example, the biochemical properties of two members of the Arabidopsis gene family have been compared, and it has been reported that MAP65-1 and MAP65-6 have different effects on microtubule polymerization and bundling (Mao et al., 2005b). Moreover, studies using GFP fusions to MAP65s reveal differences in their intracellular localization (VanDamme et al., 2004a). Analysis of mutants has only identified a phenotype for one member of the Arabidopsis MAP65 gene family so far, mutations in MAP65-3. The pleiade/MAP65-3 mutants have either an absent or a modified microtubule binding domain, which results in abnormal phragmoplast formation and incomplete cytokinesis with the accumulation of multinucleate cells, resulting in abnormal root development (Muller et al., 2004, Smertenko et al., 2004). Another allele of pleiade/MAP65-3 has been identified, and it has been shown that this is resistant to nematode invasion (Caillaud et al., 2008).
Here, further analysis of the primary structure of Arabidopsis MAP65 proteins has identified five phylogenetic clades that are also conserved in the monocot plant rice (Oryza sativa). We have compared the localization and biochemical activities of at least one member from each clade and demonstrated that three of them, MAP65-1, MAP65-3, and MAP65-5, have tight association with the phragmoplast midzone. These isotypes have different dynamics of association with the midzone and cortical microtubules in vivo, which correlates with the ability of the protein to promote microtubule polymerization and is determined by the isotype-specific C-terminal microtubule binding domain MTB2.
RESULTS
Sequence Analysis of MAP65-Like Proteins
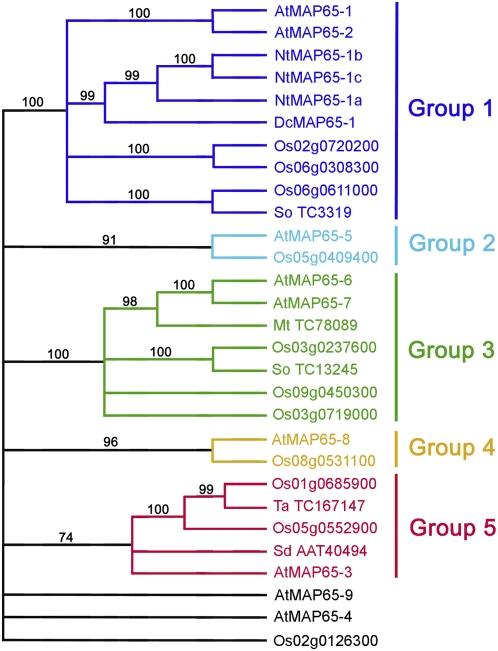
The Arabidopsis MAP65 gene family contains nine members that share identities between 28% (MAP65-4 and MAP65-8) and 78% (MAP65-1 and MAP65-2). On the phylogenetic tree of Hussey et al. (2002), MAP65-1 and MAP65-2 constitute a separate clade, as do MAP65-6 and MAP65-7, while the remaining five proteins had no close partners within the same species. A similar phylogenetic arrangement of proteins could be generated for the rice MAP-65 family. When both gene families were combined in one phylodendrogram (Figure 1), they formed five distinct clades with a high statistical probability (bootstrap values > 70%), and these clades were named groups 1 to 5. Each group included at least one Arabidopsis and one rice gene, indicating that there is higher conservation of the protein primary structure between MAP65 homologs from different species within a group than between proteins within the same species. Group 1 includes the previously published tobacco MAP65-1a, -b, and -c isotypes and carrot MAP65-1 sequences (Smertenko et al., 2000, 2004; Chan et al., 2003). Three proteins, MAP65-4, MAP65-9, and Os 02d0126300, did not fall into any group (bootstrap values < 70%) and therefore represent separate branches on the tree. Randomly chosen MAP65 homologs from other plant genomes did fall into one or more of groups 1 to 5, further indicating the conservation of theses groups across the plant kingdom. MAP65 homologs from vertebrates, invertebrates, and fungi did not fall into any of the five groups. As monocotyledons and dicotyledons diverged 150 million years ago, these data indicate that in the evolution of flowering plants there has been a functional constraint on the divergence of the primary structure of MAP65 isotypes.
Figure 1.
Phylodendrogram of MAP65 Gene Families from Arabidopsis and Rice.
The tree was rooted using sequences of animal and fungal MAP65 homologs (not shown on the tree). At, Arabidopsis thaliana; Dc, Daucus carota; Mt, Medicago truncatula; Nt, Nicotiana tabacum; Os, Oryza sativa; Sd, Solanum demissum; So, Saccharum officinarum; Ta, Triticum aestivum. Numbers represent GenBank accession numbers for the corresponding genes. [See online article for color version of this figure.]
Anti-MAP65 Antibodies
In order to address the question of any functional specification for each of the five Arabidopsis MAP65 groups, we first determined the intracellular localization of individual isotypes. We raised polyclonal antisera in mice against full-length MAP65-5 and MAP65-6 and fragments of MAP65-3 (N-terminal fragment), -4, -8, and -9 selected to be specific for each isotype (see Supplemental Figure 1A online). Antibodies against MAP65-1 were already in hand (Smertenko et al., 2004). The specificity of the antibodies for each isotype was tested by immunoblotting a total protein extract from Arabidopsis tissue culture cells and purified recombinant MAP65 isotypes. Each antibody recognized a band similar to the predicted molecular weight of the corresponding MAP65 and also the corresponding recombinant protein (see Supplemental Figures 1B and 1C and Supplemental Table 1 online). The antibody raised against MAP65-1 cross-reacted with the MAP65-2 recombinant protein as expected, as these are so similar in primary sequence, while anti-MAP65-4 and anti-MAP65-8 did not cross-react with any of the recombinant proteins used but produced a specific signal with the total protein extract.
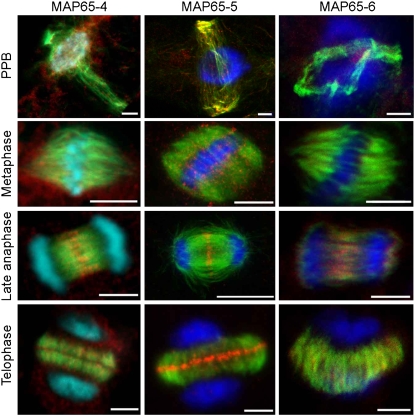
The results of the immunolocalization of MAP65 isotypes in Arabidopsis tissue culture cells are summarized in Table 1 and shown in detail for MAP65-4, MAP65-5, and MAP65-6 in Supplemental Figures 2 to 4 online (the localization of MAP65-1 and MAP65-3 is described in Smertenko et al., 2004 and Müller et al., 2004, respectively). Comparison of the staining patterns revealed that the major difference between the isotypes was at the cell division midzone. While MAP65-1 (Smertenko et al., 2004), MAP65-3 (Muller et al., 2004), and MAP65-5 concentrated at the midzones of the spindle during anaphase B and the phragmoplast, MAP65-4 and MAP65-6 concentrated at the anaphase spindle midzone but showed a more dispersed localization along the phragmoplast microtubules (Figure 2; Table 1; see Supplemental Figures 2 to 4 online). MAP65-6 was occasionally observed at the midzone in the expanding edge of the phragmoplast (see Supplemental Figure 4 online). A speckled staining particularly in metaphase cells was also apparent with all antibodies. This may reflect either a background staining or cytoplasmic MAP65 proteins and/or complexes not bound to microtubules, perhaps as a consequence of their phosphorylation state (Mao et al. 2005a; Sasabe et al., 2006; Smertenko et al., 2006). Out of all isotypes, MAP65-4 has the strongest accumulation around the nucleus during preprophase band formation (Figure 2). We also compared the localization of the MAP65s during interphase using protoplasts isolated from Arabidopsis tissue culture cells. Double staining with anti-MAP65-1 and anti-MAP65-5 demonstrated that both proteins decorated the same microtubules (see Supplemental Figure 5A online). Anti-MAP65-6 stained short microtubule fragments in a punctate manner, along microtubules that also stained with anti-MAP65-1 (see Supplemental Figure 5B online). Anti-MAP65-3, which was raised against the N-terminal fragment, occasionally stained cortical microtubules and the metaphase spindle as well as the cell division midzone (see Supplemental Figures 5C and 5D online); the cortical microtubule and metaphase spindle staining was not seen previously using an antibody raised to the C-terminal part of MAP65-3 (Muller et al., 2004). The anti-MAP65-8 and -9 antibodies did not stain any microtubule structures.
