Abstract
We isolated a total of 3 × 108 guard cell protoplasts from 22,000 Arabidopsis thaliana plants and identified 1734 unique proteins using three complementary proteomic methods: protein spot identification from broad and narrow pH range two-dimensional (2D) gels, and 2D liquid chromatography–matrix assisted laser desorption/ionization multidimensional protein identification technology. This extensive single-cell-type proteome includes 336 proteins not previously represented in transcriptome analyses of guard cells and 52 proteins classified as signaling proteins by Gene Ontology analysis, of which only two have been previously assessed in the context of guard cell function. THIOGLUCOSIDE GLUCOHYDROLASE1 (TGG1), a myrosinase that catalyzes the production of toxic isothiocyanates from glucosinolates, showed striking abundance in the guard cell proteome. tgg1 mutants were hyposensitive to abscisic acid (ABA) inhibition of guard cell inward K+ channels and stomatal opening, revealing that the glucosinolate-myrosinase system, previously identified as a defense against biotic invaders, is required for key ABA responses of guard cells. Our results also suggest a mechanism whereby exposure to abiotic stresses may enhance plant defense against subsequent biotic stressors and exemplify how enhanced knowledge of the signaling networks of a specific cell type can be gained by proteomics approaches.
INTRODUCTION
Multicellular organisms develop specialized cell types, each with unique functions with regard to its specific role in the organism. The importance of single-cell-type transcriptomics studies in elucidating the functions of specialized cell types is uncontested (Dinneny et al., 2008). Single-cell-type proteomics studies are also essential to unravel the functions of specialized cells, particularly for cell types, like the guard cell (GC), where essential responses to stimuli can occur within seconds (Assmann and Grantz, 1990) and thus are unlikely to be mediated by transcriptomic changes. However, there have been very few single-cell-type proteomics studies in either plant or metazoan systems to date, in part owing to the greater complexity of the proteome and the greater technical challenges of proteomic methodologies (Tyers and Mann, 2003).
The most common subjects for single-cell-type proteomic studies have been cultured mammalian cell lines, where material is not limiting for proteomic analyses (Schirle et al., 2003; Diks and Peppelenbosch, 2004). In addition, studies have been done on human red blood cells (Pasini et al., 2006) and mouse red blood cells (Pasini et al., 2008), where 593 and 668 proteins were identified. By contrast, for plant cells, only the proteomes of Arabidopsis thaliana and tobacco (Nicotiana tabacum) trichomes (Wienkoop et al., 2004; Amme et al., 2005), Arabidopsis epidermal cells (Wienkoop et al., 2004), and soybean (Glycine max) root hairs (Wan et al., 2005) have been assessed, and in each case only a handful of proteins were identified: 63 from Arabidopsis trichomes, 35 from tobacco trichomes, 26 from Arabidopsis epidermal cells, and 36 from root hairs. Arabidopsis pollen (a tricell microspore) has been more widely studied using proteomics because of the relative ease of isolation; however, only 135 (Holmes-Davis et al., 2005) and 121 (Noir et al., 2005) proteins have been identified from these studies. In contrast with previously published studies that reported at most hundreds of proteins in single-cell proteomes from a multicellular organism, here we report the identification of 1734 proteins in the GC proteome of Arabidopsis.
Carbon dioxide (CO2) uptake for photosynthesis, as well as water vapor loss, occur through stomatal pores and are controlled by GC movements, regulated by changes in the turgor pressure and volume of GCs. The study of GC function is particularly topical given that climate change models predict that global warming will result in more frequent and more severe droughts (Breshears et al., 2005; Schroter et al., 2005) and that stomata have been implicated in physiological forcing of the global water cycle (Hetherington and Woodward, 2003; Betts et al., 2007). In addition to the central importance of GC function to terrestrial vegetation, the GC system has also become a model system in plant cell biology. GCs respond autonomously, directly, rapidly, and reversibly to diverse environmental cues, including light, CO2, oxidative stress, humidity, and pathogens (Blatt, 2000; Fan et al., 2004, 2008; Israelsson et al., 2006; Pandey et al., 2007; Shimazaki et al., 2007; Acharya and Assmann, 2008). Stomatal apertures can be easily observed under the microscope, and direct measurement of ion channel and pump activity can be attained by electrophysiological assays. Ions (Ca2+, K+, Cl−, malate2−, and NO3−), signaling elements (e.g., G-proteins, phospholipase C [PLC], phospholipase D [PLD], inositol 1,4,5-trisphosphate, nitric oxide [NO], and reactive oxygen species [ROS]), and plant hormones (e.g., abscisic acid [ABA], auxin, and ethylene) regulate stomatal movements. With the advent of systems biology techniques, a dynamic model for induction of stomatal closure by the drought and stress hormone, ABA, has been developed (Li et al., 2006).
However, despite these achievements, basic questions remain unanswered: Are there proteins or sets of proteins that are preferentially expressed in GCs? How does the GC proteome compare with that of other plant cell types, and, importantly, how can proteomics inform studies of GC signaling and reveal new functions and relationships? These questions are addressed by this study.
RESULTS
Proteomic Methods and GC Proteins
A major challenge in single-cell-type proteomics is obtaining a sufficient quantity of highly pure cells. For GCs, as for some other plant cell types (Birnbaum et al., 2003), such purity can only be achieved by isolation of protoplasts (Figure 1A). Previous studies have validated that GC protoplasts (GCPs) retain key physiological responses present in GC in situ, including responsiveness to environmental signals such as light, ABA, and CO2. To obtain enough GCPs for our proteomic study, we adapted our preparation method (Pandey et al., 2002) by increasing the plant material from 100 to 300 leaves per isolation and performed a total of ∼100 GCP isolations.
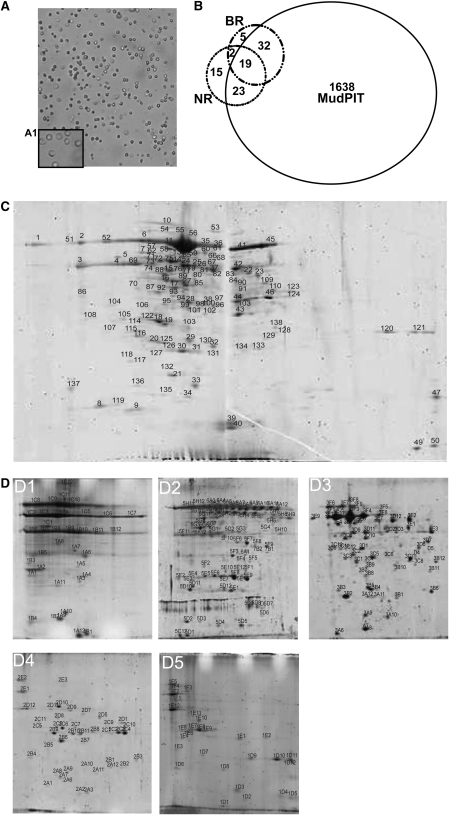
Figure 1.
Three Proteomic Methods Identified 1734 Unique Proteins of the Arabidopsis GC Proteome.
(A) Images showing high-purity GCP preparations (×100; inset magnification ×400).
(B) A total of 1712, 58, and 59 unique proteins were identified from 2D LC-MALDI MudPIT, BR, and NR methods, respectively; 19 proteins were identified by all three methods. For each method, two independent biological samples were analyzed.
(C) A 2D gel image from the broad pH range method. The first dimension was run using a 24-cm, pH 3 to 10 IPG strip. In total, 138 protein spots were detected via Coomassie blue staining. Twelve spots were identified as TGG1. Identifications of numbered spots can be found in Supplemental Table 5 online.
(D) 2D gel images from the narrow pH range method. Proteins were first fractionated into five fractions, and each protein fraction was separated on a narrow pH range IPG strip. From B1 to B5, the pH ranges are 3 to 6, 4.5 to 5.5, 5.3 to 6.3, 6.1 to 7.1, and 6 to 10 respectively. Thirty-seven spots were identified as TGG1. Identifications of numbered spots can be found in Supplemental Table 5 online.
GC proteins were isolated from GCPs and subjected to three complementary proteomics methods (Figure 1B): broad pH range two-dimensional (2D) gels (BR; Figure 1C), narrow pH range 2D gels (NR; Figure 1D), and 2D liquid chromatography–matrix assisted laser desorption/ionization multidimensional protein identification technology (2D LC-MALDI MudPIT). Gel-based and gel-free methods both have advantages and drawbacks. Gel-based methods can simultaneously separate and visualize hundreds of proteins on a single gel but have poor resolving power for membrane and low abundance proteins (Roe and Griffin, 2006). Gel-free methods have greater dynamic range in peptide identification compared with gel-based methods; however, these methods are biased against identification of small proteins (Baerenfaller et al., 2008).
A total of 1712 unique proteins (see Supplemental Table 1 online) was identified from two independent biological samples of GCP subjected to LC-MALDI MudPIT using Mascot with global false discovery rate (FDR) < 0.015 and Protein Pilot with local FDR < 0.05 (see Methods). Protein identification acceptance criteria were confidence interval (CI) ≥ 98% for proteins identified from multiple peptides and CI ≥ 99.9% for proteins identified from a single peptide. In total, 1489 proteins were identified by multiple peptides (nonbold proteins in Supplemental Table 2 online), 19 proteins were identified from a single peptide but from two independent replicates (bold proteins in Supplemental Table 2 online), and 204 unique proteins were identified from single peptides and from only one MudPIT replicate (peptide sequences are provided in Supplemental Table 3 online). For any of the 204 singletons that appear in Tables 2 or 3 (discussed below), protein identity was verified by manual inspection of spectra (see Supplemental Figure 3 online for these spectra). There were 13 peptides that could be assigned to a small group of homologous proteins but could not be assigned unambiguously to one particular protein within that group (peptide sequences are provided in Supplemental Table 3 online).
Table 2.
