Abstract
The translationally controlled tumor protein (TCTP) is an important component of the TOR (target of rapamycin) signaling pathway, the major regulator of cell growth in animals and fungi. TCTP acts as the guanine nucleotide exchange factor of the Ras GTPase Rheb that controls TOR activity in Drosophila melanogaster. We therefore examined the role of Arabidopsis thaliana TCTP in planta. Plant TCTPs exhibit distinct sequence differences from nonplant homologs but share the key GTPase binding surface. Green fluorescent protein reporter lines show that Arabidopsis TCTP is expressed throughout plant tissues and developmental stages with increased expression in meristematic and expanding cells. Knockout of TCTP leads to a male gametophytic phenotype with normal pollen formation and germination but impaired pollen tube growth. Silencing of TCTP by RNA interference slows vegetative growth; leaf expansion is reduced because of smaller cell size, lateral root formation is reduced, and root hair development is impaired. Furthermore, these lines show decreased sensitivity to an exogenously applied auxin analog and have elevated levels of endogenous auxin. These results identify TCTP as an important regulator of growth in plants and imply a function of plant TCTP as a mediator of TOR activity similar to that known in nonplant systems.
INTRODUCTION
The translationally controlled tumor protein (TCTP; also called p21, p23, histamine releasing factor and fortilin; Bommer and Thiele, 2004) is a ubiquitously expressed and distributed protein in eukaryotes. Initially identified in mammalian tumor cells and found to be controlled posttranscriptionally (Chitpatima et al., 1988), its molecular function has remained elusive. A large number of studies in various organisms have related TCTP to diverse cellular processes, for example, apoptosis, microtubule organization, or ion homeostasis, and several interacting proteins (e.g., polo kinase, tubulin, and Na+/K+-ATPase) were identified (for review, see Bommer and Thiele, 2004). Several studies in yeast and human cell lines suggest a role of TCTP in the regulation of cell growth and the control of the cell cycle through TCTP phosphorylation by polo kinase and its interaction with regulatory proteins, such as the myeloid cell leukemia protein MCL1 (Gachet et al., 1999; Li et al., 2001; Yarm, 2002). Overexpression or silencing of TCTP leads to alterations in cell cycle progression and growth defects (Tuynder et al., 2002; Liu et al., 2005). Knockout of the TCTP gene in Caenorhabditis elegans caused slow growth, progeny sterility, and embryo lethality, whereas yeast cells are viable but with reduced fitness (Rual et al., 2004; Deutschbauer et al., 2005; Sonnichsen et al., 2005). Most recently it has been shown that TCTP controls cell proliferation by inhibition of apoptosis in mice, and knockout of the gene leads to an embryo-lethal phenotype (Chen et al., 2007).
A hint for TCTP function comes from the elucidation of the Schizosaccharomyces pombe TCTP protein structure, which revealed similarity to the mammalian/dominant suppressor of sec4 (MSS4/DSS4) protein family known to bind to the Rab subfamily of Ras GTPases. It has been proposed that in this interaction TCTP acts either as a guanine nucleotide exchange factor (GEF) or guanine nucleotide-free chaperone (Boguski and McCormick, 1993; Nuoffer et al., 1997; Pai, 1998; Thaw et al., 2001; Itzen et al., 2006). By contrast, another study identified human TCTP as a guanine nucleotide dissociation inhibitor of the translation elongation factor eEF1A (Cans et al., 2003). Thus, although the molecular function of TCTP cannot be unequivocally established from these studies, it seems clear that TCTP is a modulator of GTPase activity. As GTPases act as molecular switches for a vast number of cellular processes in all eukaryotes (Vernoud et al., 2003), this GTPase-regulating property might explain TCTP's involvement in seemingly unrelated cellular processes. Importantly, it has been found recently that Drosophila melanogaster TCTP activates the Ras GTPase dRheb via a GEF activity, and silencing of TCTP expression by RNA interference (RNAi) led to reduced cell size and number (Hsu et al., 2007). In this interaction, dRheb and TCTP are regulatory factors of the TOR (target of rapamycin) Ser/Thr kinase, which regulates cell growth in response to external (e.g., hormones) and internal (e.g., amino acid levels and energy status) stimuli as has been shown for many nonplant eukaryotes, such as yeast, Drosophila, C. elegans, and human (for review, see Wullschleger et al., 2006).
In nonplant eukaryotes, TOR forms two multiprotein complexes (TORC1 and TORC2) with only partly overlapping components and functions. TORC1 controls overall cell growth through regulation of protein synthesis, ribosome biogenesis, transcription, and autophagy by phosphorylation of downstream targets (e.g., S6 kinase and eIF4E binding protein). Only TORC1 is sensitive to inhibition by a rapamycin/FKBP12 (member of the FK506 binding proteins belonging to the peptidyl-prolyl cis/trans-isomerases) complex that mimics the endogenous inhibitor FKBP38 (Bai et al., 2007). TORC2, on the other hand, determines the site of cellular growth and thereby also cell shape by activation of the Rho1 GTPase and altering actin cytoskeleton organization (Jacinto et al., 2004; Sarbassov et al., 2004; Lee et al., 2005). A similar regulatory pathway appears to also exist in plants where few homologous members of the network, such as TOR, S6 kinase, and RAPTOR, have been identified (Menand et al., 2002; Mahfouz et al., 2006). Furthermore, as in other nonplant eukaryotes, silencing of Arabidopsis thaliana TOR leads to a reduction of cell size and subsequently organ size as a consequence of reduced translational activity (Deprost et al., 2007). Interestingly, higher plants are not sensitive to rapamycin, but expression of the yeast FKBP12 in Arabidopsis leads to rapamycin susceptibility, demonstrating that a similar control of TOR by plant FKBPs and Rheb homologs exists (Geisler and Bailly, 2007; Sormani et al., 2007).
As for TCTP, knowledge of its function in plants is very limited. Publicly available databases show that the expression of TCTP in Arabidopsis is ubiquitous and high in most tissues as found for its nonplant homologs (Schmid et al., 2005). A first report associated TCTP to cell division in the root cap of Pisum sativum (Woo and Hawes, 1997). Several later studies found changes in TCTP transcript or protein levels under diverse physiological conditions, such as light, aluminum stress, cold stress, Agrobacterium tumefaciens–mediated transformation, egg cell fertilization, or water deficit (Sage-Ono et al., 1998; Ermolayev et al., 2003; Lee and Lee, 2003; Veena et al., 2003; Sprunck et al., 2005; Vincent et al., 2007). However, the significance of these changes in TCTP transcript or protein levels has not been analyzed, and the molecular function of TCTP in plants is still not established. The only functional insight from the published literature is a likely involvement of TCTP in long-distance movement of phloem proteins (Aoki et al., 2005).
Here, we report detailed characterization of a plant TCTP. The Arabidopsis TCTP is shown to be more highly expressed in physiologically active and proliferating tissues as would be expected for a regulatory protein of the TOR pathway. Analysis of a T-DNA insertion line demonstrates that TCTP knockout causes a male gametophytic phenotype due to impaired pollen tube growth. TCTP silencing induces slower vegetative development and inhibits leaf expansion due to reduced cell size. Inhibition of TCTP also alters root development. More specifically, we reveal a role of TCTP in root hair development and lateral root formation, as well as an effect on auxin homeostasis. These results are consistent with a role of TCTP as an activator of a yet to be identified plant Rheb GTPase homolog and ultimately plant TOR, with auxin as a possible signaling molecule in the pathway.
RESULTS
Sequence Comparison and Structural Modeling
Searches of nucleotide and protein databases for sequences similar to well-characterized TCTPs (human, Drosophila, yeast, and C. elegans) revealed TCTP homologs in a large number of plant species. Protein sequence comparison of plant and nonplant TCTPs showed amino acid identities in the range of 40% and similarities of ∼60% using the PAM250 matrix (Dayhoff et al., 1978). Among plant TCTPs only, these values rise to 75 and 90%, respectively. These data indicate that plant and nonplant TCTPs diverged early, a conclusion also supported by our phylogenetic analysis (see Supplemental Figure 1 and Supplemental Table 1 online). The phylogenetic tree shows the divergence of a nonplant from the plant TCTP branch, and low bootstrap values within the plant cluster indicate the high similarity of plant TCTP sequences. In contrast with nonplant species where TCTP is usually encoded by a single functional gene (Thiele et al., 2000), we were able to identify several different cDNAs in many plant species indicative of several TCTP genes in these species. This might point toward a specialization of TCTP function within plants, although the genomes of Arabidopsis (see below) and rice (Oryza sativa) contain only one functional TCTP gene.
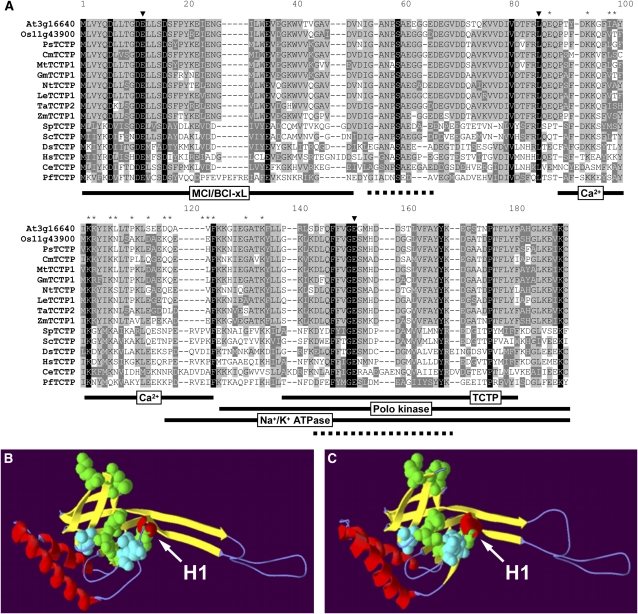
Several nonplant TCTPs have been characterized, leading to the identification of functional domains for interaction with other proteins, such as the cell cycle controlling polo kinase (Yarm, 2002), the antiapoptotic protein MCL/BCL (Yang et al., 2005), and a Na+/K+ ATPase (Jung et al., 2004). A Ca2+ binding domain and a TCTP self-interaction domain have also been identified (Kim et al., 2000; Yoon et al., 2000). Interestingly, in plant TCTP proteins these domains are not conserved, with many nonconservative amino acid substitutions making it unlikely that these regions have similar functions (Figure 1). Consistent with this, homologs of possible interacting partners (e.g., polo kinase and MCL) have not been identified in plant genomes. By contrast, the amino acid triad E12 L85 E152, characterized as an interaction surface for binding of Rab GTPases in the structurally related MSS4/DSS4 protein family (Thaw et al., 2001), together with surrounding amino acids, is well conserved (Figure 1). To investigate the possibility that the plant TCTP protein might have a similar function as TCTP in other organisms, we used homology modeling to obtain a structure prediction for the Arabidopsis TCTP protein At3g16640 (Figures 1B and 1C). This model shows high similarity to the known structure of the human TCTP protein with a nearly identical spatial organization of α-helices and β-sheets. Amino acids that are conserved in all TCTPs together with the H1 helix are clustered around the GTPase interaction surface. As the structural similarities between the MSS4/DSS4 and the TCTP protein family, although not paralleled in overall sequence similarity, have been linked to a GTPase binding property (Thaw et al., 2001; Itzen et al., 2006), our results strongly suggest a similar role of TCTP in mediating the activity of a plant Rheb GTPase. Identification of such a homolog is hampered by the absence of the Ras GTPase family in plants whose function has likely been adopted by a different class of plant GTPases (Vernoud et al., 2003). The closest Rheb GTPase plant homolog we could identify through sequence comparison is Ara6, which is involved in endosomal trafficking (Ueda et al., 2001).