Table 1.
Localization of MAP65 Proteins in Microtubules Arrays
| Protein | Cortical Microtubules | Preprophase Band | Metaphase | Anaphase | Phragmoplast | Phragmoplast Midzone |
|---|---|---|---|---|---|---|
| MAP65-1/2 | + | + | − | + | + | + |
| MAP65-3 | ± | + | − | + | + | + |
| MAP65-4 | − | ± | − | + | + | − |
| MAP65-5 | + | + | − | + | + | + |
| MAP65-6 | − | + | − | + | + | − |
| MAP65-8 | − | − | − | − | − | − |
| MAP65-9 | − | − | − | − | − | − |
− indicates no detectable staining, + indicates strong staining, and ± indicates weak staining.
Figure 2.
Immunolocalization of MAP65-4, MAP65-5, and MAP65-6 in the Preprophase Band (PPB), Mitotic Spindle (Metaphase), Late Anaphase Spindle (Late Anaphase), and Phragmoplast (Telophase) in Arabidopsis Suspension Culture Cells.
Microtubules are in green, MAP65 isotypes in red, DNA in blue, and colocalization of microtubules and MAP65s in yellow. Note that out of three antibodies shown, only MAP65-4 produces a strong signal around the nucleus at the site of the spindle formation during the preprophase band stage, while only MAP65-5 shows strong concentration at the phragmoplast midzone during telophase. Bars = 5 μm.
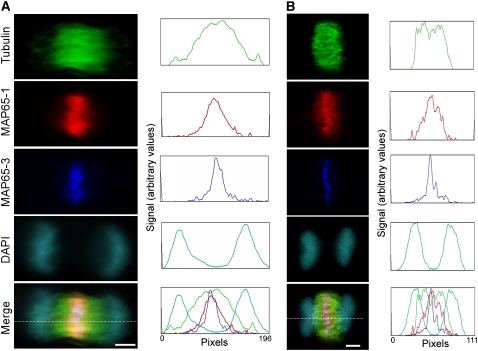
Interestingly, anti-MAP65-3 stained a very narrow line in the phragmoplast midzone compared with MAP65-1. In cells triple stained for tubulin, MAP65-1, and MAP65-3 (Figure 3), the localization of both isotypes in anaphase was similar, but during telophase the anti-MAP65-1–stained zone was almost two times wider than that stained using anti-MAP65-3, suggesting that MAP65-3 is more specifically associated with the phragmoplast midzone.
Figure 3.
Comparison of MAP65-1 and MAP65-3 Immunolocalization in the Phragmoplast Midzone in Arabidopsis Suspension Culture Cells.
Single confocal Z-sections (1 μM thick) show early telophase (A) and late telophase (B) cells stained for tubulin (green), MAP65-1 (red), MAP65-3 (blue), and DNA (cyan), with corresponding optical density plots recorded along the lines indicated in the merged image panels. DAPI, 4′,6-diamidino-2-phenylindole. Bars = 5 μm.
Dynamics of MAP65 Binding to the Phragmoplast Midzone
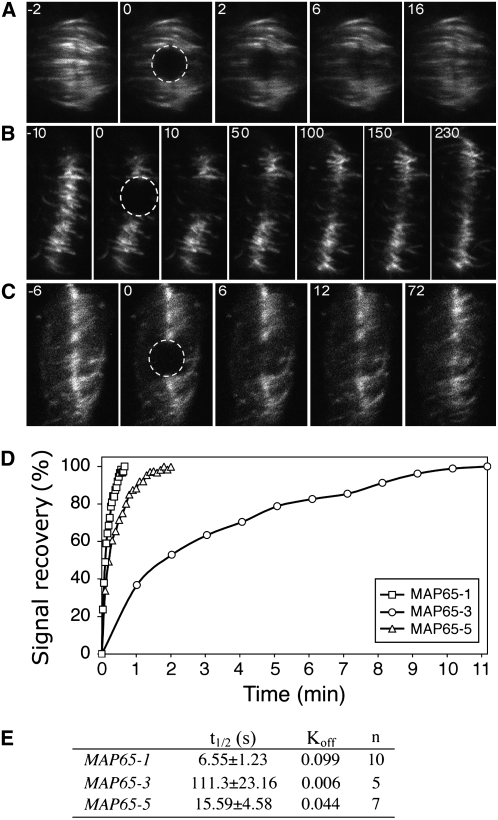
The differences in the localization between MAP65-1 and MAP65-3 at the phragmoplast midzone prompted a more detailed analysis of their association with microtubules in this region as well as other isotypes that located to this site, specifically MAP65-5. Previously, we noted that in At MAP65-1:GFP–expressing BY-2 cell lines, the GFP fluorescence concentrated at the midzone of the early phragmoplast, and as the phragmoplast expanded the margin of fluorescence was broadened (Chang et al., 2005). By contrast, in BY-2 cell lines expressing At MAP65-3:GFP and also At MAP65-5:GFP, the fluorescence stayed at the midzone throughout phragmoplast formation (Van Damme et al., 2004a). Using these three cell lines, we tested whether each of these isotypes had a more dynamic association with microtubules in the phragmoplast. To do this, we performed a FRAP analysis of BY-2 cell lines expressing C-terminal GFP fusions of each isotype under the control of the 35S promoter. For MAP65-1:GFP and MAP65-5:GFP, fluorescence intensity in the photobleached area recovered at least five times faster than in MAP65-3 (Figure 4). The mean value of replicates for each GFP chimera is plotted in Figure 4D. Fitting the curves into a single exponential equation generated t1/2 values (the time taken for 50% of the signal to recover) of 6.55 ± 0.86 s for MAP65-1 (n = 10), 15.94 ± 4.58 s for MAP65-5 (n = 7), and 111.30 ± 23.16 s for MAP65-3 (n = 7) (Figure 4E). Therefore, the more compact immunostaining of MAP65-3 in the phragmoplast midzone in Figure 3 correlates with a slower turnover compared with MAP65-1. These data suggest that functional specialization of MAP65 isotypes has generated an array of proteins with different dynamics at the division midzone, where they bundle microtubules with different efficacy.
Figure 4.
Analysis of MAP65 Turnover in the Phragmoplast Midzone.
(A) to (C) Sequences of images taken from the FRAP experiments in BY-2 cells expressing GFP fusions of MAP65-1 (A), MAP65-3 (B), and MAP65-5 (C). The number on each image corresponds to the time point in the experiment, where 0 is the image taken after completion of the photobleaching phase. The bleached area is circled; the signal in this area was measured and is shown in (D).