Fifty-Three Proteins Were Identified in Our GC Proteome and Not in Other Proteomes
| Ten Proteins Have Been Shown to Function in Other Tissues by Mutant Analysis | ||
|---|---|---|
| At1g07780 | PAI1 | Melquist and Bender (2004) |
| At1g48270 | GCR1 | Pandey and Assmann (2004) |
| At1g65860 | A FMO family protein | Hansen et al. (2007) |
| At2g29980 | FAD3 | Browse et al. (1993) |
| At2g46800 | At-MTP1 | Desbrosses-Fonrouge et al. (2005) |
| At3g52590 | UBQ1 | Johnson et al. (2004) |
| At3g54560 | HTA11 | March-Diaz et al. (2008) |
| At3g55830 | EPC1 | Bown et al. (2007) |
| At5g04110 | DNA topoisomerase II family protein | Wall et al. (2004) |
| At5g59820 | RHL41 | Davletova et al. (2005) |
| Forty-Three Proteins Have Not Been Studied | |
|---|---|
| At1g06030 | pfkB-type carbohydrate kinase family protein |
| At1g06630 | F-box family protein |
| At1g07350 | Transformer Ser/Arg-rich ribonucleoprotein, putative |
| At1g08135 | CHX6B |
| At1g10770 | Invertase/pectin methylesterase inhibitor family protein |
| At1g16140 | WAKL3, similar to wall-associated kinase protein family |
| At1g21540 | AMP binding protein, putative |
| At1g32120 | Unknown protein |
| At1g50700 | CPK33 (calcium-dependent protein kinase 33) |
| At1g51490 | Glycosyl hydrolase family 1 protein |
| At1g53290 | Galactosyltransferase family protein |
| At1g57620 | Emp24/gp25L/p24 family protein |
| At1g63310 | Unknown protein |
| At1g67580 | Protein kinase family protein |
| At1g73670 | At-MPK15 |
| At2g02540 | At-HB21/ZFHD4 |
| At2g46320 | Mitochondrial substrate carrier family protein |
| At3g05030 | NHX2 |
| At3g06410 | Nucleic acid binding |
| At3g17070 | Peroxidase, putative |
| At3g26430 | GDSL-motif lipase/hydrolase family protein |
| At3g26740 | CCL (CCR-LIKE) |
| At3g28530 | UDP-glucose 4-epimerase |
| At3g42170 | Transposase-like gene |
| At3g42950 | Glycoside hydrolase family 28 protein |
| At4g02420 | Lectin protein kinase, putative |
| At4g05590 | Unknown protein |
| At4g11660 | At-HSFB2B |
| At4g16590 | At-CSLA01 |
| At4g17100 | Ser protease protein-related |
| At4g23900 | NDK4 |
| At4g28650 | Leu-rich repeat transmembrane protein kinase, putative |
| At4g31130 | Unknown protein |
| At5g15210 | ZFHD3 |
| At5g50230 | Nucleotide binding |
| At5g53320 | Leu-rich repeat transmembrane protein kinase, putative |
| At5g55040 | DNA binding bromodomain-containing protein |
| At5g58340 | DNA binding |
| At5g60720 | Unknown protein |
| At5g62020 | At-HSFB2A |
| AtCg00860 | Unknown protein |
| AtCg01000 | Unknown protein |
| AtMg00480 | Subunit 8 of the mitochondrial F0 ATP synthase complex |
Proteins in italics were identified by a single peptide from one MudPIT replicate. Corresponding peptide sequences and spectra for these proteins are provided in Supplemental Table 3 and Supplemental Figure 3 online, respectively.
Table 3.
Identified GC Signaling Proteins in Our GC Proteome
| AGI | Name | AGI | Name |
|---|---|---|---|
| Twelve Proteins Previously Shown to Have a Role in GC Function Were Identified in Our GC Proteome | |||
| At1g11260 | STP1 | At3g45780 | PHOT1 |
| At1g37130 | NIA2 | At3g53720 | CHX20 |
| At1g48270 | GCR1 | At4g18290 | KAT2 |
| At2g18960 | OST2 | At4g23650 | CPK3 or CDPK6 |
| At2g21660 | GRP7 | At5g23060 | CAS |
| At3g15730 | PLDα1 | At5g58140 | PHOT2 |
| AGI | Name |
|---|---|
| Fifty-Two Proteins in Our GC Proteome Are Predicted to Be Signaling Proteins by GO Software | |
| At1g05810 | ARA-1 |
| At1g06840 | Leu-rich repeat transmembrane protein kinase |
| At1g09100 | 26S protease regulatory subunit 6A |
| At1g35160 | GF14/GRF4 |
| At1g52280 | Ras-related GTP binding protein |
| At1g71860 | PTP1 |
| At1g73670 | At-MPK15 |
| At1g75640 | Leu-rich repeat family protein |
| At1g76030 | Vacuolar ATP synthase subunit B |
| At1g78580 | At-TPS1 |
| At2g04880 | ZAP1 |
| At2g16600 | ROC3 |
| At2g19860 | At-HXK2 |
| At2g20610 | SUR1 |
| At2g21880 | At-RABG2 |
| At2g25170 | PKL |
| At2g26730 | Leu-rich repeat transmembrane protein kinase |
| At2g32410 | AXL |
| At2g36830 | γ-TIP |
| At2g37970 | SOUL heme-binding family protein |
| At2g44050 | COS1 |
| At2g46070 | At-MPK12 |
| At3g02880 | Leu-rich repeat transmembrane protein kinase |
| At3g08510 | At-PLC2 |
| At3g08680 | Leu-rich repeat transmembrane protein kinase |
| At3g15730 | PLDα1 |
| At3g18040 | At-MPK9 |
| At3g18820 | Ras-related GTP-binding protein |
| At3g46060 | ARA3 |
| At3g53020 | STV1 |
| At3g55020 | RabGAP |
| At3g62030 | ROC4 |
| At4g01370 | At-MPK4 |
| At4g02080 | SAR1A |
| At4g03550 | At-GSL5 |
| At4g15900 | PRL1 |
| At4g18430 | Ras-related GTP binding protein |
| At4g23650 | CPK3 or CDPK6 |
| At4g28650 | Leu-rich repeat transmembrane protein kinase |
| At4g29810 | At-MKK2 |
| At4g33680 | AGD2 |
| At4g34870 | ROC5 |
| At4g38690 | 1-Phosphatidylinositol phosphodiesterase-related |
| At4g38740 | ROC1 |
| At5g16590 | Leu-rich repeat transmembrane protein kinase |
| At5g19390 | Similar to pleckstrin homology domain-containing protein |
| At5g39500 | Pattern formation protein |
| At5g53320 | Leu-rich repeat transmembrane protein kinase |
| At5g58440 | Phox domain–containing protein |
| At5g60600 | GCPE |
| At5g63310 | NDPK2 |
| At5g67030 | ABA1 |
Proteins in boldface were shown to play a role in GC function in previous studies. Proteins in italics were identified by a single peptide from one MudPIT replicate. Corresponding peptide sequences and spectra for these proteins are provided in Supplemental Table 3 and Supplemental Figure 3 online, respectively.
Since no single proteomic method is able to provide a comprehensive picture of the proteome (Kleffmann et al., 2007), we also pursued protein identifications using two gel-based methods that can provide information complementary to gel-free methods such as LC-MALDI MudPIT. For these methods, stained spots were in-gel digested by trypsin, identified by MALDI-tandem time-of-flight (TOF/TOF), and analyzed with Mascot software (see Methods). In two independent biological replicates of BR 2D gels, 138 spots were consistently detected, and from these, 85 spots, representing 58 unique proteins, were accepted as identified, meeting a very stringent acceptance criterion of requiring CI > 99.5% for identification.
To identify a greater number of proteins, GCP proteins were first prefractionated into five fractions with pH ranges 3 to 4.6, 4.6 to 5.4, 5.4 to 6.2, 6.2 to 7.0, and 7.0 to 10.0 using an isoelectric focusing (IEF) fractionator. Each fraction was applied to a corresponding single pH 2D gel (see Methods). A total of 250 spots was consistently detected from two biological replicates, from which 120 spots were identified with CI > 99.5%; these spots represent 59 unique proteins. By far the largest protein spot on either BR or NR 2D gels was the myrosinase THIOGLUCOSIDE GLUCOHYDROLASE1 (TGG1) (Figure 1C, spot 12, and Figure 1D3, spot 3E12). TGG1 was also detected in 11 and 36 more spots on the BR and NR 2D gels, respectively (see Supplemental Tables 4 and 5 online). Fourteen and 19 proteins were identified in multiple spots on the BR and NR 2D gels, respectively, representing a total of 28 unique proteins (see Supplemental Table 5 online).
In total, 1734 unique Arabidopsis GC proteins were identified from the combined application of all methods (see Figure 1B for summary; see Supplemental Tables 1 and 4 online for all identified GC proteins and Supplemental Table 6 online for the proteins identified from any two of the three methods). Of these proteins, 336 proteins are not represented in previously reported GC transcriptomes (Leonhardt et al., 2004; Yang et al., 2008), illustrating the complementarity of proteomic and transcriptomic approaches.
Global Bioinformatic Analyses of the GC Proteome
Enrichment of Gene Ontology (GO) Biological Process (http://www.Arabidopsis.org/tools/bulk/go/index.jsp) categories in our GC proteome compared with the conceptually predicted whole Arabidopsis proteome was determined using the topGO package (Alexa et al., 2006; Baerenfaller et al., 2008). The numbers of proteins present in the published pavement epidermal cell and trichome proteomes are too low for valid topGO analysis, but it was possible to perform topGO analysis of the leaf proteome (Lee et al., 2007) for comparison to the GC proteome. Out of the eight categories most significantly enriched in the GC proteome compared with the entire (conceptual) proteome, four categories (response to cold, translation, protein folding, and photosynthesis–general) were also among the eight most significantly enriched in the leaf proteome (Table 1). However, the other four categories (glycolysis, photosynthesis–light reactions, fatty acid biosynthetic process, and amino acid metabolic process) were present in the top GC hits but absent from the top leaf hits (Table 1), suggestive of particular roles in GC (see Discussion).
Table 1.