Figure 1.
Sequence Comparison and Structural Modeling of TCTP.
(A) The amino acid sequence alignment was performed using the ClustalX1.8b software on representative plant and nonplant sequences (see Supplemental Figure 1 and Supplemental Table 1 online for phylogenetic tree and sequence accessions, respectively). Positions with strictly conserved amino acids are highlighted in black, conserved substitutions in dark gray, and blocks of similar residues in light gray. Triangles and asterisks indicate residues of the Rab GTPase binding triade (Thaw et al., 2001) and the tubulin binding domain (Gachet et al., 1999), respectively. Domains identified for nonplant TCTPs (MCL/BCL-xL interaction, Yang et al., 2005; polo kinase interaction, Yarm, 2002; Na+/K+ ATPase interaction, Jung et al., 2004; Ca2+ binding, Kim et al., 2000; TCTP self interaction, Yoon et al., 2000) are indicated by black lines and the TCTP signature (Interpro accession number IPR001983) by a dotted line. The structure of the Arabidopsis TCTP protein (At3g16640) was modeled using the known structure of the human TCTP (PDB ID 2 hr9, http://www.pdb.org) as a template on the Swiss-Model server (http://swissmodel.expasy.org/; Guex and Peitsch, 1997).
(B) and (C) The model obtained for the Arabidopsis protein (B) shows high similarity when compared with the structure of the human protein (C). Most of the absolutely conserved amino acids in all TCTP sequences as identified in the sequence alignment are clustered around the Rab GTPase interaction surface (Thaw et al., 2001) and are shown as calottes. These residues are in green (Met-1, Asp-6, Asp-11, Asp-16, Val-80, and Gly-151) and the core motif in cyan (Glu-12, Leu-85, and Glu-152). The conserved helix (H1) of this region is marked by an arrow. Helices, β-sheets, and coil regions of the two structures are represented in red, yellow, and blue, respectively.
Expression Analysis Reveals Ubiquitous Expression of TCTP
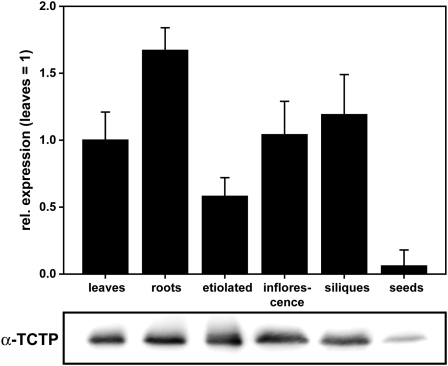
To gain insights into TCTP function in plants, we chose to analyze the Arabidopsis TCTP protein (At3g16640). Although a second isogene (At3g05540) is present in the Arabidopsis genome, there is no indication of its expression in the publicly available expression databases and not a single EST entry in the nucleotide databases. To verify this, we performed quantitative RT-PCR (qRT-PCR) analyses for a range of tissues and no At3g05540 transcript could be detected. Also, analysis of a homozygous T-DNA insertion line (SALK_010334) showed no obvious phenotype, making it highly likely that this gene is a nonfunctional pseudogene. Interestingly, in many nonplant genomes, pseudogenes of TCTP are also present (e.g., in rabbit, 10 to 15 pseudogenes are predicted) (Thiele et al., 2000). By contrast, expression of the TCTP gene At3g16640, as revealed by qRT-PCR, was very high, as anticipated from available plant microarray databases and EST clone numbers. Immunoblot analyses corroborated these results (Figure 2). With the exception of mature seeds, where gene expression was approximately 10 times below the other analyzed tissues, we found only minor differences in expression on transcript as well as protein levels among vegetative (leaves and roots) and reproductive (inflorescences and siliques) organs, indicating an association of TCTP expression with all physiologically active tissues. Transcript and protein levels in etiolated seedlings were comparable to those measured in tissues from light-grown seedlings, pointing toward a light-independent expression of TCTP.
Figure 2.
Transcript and Protein Expression Analysis.
Expression of TCTP was analyzed at the transcript level by qRT-PCR (top panel, mRNA expression levels relative to leaves and first normalized against reference genes, three biological replicates, given as means ± se; see Methods) and on the protein level by immunoblotting with a polyclonal antibody to At3g16640 TCTP protein (bottom panel, 10 μg protein loaded per lane; for TCTP antiserum specificity, see Figure 5). Differences in gene expression levels among tissues (leaves and roots: 2-week-old plants grown in plates; etiolated: grown in plates for 5 d in the dark; inflorescence: fully open flowers stage 12; siliques: fully developed green siliques, seed: mature dry seeds) were below twofold except for mature seeds that had 10-fold lower transcript levels than leaves. Protein levels (same tissues as above) rank according to mRNA levels.
TCTP Is Highly Expressed in Young Proliferating Tissues
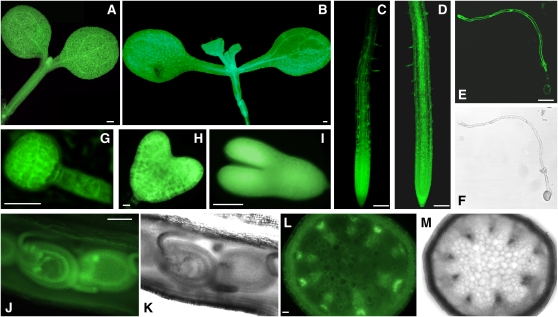
To allow for a more detailed analysis of TCTP expression, we generated TCTP-GFP reporter lines through transformation with a construct carrying a translational fusion of the full genomic TCTP region and the green fluorescent protein (GFP). This construct contained an ∼1-kb upstream region of the TCTP open reading frame, including the first two introns of the neighboring gene PRL2 (PP1/PP2A phosphatases pleiotropic regulator, At3g16650). As there is only a small intergenic region of 300 bp between TCTP and PRL2, this region therefore has to drive the expression of both genes in opposite directions. We screened 20 transformed lines for GFP expression and chose three representative homozygous single insert lines for further analyses. As expected from our expression analyses by qRT-PCR and immunoblots, GFP fluorescence was high in all tissues of young seedlings. In leaves, expression was visible in all cell types with a slightly increased GFP fluorescence associated with the vasculature (Figure 3A). This was also the case in the hypocotyl and root. In the latter, GFP fluorescence was significantly enhanced towards the root tip in the meristematic and division zones when compared with a 35S-GFP control line (Figures 3C and 3D). GFP fluorescence was also detectable throughout the elongating tube of germinating pollen (Figures 3E and 3F) and very high during early embryo development, already detectable at the globular stage and remaining high through later stages of embryogenesis (Figures 3G to 3I). In addition, TCTP expression was observable in all cell types of developing seeds but was especially high in the inner integument (Figures 3J and 3K). GFP fluorescence, although less intense, was also present in mature leaves. Although in cross sections of the stem a background expression in parenchymatic tissue was visible, a higher GFP fluorescence was associated with the vascular tissues and especially with the phloem (Figures 3L and 3M). This phloem localization is in agreement with a proposed role of TCTP in the differential translocation of phloem proteins suggested for the TCTP from cucumber (Cucumis sativus) (Aoki et al., 2005). Overall, these results from the GFP reporter lines demonstrate the high and ubiquitous expression of TCTP in plant tissues with a higher expression associated with actively dividing and differentiating cell types, such as the meristematic and division zones of the root or the embryo.
Figure 3.
TCTP Expression Analysis Using GFP Reporter Lines.
TCTP-GFP fusion protein expression under the control of the TCTP promoter in representative transgenic lines carrying the genomic TCTP-GFP construct is visualized by fluorescence imaging. TCTP expression as indicated by GFP fluorescence is ubiquitous throughout tissues and developmental stages. GFP imaging of TCTP reporter lines by epifluorescence microscopy (except for confocal images in [C] and [D]): cotyledons and hypocotyl of a young seedling (A), root (C), germinating pollen ([E] and [F]), embryo ([G], globular stage; [H], heart stage; [I], young embryo), and in cross sections of silique ([J] and [K]) and stem ([L] and [M]). For comparison, images captured for a 35S-GPF line are shown for seedling (B) and root (D). Bars = 100 μm in (A), (B), (J), and (L), 50 μm in (C) to (E) and (I), and 10 μm in (G) and (H).