(D) Rates of fluorescence signal recovery after photobleaching for each chimeric protein. The signal is expressed as a percentage of the original value read before the photobleaching phase.
(E) Table summarizing the FRAP measurements of MAP65-1:GFP, MAP65-3:GFP, and MAP65-5:GFP within the phragmoplast. The recovery rate of the fluorescence signal (t1/2) and the first-order rate constant (Koff) were calculated in n number of cells.
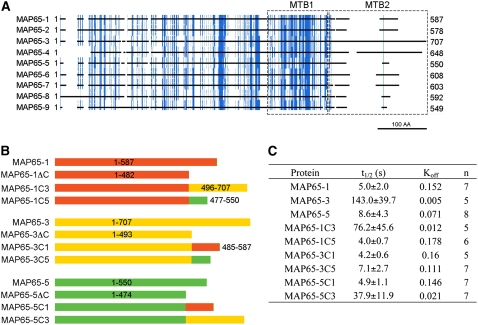
The C-Terminal Nonconserved Region Determines the Turnover of MAP65 on Microtubules
The microtubule binding domain of MAP65-1 is located in the C-terminal half of the protein and composed of two regions. One is more distal to the C terminus (MTB1; Figure 5A), conserved not just among At MAP65 isotypes and MAP65s from other plant species but also in MAP65 homologs in animals and fungi (Schuyler et al., 2003). Mutation in this region significantly reduces, but does not abolish, the ability of MAP65 to bind microtubules (Smertenko et al., 2004). The second region (MTB2; Figure 5A) is not well conserved between MAP65-like proteins from different phyla or between the plant group 1 to 5 MAP65 isotypes. This is in spite of the fact that this region harbors the phosphorylation sites that regulate MAP65-1's ability to bind microtubules. However, the phylodendrogram of MTB2 regions from MAP65 proteins (see Supplemental Figure 6 and Supplemental Data Set 2 online) for the most part retains the same phylogenetic clades as shown in Figure 1 constructed using the full-length sequences, indicating that there is considerable identity in this region between MAP65s within the same group but not between groups. MTB2 is basic in MAP65-1, with a predicted pI of 10.55. Other members of the gene family are similar in this respect, with pI values between 9.93 for MAP65-4 and 10.93 for MAP65-8 (see Supplemental Table 1 online). As the C-terminal variable region of tubulin, a known interaction site of MAPs, has an acidic pI, interaction of MTB2 with microtubules could be charge-dependent. Indeed, phosphorylation of MTB2, which reduces the pI, also reduces the ability of MAP65-1 to bind microtubules (Smertenko et al., 2006). Perhaps it is the variable MTB2 that gives the isotypes their specific properties.
Figure 5.
The MTB2 Region Modulates the Turnover of MAP65 on Microtubules.
(A) Diagram showing identities among At MAP65 amino acid sequences. The sequences were aligned, and residues conserved in more than five proteins were colored in blue, with the protein-coding sequence represented by black lines with breaks where there are gaps in the alignment. Microtubule binding regions are boxed and labeled as MTB1 and MTB2. The scale bar corresponds to 100 amino acids.
(B) Diagram of the constructed chimeric proteins. MAP65-1 and its fragments are colored in red, MAP65-3 and its fragments are colored in yellow, and MAP65-5 and its fragments are colored in green.
(C) Table summarizing the FRAP measurements for MAP65-1, MAP65-3, and MAP65-5 and their chimeras as shown in (B) on microtubules of N. benthamiana leaf epidermal cells. The recovery rate of the GFP signal (t1/2) and Koff were calculated in n number of cells.
To find out whether the MTB2 region determines the turnover rate of the MAP65s on microtubules, we constructed a series of GFP translational fusions with wild-type MAP65-1, -3, and -5 and also with the same proteins in which the MTB2 region had been swapped, as shown Figure 5B. The resulting nine constructs were transiently expressed in Nicotiana benthamiana leaf epidermal cells, and the t1/2 values of the GFP fusion proteins were measured by FRAP. In a similar fashion to the turnover rate on the cortical microtubule array of BY-2 cells, MAP65-3 had a much slower turnover (143 ± 39.7 s; Figure 5C) than At MAP65-1 (5.0 ± 2.0 s) and MAP65-5 (8.6 ± 4.3 s). It should be noted here that transfection of N. benthamiana leaf epidermal cells occurs with variable efficiency, so that the level of protein expression differs among neighboring cells. We assessed whether the t1/2 would be different in cells with higher or lower expression of the protein and found that the variation was indeed within the margin of sd; therefore, we conclude that the t1/2 does not depend on the amount of protein but on its intrinsic properties. Expression of the chimeras in which MTB2 of MAP65-3 is fused to the N terminus of MAP65-1 (MAP65-1C3) or MAP65-5 (MAP65-5C3) resulted in 15-fold (76.2 ± 45.6 s) and 4.5-fold (37.9 ± 11.9 s) increases in the t1/2 compared with wild-type MAP65-1 and MAP65-5, respectively (Figures 5B and 5C). Correspondingly, similar experiments in which the MTB2 of MAP65-1 (MAP65-3C1) or MAP65-5 (MAP65-3C5) was fused to the N terminus of MAP65-3 decreased the t1/2 by 34-fold and 20-fold compared with the wild-type MAP65-3, respectively (Figure 5C). Moreover, the MTB2 of MAP65-5 when fused to the N terminus of MAP65-1 (MAP65-1C5) had a similar t1/2 to wild-type MAP65-1, while the MTB2 of MAP65-1 fused to the N terminus of MAP65-5 (MAP65-5C1) increased the t1/2 compared with wild-type MAP65-5 by twofold. We can conclude that the MTB2 region plays a significant role in determining the binding energy of MAP65s to microtubules.
MTB2 Modulates the Effect of MAP65 on Microtubule Polymerization
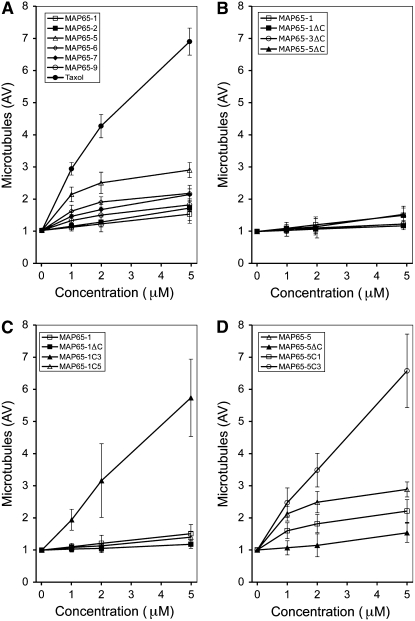
From the FRAP analyses, we have shown that the MAP65 isotypes have distinct affinities for microtubules and that this is influenced by the nature of the MTB2 region; however, it remains to be determined how these different affinities affect their molecular function. To address this question, we generated recombinant proteins and used these in microtubule polymerization assays in vitro. We were able to express and purify MAP65-1 and MAP65-5 and compare their effects on microtubule polymerization (Figure 6A). Several attempts were made to produce recombinant MAP65-3 in bacterial, insect cell, and yeast expression systems, but in all cases the protein could not be sufficiently concentrated for microtubule polymerization assays because of its low solubility under conditions for microtubule polymerization. Equal concentrations of tubulin were mixed either with microtubule stabilization buffer or microtubule stabilization buffer containing the MAP65 protein supplemented with 1 mM GTP and polymerized for 40 min. Subsequently, microtubules were centrifuged and separated on an SDS-PAGE gel, and the amount of tubulin in the pellet was quantified. We have shown previously that MAP65-1 has a minute effect on microtubule polymerization in comparison with taxol, a known inducer of microtubule polymerization (Smertenko et al., 2004). In comparison with MAP65-5, 5 μM MAP65-1 increased the amount of tubulin in the pellet by 1.7-fold compared with the control, while the same concentration of MAP65-5 increased this by 4.1-fold. The effect of MAP65-5 on microtubule polymerization was much less significant than the effect of taxol (Figure 6A). Therefore, compared with MAP65-1, the slower turnover of MAP65-5 on microtubules in vivo reflects its stronger effect on microtubule polymerization in vitro.
