Abstract
Pollen tubes must navigate through different female tissues to deliver sperm to the embryo sac for fertilization. Protein disulfide isomerases play important roles in the maturation of secreted or plasma membrane proteins. Here, we show that certain T-DNA insertions in Arabidopsis thaliana PDIL2-1, a protein disulfide isomerase (PDI), have reduced seed set, due to delays in embryo sac maturation. Reciprocal crosses indicate that these mutations acted sporophytically, and aniline blue staining and scanning electron microscopy showed that funicular and micropylar pollen tube guidance were disrupted. A PDIL2-1-yellow fluorescent protein fusion was mainly localized in the endoplasmic reticulum and was expressed in all tissues examined. In ovules, expression in integument tissues was much higher in the micropylar region in later developmental stages, but there was no expression in embryo sacs. We show that reduced seed set occurred when another copy of full-length PDIL2-1 or when enzymatically active truncated versions were expressed, but not when an enzymatically inactive version was expressed, indicating that these T-DNA insertion lines are gain-of-function mutants. Our results suggest that these truncated versions of PDIL2-1 function in sporophytic tissues to affect ovule structure and impede embryo sac development, thereby disrupting pollen tube guidance.
INTRODUCTION
In flowering plants, fertilization requires guidance of pollen tubes to deliver the sperm cells to the female gametophyte (Johnson and Preuss, 2002; Weterings and Russell, 2004). Pollen tube guidance can be divided into two phases, sporophytic and gametophytic (Ray et al., 1997; Higashiyama et al., 2003). In the sporophytic phase, when the tube grows in the transmitting tissues and on the ovary wall, tube growth is governed by extracellular matrix components and arabinogalactan proteins (Johnson and Preuss, 2002; Sanchez et al., 2004) and a signal from the female gametophyte may not be required (Hulskamp et al., 1995). The gametophytic phase, after the tube appears on the placental surface in the ovary, can be further divided into funicular guidance (Shimizu and Okada, 2000) and micropylar guidance (Higashiyama et al., 1998, 2001). Sporophytic tissues play some role in funicular guidance (Baker et al., 1997; Palanivelu et al., 2003), while the synergid cells (Higashiyama et al., 2001; Punwani et al., 2007) and central cell (Chen et al., 2007) are required for micropylar guidance.
The female gametophyte is embedded in sporophytic tissues and its development depends on them (Yang and Sundaresan, 2000; Yadegari and Drews, 2004). Missing or abnormal integuments result in meiotic arrest, which is supported by observations on the aintegumenta and bel1 mutants (Reiser et al., 1995; Klucher et al., 1996). However, in the sterile apetala mutant, megasporogenesis is arrested, although the development of the integuments are normal (Byzova et al., 1999). How sporophytic tissues influence female gametophyte development is still elusive (Yang and Sundaresan, 2000; Yadegari and Drews, 2004).
Protein disulfide isomerases (PDIs) catalyze disulfide bond formation, reduction, or isomerization, all of which are important for the maturation and proper folding of secreted or plasma membrane proteins (Wilkinson and Gilbert, 2004; Gruber et al., 2006). Typically, PDIs consist of five domains (a, b, b′, a′, and c). Domains a and a′ are similar to thioredoxin, and each contains an active site (Wilkinson and Gilbert, 2004). The noncatalytic domains b and b′ are similar in sequence to each other but not to thioredoxin, and domain c is acidic. The b′ domain contains the high affinity substrate binding site, but the a and a′ domains might contain low affinity binding sites (Darby et al., 1998; Klappa et al., 2000; Pirneskoski et al., 2004). Most PDIs also have a KDEL sequence at the C terminus, which serves as an endoplasmic reticulum (ER) retention signal.
Genomic database searches and phylogenetic analysis showed that there are 22 PDI-like (PDIL) genes in Arabidopsis thaliana, resolved into 10 groups (Houston et al., 2005). Group IV proteins have no KDEL sequence, two active thioredoxin domains, and a long C terminus called the D domain. In Dictyostelium, one D domain–containing PDI (PDI-D) without a KDEL sequence was nonetheless localized in the ER (Monnat et al., 1997), and it was suggested that this localization might be mediated by the C terminus of the D domain (Monnat et al., 2000). Expression of the Arabidopsis group IV genes was induced by ER stress, suggesting that group IV proteins might function in quality control of proteins in the ER (Martinez and Chrispeels, 2003; Kamauchi et al., 2005).
Here, we report that gain-of-function mutations in PDIL2-1 cause a delay in embryo sac maturation and thereby disrupt pollen tube guidance in Arabidopsis. PDIL2-1 is a functional PDI that is localized in the ER and is highly expressed in the micropylar region of the ovule. Our results suggest that truncated versions of PDIL2-1 that are enzymatically active act in sporophytic tissues to disrupt complete development of the female gametophyte and ovule structures, thereby disturbing pollen tube guidance.