Disruption of the TCTP Gene Causes a Male Gametophytic Phenotype
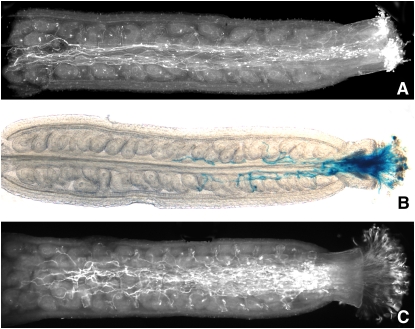
To facilitate the analysis of TCTP function, we tried to identify a T-DNA insertion line from the available Arabidopsis mutant collections. Although we were able to find an insertion line in the ABRC collection (SAIL_28_C03) and to confirm the presence of a single T-DNA insertion in the genome within exon 4 of the TCTP gene in heterozygous plants (see Supplemental Figure 2 online), we were unable to recover plants homozygous for the mutant allele. After selfing of heterozygous (TCTP/tctp) plants and analysis of their progeny by PCR genotyping or screening for T-DNA integration by Basta selection, we could only obtain wild-type and heterozygous plants (Table 1). While the observed non-Mendelian 1:1 segregation pattern was indicative of a gametophytic mutation, we first ensured that a homozygous tctp/tctp genotype did not result in an embryo lethal phenotype that would lead to a theoretical 3:1 ratio of normal to aborted seeds. Inspection of siliques from selfed heterozygous plants showed no signs of seed abortion and embryo lethality caused by the T-DNA insertion (see Supplemental Figure 3 online). We therefore reasoned that the tctp knockout might cause a gametophytic mutation leading to an aberrant transmission of the tctp mutant allele through either the male or female gametophyte, or both (Johnson-Brousseau and McCormick, 2004). To test this hypothesis, we performed reciprocal crosses between wild-type (TCTP/TCTP) and heterozygous (TCTP/tctp) plants and screened them for transmission of the tctp mutant allele through the male or female gametophyte using Basta selection. When fertilizing heterozygous plants with wild-type pollen, we observed no significant alteration in the transmission of the mutant allele. By contrast, the reciprocal cross gave almost no transmission of the mutant allele through the male gametophyte (Table 2). This result indicated an essential function of TCTP in pollen development or the fertilization process. Accordingly, we were able to demonstrate TCTP expression in pollen tubes in TCTP-GFP reporter lines (Figures 3E and 3F). We then examined the pollen phenotype (see Supplemental Figure 4 online), taking advantage of the qrt mutant background of the SAIL T-DNA collection in which the pollen grains stay together as tetrads after meiosis (Preuss et al., 1994; Sessions et al., 2002). Therefore, phenotypic changes caused by the tctp allele should be visible in two pollen grains of each tetrad, while the other two carry the wild-type allele. Alexander staining to test for pollen viability revealed no difference between the heterozygous (TCTP/tctp) line and a control line from the same SAIL collection, with each grain within a tetrad being viable. After 4',6-diamidino-2-phenylindole staining, pollen from both lines showed the typical fluorescence from the two sperm nuclei and the vegetative nucleus in each grain of a tetrad. Additionally, in germination assays, pollen tetrads from both lines frequently gave rise to three or four pollen tubes with similar final lengths in vitro, although the qrt mutation negatively affects overall germination rates and therefore hampers analysis (Boavida and McCormick, 2007). As the SAIL T-DNA construct also carries a β-glucuronidase (GUS) coding sequence under the control of the pollen-specific LAT52 promoter, we further conducted in vivo pollen germination assays (Figure 4). Our results show that pollen carrying the T-DNA insertion in the TCTP gene has a competitive disadvantage against wild-type pollen. Whereas a high number of wild-type pollen tubes reached the base of the pistil and targeted ovules, only a few tubes of tctp pollen grew past the stigma and into the ovary; they remained short and rarely targeted ovules. This pollen tube growth phenotype explains the reduced transmission of the tctp allele through the male gametophyte. We therefore conclude that TCTP is not essential for early pollen development, from the formation of the pollen mother cell to germination, but tctp knockout impedes the growth of the pollen tube, thereby reducing its competitiveness against wild-type pollen.
Table 1.
Selfed Progeny Segregation of TCTP/tctp Plants Determined by PCR and Basta Screening
| Heterozygous | Homozygous | Wild Type | χ2 (2:1:1/3:1) | χ2 (1:1) | |
|---|---|---|---|---|---|
| PCR | 52 | 0 | 53 | 53.51a | 0.01b |
| Basta | 613c | 649 | 472.48b | 1.06b | |
Selfing of TCTP/tctp (T-DNA insertion) lines failed to yield homozygous tctp/tctp plants but resulted in a non-Mendelian 1:1 segregation into heterozygous and wild-type plants.
χ2(2/0,001) = 13.82.
χ2(1/0,001) = 10.83.
Sum of Basta-resistant heterozygous or homozygous plants.
Table 2.
Progeny Segregation after Reciprocal Crosses of Wild-Type and TCTP/tctp Plants
| Cross (♀ × ♂) | Wild Typea | TCTP/tctpa | χ2 (1:1)b |
|---|---|---|---|
| TCTP/TCTP × TCTP/tctp | 496 (93.2%) | 36 (6.8%) | 397.74c |
| TCTP/tctp × TCTP/TCTP | 290 (53.2%) | 255 (46.8%) | 2.25d |
Transmission of the tctp mutant allele through the female gametophyte is unaffected, whereas transmission through the male gametophyte is greatly impaired.
Number of Basta-sensitive (wild type) and resistant (TCTP/tctp) plants.
χ2 values for a Mendelian 1:1 ratio.
χ2(1/0,001) = 10.823.
χ2(1/0,1) = 2.71.
Figure 4.
In Vivo Competitiveness Is Reduced in tctp Mutant Pollen.
Pollen from heterozygous (TCTP/tctp) SAIL line ([A] and [B]) and RNAi (C) plants was transferred to emasculated wild-type stigma. After 8 h, overall pollen tube growth was revealed by aniline blue staining, whereas germination of tctp mutant pollen (carrying a fusion of the pollen specific LAT52 promoter and the GUS coding sequence on the T-DNA) was specifically highlighted through GUS staining. Wild-type pollen tubes grew to the full length of the ovary and targeted ovules (A). By contrast, few pollen tubes carrying the T-DNA insertion penetrated the stigma and grew far into the ovary and very few targeted ovules (B). Aniline blue staining of pollen from RNAi lines could not reveal any difference to wild-type pollen (C). Pictures show representative images from 1 of 10 stained pistils for each genotype.
Silencing of TCTP by RNAi Leads to Retarded Growth Due to Decreased Cell Size
As the tctp knockout mutation causes a male gametophytic mutation and we were therefore unable to isolate a homozygous T-DNA insertion line, we took the alternative approach of RNAi to silence TCTP expression. Through qRT-PCR screening of 41 independent transgenic T1 lines transformed with the RNAi construct, we selected the three lines with lowest residual TCTP expression for further studies (TCTP-RNAi lines #1, #2, and #3). TCTP transcript levels in leaf tissue were only 1 to 2% of those in the wild type (see Supplemental Figure 5 online), and protein levels were reduced by at least 99% (Figure 5A). Given the very high expression of TCTP in wild-type plants, the reduction of TCTP expression below 1% of wild-type levels is quite remarkable. In contrast with the T-DNA insertion line, transmission of the RNAi construct through the male gametophyte was not affected even in the highest silenced lines, likely because the cauliflower mosaic virus (CaMV) 35S promoter driving the hairpin expression is only weakly active in pollen (Twell et al., 1989; Wilkinson et al., 1997). Accordingly, we could not observe a reduction in pollen tube growth in vivo (Figure 4C) or signs of seed abortion in siliques (see Supplemental Figure 3C online).
Figure 5.
TCTP Silencing and Overexpression.
Protein levels in RNAi lines were reduced to ∼1% of wild-type levels, whereas overexpression under the control of the CaMV 35S promoter led to an increase of ∼50%. TCTP overexpression lines were indistinguishable from the wild type, whereas RNAi lines showed retarded growth throughout development. RNAi lines also displayed significantly delayed bolting when compared with the wild type or overexpression lines, which bolted simultaneously. In parallel, the fresh weight of leaves and roots of in vitro–grown plants was significantly reduced for the silenced lines and again similar between the wild type and overexpressors.
(A) Immunodetection of TCTP protein in leaves in wild-type, RNAi, and overexpressing plants. The top panel shows ribulose-1,5-bis-phosphate carboxylase/oxygenase (RuBisCO) detection as loading control. The bottom panel shows detection of the endogenous Arabidopsis TCTP at 19 kD and the overexpressed TCTP carrying a fusion tag at 21 kD. Numbers below the immunoblot give TCTP protein levels relative to the wild type, quantified from band intensities of a representative immunoblot from three replicates.
(B) Images of representative plants for each genotype (the wild type, OX line #1, and RNAi line #1) 8, 18, and 31 d after germination (dag), respectively. Numbers below the frame give the average leaf number with sd at the time of bolting (n = 8).
(C) Fresh weights of in vitro–grown wild type, RNAi#1, and OX#1 lines 10 d after germination (means ± sd, five biological replicates of at least 15 pooled seedlings). Asterisks indicate statistically significant differences (analysis of variance [ANOVA] test, P < 0.001). [See online article for color version of this figure.]
In parallel, screening of plants transformed with a CaMV 35S-TCTP construct identified TCTP overexpressing plants (OX line #1, #2, and #3). The highest overexpressing line, OX line #1, showed a 13-fold increase in expression of TCTP at the transcript level (see Supplemental Figure 5 online) but only ∼50% increase at the protein level against the background of an already very high constitutive TCTP expression (Figure 5A). This result might indicate a posttranscriptional regulation of TCTP expression as has been shown for TCTP from mouse (Bommer and Thiele, 2004). Consequently, and in contrast with the RNAi lines (see below), we could not observe any phenotypical changes for any of our overexpression lines when compared with the wild type.
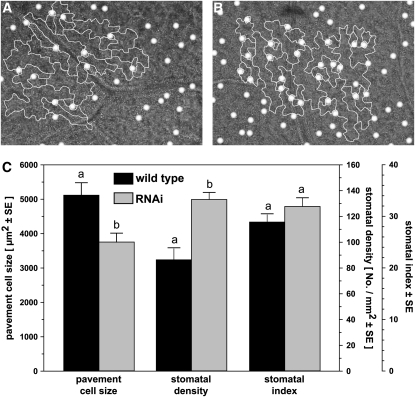
Whereas the overexpression lines were indistinguishable from the wild type, an immediately recognizable phenotype of the silencing lines was a retarded growth when grown under standard conditions on soil (Figure 5B). The growth reduction was already apparent 8 d after germination, becoming more pronounced with time, and 18 d after germination, the dwarfed phenotype of the knockdown lines was clearly visible. In addition, time of bolting was delayed when assessed on a leaf number basis. Furthermore, when grown in vitro root, and also shoot, fresh weights of knockdown lines were decreased (to 30 and 50% of the wild type, respectively; Figure 5C). This reduction in plant growth is reminiscent of a similar growth retardation observed in Arabidopsis TOR silencing lines and Drosophila TCTP RNAi lines, in both cases attributed to a reduction in cellular size (Deprost et al., 2007; Hsu et al., 2007). We therefore analyzed leaf epidermal cell size and found a reduction of pavement cell size of ∼25% in the TCTP RNAi lines (Figure 6). Stomatal density was increased in similar proportion (∼35%) so that there was no statistically significant change in the stomatal index [100 · stomatal density/(stomatal density + epidermal density)] (Figure 6C). This indicates that although the size of pavement cells is affected by TCTP silencing leading to reduced overall organ size, general epidermal development is unaltered. Comparable results have also been shown for Arabidopsis lines silenced in EBP1, a putative downstream target of TOR, as well as for potato (Solanum tuberosum) EBP1 RNAi lines where silencing of EBP1 leads to reduced growth and tuber sizes as a result of a decreased cell expansion (Horvath et al., 2006). Quantification of the EBP1 transcript levels in our TCTP RNAi lines revealed a fourfold decrease in the constitutive EBP1 expression (Figure 10D). A similar differential regulation of EBP1 has also been observed in Arabidopsis TOR knockdown lines (Deprost et al., 2007). Therefore, our results in Arabidopsis are in agreement with the TCTP function in other eukaryotes as an activator of the TOR pathway and thus cell growth.
Figure 6.
Silencing of TCTP Reduces Pavement Cell Size.
Comparison of pavement cells in the wild type and RNAi line #1 revealed that silencing of TCTP leads to a reduction in cell size of ∼25%. Stomatal density for the RNAi line was increased by 35%, but the slight increase in the stomatal index (SI) calculated from epidermal cell (E) and stomata (S) numbers per unit leaf area [SI = S*100/(S+E)] was not significant.
(A) and (B) Representative images of leaf abaxial epidermis for wild-type (A) and RNAi lines (B) with outlines of pavement cells and position of stomata highlighted.