Figure 6.
Effects of MAP65 Isotypes and Chimera Proteins on Microtubule Polymerization.
The amount of microtubule polymer was measured using a microtubule sedimentation assay in which samples contained an equal concentration of tubulin (10 μM) and increasing concentrations of the following.
(A) MAP65-1 (open squares), MAP65-2 (closed squares), MAP65-5 (open triangles), MAP65-6 (open diamonds), MAP65-7 (closed diamonds), MAP65-9 (open circles), or taxol (closed circles).
(B) MAP65-1 (open squares), MAP65-1ΔC (closed squares), MAP65-3ΔC (open circles), or MAP65-5ΔC (closed triangles).
(C) MAP65-1 (open squares), MAP65-1ΔC (closed squares), MAP65-1C3 (closed triangles), or MAP65-1C5 (open triangles).
(D) MAP65-5 (open triangles), MAP65-5ΔC (closed triangles), MAP65-5C1 (open squares), or MAP65-5C3 (open circles).
Constructs are diagrammed in Figure 5. Three independent experiments were performed for each protein, and error bars indicate sd.
The ability of MAP65-5 to promote microtubule polymerization does depend on the presence of its MTB2 region, because without this (MAP65-5ΔC) its microtubule polymerization activity is very low. In fact, MAP65-5ΔC and the similarly truncated MAP65-3 (MAP65-3ΔC) have a similar effect on microtubule polymerization as MAP65-1 and MAP65-1ΔC (Figure 6B). By substituting the MTB2 of MAP65-1 and MAP65-5 with that of MAP65-3, these proteins became more potent than their native counterparts in promoting microtubule polymerization (Figures 6C and 6D), and this correlates with the concomitant reduced turnover rate for each of the proteins in vivo (Figure 5C). Correspondingly, MAP65-5C1 (Figure 5B), composed of the MAP65-5 N terminus and the MTB2 of MAP65-1, had a reduced ability to polymerize microtubules compared with native MAP65-5 (Figure 6D).
We have also taken other members of the Arabidopsis MAP65 gene family, generated recombinant proteins from bacterially expressed cDNAs, and tested their ability to promote microtubule polymerization in vitro. MAP65-2, -6, -7, and -9 all promote microtubule polymerization to varying degrees (Figure 6A), with MAP65-2 being similar to MAP65-1 (both are in group 1) and MAP65-6 being similar to MAP65-7 (both are in group 3). These data demonstrate that conservation of sequence within a group correlates with similar effects on microtubule polymerization.
All of these data demonstrate that the MTB2 region that is conserved in the five MAP65 groups, maintained in the plant kingdom through evolution, defines the affinity of each MAP65 for microtubules and the ability of each MAP65 to promote microtubule polymerization.
DISCUSSION
Conservation of Groups of MAP65s and the At MAP65 Family
Comparison of MAP65 gene families from Arabidopsis and rice revealed that according to the primary sequence, proteins segregate into five phylogenetic clades or groups with high statistical significance, giving bootstrap values of 74% and above. Each group contains at least one Arabidopsis and one rice gene. As monocotyledons and dicotyledons diverged 150 million years ago, the maintenance of these five groups would imply some functional constraint. Despite originating from phylogenetically distinct organisms, all members of this MAP65-like family have similar predicted secondary structures. The region corresponding to amino acid residues 1 to 482 of At MAP65-1 is mainly α-helical, has a slightly acidic pI, and contains a conserved region responsible for microtubule binding (MTB1). The smaller C-terminal domain (MTB2) is predicted to be a random coil and very alkaline and is the most divergent in primary sequence between all MAP65-like proteins. Although MTB2 is highly variable in both amino acid composition and length, it is more conserved between members of each phylogenetic group than between members from different groups, underlining a functional coherence within a group. Accordingly, in vitro experiments with At MAP65-1 and -2 from group 1 and At MAP65-6 and -7 from group 3 demonstrate that members of the same group have similar effects on microtubule polymerization. However, as two Arabidopsis genes (MAP65-4 and MAP65-9) and one rice gene (Os 02g0126300) fall outside the five groups and have divergent MTB2 regions, there may be some MAP65 isotypes that function in a species-specific or genus-specific manner.
Using Genevestigator, we have analyzed available Affymetrix microarray experiments (Zimmermann et al., 2004) and extracted the transcript levels for each Arabidopsis MAP65. These values were normalized by the mean, and the sd of the signal was calculated for the different tissues and developmental stages (see Supplemental Table 2 online). The resulting numerical values represent the relative variability for each gene transcription program. MAP65-1, MAP65-4, and MAP65-7 have the lowest variability in the expression levels across tissues/developmental stages, with signal sd equal to 0.39, 0.43, and 0.31, respectively, indicating ubiquitous expression at similar levels. Other members of the gene family, on the other hand, show greater variability (signal sd for all the others is above 0.7), indicating tissue-specific enrichment for these isotypes. The highest signal sd value, at 4.65, is for pollen-specific MAP65-9, which does not fall into any of the groups, suggesting that it may perform specialized functions in pollen. In another example, MAP65-7 is ubiquitously transcribed and has a signal sd of 0.31 (see Supplemental Table 2 online), while the transcription levels of its closest homolog from group 3, MAP65-6, peak in hypocotyls, xylem, cork, and root hair zone, with signal sd of 0.76. Therefore, although the primary structure is similar within the same group, the expression programs can be different. This analysis demonstrates that each tissue expresses a representative of each of the five groups, suggesting that each isotype performs a tailored role within the cell. In this context, it is pertinent to note that the group 5 isotype MAP65-3 is essential for the completion of cytokinesis in roots (Muller et al., 2004; Caillaud et al., 2008), but this gene is expressed throughout the plant and in particular in the shoot apex. However, the pleiade/MAP65-3 mutant plants remain viable and fertile, indicating that the lack of MAP65-3 is compensated for by the expression of the other MAP65 isotypes, demonstrating that there is functional redundancy between groups.
The expression of Arabidopsis MAP65 genes changes not only during plant development but also through the cell cycle, with several MAP65 isotypes expressed at each stage. Menges et al. (2003) analyzed changes of Arabidopsis transcripts in tissue culture cells synchronized in G1/S-phases by sucrose starvation or in G2/M-phases using the DNA synthesis inhibitor aphidicolin. We have used these data for the analysis of MAP65 transcripts (see Supplemental Figure 7 online) and identified two major patterns of expression. First, some of the genes are upregulated during the G1/S transition (see Supplemental Figure 7A online): MAP65-2 (up 3.2-fold), MAP65-5 (up 3.9-fold), and MAP65-8 (up 3.4-fold). Second, some genes are upregulated toward the late G2- or M-phase (see Supplemental Figure 7B online): MAP65-2 (up 3.5-fold), MAP65-3 (up 6.4-fold), and MAP65-4 (up 6.9-fold). The increase of MAP65-3 and -4 transcript levels starts at the end of S-phase. These variations are modest compared with those of known cell cycle regulators (e.g., 14.9- and 97-fold increases for Arabidopsis cyclin A1 and cyclin B1 transcripts, respectively).