Comparison of topGO Analyses of Our GC Proteome and the Previously Identified Leaf Proteome
| Rank | GO ID | Term | Annotated | Significant | Expected | Log10(1/P Value) |
|---|---|---|---|---|---|---|
| Top Eight Biological Process GO Categories for the GC Proteome | ||||||
| 1 | GO:0009409 | Response to cold | 202 | 64 | 15.7 | 23 |
| 2 | GO:0006412 | Translation | 561 | 123 | 43.7 | 22 |
| 3 | GO:0006096 | Glycolysis | 56 | 28 | 4.4 | 17 |
| 4 | GO:0015979 | Photosynthesis | 133 | 56 | 10.4 | 14 |
| 5 | GO:0019684 | Photosynthesis, light reaction | 69 | 28 | 5.4 | 14 |
| 6 | GO:0006457 | Protein folding | 228 | 52 | 17.8 | 12 |
| 7 | GO:0006633 | Fatty acid biosynthetic process | 105 | 31 | 8.2 | 11 |
| 8 | GO:0006520 | Amino acid metabolic process | 321 | 86 | 25.2 | 10 |
| Top Eight Biological Process GO Categories for the Leaf Proteome | ||||||
| 1 | GO:0006412 | Translation | 561 | 246 | 60.6 | 30 |
| 2 | GO:0009409 | Response to cold | 202 | 83 | 21.8 | 28 |
| 3 | GO:0042254 | Ribosome biogenesis and assembly | 125 | 62 | 13.5 | 27 |
| 4 | GO:0006334 | Nucleosome assembly | 57 | 36 | 6.12 | 21 |
| 5 | GO:0015979 | Photosynthesis | 133 | 73 | 14.4 | 17 |
| 6 | GO:0006457 | Protein folding | 228 | 69 | 24.6 | 15 |
| 7 | GO:0044249 | Cellular biosynthetic process | 1347 | 441 | 145.5 | 12 |
| 8 | GO:0015986 | ATP synthesis-coupled proton transport | 32 | 19 | 3.5 | 10 |
To identify proteins in our GC proteome that may be specifically expressed or enriched in GCs, we compared our GC proteome to ∼13,000 proteins in previously identified proteomes of cell walls (Bayer et al., 2006), trichomes (Wienkoop et al., 2004), epidermal cells (Wienkoop et al., 2004), and leaves (Lee et al., 2007), as well as to the recently reported proteome map of Arabidopsis organs (Baerenfaller et al., 2008). These comparisons revealed 61 proteins that were identified in our GC proteome and not in these other proteomes (Table 2). Of these 61 proteins, we found eight that were identified in other studies in the literature: four of the eight (At3g16080, At3g18040, At3g23840, and At5g54190) were identified in studies on specific organelles (Friso et al., 2004; Heazlewood et al., 2004; Philippar et al., 2007; Carroll et al., 2008), and the other four (At1g50010, At5g08690, AtMg01190, and At5g08670) were identified by proteomic analyses of certain mutants or certain treatments, or in cultured plant cells (Rajjou et al., 2004; Dixon et al., 2005; Job et al., 2005; Sorin et al., 2006). The remaining 53 proteins may have unique roles in GC function, particularly since, to date, only 10 of these proteins have been shown to function in any other tissues by mutant analysis (Table 2). Of the proteins identified by Baerenfaller et al. (2008) as biomarkers of specific organ types, nine of these proteins (three from flowers, three from siliques, two from roots, and one from seeds) were present in our GC proteome (see Supplemental Table 7 online).
Twelve proteins previously shown to play a role in GC function were present in our GC proteome (Table 3). In addition, functional classification by GO analysis showed that 52 proteins out of our identified GC proteome are predicted as signal transduction proteins (Table 3). Of these, only two proteins, the phospholipase PLDα1 (Mishra et al., 2006) and the calcium-dependent protein kinase CPK3/CDPK6 (Mori et al., 2006), have been previously studied in the context of GC function, where they have been shown to participate in ABA signaling.
Functional Analysis of One of the Most Abundant Proteins in GCs, the Myrosinase TGG1
The plant glucosinolate-myrosinase system is known as a defense system against bacteria, pathogens, and herbivores. When tissue is damaged (e.g., by insect chewing), glucosinolates are thought to be released from the vacuole and hydrolyzed by myrosinases into a variety of toxic small molecules, including thiocyanate, isothiocyanate, and nitrile, which are active against biotic intruders (Wittstock and Halkier, 2002; Barth and Jander, 2006).
Although our gel-based analyses contributed only incrementally to the total number of GC proteins identified, these analyses revealed a remarkable abundance in GC of the myrosinase, TGG1. TGG1 was by far the largest spot on either the BR (Figure 1C, spot 12) or the NR 2D gels (Figure 1D3, spot 3E12) and was identified in multiple spots (see Supplemental Table 5 online; 12 spots on the BR and 37 spots on the NR gels), suggesting high abundance and multiple posttranslational modifications.
TGG1 protein was not identified in proteomic analyses of trichome and epidermal pavement cells (Wienkoop et al., 2004), consistent with previous reporter gene analysis demonstrating strong expression of the TGG1 gene in GC and no expression in other epidermal cell types (Husebye et al., 2002; Barth and Jander, 2006). However, the presence of the TGG1 protein in GC has not been previously shown, and no function of TGG1 in GC has been described previously. The other Arabidopsis myrosinase, TGG2, is not expressed in GC by reporter gene analysis (Barth and Jander, 2006). TGG1 and TGG2 are indistinguishable by the probes used on Affymetrix microarrays, and the only other demonstrated locus of expression of these two TGG genes is in phloem idioblasts (Husebye et al., 2002; Barth and Jander, 2006). Different masses of trypsin-digested peptides and amino acid sequences are generated from TGG1 versus the related myrosinase, TGG2, and can be detected by mass spectrometry and tandem mass spectrometry, respectively. Thus, TGG1 and TGG2 can be clearly distinguished by proteomic methods. Unlike TGG1, TGG2 was not found in any of the gel-based studies and was identified only in one replicate of the LC-MALDI MudPIT method using the Protein Pilot search algorithm. While 37 unique peptides were identified from TGG1, only two unique peptides were identified from TGG2, plus one peptide was identified that is identical in TGG1 and TGG2. These results suggest a substantially lower abundance of TGG2 in GC compared with TGG1.
TGG1 is predicted to be a cytoplasmic protein by SubLoc (http://www.bioinfo.tsinghua.edu.cn/SubLoc/), a secreted protein by Target P (http://www.cbs.dtu.dk/services/TargetP/), and a chloroplast protein by WoLF PSORT (http://wolfpsort.org/). TGG1 has been identified in proteomic studies of the chloroplast proteome (Kleffmann et al., 2004), vacuole proteome (Carter et al., 2004), and ribosome proteome (Giavalisco et al., 2005). This lack of consensus regarding the subcellular localization of TGG1 notwithstanding, previous studies by Jander and colleagues have clearly shown that degradation of glucosinolates is slower in tgg1 mutant leaves compared with the wild type (Barth and Jander, 2006).
If the primary role of myrosinases is the deterrence of herbivory, why would TGG1 expression be limited to GC and not extend throughout all cell types of the epidermal layer? The abundance of TGG1 in GCs suggested to us that TGG1 might have as yet undiscovered roles in GCs. Accordingly, we evaluated TGG1 functions in GCs using two independent tgg1 mutants, tgg1-1 and tgg1-3 (Barth and Jander, 2006; Ueda et al., 2006). tgg1-1 (SALK_130474) and tgg1-3 (SAIL_786_B08) are T-DNA insertional mutants. tgg1-3 has been shown to lack full-length TGG1 transcript by RT-PCR analysis (Barth and Jander, 2006). In in vitro assays, aboveground tissue homogenates from tgg1-1 and tgg1-3 mutants exhibit only ∼5 to 8% of wild-type levels of myrosinase activity (Barth and Jander, 2006; Ueda et al., 2006). As previously reported (Barth and Jander, 2006), these tgg1 mutants showed no obvious whole-plant phenotypes or developmental defects.
Given the roles of myrosinases in plant–herbivore interactions, we first assessed the GC response of wild-type and tgg1 mutants to a uniform wounding stimulus (Bailey et al., 2005). We found that wounding induces stomatal closure (Figure 2). tgg1 GCs showed a moderate disruption of wound-induced stomatal closure (Figure 2). Wounding induces methyl jasmonate (MJ) accumulation in leaf tissue (Maffei et al., 2007), and MJ has been reported to promote stomatal closure (Suhita et al., 2004; Munemasa et al., 2007). Therefore, we next evaluated MJ regulation of stomatal movements in Columbia (Col) and tgg1 mutants; however, we could not find any consistent effects of MJ on these responses, even in wild-type plants (see Supplemental Figure 1 online). This difference from published reports may be due to different plant growth conditions. Suhita et al. (2004) grew plants hydroponically in sand with short days (8-h photoperiod) and 300 μmol m−2s−1 light intensity, Munemasa et al. (2007) grew plants with long days (16-h photoperiod) and 8000 Lux (∼160 μmol m−2s−1) light intensity, and we grew plants in potting mix with an 8-h photoperiod and 110 μmol m−2s−1 of light (see Supplemental Methods online for details). Indeed, the effect of MJ on stomatal closure differs among published studies, consistent with the supposition that this effect may be altered by plant growth conditions; for example, the extent of MJ promotion of stomatal closure in the Suhita et al. (2004) study (∼3 μm) is greater than that reported by the Munemasa et al. (2007) study (<1 μm). It has also been reported that the sitiens mutant of tomato (Solanum lycopersicum), an ABA biosynthetic mutant, shows little MJ-induced decrease in transpiration unless ABA is exogenously supplied (Herde et al., 1997), suggesting an interplay between these two hormones in the regulation of stomatal aperture.
Figure 2.
tgg1 Mutants Are Hyposensitive to Wounding Promotion of Stomatal Closure at 5 min.
(A) At 5 min, tgg1-1 and tgg1-3 mutants are hyposensitive to wounding promotion of stomatal closure compared with Col.
(B) tgg1-1 and tgg1-3 mutants show no significant difference compared with Col in wounding inhibition of stomatal opening at 30 min and 1 h.
For (A) and (B), leaves were treated blindly and simultaneously with a slicker brush (Bailey et al., 2005) to ensure the same amounts of tiny holes were punched in all leaves. n = 4 independent experiments. At least 60 stomata were measured for each treatment per genotype per replicate. Data are presented as mean ± se. Data were analyzed by Student's t test, P < 0.05 was considered significant (*).
Because ABA regulation of stomatal movements is central to GC function, stomatal responses to ABA were evaluated in the tgg1 mutants. tgg1 mutant stomata showed wild-type responses to ABA promotion of stomatal closure (Figure 3A) but were hyposensitive to ABA inhibition of stomatal opening (Figure 3B). These results imply that TGG1 is a positive regulator in ABA inhibition of stomatal opening and thus has a heretofore unrecognized role in plant abiotic stress responses.
Figure 3.