(C) Quantification of cell size, stomatal density, and stomatal index determined for both genotypes from six independent areas per leaf and 30 cells measured within each. “a” and “b” indicate statistically significant changes (ANOVA test on raw data, P < 0.05)
Figure 10.
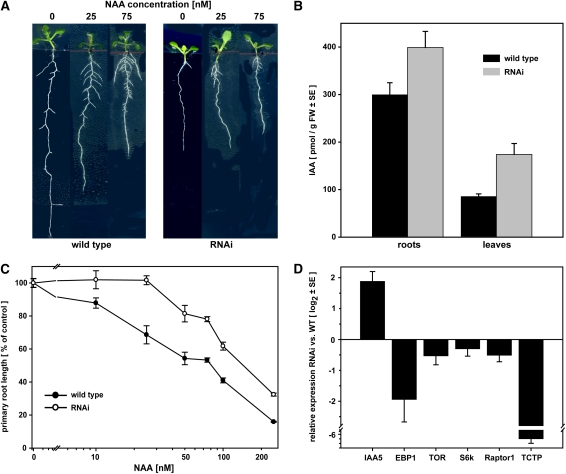
TCTP-RNAi Lines Have an Altered Auxin Homeostasis.
Treatment with increasing concentrations of NAA revealed an auxin-resistant phenotype of RNAi lines. Primary root elongation in these lines showed no response at concentrations of up to 25 nM NAA and at higher concentrations a consistently lower response than the wild type. Quantification of endogenous free IAA levels revealed increased levels in RNAi plants compared with the wild type and a constitutively higher expression of the auxin-inducible transcriptional regulator IAA5. Expression of EBP1 was reduced by approximately fourfold in the RNAi lines, and there was no significant change in the expression of other genes of the TOR pathway. [See online article for color version of this figure.]
(A) Representative images of wild-type and RNAi (RNAi#1 line) plants grown in vitro on half-strength Murashige and Skoog (MS) medium supplemented with NAA.
(B) Relative primary root length after 5 d of growth on supplemented medium (mean ± se, n = 9).
(C) Quantification of free IAA levels in RNAi and wild-type plants (mean ± se of seven biological replicates). Increases in RNAi plants are statistically significant (ANOVA test, P < 0.05).
(D) Expression analysis by qRT-PCR for IAA5, EBP1, TOR, S6k, Raptor1, and TCTP in leaf tissue. Relative expression (mean ± se) was normalized to three reference genes (APT1, PDF2, and UBC9; see Methods).
Reduced TCTP Expression Affects Root Growth and Lateral Root Formation
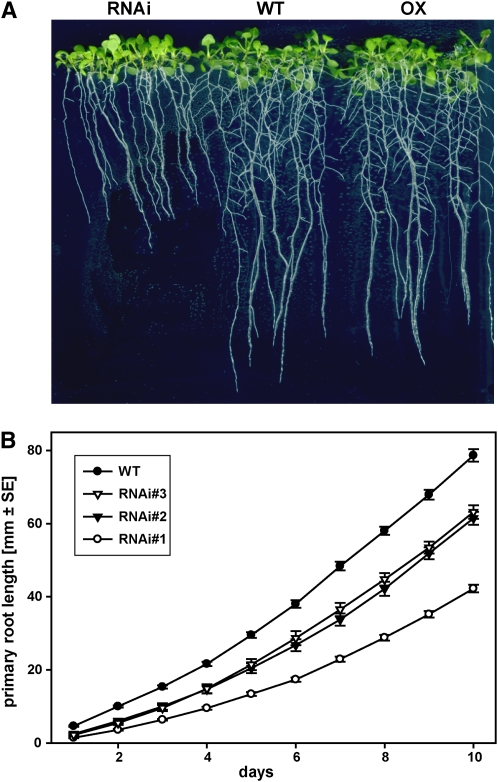
We next examined the effect of reduced TCTP levels on root growth and morphology. When grown in vitro on vertical plates, roots of RNAi lines showed significantly reduced length within a few days of germination, whereas overexpression lines were indistinguishable from the wild type (Figure 7A). Importantly, the reduction in primary root length was commensurate with TCTP silencing levels measured in the T1 generation for the three chosen RNAi lines, with the most severely silenced line having the shortest root (Figure 7B). This result points to a quantitative effect of TCTP expression on root growth. Ten days after germination the length of the primary root in RNAi line #1 was only half that measured for overexpressors or the wild type. We also quantified the number and lengths of the lateral roots. The average lateral root length and number per plant were decreased in the knockdown lines, with RNAi line #1 again showing the most severe reduction. A decreased lateral root number is consistent with a shorter overall primary root compared with the wild type (Table 3). Surprisingly, however, the lateral root density measured as the number of laterals per centimeter of primary root was reduced in the RNAi lines to between 62.5 and 75% of the wild type (i.e., the decrease in lateral root number was relatively higher than the reduction in primary root length). This decrease in lateral root density identifies the involvement of TCTP in the initiation or in the early outgrowth of lateral meristems. We therefore examined the TCTP-GFP reporter lines for TCTP expression patterns during lateral root formation in comparison to a 35S-GFP control line. GFP fluorescence in TCTP reporter lines started to increase very early at the site where the first cell divisions occur within the pericycle to initiate the lateral root primordium (Figures 8A and 8B; Laskowski et al., 1995; Malamy and Benfey, 1997). During the formation of the primordium and its penetration of the epidermis, GFP fluorescence was high throughout the forming bulge (Figures 8B to 8D). This increased GFP fluorescence then stayed with the meristematic and division zones of the developing lateral root (Figure 8E), similar to the pattern observed for the primary root. These results alongside the growth reduction seen for both the shoot and the primary root reveal an important role for TCTP in cell growth as in nonplant species (Yang et al., 2005; Hsu et al., 2007). In analogy to the role of TCTP as a GEF of the TOR activating GTPase Rheb in Drosophila, it is interesting to note that an identical phenotype with reduced shoot and root growth as a consequence of reduced cell size has also been observed for transgenic Arabidopsis lines silenced in TOR (Deprost et al., 2007).
Figure 7.
Effect of Altered TCTP Expression on Growth and Root Development.
RNAi lines showed retarded growth of the primary root, whereas overexpressors and wild-type plants were not visually different. RNAi lines developed fewer lateral roots (see Table 3 for quantitative data). Comparison of the three most severely silenced lines showed a quantitative effect of TCTP silencing on root growth with RNAi#1 having the slowest growing primary root. [See online article for color version of this figure.]
(A) Representative image of wild-type, TCTP RNAi, and TCTP overexpressing seedlings (line RNAi#1 and OX#1, respectively) grown on agar in vertical plates.
(B) Time course of primary root growth for three RNAi lines and the wild type (given as means ± se, n > 12 for each genotype).
Table 3.
Lateral Root Development in Wild-Type and TCTP-RNAi Plants
| Line | Lengtha | Numberb | Density (%)c |
|---|---|---|---|
| Wild type | 3.67 ± 0.28d | 18.9 ± 1.1d | 2.4 ± 0.1d (100%) |
| Line #1 | 1.81 ± 0.29e | 6.3 ± 0.5e | 1.5 ± 0.1e (62.5%) |
| Line #2 | 1.96 ± 0.27e | 10.3 ± 1.0f | 1.7 ± 0.1ef (70.8%) |
| Line #3 | 2.28 ± 0.25e | 11.7 ± 1.1f | 1.8 ± 0.2f (75%) |
For each genotype at least 10 seedlings have been analyzed. Superscript letters (d to f) indicate significant difference between lines in ANOVA test (P < 0.05).
Average lateral root length (mm ± se, 12 d after germination).
Average lateral root number (>100 μm, 12d after germination) per seedling (number ± se).
Lateral root density (number/centimeter primary root ± se).
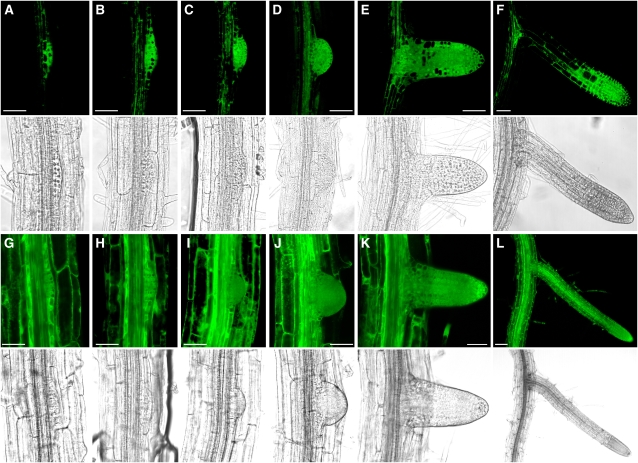
Figure 8.
GFP Fluorescence during Lateral Root Formation in TCTP-GFP Reporter Lines.
Confocal laser scanning microscopy of lateral root formation in TCTP-GFP reporter lines ([A] to [F]) in comparison to a 35S-GFP control line ([G] to [L]). GFP fluorescence for the TCTP line becomes visible early in root primordia initiation when first cell divisions occur in the pericycle and then remains strong in the actively dividing tissue layers. For the 35S-GFP control line, fluorescence in the lateral root primordia is comparable to surrounding tissues and is detectable throughout the developing lateral root. Top panels in (A) to (L) show GFP fluorescence; bottom panels correspond to transmission images. Bars = 50 μm.
(A), (B), (G), and (H) Lateral root initiation after early cell divisions in the pericycle.
(C) and (I) Later stage of lateral root primordium development, penetration of endodermal layer.
(D) and (J) Primordium reaches the epidermis.
(E) and (K) Lateral root after emergence.
(F) and (L) later stages of lateral root formation.
TCTP Silencing Leads to Altered Root Hair Morphology
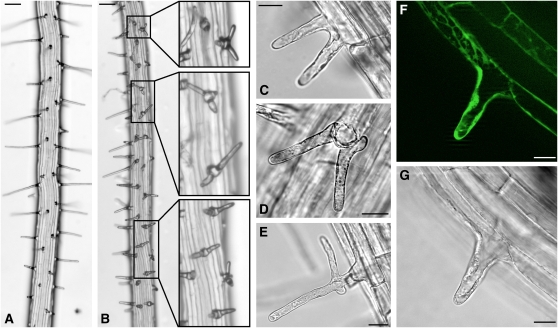
During the examination of roots of the TCTP RNAi lines, we observed an alteration of root hair morphology, especially in the most severely silenced line (RNAi line#1). In contrast with the wild type, where root hair shape and patterning are very organized (Grierson and Schiefelbein, 2002), roots of knockdown lines showed patches with disturbed root hair patterning. These patches were spread across the primary root and interspersed with regions of apparently unaltered development (Figure 9B). The position of root hair emergence close to the lower end of the trichoblast was not altered in our TCTP silenced lines when compared with the wild type, but root hairs formed from an initial central bulge whose outgrowth was interrupted and followed by the emergence of two to three additional root hair tips (Figures 9B to 9D). These tips remained much shorter than wild-type root hairs. In some cases, root hairs started to develop normally with one growing primary tip, but then additional tips started to grow at the base (Figure 9E). These root hair phenotypes suggest a role of TCTP for normal polarized tip growth; depletion of the TCTP protein in the RNAi lines leads to depolarized and branched root hair growth. Examination of root hairs in the TCTP-GFP reporter lines showed a high GFP signal in trichoblasts (Figures 9F and 9G), further underpinning a role for TCTP in root hair formation.