Differential Localization of At MAP65 Isotypes
Immunolocalization of the MAP65 isotypes in Arabidopsis tissue culture cells showed a differential localization to microtubule arrays. The data show differences from the published localizations, where GFP chimeras were expressed under the control of the strong cauliflower mosaic virus 35S promoter in BY-2 cells (Van Damme et al., 2004a). Principally, in other studies, At MAP65-4:GFP was observed to bind the metaphase spindle, At MAP65-8:GFP bound cortical microtubules, and At MAP65-6:GFP bound vesicle-like structures. These features were not observed in our immunolocalization studies. These differences could occur because of the limitations in the experimental procedures used. (1) The interaction of a MAP65 isotype with microtubules in the wild-type cells can be downregulated at a particular cell cycle or developmental stage by posttranslational modification, which is then overcome in the overexpressing lines. (2) In the case of MAP65-8, where we see no colocalization to microtubules, it could be that this isotype, although expressed, is not utilized in microtubule arrays in these cells. The same could be true for MAP65-9, which is specific to stamens and pollen. This phenomenon has also been seen in the case of maize β-tubulins, where β2-tubulin is transcribed in many tissues but utilized only in male meiocytes (Eun and Wick, 1998). (3) The amount of protein is below the sensitivity of the immunofluorescence technique. (4) The epitope for the antibodies are not exposed at these sites. (5) The overexpression of particular MAP65s might cause the isotypes to locate to structures not normally bound by the isotype. (6) The overexpression of a particular MAP65 isotype causes the downregulation of its counterpart or other isotypes, allowing the transgene to function where it is not normally required upon selection of stable lines. This, for example, is known to occur upon overexpression of tubulin isotypes in plant cells (Anthony and Hussey, 1998). Although the immunolocalization and expression of GFP chimeras in some cases yield different data, both approaches show that there is differential localization of MAP65 isotypes on microtubule arrays and demonstrate that each microtubule array is constructed using several isotypes. Moreover, it appears that while most isotypes localize to the anaphase midzone, MAP65-1, MAP65-3, and MAP65-5 are also more prevalent in the phragmoplast midzone. The remaining sections of this discussion focus on this observation.
Turnover of MAP65-1, -3, and -5 on Microtubules
We have shown previously that the interaction of MAP65-1 with microtubules in vivo is highly dynamic, more dynamic even than the turnover of tubulin in microtubules themselves (Chang et al., 2005). This led us to suggest that MAP65-1 is responsible for the cross-linking of microtubules in dynamic microtubule arrays that need to retain their spatial organization (Chang et al., 2005). The rate of turnover of MAP65-1 was similar in all microtubule arrays, including the cortical array and the phragmoplast, indicating that this isotype performs the same dynamic role in the different microtubule organizations. A striking feature of the different MAP65 isotype localizations shown here is that differences are apparent between the midzone-localized MAP65-1 and MAP65-3 isotypes. In analyzing these differences, we have shown that the turnover of MAP65-3 is far slower than that of MAP65-1. These data suggest that MAP65-3 performs a different role than MAP65-1. However, there must be another MAP65 that can perform the role of MAP65-3 in plants, as the pleiade mutants are not defective in cytokinesis in all tissues in which it is expressed, indicating an overlapping or redundant function with other MAP65 isotypes.
It is interesting that in S. cerevisiae and Schizosaccharomyces pombe, Ase1p has a long turnover (6 to 7 min) (Schuyler et al., 2003, Loiodice et al., 2005) at the anaphase spindle midzone, and in this respect it is perhaps more functionally related to At MAP65-3 than to At MAP65-1. Moreover, the slower turnover of MAP65-3 at the midzone would suggest that MAP65-3 either has a higher affinity for midzone microtubules or that it complexes with other proteins at this site. Several human PRC1 or yeast Ase1p interactors have been identified (e.g., Kurasawa et al. [2004] have shown that PRC1 binds to the midzone kinesin Kif4, and Bratman and Chang [2007] have shown an interaction of Ase1p with CLASP), and although these complexes are important for the normal function of the midzone, their effect on the turnover rates or MAP65 function is unknown. So far, no MAP65-interacting proteins have been identified, and pull-down assays using MAP65-1–coated beads in plant cell extracts only detected MAP65 isotypes, indicating the ability to form dimers (Smertenko et al., 2004).
The region of greatest heterogeneity between the MAP65 isotypes is in the second microtubule binding region (MTB2). Moreover, it is the MTB2 region that is conserved within a group but divergent across groups, indicating that MTB2 may be responsible for specifying MAP65 isotype properties and function. In this context, the differences we have seen in the turnover rates of MAP65-1, -3, and -5 may be due to the differences in the MTB2 region. In addressing this hypothesis, we made six chimeric proteins in which MTB2 was swapped between the three isotypes. These were fused to GFP and transiently expressed in N. benthamiana leaves, and the turnover rates on microtubules were measured. The MTB2 of MAP65-3 reduced the turnover rate of MAP65-1 and MAP65-5, while MTB2 of MAP65-1 caused a faster turnover of MAP65-3 and -5, indicating that MTB2 has a major influence on the properties of an isotype and in particular its affinity to the microtubule.
MTB2 is the region that harbors the phosphorylation sites that are known to control the activity of MAP65s (Sasabe et al., 2006; Smertenko et al., 2006). MAP65-1 has nine potential phosphorylation sites in this region, all of which can be phosphorylated by extracts from metaphase-arrested plant cells. Phosphorylation sites include cyclin-dependent kinase sites, aurora B kinase sites, mitogen-activated protein kinase, and casein kinase sites. The phosphorylation sites are conserved within each phylogenetic group but not between them. This raises the possibility that each group might be controlled by a unique mechanism that depends on the combined activity of varied protein kinase pathways. The amount of phosphorylation determines the ability of MAP65-1 to bind microtubules. It has been suggested that phosphorylation of S. pombe Ase1p, which is more related in its turnover rates to MAP65-3, strengthens its microtubule binding in the nucleus (Loiodice et al., 2005). Conceivably, the phosphorylation status of MAP65-3 in the phragmoplast midzone can modulate its affinity for microtubules or other proteins located in this region, resulting in the slower turnover rate.
Effects of MAP65s on Microtubule Polymerization in Vitro
Most data on the biochemical characterization of MAP65s has been generated using group 1 MAP65s. The tobacco MAP65-1b does not appear to promote microtubule polymerization (Wicker-Planquart et al., 2004). At MAP65-1 has been interpreted as having an insignificant effect on microtubule polymerization compared with taxol (Smertenko et al., 2004), but in another experiment a small increase in polymerization compared with tubulin alone was interpreted as having a significant effect on microtubule polymerization (Mao et al., 2005b). These differences in interpretation can be unified by stating that MAP65-1 has a small effect on polymerization when compared with known enhancers of microtubule polymerization. The significance of such a small promotion of microtubule polymerization by MAP65-1 in vivo is unknown. MAP65-5 has a more prominent effect on microtubule polymerization, having a threefold greater effect than MAP65-1. Removing the MTB2 region from MAP65-3 and MAP65-5 reduced the polymerization to the level of MAP65-1 itself. Swapping the MTB2 regions between isotypes demonstrated that the MTB2 region of MAP65-5 could enhance the microtubule polymerization property of MAP65-1 and that the MTB2 region of MAP65-1 could reduce the same property of MAP65-5. Although we could not make recombinant MAP65-3, by swapping the MTB2 region of MAP65-3 into At MAP65-1 or -5 we were able to significantly enhance their polymerization abilities. As the MTB2 region appears to define the properties of MAP65 in terms of their ability to promote microtubule polymerization, then by correlation we can suggest that MAP65-3 can promote microtubule polymerization more efficiently than MAP65-1 or -5. Similarly, it would appear that there is a correlation between the ability to promote microtubule polymerization and the speed of turnover on microtubules in vivo, where a greater ability to promote polymerization (e.g., for MAP65-3, MAP65-1C3, or MAP65-5C3) corresponds with a slower turnover rate.