Regulatory Effects of Glucosinolates and/or Myrosinases on Arabidopsis Stomatal Apertures and GC K+in Currents of Col and tgg1 Mutants in the Presence and Absence of ABA.
(A) tgg1-1 and tgg1-3 mutants respond similarly to the wild type (Col) in ABA induction of stomatal closure.
(B) tgg1-1 and tgg1-3 mutants are hyposensitive to ABA inhibition of stomatal opening.
For (A) and (B), n = 3 experiments. At least 60 stomata were measured for each treatment per genotype per replicate. Data are mean ± se. P < 0.05 (Student's t test) was considered significant (*).
(C) Typical GC whole-cell K+ current traces of Col and tgg1 mutants under control, 50 μM glucosinolate (G), and/or 0.2 unit/mL myrosinase (M) application to the GC cytosol, with or without 50 μM ABA. Current and time scales are as shown.
(D) K+ current density (mean ± se) at −219 mV of GC whole-cell inward K+ currents.
n = 14, 7, 6, 10, 7, and 6 for Col under control, ABA, G, G+M, M, and ABA+M treatments, respectively; n = 13, 10, 11, 6, 7, and 7 for tgg1-1 under control, ABA, G, G+M, M, and ABA+M treatments, respectively; n = 16, 8, 13, 8, 6, and 12 for tgg1-3 under control, ABA, G, G+M, M, and ABA+M treatments, respectively. P ≤ 0.01 (Student's t test) was considered significant (*) for (D).
In wild-type Col plants, ABA is known to inhibit the GC K+in channels that mediate K+ uptake during stomatal opening (Schroeder et al., 1987; Schwartz et al., 1994; Wang et al., 2001) (Figures 3C and 3D). In the absence of ABA treatment, tgg1 mutant GC had similar K+in current amplitudes and kinetics as Col; however, ABA inhibition of K+in current was abolished in both independent tgg1 mutants (Figures 3C and 3D). Consistent with the fact that tgg1 mutants show a wild-type response in ABA promotion of stomatal closure, outward K+ currents, which mediate K+ efflux during stomatal closure, were statistically identical in Col versus tgg1 mutants either with or without ABA treatment (Figure 3C; see Supplemental Figure 2 online).
Alterations in the glucosinolate profiles of tgg mutants have already been characterized at the whole-leaf level, with significant increases in aliphatic and indole glucosinolates primarily observed in tgg1 tgg2 double mutants (Barth and Jander, 2006). Since myrosinases hydrolyze glucosinolates, one key question that arises is, what is functioning in the ABA inhibition of K+in channels: myrosinase itself, glucosinolates, or the hydrolyzed products of glucosinolates? As a first step toward addressing this question, glucosinolates, myrosinase, or a combination of glucosinolates and myrosinase was directly applied to the cytosol of Col and tgg1 mutant GCs via the patch pipette solution (Figures 3C and 3D). Whole-cell patch clamp data showed that glucosinolate administration resulted in inhibition of K+in channels in Col GCs but not in GCs of tgg1 mutants, indicating that the glucosinolates themselves do not suffice to inhibit channel activity. By contrast, coadministration of glucosinolates and myrosinase resulted in a similar extent of inhibition of K+in channels in both Col and tgg1 mutants, suggesting that it is the hydrolyzed products of glucosinolates that evoke ion channel inhibition. However, myrosinase addition alone had no effect on K+in channels in either Col or tgg1 mutants, suggesting that, in the absence of an appropriate triggering event, glucosinolate substrates are not available for myrosinase action. Most importantly, even though myrosinase application alone had no effect on K+in currents, application of myrosinase restored K+in sensitivity to ABA in tgg1 GCs (Figures 3C and 3D). These results indicate that the hydrolysis of glucosinolates by myrosinases is induced in some manner by ABA in Arabidopsis GCs (see Figure 4 and Discussion) and is a necessary component of ABA-mediated K+in channel inhibition.
Figure 4.
Speculative Model of Interactions between Glucosinolates, Myrosinase (TGG1), and K+ Channels in GC ABA Signaling.
Left and right GCs indicate events before and after, respectively, activation of signaling pathways. Abiotic stresses increase ABA delivery to GCs. ABA either repartitions glucosinolates (G) from the vacuole to the cytosol (arrow 1) or enhances myrosinase (M) activity or substrate affinity (arrow 2). Inhibition of inward K+ channel (K+in) activity might result from channel modification by the resultant hydrolyzed products of glucosinolates (e.g., isothiocyanates [ITC]), as in mammalian cells (arrow 3) or might result from decreased ROS scavenging (arrow 4) by ascorbate (Asc). Hydrolyzed products of glucosinolates may diffuse through the stomatal pore (shaded arrows) and thus deter biotic invaders. Reported biotic (wound) induced increases in ABA (Schmelz et al., 2003) may also provide positive feedback to the glucosinolate-myrosinase system.
DISCUSSION
Proteomics, an important postgenomic approach, has been applied to many fields [e.g., identification of protein expression profile changes under stress conditions (Hashimoto and Komatsu, 2007), analysis of posttranslational modifications (Kwon et al., 2006), and determination of protein–protein interactions (Parrish et al., 2007)]. Quantitative proteomic methods are also emerging, but such quantifications will have greatest correspondence to biologically significant cellular protein amounts in the context of single cell type proteomes, as opposed to mixed tissues or organs where the abundance of a given protein may vary greatly among the different cell types present and thus mask abundance, or changes in abundance, within any single cell type. Although fava bean (Vicia faba) GCs were used as material for an in-gel kinase assay approximately a decade ago, leading to identification of a Ca2+-independent ABA-activated protein kinase by mass spectrometry (Li and Assmann, 1996, 2000), a major bottleneck for the characterization of the GC proteome has been obtaining enough highly pure GC from a species with a sequenced genome. Here, this challenge was overcome and three proteomics methods were used to identify 1734 unique proteins of the GC proteome.
Comparison of the Three Proteomic Methods
In our study, the gel-free method (2D LC-MALDI MudPIT) yielded by far the largest number of protein identifications: 1712 proteins were identified by this method, and, of these, 1638 were not identified by either of the gel-based methods. The two gel-based methods (BR and NR) yielded similar numbers of identified proteins (58 and 59 unique proteins, respectively), with an identification rate ∼30-fold lower than that of the gel-free method. Twenty-one proteins (i.e., approximately one-third of the proteins identified by BR or NR methods) were common to both of these gel-based approaches. Eighty-eight percent of the BR proteins (51 proteins) and 71% of the NR proteins (42 proteins) were also identified by 2D LC-MALDI MudPIT analyses. Only 19 proteins (representing 33% of the BR proteins, 32% of the NR proteins, and 1% of the 2D LC-MALDI MudPIT proteins) were found from all three methods (Figure 1B; see Supplemental Table 6 online). This low overlap is consistent with observations from other organisms that have shown that multiple strategies are required to obtain high coverage of the proteome (Kleffmann et al., 2007). The limitations of gel-based methods regarding identification of basic, high molecular mass, and membrane proteins are well known (Jorrin et al., 2007). However, the gel-based methods importantly allowed recognition of the high abundance of TGG1 in GC. In addition, gel-based methods provide more reliable inference of posttranslational modifications (see Supplemental Table 5 online). Moreover, 22 proteins of the GC proteome were exclusively identified by gel-based methods, indicating that gel-free methods cannot completely replace traditional gel-based methods (Lambert et al., 2005). Although we evaluated the 22 proteins for common characteristics (e.g., molecular weight, pI, predicted subcellular localization, and the predicted trypsin-digestion patterns of these protein sequences) that might have enhanced their detection by gel-based over gel-free methods, no such characteristics could be identified.
Comparison of the GC Proteome with Other Proteomes
Although we used GCPs as starting material, 29 of the identified GC proteins were identified by a previous cell wall proteomic study (Bayer et al., 2006): these proteins may localize to multiple subcellular compartments, including both apoplastic and symplastic destinations, or be present in secretory vesicles that have not yet fused with the plasma membrane (Lee et al., 2004). Indeed, further GO analysis predicted that 23 of the 29 (79%) proteins also localize to non–cell wall subcellular compartments.
Our topGO analysis revealed Biological Processes that were enriched in the GC proteome (Table 1) relative to the entire predicted Arabidopsis proteome and to the documented leaf proteome (Lee et al., 2007). Four of the GC-enriched Biological Processes were also enriched in the published leaf proteome (Lee et al., 2007) and thus may typify leaf cell types in general but not GC in particular. The GO category “amino acid metabolic process” was also fairly highly ranked in leaves (rank = 10). More interesting are the remaining GC-enriched Biological Processes: glycolysis (rank 3 in GC but 14 in leaf), light reactions of photosynthesis (rank 5 in GC but 29 in leaf), and fatty acid biosynthesis (rank 7 in GC but 142 in leaf).
Stomatal movement is estimated to have a high energetic requirement (Assmann and Zeiger, 1987), consistent with the topGO prediction of the importance of glycolysis and photophosphorylation to this cell type. Indeed, biochemical assays have shown disproportionately high rates of photophosphorylation in GC relative to their very low rates of carbon fixation (Shimazaki et al., 2007), and red light–stimulated stomatal opening is known to be inhibited by photosynthetic inhibitors, such as DCMU (Schwartz and Zeiger, 1984). The enrichment in the GC proteome of proteins involved in fatty acid biosynthesis may reflect not only the importance of lipids to cuticle formation (Samuels et al., 2008), but also the importance of lipids and lipid metabolites as signaling entities in GC. For example, the guard cell–specific HIGH CARBON DIOXIDE (HIC) gene encodes a likely 3-keto acyl CoA synthase, involved in the synthesis of very-long-chain fatty acids, yet hic mutants also show a dramatic CO2-dependent alteration in GC production (Gray et al., 2000). Other lipid-related molecules with demonstrated roles in GC related to ABA signaling include phosphatidic acid (Jacob et al., 1999; Zhang et al., 2005), sphinogosine-1-phosphate (Ng et al., 2001; Coursol et al., 2003), and inositol phosphates (Lee et al., 1996; Jung et al., 2002; Hunt et al., 2003). Our topGO analysis of the GC proteome suggests that additional study of mutants in enzymes related to fatty acid synthesis may uncover GC-related phenotypes.