Figure 9.
Root Hair Development in RNAi Lines Is Impaired and Depolarized.
Shown are images for RNAi line #1, which showed the most severely impaired root hair development. Root hairs exhibited altered phenotypes in patches along the root axes ([B], insets show magnifications) when compared with the wild type (A). Frequently, root hairs branched from the flanks of a central bulge ([C], side view; [D], view from the top). In some cases, root hairs were depolarized with additional tips growing from the base (E). GFP fluorescence in the TCTP-GFP reporter lines was readily observable in root hair cells ([F], GFP fluorescence; [G], transmission image, confocal images). Bars = 100 μm in (A) and (B) and 20 μm in (C) to (G). [See online article for color version of this figure.]
TCTP Silencing Affects the Auxin Homeostasis
The plant hormone auxin is a key regulator of lateral root formation (De Smet et al., 2006; Fukaki et al., 2007), suggesting that an impaired auxin homeostasis could be the cause of the reduction in primary root growth and lateral root density observed in our TCTP RNAi lines. To investigate this, wild-type and RNAi plants were grown on medium supplemented with the auxin analog 1-naphthalacetic acid (NAA). Wild-type plants showed the expected reduction in primary root growth and induction of lateral root formation with increasing concentrations of NAA as the classical responses in this bioassay (Figure 10A; Zimmerman and Hitchcock, 1942). By contrast, reduction of primary root growth in knockdown lines only occurred at high NAA concentrations and even then remained mild. Response curve analyses revealed that RNAi lines did not respond to concentrations of up to 25 nM NAA when the wild type already showed a reduction in primary root length of ∼25%. The lag in response was sustained over the higher NAA concentrations (Figure 10B). Quantification of endogenous indole-3-acetic acid (IAA) levels in RNAi line #1 revealed significantly higher amounts compared with those in the wild type, with 2-fold and 1.3-fold increases in leaves and roots, respectively (Figure 10C). These changes in IAA levels are well in the range observed for known auxin-overproducing transgenic and mutant lines in which shorter roots and a stunted growth similar to that of our TCTP RNAi plants have been found (Boerjan et al., 1995; King et al., 1995; Barlier et al., 2000; Zhao et al., 2001, 2002). The observed reduced sensitivity to exogenously applied NAA might therefore be caused by the already higher endogenous IAA levels. Interestingly, the TCTP silencing lines do not show elongated hypocotyls characteristic of auxin-overproducing Arabidopsis plants. Hypocotyl lengths of 6-d-old wild-type and TCTP silencing plants were 2.4 ± 0.3 mm and 2.3 ± 0.3 mm, respectively. As over-production of IAA leads to an increased expression of auxin-inducible genes such as the AUX/IAA family of auxin-regulated transcriptional regulators (Zhao et al., 2002), we tested the constitutive expression levels of the IAA5 gene in the RNAi line #1. Results obtained by qRT-PCR show that IAA5 expression is fourfold higher in the RNAi line when compared with the wild type in agreement with the higher endogenous IAA levels (Figure 10D). There was no significant change in the expression of other genes of the TOR pathway, such as TOR, S6 kinase, and Raptor1, although all genes showed a slight downregulation. These results demonstrate an involvement of auxin in the plant TCTP and TOR-dependent pathway of cell growth regulation.
DISCUSSION
The role of TCTP in eukaryotic organisms has been obscure for a long time, although expression of the protein is ubiquitous and high in most tissues. The difficulty in determining TCTP function has been caused by the apparent involvement of TCTP in multiple seemingly unrelated cellular processes, such as IgE binding, microtubule organization, or ion homeostasis. The diversity of potential interacting proteins, identified by the yeast two-hybrid system or coimmunoprecipitation, increased this confusion even further, especially as evidence for the biological relevance of many of these interactions, sometimes even their verification by alternative methods, is still missing (Bommer and Thiele, 2004).
Here, using Arabidopsis as a model species, we report a detailed characterization of a plant TCTP. Our sequence analyses reveal limited sequence identity between plant TCTP proteins and their homologs in other Eukaryota, making predictions of functional domains difficult. So far none has been experimentally identified for a plant TCTP. This lack of sequence conservation is not surprising as many interactors of nonplant TCTPs are regulators of cell division specific to animals and fungi (Lee and Amon, 2003; Ohkura, 2003; Yang-Yen, 2006). The different mechanism by which cell division proceeds in plants has recruited a different set of regulatory proteins during evolution, and consistently TCTP-interacting proteins, such as MCL1 or polo kinase, have not been found in plants (Barr and Gruneberg, 2007). Nevertheless, our structural model for the Arabidopsis TCTP protein shows high similarity to the human protein. TCTP has been identified as a modulator of GTPase activity, for example, of the human elongation factor eEF1a and the Rheb GTPase in Drosophila (Cans et al., 2003; Hsu et al., 2007). It is remarkable that the GTPase interaction surface built by amino acids strictly conserved in all TCTPs is almost identical in the human TCTP and our Arabidopsis protein model. From this it appears highly likely that plant TCTP, despite having evolved to interact with plant-specific effectors, has retained its GTPase binding property and therefore GTPase activity–regulating function.
A deeper understanding of TCTP function has been hampered by the lack of viable mutant lines in multicellular organisms. In mouse, Drosophila, and C. elegans the knockout of the TCTP gene is lethal at an early embryonic stage, whereas heterozygous mutants show a wild type–like phenotype (Sonnichsen et al., 2005; Chen et al., 2007). Surprisingly, our results show that in Arabidopsis a TCTP knockout through T-DNA insertion results in a male gametophytic mutation preventing the generation of a homozygous mutant line. This is caused by a reduced competitiveness of pollen carrying the tctp mutant allele as a result of a slower and prematurely arrested growth of the pollen tubes within the ovary. As TCTP is a putative upstream regulator of the cell growth–regulating TOR network, it is not totally surprising to observe such a growth reduction in one of the fastest enlarging cell types in plants (Cheung and Wu, 2008). With a function in the modulation of GTPase activity implied by our structural analysis, it is intriguing to speculate that TCTP might act as a regulator of a Rab or Rho GTPase involved in pollen tube growth. Members of both families and their regulatory interactors have been shown to have important functions for pollen development (Kost et al., 1999; Li et al., 1999; Fu et al., 2001; Cheung et al., 2002; Chen et al., 2003; de Graaf et al., 2005; Gu et al., 2005; Klahre et al., 2006; Klahre and Kost, 2006). Interestingly, pollen of the rop2 GTPase mutant in maize (Zea mays) has a similar reduction in competitiveness as the TCTP knockout line (Arthur et al., 2003). Members of the same GTPase families have also been implicated in the regulation of polarized tip growth in root hairs (Molendijk et al., 2001; Preuss et al., 2004), therefore suggesting a similar function for TCTP in root hair development as proposed for pollen development. Overexpression of the ROP2 GTPase causes the formation of multiple tips on the same cell or hairs with several tips (Jones et al., 2002). The scn1 mutant shows initiation of several bulges from the same trichoblast and multiple tips growing from the same hair, whereas mutants of the ARF GAP protein RPA have branched hairs (Carol et al., 2005; Song et al., 2006). In addition to this proposed regulation of GTPase activity by TCTP to control pollen and root hair development, the TORC2 complex in yeast and Dictyostelium controls the organization of the actin cytoskeleton to maintain polarized growth (Jacinto et al., 2004; Sarbassov et al., 2004; Lee et al., 2005), a central process in tip growth of pollen tubes and root hairs in plants (Kost et al., 1998; Gibbon et al., 1999; Baluska et al., 2000; Fu et al., 2001; Chen et al., 2002; Gilliland et al., 2002; Jones et al., 2002; Ringli et al., 2002). Therefore, TCTP might also control spatial growth in pollen tubes or root hairs via the TORC2 signaling branch (see Supplemental Figure 6 online).
The question of how eukaryotic cell growth (i.e., increase in cytoplasm mass) and cell proliferation (i.e., increase in cell number) are regulated in their temporal and spatial patterns by internal and external cues is still a matter of much debate in animals (Jorgensen and Tyers, 2004; Cook and Tyers, 2007) and in plants (Sugimoto-Shirasu and Roberts, 2003; Ingram and Waites, 2006; Tsukaya, 2006). Currently, it is not clear how cell growth and division are coordinated to organ growth and how these processes shape the development of the plant (Ingram and Waites, 2006; John and Qi, 2008). Nevertheless, over the past years there has been wide agreement that the TOR signaling network is of central regulatory significance for the control of cell growth in yeast and animals (see Supplemental Figure 6 online; Schmelzle and Hall, 2000; De Virgilio and Loewith, 2006; Wullschleger et al., 2006). Recently, the importance of TOR and downstream targets, such as S6 kinase, has also become apparent in plants (Menand et al., 2002; Turck et al., 2004; Mahfouz et al., 2006; Deprost et al., 2007; Sormani et al., 2007). The implication of TCTP in the control of cellular growth through positive regulation of the GTPase Rheb and thereby TOR has recently been established in Drosophila (Hsu et al., 2007). The analysis of Arabidopsis TCTP RNAi lines in this study suggests a function of TCTP upstream of TOR in plants as in other eukaryotes (see Supplemental Figure 6 online). Consistent with that is the similar growth phenotype reported for Arabidopsis plants silenced for TOR (Deprost et al., 2007) compared with the phenotype we identify in our TCTP RNAi lines. In both cases, lines show retarded vegetative growth that is caused by a reduction in cellular growth as is exemplified by the smaller pavement cell size in the leaf epidermis. The increased TCTP expression at the initiation site of the nascent lateral roots as revealed through the analysis of our TCTP-GFP reporter lines associates TCTP to the establishment of the lateral root meristem. In this process, the founder cells at the protoxylem pole of the pericycle become meristematic with the development of a dense cytoplasm and ribosome biogenesis (Parizot et al., 2008), which have been identified as key downstream processes activated through TOR signaling in other organisms. Thus, an increased expression of TCTP in these founder cells and in the derived meristematic tissue is in agreement with the proposed function of TCTP as an upstream activator of the TOR pathway to drive increases in cytoplasmic content. Consistent with a plant TOR signaling network of comparable function to that in nonplant eukaryotes is the fact that expression of the yeast FKBP12 in rapamycin-insensitive Arabidopsis plants results in rapamycin susceptibility. The FKBP12-rapamycin complex mimics the endogenous TOR inhibitor FKBP38 (Bai et al., 2007), and treatment of these Arabidopsis transgenics with rapamycin leads to a reduction of primary root growth as in our TCTP silencing lines (Sormani et al., 2007). Therefore, it appears that rapamycin and TCTP knockdown result in a downregulation of TOR activity with comparable effects on primary root growth. Similarly, in the green algae Chlamydomonas reinhardtii, binding of rapamycin to a sensitive FKBP12 homolog results in a reduction of growth (Crespo et al., 2005). This shows that although higher plants are not susceptible to rapamycin, most likely because their FKBP12 protein cannot bind rapamycin, a regulation of plant TOR by a FKBP38 homolog must exist. Conversely, overexpression of Arabidopsis TOR led to an increase in cell size and subsequent plant growth (Deprost et al., 2007). Interestingly, this was not observed in our TCTP-overexpressing plants, which showed no detectable phenotypic differences to the wild type. Overexpression of TCTP in Drosophila did not result in overgrowth phenotypes either (Hsu et al., 2007), indicating that elevated TCTP protein levels are not resulting in increased activity of downstream targets and therefore growth promotion.