The exact mechanism of the promotion of microtubule polymerization by MAP65 is unknown, but two hypotheses can be brought forward. The binding energy of a MAP to a microtubule can enhance the lateral or longitudinal interactions of the tubulin dimers and in this way stabilize the microtubule lattice. Alternatively, bridges between microtubules created by a MAP can counteract the tension imposed on the microtubule lattice by the curved conformation of GDP-tubulin. In both cases, we can surmise that longer association of MAP65 with microtubules can result in increased microtubule polymerization. A similar correlation between turnover rate and polymerization activity has been reported for other MAPs in animals. The turnover of MAP2 and MAP4 (Olmsted et al., 1989), which can promote the polymerization of microtubules, is slower then the turnover of ensconsin, which has no effect on microtubule polymerization (Bulinski et al., 2001). This, however, would only seem to apply for structural proteins, as regulatory proteins like EB1 bind the plus ends of microtubules and have a fast turnover (3.6 s) but still can promote microtubule polymerization by suppressing catastrophes and reducing the depolymerization rate (Dragestein et al., 2008).
METHODS
Plant and Tissue Culture
Arabidopsis thaliana and tobacco (Nicotiana tabacum) BY-2 cells were grown as described by Smertenko et al. (2004).
Phylogenetic and Sequence Analysis
Nine protein sequences encoded by the Arabidopsis MAP65 gene family (Hussey et al., 2002) were analyzed with published MAP65 sequences from tobacco and carrot (Daucus carota), rice (Oryza sativa), and several MAP65 homologs from Medicago truncatula, Saccharum officinarum, Triticum aestivum, and Solanum demissum. The sequences were aligned using the ClustalX software package (Thompson et al., 1997), and the phylodendrogram was constructed using the parsimony algorithm, the bootstrap resampling method, and full heuristic search using PAUP 4.0 software (Sinauer Associates). Bootstrap values were calculated from 1000 replicates of the tree, and the groups with bootstrap scores below 60 were not retained in the tree. Animal and fungal homologs of MAP65 were used as an outgroup (see Supplemental Data Set 1 online). GenBank accession numbers for rice, M. truncatula, S. officinarum, T. aestivum, and S. demissum MAP-65 homologs are indicated on the phylodendrogram. Transcription profiles were analyzed using Genevestigator (Zimmermann et al., 2004; https://www.genevestigator.ethz.ch). Molecular weight and pI values were estimated using the EXPASY pI tool (http://expasy.org/cgi-bin/pi_tool).
Antibodies and Immunostaining
Antiserum was raised in 4- to 8-week-old mice against full-length MAP65-5 and -6 proteins or fragments corresponding to amino acid residues 94 to 176 and 556 to 631 of MAP65-3, amino acid residues 466 to 534 of MAP65-4, amino acid residues 519 to 590 of MAP65-8, and amino acid residues 48 to 158 of MAP65-9 (see Supplemental Figure 1A online). DNA fragments corresponding to each antigen were amplified by RT-PCR using specific primers containing restriction endonuclease sites added to their 5′ ends: NdeI and XhoI for MAP65-4, -6, -8, and -9 and NheI and XhoI for MAP65-5. PCR fragments were cloned into the pGEMT vector (Promega), sequenced, excised with the respective endonucleases, and cloned into the corresponding sites of the pET28a vector (Novagen). Antigens were expressed as recombinant proteins containing an N-terminal His tag in Escherichia coli strain DH3(BL21)pLysS or Rosetta II (Novagen). Total bacterial protein was extracted using sonication, and the recombinant proteins were purified under denaturing conditions in urea-containing buffers on nickel-nitrilotriacetic acid agarose columns (Qiagen) according to the manufacturer's recommendations. Purified proteins were dialyzed against PBS supplemented with 20% glycerol overnight at 4°C, and the final concentration was adjusted to 1 mg/mL. Fifty micrograms of recombinant protein was used for each boost, and a total of four boosts over 2 months were performed. Antiserum was collected 10 d after the final boost and tested by immunoblotting as described elsewhere (Smertenko et al., 2004).
For immunostaining, Arabidopsis 3- to 5-d-old suspension culture cells were separated from the tissue culture medium using 50-mesh nylon cloth and fixed for 30 min at room temperature with 4% paraformaldehyde in 0.1 M PIPES, pH 6.8, 5 mM EGTA, 2 mM MgCl2, and 0.4% Triton X-100. The fixative was washed away with PBS, and cells were treated for 5 min at room temperature with a solution of 0.8% Macerozyme R-10 and 0.2% Pectolyase Y-23 in 0.4 M mannitol, 5 mM EGTA, 15 mM MES, pH 5.0, 1 mM phenylmethylsulfonyl fluoride, 10 μg/mL leupeptin, and 10 μg/mL pepstatin A. Afterward, cells were washed in PBS and attached to poly-l-Lys–coated cover slips. The cover slips were then incubated for 30 min in 1% (w/v) BSA in PBS and incubated for 1 h with primary antibodies. The primary antibodies used were rabbit polyclonal anti-At MAP65-1 (Smertenko et al., 2004) diluted 1:500, mouse polyclonal anti MAP65-3, -4, -5, -6, -8, and -9 diluted 1:500, and rat monoclonal anti-tubulin YL1/2 diluted 1:100 (Serotec). The specimens were then washed three times for 10 min each in PBS and incubated for 1 h with secondary antibodies (Jackson ImmunoResearch) diluted 1:100 in the following combinations. For double immunolabeling, anti-mouse TRITC (minimal cross-reaction to rat and other IgGs; catalog number 715-025-151) and anti-rat fluorescein isothiocyanate (minimal cross-reaction to mouse and other IgGs; catalog number 112-095-167) conjugates were used, and for triple labeling, anti-rat fluorescein isothiocyanate (as above), anti-mouse Cy5 (minimal cross-reaction to rat and other IgGs; catalog number 715-175-150), and anti-rabbit TRITC (minimal cross-reaction to rat, mouse, and other IgGs; catalog number 711-026-152) were used. After incubation with secondary antibodies, the specimens were washed two times for 10 min each in PBS and 10 min in PBS supplemented with 0.5 μg/mL 4′,6-diamidino-2-phenylindole, mounted in Vectashield (Vector Laboratories) mounting medium, and examined with a Zeiss 510 confocal microscope. Images were quantified using NIH Image software (http://rsb.info.nih.gov). As negative controls, each sample was incubated with one of the primary antibodies followed by a mixture of all secondary conjugates used in the experiment. No detectable signal was recorded on the fluorochrome channels where the corresponding primary antibody had not been applied.