Of the 53 proteins that were identified in our GC proteome (Table 2) but not in other known proteomes, some may be specific to GCs and thus can be considered as candidate GC biomarkers. In this regard, it would be of particular interest to characterize the seven proteins of unknown function in this set (Table 2). Others may be more abundant in GCs than elsewhere; thus, we succeeded in identifying these proteins as part of the GC proteome while they were missed in other proteomic analyses (e.g., the likely G-protein coupled receptor, GCR1, which confers ABA hypersensitivity to both stomatal movements and root growth) (Pandey and Assmann, 2004). Finally, some proteins may be specifically present in GC plus a few other specialized cell types and thus missed in proteome analyses of whole organs. Conversely, our identification in the GC proteome of proteins previously thought to be biomarkers for specific organs (see Supplemental Table 7 online), including roots and seeds, which lack GC, indicates the importance of single-cell-type proteome analysis in determinations of protein distribution.
The GC Proteome and GC Signaling
We identified 67 proteins from the literature as previously shown to function in mature Arabidopsis GCs/GCPs (see Supplemental Table 8 online). Of these, 51 participate in GC responses to ABA, light, and/or CO2, while six of the remaining 16 proteins function in solute transport. Twelve of the 67 proteins were present in our GC proteome (Table 3); the other 55 may be low abundance proteins or induced under specific conditions. The 12 identified proteins are involved in light (PHOT1, PHOT2, and CHX20) and ABA (CPK3, GCR1, GRP7, OST2, NIA2, and PLDα1) signaling and in solute transport (CHX20, STP1, KAT2, and OST2) (Table 3).
GO analysis categorizes 52 proteins in our GC proteome as signal transduction proteins (Table 3). Of these, 50 have yet to be studied in the context of GC function, highlighting the usefulness of proteome analysis in identifying targets for further functional analyses. Thirteen are protein kinases, including one CDPK (CPK3/CDPK6), seven LRR protein kinases, and five MAP kinases; four of these kinases (At1g73670, At3g18040, At4g28650, and At5g53320) are also in the list of proteins only found to date in the GC proteome (Table 2). This information strongly suggests that phosphorylation is one of the main posttranslational modifications in GCs, consistent with previous demonstrations of the importance of phosphorylation to light and ABA signaling in GCs (Kinoshita et al., 1993; Li and Assmann, 1996, 2000; Li et al., 2000; Mustilli et al., 2002; Kinoshita et al., 2003; Sokolovski et al., 2005). Five of the 52 proteins are involved in auxin signaling, suggesting that auxin may be more important in GC physiology than previously recognized (Acharya and Assmann, 2008).
TGG1 Function in GCs
While many interesting candidates for further downstream analysis were revealed in our GC proteome analysis, we chose to perform an in-depth functional study of one protein. TGG1 was chosen due to its superabundance in GC: TGG1 comprises 40 to 50% of the total protein identified on 2D gels and was identified by >30 unique peptides (not shared with TGG2) in our MudPIT analysis. However, no roles for TGG1 in GCs had been previously demonstrated.
We examined the effect of wounding on stomatal apertures and found that wounding of the epidermis did stimulate a stomatal response and that TGG1 seemed to participate in this effect (Figure 2). However, TGG1 appeared to play a more essential role in GC ABA signaling: tgg1 mutant plants lacking this enzyme were unresponsive to ABA inhibition of stomatal opening and K+in channel regulation. Intracellular application of myrosinase alone did not restore K+in channel inhibition in tgg1 GCs or cause channel inhibition in wild-type GCs, yet application of myrosinase in the presence of ABA restored channel inhibition to the tgg1 mutants.
Our electrophysiological results are consistent with the following scenario (Figure 4): (1) myrosinase and its substrates, the glucosinolates, are localized in distinct subcellular compartments in GCs, as has been proposed for other cell types (Grubb and Abel, 2006). (2) ABA induces the relocalization of glucosinolates to the cytosol (arrow 1 in Figure 4). (3) The hydrolysis of available glucosinolates by TGG1 leads to inhibition of K+in channels in GCs, and this is one component of ABA inhibition of stomatal opening. However, we note that there are also alternative interpretations that are consistent with our data (e.g., ABA signaling might somehow increase the activity of myrosinase or its affinity for its substrate) (arrow 2 in Figure 4).
ABA regulation of glucosinolate compartmentalization and thus availability for hydrolysis by myrosinases, as hypothesized here, could help to explain nondefensive developmental changes in glucosinolate concentrations (Petersen et al., 2002; Brown et al., 2003) that occur in the absence of the tissue disruption that brings substrate and enzyme together during herbivory. Such a phenomenon could also provide a mechanism whereby exposure to abiotic stress could strengthen defenses against subsequent biotic stressors. In addition, given the mechanisms described here, biotic (wound)-induced increases in ABA, as reported to occur in leaf tissue (Schmelz et al., 2003), might also provide positive feedback to the glucosinolate-myrosinase defense pathway (Figure 4), priming plant defense mechanisms against abiotic and biotic (Beckers and Conrath, 2007) stressors.
Alteration in membrane potential is a rapid response to wounding (Maffei et al., 2007), and our observations suggest that K+ channel regulation may contribute to these early electrical events. Since application of glucosinolates to wild-type GCs, or application of glucosinolates plus myrosinase to tgg1 mutant GCs, also inhibits the K+in channels, we infer that it is the reaction catalyzed by myrosinase (e.g., the hydrolyzed products of glucosinolates) that evoke channel inhibition. Because the inward K+ channels of GCs are, on a sequence homology basis, most similar to metazoan Shaker channels (Pilot et al., 2003; Pandey et al., 2007), animal Shaker K+ channels may also be targets for these plant secondary compounds, which are known to have both toxic and anticarcinogenic effects in mammals (Halkier and Gershenzon, 2006). Preharvest growth conditions, harvesting processes, and storage conditions all can affect plant glucosinolate concentrations (Johnson, 2002). The implication from our data that ABA may regulate glucosinolate sequestration and stability may suggest modifications to extant agronomic protocols, with implications for food quality.
Our results (Figures 3C and 3D) demonstrate an interconnection between the products of myrosinase activity and K+ channel regulation, but further research will required to determine the exact mechanistic basis of this response. It may be relevant that recent studies have shown that isothiocyanates alter activity of the mammalian pain-sensing TRP1A channels by covalent modification at Cys residues (Hinman et al., 2006). Alternatively, or in addition, the fact that myrosinase binds the antioxidant ascorbic acid and can catalyze formation of a condensation product of ascorbic acid with methylindoles (Burmeister et al., 2000) may suggest that myrosinase activity promotes ABA signaling via decreasing the ability of the GC to scavenge ROS. ROS elevation is a key signal transduction element in ABA signaling (Pei et al., 2000; Kwak et al., 2003), and plants engineered for increased ascorbate levels exhibit decreased levels of ROS and decreased GC responsiveness to ABA (Chen and Gallie, 2004).
While it has recently been demonstrated that small molecules, such as NO and ROS, are shared between ABA and defense signaling pathways, including in GCs (Melotto et al., 2006; Ali et al., 2007; Zhang et al., 2008), enzymes of secondary metabolism, such as TGG1, have not previously been implicated in ABA signaling. The interconnection discovered here between ABA and the glucosinolate-based biotic defense mechanism suggests a mechanism whereby exposure to abiotic stresses may enhance plant defense against subsequent biotic invaders. One general property of the hydrolyzed products of glucosinolates is volatility (Yan and Chen, 2007). Speculatively, the localized and extremely high abundance of TGG1 in GC might facilitate evaporation of the hydrolyzed products from stomatal pores and thus maximize both deterrence of would-be herbivores and attraction of their parasites and predators (Bradburne and Mithen, 2000) as well as possibly initiate between- or within-plant defense signaling mechanisms (Baldwin et al., 2006; Frost et al., 2007).
In conclusion, the GC is a model system in plant cell biology, and the discovered GC proteome can be used to inform the reconstruction of GC signaling networks in silico (Li et al., 2006) and in planta. Assessment of candidate proteins identified by our GC proteomic analysis also has the potential to enhance our understanding of how plants interact with the local climate and biotic environment. In particular, demonstration of TGG1 involvement in ABA signaling demonstrates novel roles for this known enzyme and highlights an interplay between biotic and abiotic stress responses in plants. The strategy applied here, beginning with global protein identification within a single cell type and ending with discovery of novel signaling pathways by functional analysis of protein candidates identified from proteomic analyses, will be a powerful approach for future single cell type studies in both plants and metazoans.
METHODS
Plant Material and GCP Isolation and Purity
Healthy rosette leaves harvested from 5-week-old plants were the tissue source for GCP isolation. Fifty million GCPs were used per independent replicate for the BR (pH 3 to 10) gel-based proteomics study, the NR gel-based study, and the LC-MALDI MudPIT run. Two replicates were performed for each method, and ∼22,000 Wassilewskija Arabidopsis thaliana plants were used, yielding ∼3 × 108 GCPs. All plants were grown in the same growth chamber (see Supplemental Methods online for details).
GCPs were isolated using the same-day two-step enzyme digestion method (Pandey et al., 2002). Epidermal peels are first obtained by blending Arabidopsis leaves in buffer. Epidermal cell protoplasts are then released from the epidermal peels by enzymatic digestion of their cell walls in the first enzyme digestion step, while the thicker-walled GCs remain attached to the cuticle. In the second enzyme digestion step, GCPs are released by further cell wall digestion. The main contamination for GCP isolation comes from mesophyll cell protoplasts, so we discarded any GCP isolations with >1% mesophyll cell protoplast contamination, as quantified by observing under the microscope and counting numbers of GCPs versus mesophyll cell protoplasts. After obtaining the GC proteome, we further evaluated the issue of contamination by comparing the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) protein amount in leaves and GCP. Rubisco is a key enzyme in the Calvin cycle and accounts for >50% of the total leaf protein (Evans, 1989). However, in our proteomic studies, the most abundant protein spot on 2D gels is TGG1 in both BR and NR methods, and Rubisco protein spots are hardly seen (Figures 1C and 1D; see Supplemental Table 4 online). This result not only indicates that the protein contamination from chloroplasts and chloroplast-rich mesophyll cells is low, but also is consistent with previous indications that photosynthethic carbon fixation is not a major function of GC chloroplasts (Shimazaki, 1989).