For several other genes of the TOR pathway, a similar growth inhibition in the corresponding mutant or silencing lines has been observed. Arabidopsis knockdown lines for the epidermal growth factor receptor binding protein (ErbB-3) homolog, EBP1, a protein involved in ribosome function and translation efficiency, showed similar phenotypes as TCTP or TOR RNAi lines with alterations in cell sizes (Horvath et al., 2006). As EBP1 is differentially regulated in TOR RNAi lines, it has been suggested that EBP1 is a downstream target of TOR (Horvath et al., 2006; Deprost et al., 2007). The downregulation of EBP1 in our TCTP silencing lines is slightly more pronounced than in TOR RNAi lines and in agreement with its predicted function as a target of the TOR signaling pathway similar to S6 kinase. Mutants for members of the TORC1 complex, Raptor1 and Raptor2, also revealed the importance of these complexes for the control of vegetative growth (Anderson et al., 2005; Deprost et al., 2005).
The effect of TCTP silencing on auxin homeostasis that we identify here is in line with other evidence connecting the TOR pathway with auxin signaling. An involvement of auxin in the function of several of the few known plant homologs of the TOR protein network has been found. Activation of the Arabidopsis S6 kinase has been shown to be at least partly dependent on auxin, leading to the phosphorylation of the downstream target ribosomal S6 protein (Turck et al., 2004). Furthermore, the Arabidopsis homolog of FKBP38, FKBP42 (TWISTED DWARF1/ULTRACURVATA2), has been identified as a positive regulator of PGP1-mediated auxin transport (Perez-Perez et al., 2004; Bouchard et al., 2006), and the EBP1 protein, a putative downstream target of TOR, is stabilized by auxin thereby preventing its degradation and maintaining its growth stimulating activity (Horvath et al., 2006). The increase in IAA levels measured in our knockdown plants, corroborated by the higher constitutive expression of the auxin-inducible transcriptional regulator IAA5 similar to that described in auxin-overproducing plants (Zhao et al., 2002), is in agreement with auxin being a central hormone in the control of cellular growth. From our results, however, we cannot separate whether auxin is integral to the TOR pathway, either as an upstream or a downstream signal, or if auxin acts in parallel to that pathway with both signaling cascades converging on the same downstream targets (see Supplemental Figure 6 online). The fact that an increase in hypocotyl length typically observed in auxin-overproducing plants could not be detected in our TCTP RNAi lines suggests that the increased auxin levels present in our lines cannot overrule the growth inhibition by TCTP silencing. Furthermore, exogenous auxin application or endogenous overproduction in Arabidopsis mutants, such as rty and sur1, leads to increased formation of lateral roots (Zimmerman and Hitchcock, 1942; Boerjan et al., 1995; King et al., 1995). This response could not be observed in our TCTP knockdown lines, although our GFP reporter lines highlight an involvement of TCTP in lateral root formation, which might also reflect an overriding inhibition through decreased TCTP levels. It therefore would seem likely that in an integral model auxin is upstream of TCTP and TOR. If, on the other hand, auxin acts in parallel to the TOR pathway in regulating cellular growth and root development, then there has to be a feedback mechanism that monitors cellular growth and, if enlargement is repressed under otherwise growth favoring conditions as is the case in our TCTP RNAi lines, adjusts auxin homeostasis to support growth.
This study establishes the crucial function of TCTP for the control of cellular growth in plants and its translation into organ growth, as is known of its homologs in nonplant systems. Several studies in plants have provided evidence that differential expression of TCTP occurs under abiotic stresses, such as aluminum exposure, cold, salinity, and water limitation (Ermolayev et al., 2003; Lee and Lee, 2003; Vincent et al., 2007). A role for TOR and its downstream target S6 kinase has also been identified in the response to osmotic stress, and increased TOR expression leads to increased seed number (Deprost et al., 2007). This is reminiscent of the involvement of the TOR signaling pathway in the response to stresses such as hypoxia, low energy status, or amino acid deprivation found in nonplant eukaryotes. Experiments are underway to elucidate the functional bases of these observations and what role TCTP plays in interconnecting growth and the response to environmental stresses or changing conditions in plants.
METHODS
Plant Material and Growth
All seeds (Arabidopsis thaliana ecotype Columbia-0) were sterilized by treatment with 70% ethanol for 2 min, 5% sodium hypochlorite for 5 min, and several washes with sterile water. For growth on vertical plates, seeds were sown on half-strength MS medium (0.5 MS salts, 1% sucrose, and 0.8% micro agar [Duchefa]) in 15-mm square plates sealed with microporous tape (3M) in growth chambers under short-day conditions (10 h light/14 dark, 21°C/19°C, 120 μE m−2 s−1). For auxin treatments, seeds were sown on half-strength MS medium supplemented with NAA at various concentrations made up from a 0.1 M stock solution in 1 M NaOH. Plants grown on soil were first germinated on half-strength MS medium for 5 d, and then seedlings of even sizes were transferred to soil and grown under long-day conditions (16 h light/8 h dark, 21°C/19°C and 120 μE m−2 s−1). For Basta selection, plants were grown on half-strength MS medium as above supplemented with 10 μg/mL phosphinotricin. Crosses were performed as described (Weigel and Glazebrook, 2002). TCTP T-DNA insertion lines SAIL_28_C03 and SALK_010334 were obtained from the ABRC collection.
Pollen Germination and Staining
For in vitro germination assays, pollen was harvested from flowers at stage 12 (i.e., on the day the flower opened) and transferred to agarose pads (1% agarose in 0.01% boric acid, 5 mM CaCl2, 5 mM KCl, 1mM MgSO4, and 10% sucrose, pH 7.5) on microscopy slides for germination at 22°C for 16 h in the dark. For Alexander and 4',6-diamidino-2-phenylindole staining, pollen grains were directly mounted in staining solution before microscopy (Boavida and McCormick, 2007). In vivo germination was performed as follows: wild-type flowers were emasculated, and the next day pollen from heterozygous (TCTP/tctp) or RNAi plants was transferred to the stigmata, and after 8 h, pistils were harvested for staining. For GUS staining, pistils were vacuum infiltrated with staining solution (Weigel and Glazebrook, 2002), incubated at 37°C overnight, fixed in ethanol:acetic acid (9:1), and cleared with chloral hydrate/glycerol/water (8:1:2) solution (Berleth and Jurgens, 1993). Aniline blue staining was performed as described (Preuss et al., 1993).
Molecular Techniques and Construct Design
General molecular techniques were performed as described (Sambrook et al., 1989). For GFP reporter lines, the genomic region between the first two introns of the 5′ upstream gene (At3g16640, PRL2) and the TCTP stop codon were PCR amplified from genomic DNA (primer gTCTPfor 5′-GGATCCTTTCACTGTTATGGTCAGCAATACGTGTAG-3′ and gTCTPrev 5′-AGCACTTGACCTCCTTCAAACCATG-3′) and cloned into the pENTR1A vector (Invitrogen) using BamHI and EcoRV sites. This genomic fragment was then transferred into the vector pMDC107 (Curtis and Grossniklaus, 2003) by LR recombinase reaction according to the recommendations of the manufacturer. For the TCTP RNAi construct, a DNA fragment of the TCTP cDNA was amplified by PCR (forward primer 5′-ATGTTGGTGTACCAAGATCTTCTCAC-3′; reverse primer 5′-AGCACTTGACCTCCTTCAAACC-3′), cloned blunt end into the vector pENTR1A cut with XmnI and EcoRV, and transferred into the vector pHELLSGATE8 (Helliwell et al., 2002) by LR recombinase reaction. For the TCTP overexpression construct, the coding region was amplified by PCR from the cDNA (forward primer 5′-AACCATGGTGGTGTACCAAGATCTTCTCACC-3′; reverse primer 5′-ATCTGCACTTGACCTCCTTCAAACC-3′), cloned into the vector pENTR1A cut with NcoI and EcoRV, and transferred into the vector pMDC32 (Curtis and Grossniklaus, 2003) by LR recombination. The binary vectors were introduced into the Agrobacterium tumefaciens strain AGL1, and plants were transformed using the floral dip method (Clough and Bent, 1998). Genotyping and characterization of the T-DNA insertion site of the SAIL line by PCR was performed using primers specific for the left and right borders of the T-DNA and TCTP, respectively (SAIL-LB primer 5′-ATAGCCTTGCTTCCTATTATATCTTCCCA-3′; SAIL-RB primer 5′-TAAATTATCGCGCGCGGTGTCA-3′; 5′-TCTP-SAIL1 primer 5′- AGATATCACACCAAATAACACAAAAAGTAACG-3′; TCTP-SAIL2 primer 5′-GACACCTTCAGACTTCAGGAGCAAC-3′) and genomic DNA prepared as described (Xin et al., 2003).
Expression Analysis by qRT-PCR
Expression of target genes was quantified as described (Jost et al., 2007). Briefly, mRNA from extracts of 50 mg of plant tissue was captured and transcribed into cDNA on oligo(dT)-coated magnetic beads. Subsequently, 0.5 ng of bead-bound cDNA was used to perform qPCR in a total volume of 10 μL using the Power Sybr Green PCR Master Mix on an ABI 7900HT fast real-time instrument (at the Australian Cancer Research Foundation Biomolecular Resource Facility, John Curtin School of Medical Research, Australian National University) according to the manufacturer's instructions (Applied Biosystems). Relative quantification of expression was calculated after data analysis using SDS2.2 software (Applied Biosystems) by the comparative ΔΔCT method (Livak and Schmittgen, 2001) with APT1, PDF2, and UBC9 as reference genes. For evaluation of reference genes, normalization and determination of amplification efficiencies the geNORM and LinRegPCR algorithms, respectively, were used (Vandesompele et al., 2002; Ramakers et al., 2003). For primer sequences, see Supplemental Table 2 online.