Electrophoresis and Immunoblotting
Total protein was extracted from 5-d-old Arabidopsis tissue culture cells by precipitation with trichloroacetic acid. Protein extract (∼50 μg) was separated on 7.5% SDS-PAGE gels and transferred onto pure nitrocellulose membrane. The membrane was cut into strips, and each strip was separately washed in 2× TBST supplemented with 5% (w/v) fat-free milk powder for 20 min and incubated for 1 h at room temperature with primary antibody diluted 1:1000 in the same buffer. Afterward, the strips were washed three times for 10 min each in TBST and incubated for 40 min with secondary anti-mouse or anti-rabbit horseradish peroxidase conjugates (GE Healthcare) diluted 1:3000. Unbound secondary antibody was washed off in TBST (three times for 5 min each), and the signal was developed using the ECL reagent (GE Healthcare).
Live Cell Imaging
Tobacco strain BY-2 cell lines expressing MAP65:GFP fusion proteins were kindly donated by D. Geelen and grown as described by VanDamme et al. (2004a). For imaging, 3- to 5-d-old cultures were used. Cells were mounted in 2% low-gelling point agarose and observed on a Zeiss 510 confocal scanning microscope. Photobleaching for the FRAP analysis was done using a 488-nm argon laser line for 20 iterations at 80% strength. The numerical data were exported to Excel and analyzed as described by Chang et al. (2005).
For infiltration into Nicotiana benthamiana leaves, full-length and chimeric proteins were inserted into pMDC43 binary vectors (Curtis and Grossniklaus, 2003) using Gateway technology and transformed into chemical-competent cells of Agrobacterium tumefaciens strain GV3101. Three mature leaves from different 7- to 10-week-old plants were infiltrated as described by Voinnet et al. (2003) and imaged after 2 to 3 d. The FRAP analysis was performed as described above. Different leaves as well as cells expressing different levels of protein had similar turnover rates for the corresponding proteins.
Recombinant Protein Expression
MAP65-1 was expressed as described by Smertenko et al. (2004). The MAP65-5 coding sequence was amplified from Arabidopsis mRNA by RT-PCR, cloned into pGEMT (Promega) vector, and then cloned into NheI/BamHI sites of pET28a (Novagen) vector. The MAP65-6 coding sequence was isolated from Arabidopsis cDNA library CD4-15 using EST 159E10T7 (both were kindly provided by the ABRC) as a probe and cloned into NdeI/XhoI sites of pET28a. Full-length cDNA clones of At MAP65-2 (clone pda08789) and MAP65-7 (clone pda09605) were obtained from the RIKEN Bioresource Center and cloned into NdeI/SalI and NdeI/XhoI sites of pET28a, respectively. Full-length MAP65-9 cDNA clone (DQ056733) was obtained from the ABRC and cloned into NdeI/XhoI sites of pET28a.
For swapping the MTB2 regions between isotypes, coding sequences corresponding to amino acids 483 to 484 of MAP65-1, amino acids 484 to 485 of MAP65-3, and amino acids 511 to 512 of MAP65-5 were modified to introduce a SacII restriction site. Afterward, the SacII site was used to join the corresponding N-terminal and MTB2 fragments as shown in Figure 6B. The introduction of a SacII site changed the Gln and Lys residues within the random coil region to Pro and Arg. These substitutions do not change the pI of the protein or affect predicted secondary structure. The resulting MAP65-1ΔC, MAP65-3ΔC, MAP65-1C5, MAP65-1C3, MAP65-3C1, and MAP65-3C5 cDNAs were cloned into NdeI/XhoI sites of pET28a, and MAP65-5ΔC, MAP65-5C1, and MAP65-5C3 were cloned into NheI/XhoI sites of pET28a. The sequences of the clones were verified by sequencing.
Recombinant proteins were produced in E. coli strain BL21(DE3)pLysS or Rosetta II and purified using nickel-affinity chromatography as described elsewhere (Smertenko et al., 2004).
Microtubule Polymerization Assay
Microtubule polymerization assays were performed as described by Smertenko et al. (2004).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5g55230 (MAP65-1), At4g26760 (MAP65-2), At5g51600 (MAP65-3), At3g60840 (MAP65-4), At2g38720 (MAP65-5), At2g01910 (MAP65-6), At1g14690 (MAP65-7), At1g27920 (MAP65-8), At5g62250 (MAP65-9), CAC17794 (tobacco MAP65-1a), CAC17795 (tobacco MAP65-1b), CAC17796 (tobacco MAP65-1c), and CAD58680 (carrot MAP65-1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of Antibodies against the MAP65 Isotypes.
Supplemental Figure 2. Immunolocalization of MAP65-4 in Microtubule Arrays of Arabidopsis Tissue Culture Cells.
Supplemental Figure 3. Immunolocalization of MAP65-5 in Microtubule Arrays of Arabidopsis Tissue Culture Cells.
Supplemental Figure 4. Immunolocalization of MAP65-6 in Microtubule Arrays of Arabidopsis Tissue Culture Cells.
Supplemental Figure 5. Immunolocalization of MAP65 Isotypes in Arabidopsis Protoplasts and Tissue Culture Cells.
Supplemental Figure 6. Phylodendrogram of MTB2 Regions of MAP65 Genes.
Supplemental Figure 7. Cell Cycle-Dependent Changes in the Levels of MAP65 Gene Transcription.
Supplemental Figure 8. Transcription of MAP65 Genes in Tissues and Organs of Arabidopsis Plants.
Supplemental Table 1. Characteristics of At MAP65 Protein Sequences.
Supplemental Table 2. Analysis of At MAP65 Transcription Programs Using Affymetrix Microarray Data.
Supplemental Table 3. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Corresponding to Figure 1.
Supplemental Data Set 2. Text File of the Alignment Corresponding to Supplemental Figure 6 online.
Supplementary Material
Acknowledgments
We thank D. Geelen for providing us with BY-2 cell lines expressing At MAP65-1:GFP, At MAP65-3:GFP, and At MAP65-5:GFP; Safina Khan for cloning At MAP65-5; Alison Ritchie for technical assistance in raising antibodies; and the ABRC and the RIKEN Bioresource Center for cDNA clones and libraries. This work was supported by the Biotechnology and Biological Sciences Research Council (Grant G12762).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Patrick J. Hussey (p.j.hussey@durham.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Anthony, R.G., and Hussey, P.J. (1998). Suppression of endogenous alpha and beta tubulin synthesis in transgenic maize calli overexpressing alpha and beta tubulins. Plant J. 16 297–304. [DOI] [PubMed] [Google Scholar]
- Anthony, R.G., and Hussey, P.J. (1999). Dinitroaniline herbicide resistance and the microtubule cytoskeleton. Trends Plant Sci. 4 112–116. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Wilson, J.E., Cork, A., and Williamson, R.E. (1994). Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 35 935–942. [PubMed] [Google Scholar]
- Bayley, P.M., Sharma, K.K., and Martin, S.R. (1994). Microtubule dynamics in vitro. In Microtubules, J.S. Hyams and C.W. Lloyd, eds (New York: Wiley-Liss), pp. 111–137.