Protein Extraction and Separation for Mass Spectrometry Analysis
For the BR analysis, proteins were extracted (see Supplemental Methods online) from ∼50 million GCP. One milligram of total protein was cup-loaded onto a pH 3 to 10, 24-cm IPG strip (Bio-Rad) that was rehydrated overnight with 450 μL rehydration solution (8 M urea, 2% CHAPS, 2% IPG buffer, 20 mM DTT, and 0.001% bromophenol blue). IPG strips were run in a Multiphor II system (Pharmacia Biotech) at 500 V (gradient) for 1 min, 3500 V (gradient) for 1.5 h, and 3500 V for 10.5 h. The second dimension was run in a Protean II Xi cell (Bio-Rad) at 45 mA for 4.5 h. Gels were stained with Colloidal Coomassie Blue.
For the NR analysis, ∼50 million GCPs per sample were ground into fine powder under liquid nitrogen, and proteins were TCA precipitated. One milligram of total protein per replicate was prefractioned into five different pH ranges by six pH discs (3, 4.6, 5.4, 6.2, 7.0, and 10.0) using an IEF fractionator (Invitrogen; see Supplemental Methods online for details). Fractions were then separated on IPG strips of the corresponding pH range. IPG strips were run at 175 V (gradient) for 15 min, 2000 V (gradient) for 1 h, and 2000 V for 6 h. The second dimension was run in a miniprotean cell system (Bio-Rad) at 100 V for 10 min and then 200 V for 45 min. Gels were stained with Sypro-Ruby (Molecular Probes/Invitrogen).
For 2D LC-MALDI MudPIT analyses, total protein from ∼50 million GCP per sample was extracted as for IEF fractionation. Proteins were in-solution digested according to Adachi et al. (2006), and then trypsin-digested peptides were separated using two sequential separation methods, strong cation exchange and C18 nanoflow chromatography. See Supplemental Methods online for detailed separation methods.
Spot Cutting, Trypsin Digestion, and Spotting on MALDI Plates
All visible spots in both BR 2D gels were cut manually, and all spots in both NR 2D gels were cut using a spot-cutter (Bio-Rad EXQuest spot cutter). All spots were digested with trypsin (Promega Sequencing Grade) according to http://www.hmc.psu.edu/core/proteins_MassSpec/MassSpec/sampleprep.htm, desalted with SCX Ziptips (Millipore), and then spotted on MALDI plates. After the samples were dried, each spot was overlaid by 0.6 μL of matrix solution (5 mg/mL of a-cyano-4-hydroxycinnamic acid, 2 mg/mL of ammonium phosphate, 0.1% trifluoroacetic acid, and 50% acetonitrile).
Mass Spectrometry and Data Analysis
All peptides were analyzed using a 4700 or 4800 proteomic analyzer MALDI-TOF/TOF tandem system (Applied Biosysems). Two different software packages were used: GPS Explorer (Applied Biosystems/MDS Sciex), using as an underlying search algorithm a locally installed copy of the Mascot software programs, version 2.1 (Matrix Science; http://www.matrixscience.com), or Protein Pilot software version 2.0 (Applied Biosystems/MDS Sciex), using the Paragon algorithm (Shilov et al., 2007) for searching and the ProFound algorithm for protein inference and grouping from tandem mass spectrometry (MS/MS) spectral/peptide data.
All MS and MS/MS data obtained from gel-based methods were analyzed using GPS Explorer (Applied Biosystems). Candidate protein IDs from individual gel spots were accepted if they had a GPS Explorer protein CI > 99.5% (equivalent to a Mascot Score of P < 0.005). MS/MS data from 2D LC-MALDI MudPIT experiments were analyzed using both Mascot and Protein Pilot software version 2.0. For both algorithms, protein identification acceptance criteria were CI ≥ 98% (equal to a Protein Pilot unused score of 1.7) for proteins identified with multiple peptides and CI ≥ 99.9% for proteins detected from a single peptide, plus acceptable estimated FDRs (see Supplemental Methods online for details). In addition to the much more stringent CI requirements for acceptance of protein identifications that were identified solely from single peptide sequences from one biological replicate of MudPIT (requirements whose stringency guaranteed almost complete presence of appropriate B- and Y-ions), all such identifications in Tables 2 and 3 were further verified by manual analysis of spectra (see Supplemental Figure 3 online) to verify the lack of significant unmatched MS/MS peaks, the presence of strong peaks representing fragmentation at expected favored residues (e.g., after Asp and before Pro residues), and the presence of expected immonium ions from sequences containing amino acid residues expected to give strong immonium ion signals (e.g., a 110 m/z peak from His-containing peptides, an 86 m/z peak from Iso/Leu-containing peptides, etc.).
Stomatal Aperture Measurement
tgg1 mutants were generously provided by Georg Jander, Cornell University. Stomatal aperture measurements were basically performed as previously described (Fan et al., 2008). Leaves were harvested from 5-week-old healthy Arabidopsis plants just before initiation of the photoperiod in the growth chambers for stomatal opening assays and after the lights had been on for 5 min for stomatal closure measurements. Excised leaves were placed abaxial side down in a 6-well Petri dish containing 5 mL of solution in each well. The solution for assays of stomatal opening was 10 mM KCl, 7.5 mM IDA, and 10 mM MES, pH 6.15, with KOH. The solution for assays of stomatal closure was 20 mM KCl, 5 mM MES, and 1 mM CaCl2, pH 6.15, with KOH.
For wounding induction of stomatal closure, excised leaves in closure solution were first put under light (200 μmol m−2s−1) for 3 h to open stomata. For wounding inhibition of stomatal opening experiments, excised leaves in opening solution were first put under darkness for 1 h to close stomata. Next, leaves were treated with a slicker brush at the same time to ensure that the same extent of wounding treatment was administered to all the leaves (Bailey et al., 2005). Leaves were immediately returned to the dishes and put under light, and stomatal aperture measurements were taken at time points as indicated in Figure 2.
For stomatal opening experiments with MJ or ABA, the dish containing excised leaves was placed in darkness for 2 h to promote stomatal closure. For assays of stomatal closure, the leaves were placed under light (200 μmol m−2s−1) for 2 h to induce stomatal opening. Five microliters of 50 mM MJ (Sigma-Aldrich), 50 mM ABA (A.G. Scientific) (50 μM final concentration), DMSO (solvent control for MJ), or 100% ethanol (solvent control for ABA) was then added in each well for treatment or control, respectively, and leaves were further left under light for 2 more h for both treatment and solvent controls.
Four independent replicates were performed for the wounding experiment; eight and 10 independent replicates were performed for MJ inhibition of opening and promotion of closure experiments, respectively, and three replicates were performed for ABA regulation of stomatal aperture experiments in tgg1 mutants. Epidermal peels were prepared and 10 epidermal images were photographed per leaf. At least 50 stomatal apertures were measured per leaf. All stomatal apertures were measured using free access Image J software, version 1.34s.
Electrophysiology
Arabidopsis GCP isolation and standard whole-cell K+ recording were as previously described (Wang et al., 2001; Coursol et al., 2003). For ABA treatment, 50 μM ABA was added in basic solution for ≥1.5 h pretreatment of GCP, and the same concentration of ABA was also present in the bath solution during patch clamping. For glucosinolate and myrosinase treatments, final concentrations of 50 μM total glucosinolates (Sigma-Aldrich) and/or 0.2 units/mL myrosinase (Sigma-Aldrich) were added (from 50 mM and 50 units/mL stock solution for glucosinolates and myrosinase, respectively) into the pipette solution immediately before the start of the experiment. Glucosinolates were extracted according to the protocol provided (Sigma-Aldrich). K+ current magnitudes were compared with Student's t test; results with P ≤ 0.01 were considered significantly different.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession number: TGG1 (At5g26000, P37702).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MJ Does Not Affect Stomatal Movements.
Supplemental Figure 2. I/V Curves of Time-Activated Whole-Cell K+ Currents of Col and tgg1 Mutant Guard Cells.
Supplemental Figure 3. Spectra of Proteins Listed in Tables 2 and 3 That Were Identified by Single Peptides from One MudPIT Experiment.
Supplemental Table 1. Proteins Identified in our GC Proteome by the MudPIT Method.
Supplemental Table 2. Proteins Identified by Multiple Peptides and Proteins Identified by Single Peptides but from Both Replicates of the MudPIT Method.
Supplemental Table 3. Peptide Sequences for Proteins Identified by a Single Peptide and in One Replicate of the MudPIT Method.
Supplemental Table 4. Proteins Identified in the GC Proteome by Gel-Based Methods.
Supplemental Table 5. Twenty-Eight Proteins Were Identified in Multiple Spots from the Gel-Based Methods.
Supplemental Table 6. GC Proteins Identified by Any Two Proteomic Methods.
Supplemental Table 7. Nine Previously Identified Organ Biomarker Proteins Were Identified in Our GC Proteome.
Supplemental Table 8. Sixty-Seven Proteins Demonstrated to Function in Mature Arabidopsis Guard Cells Based on Published Literature.
Supplemental Methods.
Supplementary Material
Acknowledgments
We acknowledge the College of Medicine Mass Spectromety and Proteomics Facility and the Proteomics and Mass Spectrometry Core Facility at Penn State University. We thank Dr. Georg Jander for tgg mutant seeds and Drs. Hong Ma, Daniel Jones, and Jiaxu Li for comments on the manuscript. This work was supported by National Science Foundation Grants MCB-0209694 and 6-2066-01 and USDA Grant 2006-35100-17254 to S.M.A.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sarah M. Assmann (sma3@psu.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Acharya, B.R., and Assmann, S.M. (November 25, 2008). Hormone interactions in stomatal function. Plant Mol. Biol. 25 http://dx.doi.org/10.1007/s11103-008-9427-0. [DOI] [PubMed]
- Adachi, J., Kumar, C., Zhang, Y., Olsen, J.V., and Mann, M. (2006). The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 7 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa, A., Rahnenfuhrer, J., and Lengauer, T. (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22 1600–1607. [DOI] [PubMed] [Google Scholar]
- Ali, R., Ma, W., Lemtiri-Chlieh, F., Tsaltas, D., Leng, Q., von Bodman, S., and Berkowitz, G.A. (2007). Death don't have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19 1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amme, S., Rutten, T., Melzer, M., Sonsmann, G., Vissers, J.P., Schlesier, B., and Mock, H.P. (2005). A proteome approach defines protective functions of tobacco leaf trichomes. Proteomics 5 2508–2518. [DOI] [PubMed] [Google Scholar]
- Assmann, S.M., and Grantz, D.A. (1990). Stomatal response to humidity in sugarcane and soybean: Effect of vapour pressure difference on the kinetics of the blue light response. Plant Cell Environ. 13 163–169. [Google Scholar]
- Assmann, S.M., and Zeiger, E. (1987). Guard cell bioenergetics. In Stomatal Function, E. Zeiger, G. Farquhar, and I. Cowan, eds (Stanford, CA: Stanford University Press), pp. 163–194.