Antibody Production
TCTP was expressed as a 6xHis fusion protein after PCR amplification of the open reading frame and cloning into NcoI/BamHI sites of the vector pET28. The construct was transformed into BL21(DE3) cells (Novagen). After growth of the bacteria at 37°C and 220 rpm to an OD600 of 0.5, isopropylthio-β-galactoside was added to a final concentration of 1 mM and the bacteria grown further for 4 h. The bacteria were harvested by centrifugation, resuspended in 10 mL buffer (50 mM Tris, 300 mM NaCl, 10 mM β-mercaptoethanol, and 0.1 mM PMSF) + 20 mM imidazole and disrupted by ultrasonication. After centrifugation, the supernatant was applied to a HiTrap affinity purification column (Amersham) according to the manufacturer's protocol (consecutive washes with 5 mL buffer + 20 and 50 mM imidazole, respectively; elution with 5 mL buffer + 250 mM imidazole). The buffer with the highly purified TCTP protein was exchanged to 1× PBS using a PD10 column (Amersham). This antigen solution (4 mg/mL protein) was injected into rabbits using standard protocols for antibody production (Sambrook et al., 1989), and the resulting antiserum was used for immunoblotting.
Protein Extraction and Immunoblotting
For protein extraction, plant tissues were ground in extraction buffer (50 mM Tris, 300 mM NaCl, 0.1 mM PMSF, and 10 mM β-mercaptoethanol), and after centrifugation (14,000 rpm, 20 min, 4°C), the supernatant was denatured (95°C, 3 min) after addition of 2× sample buffer (Laemmli, 1970). Protein concentrations were determined using the Bradford assay system (Bio-Rad). For immunoblotting, plant proteins were separated on 12.5% SDS-PAGE gels and transferred to nitrocellulose membranes using a protean III system according to the manufacturer (Bio-Rad). Anti-TCTP primary antibody and anti-rabbit IgG secondary antibody (Promega) were used in 1:5000 and 1:10,000 dilutions, respectively. Chemiluminescent detection and quantification were performed using the Western-Star Immunodetection System according to the manufacture (Applied Biosystems) and a VersaDoc MP 4000 CCD detector system with Quantity One software (Bio-Rad).
IAA Determination
For determination of endogenous IAA levels, plants were grown in vitro as described above. Approximately 100 mg of leaf or root tissue were extracted with 1 mL methanol at 70°C for 1 h with 30 pmol [2H]2-IAA as an internal standard. After centrifugation to remove cellular debris, the supernatant was dried in a speed vac and free IAA quantified by a gas chromatography–mass spectrometry technique as described (Müller et al., 2002).
Microscopy
For determination of epidermal anatomy parameters, six small areas selected in the central part of the blades beside the major vein were cut out, cleared overnight in 85% lactic acid, and mounted for microscopy for imaging by light microscopy. Measurements of cell sizes were determined from these images using Zeiss LSM software (Zeiss) and the appropriate pixel to distance conversion. GFP confocal microscopy was performed with a Leica SP2 confocal laser scanning microscope (488-nm laser excitation, 510- to 560-nm emission). For epifluorescence microscopy, a Zeiss Axioplan2 microscope fitted with an Apotome imaging system (Zeiss) was used. Images were processed using Photoshop 7.0 software (Adobe).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TCTP, At3g16640 and At3g05540; PRL2, At3g16650; TOR, At1g50030; EBP1/AtG2, At3g51800; S6 kinase/S6k1, At3g08730; RAPTOR1, At3g08850; IAA5, At1g15580; APT1, At1g27450; PDF2/PP2AA3, At1g13320; UBC9, At4g27960.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogentic Tree for Plant and Selected Nonplant TCTPs.
Supplemental Figure 2. Characterization of TCTP T-DNA Insertion Line SAIL_28_C03.
Supplemental Figure 3. Seed Phenotype in Developing Siliques.
Supplemental Figure 4. Analysis of TCTP Pollen Phenotype.
Supplemental Figure 5. Screening of RNAi and Overexpression Lines.
Supplemental Figure 6. Comparison of TOR Pathway in Plants and Other Eukaryotes.
Supplemental Table 1. Plant and Nonplant TCTP Sequences Used in Phylogenetic Analysis.
Supplemental Table 2. Primers Used for Quantitative RT-PCR.
Supplemental Data Set 1. Text File of TCTP Protein Alignments Corresponding to the Phylogenetic Tree Shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Jan Elliot for antibody production, the Australian National University electron microscopy unit for providing microscopy facilities, and Allan Lohe for help with the phenotyping of gametophytic development. We also acknowledge Peter Waterhouse and the ABRC for providing the vector pHELLSGATE8 and the T-DNA insertion lines, respectively. This work has been funded by the Australian National University and the Grains Research and Development Corporation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Josette Masle (josette.masle@anu.edu.au).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Anderson, G.H., Veit, B., and Hanson, M.R. (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, K., Suzui, N., Fujimaki, S., Dohmae, N., Yonekura-Sakakibara, K., Fujiwara, T., Hayashi, H., Yamaya, T., and Sakakibara, H. (2005). Destination-selective long-distance movement of phloem proteins. Plant Cell 17 1801–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur, K.M., Vejlupkova, Z., Meeley, R.B., and Fowler, J.E. (2003). Maize ROP2 GTPase provides a competitive advantage to the male gametophyte. Genetics 165 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X., Ma, D., Liu, A., Shen, X., Wang, Q.J., Liu, Y., and Jiang, Y. (2007). Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science 318 977–980. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Salaj, J., Mathur, J., Braun, M., Jasper, F., Samaj, J., Chua, N.H., Barlow, P.W., and Volkmann, D. (2000). Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 227 618–632. [DOI] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, F.A., and Gruneberg, U. (2007). Cytokinesis: Placing and making the final cut. Cell 131 847–860. [DOI] [PubMed] [Google Scholar]
- Berleth, T., and Jurgens, G. (1993). The role of the monopteros gene in organizing the basal body region of the Arabidopsis embryo. Development 118 575–587. [Google Scholar]
- Boavida, L.C., and McCormick, S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52 570–582. [DOI] [PubMed] [Google Scholar]
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Van Onckelen, H., Van Montagu, M., and Inze, D. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski, M.S., and McCormick, F. (1993). Proteins regulating Ras and its relatives. Nature 366 643–654. [DOI] [PubMed] [Google Scholar]
- Bommer, U.A., and Thiele, B.J. (2004). The translationally controlled tumour protein (TCTP). Int. J. Biochem. Cell Biol. 36 379–385. [DOI] [PubMed] [Google Scholar]
- Bouchard, R., Bailly, A., Blakeslee, J.J., Oehring, S.C., Vincenzetti, V., Lee, O.R., Paponov, I., Palme, K., Mancuso, S., Murphy, A.S., Schulz, B., and Geisler, M. (2006). Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J. Biol. Chem. 281 30603–30612. [DOI] [PubMed] [Google Scholar]
- Cans, C., et al. (2003). Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc. Natl. Acad. Sci. USA 100 13892–13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol, R.J., Takeda, S., Linstead, P., Durrant, M.C., Kakesova, H., Derbyshire, P., Drea, S., Zarsky, V., and Dolan, L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438 1013–1016. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Cheung, A.Y., and Wu, H.M. (2003). Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 15 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y., Wong, E.I., Vidali, L., Estavillo, A., Hepler, P.K., Wu, H.M., and Cheung, A.Y. (2002). The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Wu, P.-H., Chou, C.-H., Yan, Y.-T., Liu, H., Weng, S.-Y., and Yang-Yen, H.-F. (2007). A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type–specific manner. Mol. Cell. Biol. 18 2525–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., Chen, C.Y., Glaven, R.H., de Graaf, B.H., Vidali, L., Hepler, P.K., and Wu, H.M. (2002). Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14 945–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., and Wu, H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59 547–572. [DOI] [PubMed] [Google Scholar]
- Chitpatima, S.T., Makrides, S., Bandyopadhyay, R., and Brawerman, G. (1988). Nucleotide sequence of a major messenger-RNA for a 21 kilodalton polypeptide that is under translational control in mouse tumor cells. Nucleic Acids Res. 16 2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cook, M., and Tyers, M. (2007). Size control goes global. Curr. Opin. Biotechnol. 18 341–350. [DOI] [PubMed] [Google Scholar]
- Crespo, J.L., Diaz-Troya, S., and Florencio, F.J. (2005). Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 139 1736–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff, M.O., Schwartz, R.M., and Orcutt, B.C. (1978). A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure, M.O. Dayhoff, ed (Washington DC: National Biomedical Research Foundation), pp. 345–352.
- de Graaf, B.H., Cheung, A.Y., Andreyeva, T., Levasseur, K., Kieliszewski, M., and Wu, H.M. (2005). Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused pollen tube growth in tobacco. Plant Cell 17 2564–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost, D., Truong, H.N., Robaglia, C., and Meyer, C. (2005). An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 326 844–850. [DOI] [PubMed] [Google Scholar]
- Deprost, D., Yao, L., Sormani, R., Moreau, M., Leterreux, G., Nicolai, M., Bedu, M., Robaglia, C., and Meyer, C. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet, I., Vanneste, S., Inze, D., and Beeckman, T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60 871–887. [DOI] [PubMed] [Google Scholar]
- Deutschbauer, A.M., Jaramillo, D.F., Proctor, M., Kumm, J., Hillenmeyer, M.E., Davis, R.W., Nislow, C., and Giaever, G. (2005). Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio, C., and Loewith, R. (2006). Cell growth control: Little eukaryotes make big contributions. Oncogene 25 6392–6415. [DOI] [PubMed] [Google Scholar]
- Ermolayev, V., Weschke, W., and Manteuffel, R. (2003). Comparison of Al-induced gene expression in sensitive and tolerant soybean cultivars. J. Exp. Bot. 54 2745–2756. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki, H., Okushima, Y., and Tasaka, M. (2007). Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 256 111–137. [DOI] [PubMed] [Google Scholar]
- Gachet, Y., Tournier, S., Lee, M., Lazaris-Karatzas, A., Poulton, T., and Bommer, U.A. (1999). The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J. Cell Sci. 112 1257–1271. [DOI] [PubMed] [Google Scholar]
- Geisler, M., and Bailly, A. (2007). Tete-a-tete: The function of FKBPs in plant development. Trends Plant Sci. 12 465–473. [DOI] [PubMed] [Google Scholar]
- Gibbon, B.C., Kovar, D.R., and Staiger, C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11 2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland, L.U., Kandasamy, M.K., Pawloski, L.C., and Meagher, R.B. (2002). Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol. 130 2199–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson, C.S., and Schiefelbein, J. (2002). Root hairs. In The Arabidopsis Book, C. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/, www.aspb.org/publications/arabidopsis/.