- Bratman, S.V., and Chang, F. (2007). Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell 13 812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulinski, J.C., Odde, D.J., Howell, B.J., Salmon, T.D., and Waterman-Storer, C.M. (2001). Rapid dynamics of the microtubule binding of ensconsin in vivo. J. Cell Sci. 114 3885–3897. [DOI] [PubMed] [Google Scholar]
- Caillaud, M.C., Lecomte, P., Jammes, F., Quentin, M., Pagnotta, S., Andrio, E., Engler, J.D., Marfaing, N., Gounon, P., Abad, P., and Favery, B. (2008). MAP65-3 microtubule-associated protein is essential for nematode-induced giant cell ontogenesis in Arabidopsis. Plant Cell 20 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Jensen, C.G., Jensen, L.C.W., Bush, M., and Lloyd, C.W. (1999). The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. USA 96 14931–14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Mao, G.J., Smertenko, A., Hussey, P.J., Naldrett, M., Bottrill, A., and Lloyd, C.W. (2003). Identification of a MAP65 isoform involved in directional expansion of plant cells. FEBS Lett. 534 161–163. [DOI] [PubMed] [Google Scholar]
- Chang, H.-Y., Smertenko, A.P., Igarashi, H., Dixon, D.P., and Hussey, P.J. (2005). Dynamic interaction of NtMAP65-1a with microtubules in vivo. J. Cell Sci. 118 3195–3201. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragestein, K.A., van Cappellen, W.A., van Haren, J., Tsibidis, G.D., Akhmanova, A., Knoch, T.A., Grosveld, F., and Galjart, N. (2008). Dynamic behavior of GFP-CLIP-170 reveals fast protein turnover on microtubule plus ends. J. Cell Biol. 180 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun, S.-O., and Wick, S.M. (1998). Tubulin isoform usage in maize microtubules. Protoplasma 204 235–244. [Google Scholar]
- Gardiner, J.C., Harper, J.D., Weerakoon, N.D., Collings, D.A., Ritchie, S., Gilroy, S., Cyr, R.J., and Marc, J. (2001). A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell 13 2143–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, R.H., Wick, S.M., Silflow, C.D., and Snustad, D.P. (1994). Microtubule component of the plant-cell cytoskeleton. Plant Physiol. 104 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, T. (2007). Microtubule-associated proteins in higher plants. J. Plant Res. 120 79–98. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., Hawkins, T.J., Igarashi, H., Kaloriti, D., and Smertenko, A. (2002). The plant cytoskeleton: Recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 50 915–924. [DOI] [PubMed] [Google Scholar]
- Ishida, T., Thitamadee, S., and Hashimoto, T. (2007). Twisted growth and organisation of cortical microtubules. J. Plant Res. 120 61–70. [DOI] [PubMed] [Google Scholar]
- Jiang, C.J., and Sonobe, S. (1993). Identification and preliminary characterization of a 65 kDa higher-plant microtubule-associated protein. J. Cell Sci. 105 891–901. [DOI] [PubMed] [Google Scholar]
- Jiang, W., Jimenez, G., Wells, N.J., Hope, T.J., Wahl, G.M., Hunter, T., and Fukunaga, F. (1998). PRC1: A human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell 2 877–885. [DOI] [PubMed] [Google Scholar]
- Kurasawa, Y., Earnshaw, W.C., Mochizuki, Y., Dohmae, N., and Todokoro, N. (2004). Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23 3237–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice, I., Staub, J., Setty, T.G., Nguyen, N.-P.T., Paoletti, A., and Tran, P.T. (2005). Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell 16 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, G., Chan, J., Calder, G., Doonan, J.H., and Lloyd, C.W. (2005. a). Modulated targeting of GFP-AtMAP65–1 to central microtubules during division. Plant J. 43 469–478. [DOI] [PubMed] [Google Scholar]
- Mao, T.L., Jin, L.F., Li, H., Liu, B., and Yuan, M. (2005. b). Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol. 138 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., Hennig, L., Gruissem, W., and Murrey, J.A.H. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53 423–442. [DOI] [PubMed] [Google Scholar]
- Muller, S., Smertenko, A., Wagner, V., Heinrich, M., Hussey, P.J., and Hauser, M.-T. (2004). The plant microtubule associated protein, AtMAP65–3/PLE, is essential for cytokinetic phragmoplast function. Curr. Biol. 14 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted, J.B., Stemple, D.L., Saxton, W.M., Neighbors, B.W., and McIntosh, J.R. (1989). Cell cycle-dependent changes in the dynamics of MAP2 and MAP4 in cultured cells. J. Cell Biol. 109 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia, M., and Bouchez, D. (2007). Molecular encounters at microtubule ends in the plant cell cortex. Curr. Opin. Plant Biol. 10 557–563. [DOI] [PubMed] [Google Scholar]
- Pellman, D., Bagget, M., Tu, H., and Fink, G.R. (1995). Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J. Cell Biol. 130 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin, R.M., Wang, Y., Yuen, C.Y.L., Will, J., and Masson, P.H. (2007). WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J. 49 961–971. [DOI] [PubMed] [Google Scholar]
- Sasabe, M., Soyano, S., Takahashi, Y., Sonobe, S., Igarashi, H., Itoh, T.J., Hidaka, M., and Machida, Y. (2006). Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes Dev. 20 1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler, S.C., Liu, J.Y., and Pellman, D.J. (2003). The molecular function of Ase1p: Evidence for a MAP-dependent midzone-specific spindle matrix. J. Cell Biol. 160 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook, J.C., and Kaloriti, D. (2008). Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 13 303–310. [DOI] [PubMed] [Google Scholar]
- Smertenko, A., Saleh, N., Igarashi, H., Mori, H., Hauser-Hahn, I., Jiang, C.J., Sonobe, S., Lloyd, C.W., and Hussey, P.J. (2000). A new class of microtubule-associated proteins in plants. Nat. Cell Biol. 2 750–753. [DOI] [PubMed] [Google Scholar]
- Smertenko, A.P., Chang, H.Y., Sonobe, S., Fenyk, S.I., Weingartner, M., Bogre, L., and Hussey, P.J. (2006). Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J. Cell Sci. 119 3227–3237. [DOI] [PubMed] [Google Scholar]
- Smertenko, A.P., Chang, H.Y., Wagner, V., Kaloriti, D., Fenyk, S., Sonobe, S., Lloyd, C., Hauser, M.T., and Hussey, P.J. (2004). The Arabidopsis microtubule-associated protein AtMAP65-1: Molecular analysis of its microtubule bundling activity. Plant Cell 16 2035–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375 676–677. [Google Scholar]
- Van Damme, D., Bouget, F.-Y., Van Poucke, K., Inze, D., and Geelen, D. (2004. a). Molecular dissection of plant cytokinesis and structure: A survey of GFP-tagged proteins. Plant J. 40 386–398. [DOI] [PubMed] [Google Scholar]
- Van Damme, D., Van Poucke, K., Boutant, E., Ritzenthaler, C., Inze, D., and Geelen, D. (2004. b). In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 136 3956–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Wade, R.H., and Hyman, A.A. (1997). Microtubule structure and dynamics. Curr. Opin. Cell Biol. 9 12–17. [DOI] [PubMed] [Google Scholar]
- Wicker-Planquart, C., Stoppin-Mellet, V., Blanchoin, L., and Vantard, M. (2004). Interactions of tobacco microtubule-associated protein MAP65-1b with microtubules. Plant J. 39 126–134. [DOI] [PubMed] [Google Scholar]
- Yasuhara, H., Muraoka, M., Shogaki, H., Mori, H., and Sonobe, S. (2002). TMBP200, a microtubule bundling polypeptide isolated from telophase tobacco BY-2 cells is a MOR1 homologue. Plant Cell Physiol. 43 595–603. [DOI] [PubMed] [Google Scholar]
- Yasuhara, H., Sonobe, S., and Shibaoka, H. (1993). Effects of taxol on the development of the cell plate and of the phragmoplast in tobacco BY-2 cells. Plant Cell Physiol. 34 21–29. [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 135 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.