- Baerenfaller, K., Grossmann, J., Grobei, M.A., Hull, R., Hirsch-Hoffmann, M., Yalovsky, S., Zimmermann, P., Grossniklaus, U., Gruissem, W., and Baginsky, S. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320 938–941. [DOI] [PubMed] [Google Scholar]
- Bailey, B.A., Strem, M.D., Bae, H., de Mayolo, G.A., and Guiltinan, M.J. (2005). Gene expression in leaves of Theobroma cacao in response to mechanical wounding, ethylene, and/or methyl jasmonate. Plant Sci. 168 1247–1258. [Google Scholar]
- Baldwin, I.T., Halitschke, R., Paschold, A., von Dahl, C.C., and Preston, C.A. (2006). Volatile signaling in plant-plant interactions: “Talking trees” in the genomics era. Science 311 812–815. [DOI] [PubMed] [Google Scholar]
- Barth, C., and Jander, G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46 549–562. [DOI] [PubMed] [Google Scholar]
- Bayer, E.M., Bottrill, A.R., Walshaw, J., Vigouroux, M., Naldrett, M.J., Thomas, C.L., and Maule, A.J. (2006). Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6 301–311. [DOI] [PubMed] [Google Scholar]
- Beckers, G.J., and Conrath, U. (2007). Priming for stress resistance: from the lab to the field. Curr. Opin. Plant Biol. 10 425–431. [DOI] [PubMed] [Google Scholar]
- Betts, R.A., Boucher, O., Collins, M., Cox, P.M., Falloon, P.D., Gedney, N., Hemming, D.L., Huntingford, C., Jones, C.D., Sexton, D.M., and Webb, M.J. (2007). Projected increase in continental runoff due to plant responses to increasing carbon dioxide. Nature 448 1037–1041. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blatt, M.R. (2000). Ca2+ signalling and control of guard-cell volume in stomatal movements. Curr. Opin. Plant Biol. 3 196–204. [PubMed] [Google Scholar]
- Bown, L., Kusaba, S., Goubet, F., Codrai, L., Dale, A.G., Zhang, Z., Yu, X., Morris, K., Ishii, T., Evered, C., Dupree, P., and Jackson, S. (2007). The ectopically parting cells 1-2 (epc1-2) mutant exhibits an exaggerated response to abscisic acid. J. Exp. Bot. 58 1813–1823. [DOI] [PubMed] [Google Scholar]
- Bradburne, R.P., and Mithen, R. (2000). Glucosinolate genetics and the attraction of the aphid parasitoid Diaeretiella rapae to Brassica. Proc. Biol. Sci. 267 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breshears, D.D., et al. (2005). Regional vegetation die-off in response to global-change-type drought. Proc. Natl. Acad. Sci. USA 102 15144–15148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, P.D., Tokuhisa, J.G., Reichelt, M., and Gershenzon, J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481. [DOI] [PubMed] [Google Scholar]
- Browse, J., McConn, M., James, D., Jr., and Miquel, M. (1993). Mutants of Arabidopsis deficient in the synthesis of α-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268 16345–16351. [PubMed] [Google Scholar]
- Burmeister, W.P., Cottaz, S., Rollin, P., Vasella, A., and Henrissat, B. (2000). High resolution X-ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base. J. Biol. Chem. 275 39385–39393. [DOI] [PubMed] [Google Scholar]
- Carroll, A.J., Heazlewood, J.L., Ito, J., and Millar, A.H. (2008). Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell. Proteomics 7 347–369. [DOI] [PubMed] [Google Scholar]
- Carter, C., Pan, S., Zouhar, J., Avila, E.L., Girke, T., and Raikhel, N.V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., and Gallie, D.R. (2004). The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 16 1143–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursol, S., Fan, L.M., Le Stunff, H., Spiegel, S., Gilroy, S., and Assmann, S.M. (2003). Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423 651–654. [DOI] [PubMed] [Google Scholar]
- Davletova, S., Schlauch, K., Coutu, J., and Mittler, R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses-Fonrouge, A.G., Voigt, K., Schroder, A., Arrivault, S., Thomine, S., and Kramer, U. (2005). Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 579 4165–4174. [DOI] [PubMed] [Google Scholar]
- Diks, S.H., and Peppelenbosch, M.P. (2004). Single cell proteomics for personalised medicine. Trends Mol. Med. 10 574–577. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., Long, T.A., Wang, J.Y., Jung, J.W., Mace, D., Pointer, S., Barron, C., Brady, S.M., Schiefelbein, J., and Benfey, P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320 942–945. [DOI] [PubMed] [Google Scholar]
- Dixon, D.P., Skipsey, M., Grundy, N.M., and Edwards, R. (2005). Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 138 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J.R. (1989). Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78 9–19. [DOI] [PubMed] [Google Scholar]
- Fan, L.M., Zhang, W., Chen, J.G., Taylor, J.P., Jones, A.M., and Assmann, S.M. (2008). Abscisic acid regulation of guard-cell K+ and anion channels in Gβ- and RGS-deficient Arabidopsis lines. Proc. Natl. Acad. Sci. USA 105 8476–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, L.M., Zhao, Z., and Assmann, S.M. (2004). Guard cells: A dynamic signaling model. Curr. Opin. Plant Biol. 7 537–546. [DOI] [PubMed] [Google Scholar]
- Friso, G., Giacomelli, L., Ytterberg, A.J., Peltier, J.B., Rudella, A., Sun, Q., and Wijk, K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost, C.J., Appel, H.M., Carlson, J.E., De Moraes, C.M., Mescher, M.C., and Schultz, J.C. (2007). Within-plant signalling via volatiles overcomes vascular constraints on systemic signalling and primes responses against herbivores. Ecol. Lett. 10 490–498. [DOI] [PubMed] [Google Scholar]
- Giavalisco, P., Wilson, D., Kreitler, T., Lehrach, H., Klose, J., Gobom, J., and Fucini, P. (2005). High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol. Biol. 57 577–591. [DOI] [PubMed] [Google Scholar]
- Gray, J.E., Holroyd, G.H., van der Lee, F.M., Bahrami, A.R., Sijmons, P.C., Woodward, F.I., Schuch, W., and Hetherington, A.M. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408 713–716. [DOI] [PubMed] [Google Scholar]
- Grubb, C.D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11 89–100. [DOI] [PubMed] [Google Scholar]
- Halkier, B.A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. [DOI] [PubMed] [Google Scholar]
- Hansen, B.G., Kliebenstein, D.J., and Halkier, B.A. (2007). Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 50 902–910. [DOI] [PubMed] [Google Scholar]
- Hashimoto, M., and Komatsu, S. (2007). Proteomic analysis of rice seedlings during cold stress. Proteomics 7 1293–1302. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., Tonti-Filippini, J.S., Gout, A.M., Day, D.A., Whelan, J., and Millar, A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herde, O., Pena-Cortes, H., Willmitzer, L., and Eisahn, J. (1997). Stomatal responses to jasmonic acid, linolenic acid and abscisic acid in wild-type and ABA-deficient tomato plants. Plant Cell Environ. 20 136–141. [Google Scholar]
- Hetherington, A.M., and Woodward, F.I. (2003). The role of stomata in sensing and driving environmental change. Nature 424 901–908. [DOI] [PubMed] [Google Scholar]
- Hinman, A., Chuang, H.H., Bautista, D.M., and Julius, D. (2006). TRP channel activation by reversible covalent modification. Proc. Natl. Acad. Sci. USA 103 19564–19568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes-Davis, R., Tanaka, C.K., Vensel, W.H., Hurkman, W.J., and McCormick, S. (2005). Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5 4864–4884. [DOI] [PubMed] [Google Scholar]
- Hunt, L., Mills, L.N., Pical, C., Leckie, C.P., Aitken, F.L., Kopka, J., Mueller-Roeber, B., McAinsh, M.R., Hetherington, A.M., and Gray, J.E. (2003). Phospholipase C is required for the control of stomatal aperture by ABA. Plant J. 34 47–55. [DOI] [PubMed] [Google Scholar]
- Husebye, H., Chadchawan, S., Winge, P., Thangstad, O.P., and Bones, A.M. (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsson, M., Siegel, R.S., Young, J., Hashimoto, M., Iba, K., and Schroeder, J.I. (2006). Guard cell ABA and CO2 signaling network updates and Ca2+ sensor priming hypothesis. Curr. Opin. Plant Biol. 9 654–663. [DOI] [PubMed] [Google Scholar]
- Jacob, T., Ritchie, S., Assmann, S.M., and Gilroy, S. (1999). Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 96 12192–12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job, C., Rajjou, L., Lovigny, Y., Belghazi, M., and Job, D. (2005). Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 138 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, I.T. (2002). Glucosinolates in the human diet. Bioavailability and implications for health. Phytochem. Rev. 1 183–188. [Google Scholar]
- Johnson, M.A., von Besser, K., Zhou, Q., Smith, E., Aux, G., Patton, D., Levin, J.Z., and Preuss, D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorrin, J.V., Maldonado, A.M., and Castillejo, M.A. (2007). Plant proteome analysis: A 2006 update. Proteomics 7 2947–2962. [DOI] [PubMed] [Google Scholar]
- Jung, J.Y., Kim, Y.W., Kwak, J.M., Hwang, J.U., Young, J., Schroeder, J.I., Hwang, I., and Lee, Y. (2002). Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Emi, T., Tominaga, M., Sakamoto, K., Shigenaga, A., Doi, M., and Shimazaki, K. (2003). Blue-light- and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiol. 133 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Shimazaki, K., and Nishimura, M. (1993). Phosphorylation and dephosphorylation of guard-cell proteins from Vicia faba L. in response to light and dark. Plant Physiol. 102 917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann, T., Russenberger, D., von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W., and Baginsky, S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14 354–362. [DOI] [PubMed] [Google Scholar]
- Kleffmann, T., von Zychlinski, A., Russenberger, D., Hirsch-Hoffmann, M., Gehrig, P., Gruissem, W., and Baginsky, S. (2007). Proteome dynamics during plastid differentiation in rice. Plant Physiol. 143 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.M., Leonhardt, N., Torres, M.A., Dangl, J.L., Bloom, R.E., Bodde, S., Jones, J.D., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S.J., Choi, E.Y., Choi, Y.J., Ahn, J.H., and Park, O.K. (2006). Proteomics studies of post-translational modifications in plants. J. Exp. Bot. 57 1547–1551. [DOI] [PubMed] [Google Scholar]