- Gu, Y., Fu, Y., Dowd, P., Li, S., Vernoud, V., Gilroy, S., and Yang, Z. (2005). A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell Biol. 169 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., and Peitsch, M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18 2714–2723. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Wesley, S.V., Wielopolska, A.J., and Waterhouse, P. (2002). High-throughput vectors for efficient gene silencing in plants. Funct. Plant Biol. 29 1217–1225. [DOI] [PubMed] [Google Scholar]
- Horvath, B.M., Magyar, Z., Zhang, Y., Hamburger, A.W., Bako, L., Visser, R.G., Bachem, C.W., and Bogre, L. (2006). EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 25 4909–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, Y.C., Chern, J.J., Cai, Y., Liu, M., and Choi, K.W. (2007). Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 445 785–788. [DOI] [PubMed] [Google Scholar]
- Ingram, G.C., and Waites, R. (2006). Keeping it together: Co-ordinating plant growth. Curr. Opin. Plant Biol. 9 12–20. [DOI] [PubMed] [Google Scholar]
- Itzen, A., Pylypenko, O., Goody, R.S., Alexandrov, K., and Rak, A. (2006). Nucleotide exchange via local protein unfolding–structure of Rab8 in complex with MSS4. EMBO J. 25 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto, E., Loewith, R., Schmidt, A., Lin, S., Ruegg, M.A., Hall, A., and Hall, M.N. (2004). Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6 1122–1128. [DOI] [PubMed] [Google Scholar]
- John, P.C., and Qi, R. (2008). Cell division and endoreduplication: Doubtful engines of vegetative growth. Trends Plant Sci. 13 121–127. [DOI] [PubMed] [Google Scholar]
- Johnson-Brousseau, S.A., and McCormick, S. (2004). A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 39 761–775. [DOI] [PubMed] [Google Scholar]
- Jones, M.A., Shen, J.J., Fu, Y., Li, H., Yang, Z., and Grierson, C.S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., and Tyers, M. (2004). How cells coordinate growth and division. Curr. Biol. 14 R1014–R1027. [DOI] [PubMed] [Google Scholar]
- Jost, R., Berkowitz, O., and Masle, J. (2007). Magnetic quantitative reverse transcription PCR: A high-throughput method for mRNA extraction and quantitative reverse transcription PCR. Biotechniques 43 206–211. [DOI] [PubMed] [Google Scholar]
- Jung, J., Kim, M., Kim, M.J., Kim, J., Moon, J., Lim, J.S., and Lee, K. (2004). Translationally controlled tumor protein interacts with the third cytoplasmic domain of Na,K-ATPase alpha subunit and inhibits the pump activity in HeLa cells. J. Biol. Chem. 279 49868–49875. [DOI] [PubMed] [Google Scholar]
- Kim, M., Jung, Y., Lee, K., and Kim, C. (2000). Identification of the calcium binding sites in translationally controlled tumor protein. Arch. Pharm. Res. 23 633–636. [DOI] [PubMed] [Google Scholar]
- King, J.J., Stimart, D.P., Fisher, R.H., and Bleecker, A.B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre, U., Becker, C., Schmitt, A.C., and Kost, B. (2006). Nt-RhoGDI2 regulates Rac/Rop signaling and polar cell growth in tobacco pollen tubes. Plant J. 46 1018–1031. [DOI] [PubMed] [Google Scholar]
- Klahre, U., and Kost, B. (2006). Tobacco RhoGTPase ACTIVATING PROTEIN1 spatially restricts signaling of RAC/Rop to the apex of pollen tubes. Plant Cell 18 3033–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16 393–401. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Laskowski, M.J., Williams, M.E., Nusbaum, H.C., and Sussex, I.M. (1995). Formation of lateral root meristems is a two-stage process. Development 121 3303–3310. [DOI] [PubMed] [Google Scholar]
- Lee, B.H., and Amon, A. (2003). Polo kinase–meiotic cell cycle coordinator. Cell Cycle 2 400–402. [PubMed] [Google Scholar]
- Lee, J.Y., and Lee, D.H. (2003). Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiol. 132 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Comer, F.I., Sasaki, A., McLeod, I.X., Duong, Y., Okumura, K., Yates III, J.R., Parent, C.A., and Firtel, R.A. (2005). TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell 16 4572–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Zhang, D., and Fujise, K. (2001). Characterization of fortilin, a novel antiapoptotic protein. J. Biol. Chem. 276 47542–47549. [DOI] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Peng, H.W., Cheng, Y.S., Yuan, H.S., and Yang-Yen, H.F. (2005). Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol. Cell. Biol. 25 3117–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Mahfouz, M.M., Kim, S., Delauney, A.J., and Verma, D.P. (2006). Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44. [DOI] [PubMed] [Google Scholar]
- Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., and Robaglia, C. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk, A.J., Bischoff, F., Rajendrakumar, C.S., Friml, J., Braun, M., Gilroy, S., and Palme, K. (2001). Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, A., Duchting, P., and Weiler, E.W. (2002). A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta 216 44–56. [DOI] [PubMed] [Google Scholar]
- Nuoffer, C., Wu, S.K., Dascher, C., and Balch, W.E. (1997). MSS4 does not function as an exchange factor for Rab in endoplasmic reticulum to Golgi transport. Mol. Biol. Cell 8 1305–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura, H. (2003). Phosphorylation: Polo kinase joins an elite club. Curr. Biol. 13 R912–R914. [DOI] [PubMed] [Google Scholar]
- Pai, E.F. (1998). The alpha and beta of turning on a molecular switch. Nat. Struct. Biol. 5 259–263. [DOI] [PubMed] [Google Scholar]
- Parizot, B., et al. (2008). Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol. 146 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez, J.M., Ponce, M.R., and Micol, J.L. (2004). The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol. 134 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D., Lemieux, B., Yen, G., and Davis, R.W. (1993). A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 7 974–985. [DOI] [PubMed] [Google Scholar]
- Preuss, D., Rhee, S.Y., and Davis, R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264 1458–1460. [DOI] [PubMed] [Google Scholar]
- Preuss, M.L., Serna, J., Falbel, T.G., Bednarek, S.Y., and Nielsen, E. (2004). The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16 1589–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H., and Moorman, A.F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339 62–66. [DOI] [PubMed] [Google Scholar]
- Ringli, C., Baumberger, N., Diet, A., Frey, B., and Keller, B. (2002). ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol. 129 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual, J.F., Ceron, J., Koreth, J., Hao, T., Nicot, A.S., Hirozane-Kishikawa, T., Vandenhaute, J., Orkin, S.H., Hill, D.E., van den Heuvel, S., and Vidal, M. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage-Ono, K., Ono, M., Harada, H., and Kamada, H. (1998). Dark-induced accumulation of mRNA for a homolog of translationally controlled tumor protein (TCTP) in Pharbitis. Plant Cell Physiol. 39 357–360. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sarbassov, D.D., Ali, S.M., Kim, D.H., Guertin, D.A., Latek, R.R., Erdjument-Bromage, H., Tempst, P., and Sabatini, D.M. (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14 1296–1302. [DOI] [PubMed] [Google Scholar]
- Schmelzle, T., and Hall, M.N. (2000). TOR, a central controller of cell growth. Cell 103 253–262. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.F., Yang, C.Y., Liu, J., and Yang, W.C. (2006). RPA, a class II ARFGAP protein, activates ARF1 and U5 and plays a role in root hair development in Arabidopsis. Plant Physiol. 141 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434 462–469. [DOI] [PubMed] [Google Scholar]
- Sormani, R., Yao, L., Menand, B., Ennar, N., Lecampion, C., Meyer, C., and Robaglia, C. (2007). Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunck, S., Baumann, U., Edwards, K., Langridge, P., and Dresselhaus, T. (2005). The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J. 41 660–672. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6 544–553. [DOI] [PubMed] [Google Scholar]
- Thaw, P., Baxter, N.J., Hounslow, A.M., Price, C., Waltho, J.P., and Craven, C.J. (2001). Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones. Nat. Struct. Biol. 8 701–704. [DOI] [PubMed] [Google Scholar]
- Thiele, H., Berger, M., Skalweit, A., and Thiele, B.J. (2000). Expression of the gene and processed pseudogenes encoding the human and rabbit translationally controlled tumour protein (TCTP). Eur. J. Biochem. 267 5473–5481. [DOI] [PubMed] [Google Scholar]
- Tsukaya, H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57 477–496. [DOI] [PubMed] [Google Scholar]
- Turck, F., Zilbermann, F., Kozma, S.C., Thomas, G., and Nagy, F. (2004). Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol. 134 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuynder, M., Susini, L., Prieur, S., Besse, S., Fiucci, G., Amson, R., and Telerman, A. (2002). Biological models and genes of tumor reversion: Cellular reprogramming through tpt1/TCTP and SIAH-1. Proc. Natl. Acad. Sci. USA 99 14976–14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D., Klein, T.M., Fromm, M.E., and McCormick, S. (1989). Transient expression of chimeric genes delivered into pollen by microprojectile bombardment. Plant Physiol. 91 1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., and Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veena, Jiang, H., Doerge, R.W., and Gelvin, S.B. (2003). Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 35 219–236. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A.C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, D., et al. (2007). Proteomic analysis reveals differences between Vitis vinifera L. cv. Chardonnay and cv. Cabernet Sauvignon and their responses to water deficit and salinity. J. Exp. Bot. 58 1873–1892. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Wilkinson, J.E., Twell, D., and Lindsey, K. (1997). Activities of CaMV 35S and nos promoters in pollen: Implications for field release of transgenic plants. J. Exp. Bot. 48 265–275. [Google Scholar]
- Woo, H.H., and Hawes, M.C. (1997). Cloning of genes whose expression is correlated with mitosis and localized in dividing cells in root caps of Pisum sativum L. Plant Mol. Biol. 35 1045–1051. [DOI] [PubMed] [Google Scholar]
- Wullschleger, S., Loewith, R., and Hall, M.N. (2006). TOR signaling in growth and metabolism. Cell 124 471–484. [DOI] [PubMed] [Google Scholar]
- Xin, Z., Velten, J.P., Oliver, M.J., and Burke, J.J. (2003). High-throughput DNA extraction method suitable for PCR. Biotechniques 34 820–824, 826. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Yang, F., Xiong, Z., Yan, Y., Wang, X., Nishino, M., Mirkovic, D., Nguyen, J., Wang, H., and Yang, X.F. (2005). An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene 24 4778–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang-Yen, H.F. (2006). Mcl-1: A highly regulated cell death and survival controller. J. Biomed. Sci. 13 201–204. [DOI] [PubMed] [Google Scholar]
- Yarm, F.R. (2002). Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell. Biol. 22 6209–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, T., Jung, J., Kim, M., Lee, K.M., Choi, E.C., and Lee, K. (2000). Identification of the self-interaction of rat TCTP/IgE-dependent histamine-releasing factor using yeast two-hybrid system. Arch. Biochem. Biophys. 384 379–382. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Christensen, S.K., Fankhauser, C., Cashman, J.R., Cohen, J.D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Hull, A.K., Gupta, N.R., Goss, K.A., Alonso, J., Ecker, J.R., Normanly, J., Chory, J., and Celenza, J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, P.W., and Hitchcock, A.E. (1942). Substituted phenoxy and benzoic acid growth substances and the relation of structure to physiological activity. Contrib. Boyce Thompson Instit. 12 491–496. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.