- Lambert, J.P., Ethier, M., Smith, J.C., and Figeys, D. (2005). Proteomics: From gel based to gel free. Anal. Chem. 77 3771–3788. [DOI] [PubMed] [Google Scholar]
- Lee, J., Garrett, W.M., and Cooper, B. (2007). Shotgun proteomic analysis of Arabidopsis thaliana leaves. J. Sep. Sci. 30 2225–2230. [DOI] [PubMed] [Google Scholar]
- Lee, S.-J., Saravanan, R.S., Damasceno, C.M.B., Yamane, H., Kim, B.-D., and Rose, J.K.C. (2004). Digging deeper into the plant cell wall proteome. Plant Physiol. Biochem. 42 979–988. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Choi, Y.B., Suh, S., Lee, J., Assmann, S.M., Joe, C.O., Kelleher, J.F., and Crain, R.C. (1996). Abscisic acid-induced phosphoinositide turnover in guard cell protoplasts of Vicia faba. Plant Physiol. 110 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt, N., Kwak, J.M., Robert, N., Waner, D., Leonhardt, G., and Schroeder, J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16 596–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Assmann, S.M. (1996). An abscisic acid-activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell 8 2359–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Assmann, S.M. (2000). Mass spectrometry. An essential tool in proteome analysis. Plant Physiol. 123 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wang, X.Q., Watson, M.B., and Assmann, S.M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287 300–303. [DOI] [PubMed] [Google Scholar]
- Li, S., Assmann, S.M., and Albert, R. (2006). Predicting essential components of signal transduction networks: A dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei, M.E., Mithofer, A., and Boland, W. (2007). Before gene expression: Early events in plant-insect interaction. Trends Plant Sci. 12 310–316. [DOI] [PubMed] [Google Scholar]
- March-Diaz, R., Garcia-Dominguez, M., Lozano-Juste, J., Leon, J., Florencio, F.J., and Reyes, J.C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53 475–487. [DOI] [PubMed] [Google Scholar]
- Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126 969–980. [DOI] [PubMed] [Google Scholar]
- Melquist, S., and Bender, J. (2004). An internal rearrangement in an Arabidopsis inverted repeat locus impairs DNA methylation triggered by the locus. Genetics 166 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, G., Zhang, W., Deng, F., Zhao, J., and Wang, X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312 264–266. [DOI] [PubMed] [Google Scholar]
- Mori, I.C., Murata, Y., Yang, Y., Munemasa, S., Wang, Y.F., Andreoli, S., Tiriac, H., Alonso, J.M., Harper, J.F., Ecker, J.R., Kwak, J.M., and Schroeder, J.I. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 4 e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa, S., Oda, K., Watanabe-Sugimoto, M., Nakamura, Y., Shimoishi, Y., and Murata, Y. (2007). The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol. 143 1398–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli, A.C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, C.K., Carr, K., McAinsh, M.R., Powell, B., and Hetherington, A.M. (2001). Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410 596–599. [DOI] [PubMed] [Google Scholar]
- Noir, S., Brautigam, A., Colby, T., Schmidt, J., and Panstruga, R. (2005). A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem. Biophys. Res. Commun. 337 1257–1266. [DOI] [PubMed] [Google Scholar]
- Pandey, S., and Assmann, S.M. (2004). The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S., Wang, X.Q., Coursol, S.A., and Assmann, S.M. (2002). Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytol. 153 517. [DOI] [PubMed] [Google Scholar]
- Pandey, S., Zhang, W., and Assmann, S.M. (2007). Roles of ion channels and transporters in guard cell signal transduction. FEBS Lett. 581 2325–2336. [DOI] [PubMed] [Google Scholar]
- Parrish, J.R., et al. (2007). A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 8 R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini, E.M., Kirkegaard, M., Mortensen, P., Lutz, H.U., Thomas, A.W., and Mann, M. (2006). In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood 108 791–801. [DOI] [PubMed] [Google Scholar]
- Pasini, E.M., Kirkegaard, M., Salerno, D., Mortensen, P., Mann, M., and Thomas, A.W. (2008). Deep coverage mouse red blood cell proteome: A first comparison with the human red blood cell. Mol. Cell. Proteomics 7 1317–1330. [DOI] [PubMed] [Google Scholar]
- Pei, Z.M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G.J., Grill, E., and Schroeder, J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406 731–734. [DOI] [PubMed] [Google Scholar]
- Petersen, B., Chen, S., Hansen, C., Olsen, C., and Halkier, B. (2002). Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214 562–571. [DOI] [PubMed] [Google Scholar]
- Philippar, K., Geis, T., Ilkavets, I., Oster, U., Schwenkert, S., Meurer, J., and Soll, J. (2007). Chloroplast biogenesis: the use of mutants to study the etioplast-chloroplast transition. Proc. Natl. Acad. Sci. USA 104 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot, G., Pratelli, R., Gaymard, F., Meyer, Y., and Sentenac, H. (2003). Five-group distribution of the Shaker-like K+ channel family in higher plants. J. Mol. Evol. 56 418–434. [DOI] [PubMed] [Google Scholar]
- Rajjou, L., Gallardo, K., Debeaujon, I., Vandekerckhove, J., Job, C., and Job, D. (2004). The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134 1598–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, M.R., and Griffin, T.J. (2006). Gel-free mass spectrometry-based high throughput proteomics: Tools for studying biological response of proteins and proteomes. Proteomics 6 4678–4687. [DOI] [PubMed] [Google Scholar]
- Samuels, L., Kunst, L., and Jetter, R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59 683–707. [DOI] [PubMed] [Google Scholar]
- Schirle, M., Heurtier, M.A., and Kuster, B. (2003). Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 2 1297–1305. [DOI] [PubMed] [Google Scholar]
- Schmelz, E.A., Engelberth, J., Alborn, H.T., O'Donnell, P., Sammons, M., Toshima, H., and Tumlinson III, J.H. (2003). Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. Proc. Natl. Acad. Sci. USA 100 10552–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, J.I., Raschke, K., and Neher, E. (1987). Voltage dependence of K+ channels in guard-cell protoplasts. Proc. Natl. Acad. Sci. USA 84 4108–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroter, D., et al. (2005). Ecosystem service supply and vulnerability to global change in Europe. Science 310 1333–1337. [DOI] [PubMed] [Google Scholar]
- Schwartz, A., Wu, W.H., Tucker, E.B., and Assmann, S.M. (1994). Inhibition of inward K+ channels and stomatal response by abscisic acid: An intracellular locus of phytohormone action. Proc. Natl. Acad. Sci. USA 91 4019–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, A., and Zeiger, E. (1984). Metabolic energy for stomatal opening. Roles of photophosphorylation and oxidative phosphorylation. Planta 161 129–136. [DOI] [PubMed] [Google Scholar]
- Shilov, I.V., Seymour, S.L., Patel, A.A., Loboda, A., Tang, W.H., Keating, S.P., Hunter, C.L., Nuwaysir, L.M., and Schaeffer, D.A. (2007). The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6 1638–1655. [DOI] [PubMed] [Google Scholar]
- Shimazaki, K., Doi, M., Assmann, S.M., and Kinoshita, T. (2007). Light regulation of stomatal movement. Annu. Rev. Plant Biol. 58 219–247. [DOI] [PubMed] [Google Scholar]
- Shimazaki, K.I. (1989). Ribulosebisphosphate carboxylase activity and photosynthetic O2 evolution rate in Vicia guard-cell protoplasts. Plant Physiol. 91 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolovski, S., Hills, A., Gay, R., Garcia-Mata, C., Lamattina, L., and Blatt, M.R. (2005). Protein phosphorylation is a prerequisite for intracellular Ca2+ release and ion channel control by nitric oxide and abscisic acid in guard cells. Plant J. 43 520–529. [DOI] [PubMed] [Google Scholar]
- Sorin, C., Negroni, L., Balliau, T., Corti, H., Jacquemot, M.P., Davanture, M., Sandberg, G., Zivy, M., and Bellini, C. (2006). Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 140 349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhita, D., Raghavendra, A.S., Kwak, J.M., and Vavasseur, A. (2004). Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol. 134 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H., Nishiyama, C., Shimada, T., Koumoto, Y., Hayashi, Y., Kondo, M., Takahashi, T., Ohtomo, I., Nishimura, M., and Hara-Nishimura, I. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47 164–175. [DOI] [PubMed] [Google Scholar]
- Tyers, M., and Mann, M. (2003). From genomics to proteomics. Nature 422 193–197. [DOI] [PubMed] [Google Scholar]
- Wall, M.K., Mitchenall, L.A., and Maxwell, A. (2004). Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA 101 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, J., Torres, M., Ganapathy, A., Thelen, J., DaGue, B.B., Mooney, B., Xu, D., and Stacey, G. (2005). Proteomic analysis of soybean root hairs after infection by Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 18 458–467. [DOI] [PubMed] [Google Scholar]
- Wang, X.Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292 2070–2072. [DOI] [PubMed] [Google Scholar]
- Wienkoop, S., Zoeller, D., Ebert, B., Simon-Rosin, U., Fisahn, J., Glinski, M., and Weckwerth, W. (2004). Cell-specific protein profiling in Arabidopsis thaliana trichomes: Identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry 65 1641–1649. [DOI] [PubMed] [Google Scholar]
- Wittstock, U., and Halkier, B.A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7 263–270. [DOI] [PubMed] [Google Scholar]
- Yan, X., and Chen, S. (2007). Regulation of plant glucosinolate metabolism. Planta 226 1343–1352. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Costa, A., Leonhardt, N., Siegel, R.S., and Schroeder, J.I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., He, S.-Y., and Assmann, S.M. (2008). The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. 56 984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Yu, L., Zhang, Y., and Wang, X. (2005). Phospholipase D in the signaling networks of plant response to abscisic acid and reactive oxygen species. Biochim. Biophys. Acta 1736 1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.