RESULTS
Certain T-DNA Insertions in PDIL2-1 Cause Reduced Seed Set
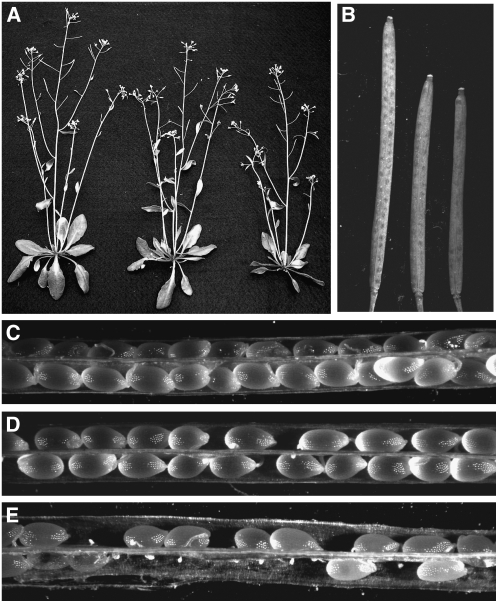
An extracellular PDI was shown to be important for gamete fusion in mammals (Ellerman et al., 2006). Analyses of EST databases obtained from egg, sperm, and embryo sac EST libraries in maize (Zea mays) (Engel et al., 2003; Yang et al., 2006) indicated that several PDI-like genes were represented in these libraries. To determine if Arabidopsis homologs of these maize PDIL proteins played any role in plant reproduction, we analyzed several Arabidopsis T-DNA insertion mutants. There were no reproductive phenotypes for the PDIL1-1, PDIL2-3, or PDIL5-3 insertion lines. However, plants with certain insertions in PDIL2-1 showed smaller siliques and reduced seed set in both heterozygous and homozygous plants (Figure 1B, 1D, and 1E), indicating the dominant effect of these insertions. Vegetative growth was normal except for the shorter stature (Figure 1A).
Figure 1.
Vegetative and Reproductive Development of Wild-Type and Line 5 Plants.
(A) Thirty-five-day-old homozygous plants (right) and heterozygous plants (middle) show a slight reduction in growth relative to wild-type plants (left).
(B) Mature siliques from the homozygous (right) and heterozygous mutant (middle) are shorter than wild-type siliques (left).
(C) Dissected mature wild-type siliques with full seed set.
(D) Siliques from the heterozygous plants with reduced seed set.
(E) Siliques from homozygous plants have severely reduced seed set and obvious undeveloped ovules. Similar results were seen with line 4.
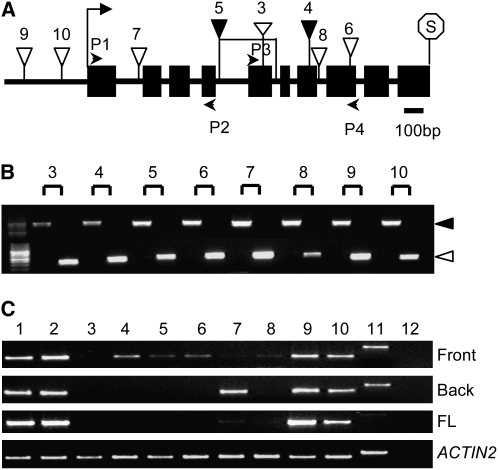
We obtained eight T-DNA insertion mutants (Figure 2A) in PDIL2-1 (Samson et al., 2002; Alonso et al., 2003). For all of these, we could identify homozygous lines (Figure 2B), but only FLAG_427G04 (line 4) and SALK_046705 (line 5) showed reduced seed set (Figure 1). To understand why this might be, we performed detailed expression and phenotypic analyses on these mutants. As shown in Figures 2A and 2C, the T-DNA insertion lines SALK_097094 and CS859015 (lines 9 and 10), located in the promoter region, had no or only a minor effect on gene expression. By contrast, in SALK_135268c and SALK_148421 (lines 7 and 8), inserted in the first or 7th introns, respectively, expression levels were significantly reduced. For three other insertion lines (i.e., FLAG_427G04, SALK_046705, and SALK_082179; lines 4, 5, and 6), we could only detect truncated transcripts. Although line 5 was predicted to be inserted in the 4th intron according to The Arabidopsis Information Resource (http://www.Arabidopsis.org), our thermal asymmetric interlaced PCR and sequencing analyses indicated that there was another insertion in the 5th intron, with an opposite orientation. There was also a 344-bp deletion between these two insertions. The FLAG_015D01 (line 3) appears to be a null, since no transcript was detected with any of the primer pairs tested.
Figure 2.
Gene Structure and T-DNA Insertion Analyses of PDIL2-1.
(A) PDIL2-1 gene structure and T-DNA insertion sites. P1, P2, P3, and P4 indicate position of primers used to check expression. Start and stop codons are noted with an arrow and octagon, respectively. Black boxes and black lines represent exons and introns, respectively.
(B) PCR identification of homozygotes in the indicated T-DNA insertion lines. For each line, the left lane is a wild-type plant having only a gene-specific band (black arrow), and the right lane is an insertion line, having only a T-DNA–specific band (white arrow).
(C) RT-PCR analyses. PDIL2-1 expression was examined using primers covering different parts of the gene (primers P1 and P2, front; or primers P3 and P4, back) or full-length (FL) cDNA. ACTIN2 expression was used as a control.
The numbers shown in (A) to (C) represent (1) Ws4 wild type, (2) Col-0 wild type, (3) FLAG_015D01, (4) FLAG_427G04, (5) SALK_046705, (6) SALK_082179, (7) SALK_135268c, (8) SALK_148421, (9) SALK_097094, (10) CS859015, (11) gDNA control, and (12) water control.
Both heterozygous and homozygous plants of line 4 and line 5 had reduced seed set (Figure 1; data not shown), somewhat more so in the homozygotes. As shown in Figure 2C, the expression level of both these insertion lines was lower than in the wild type, suggesting that the phenotype was due to the truncated protein, rather than to overexpression. The siliques of these two lines were also significantly smaller than those of the wild type (Figure 1B; data not shown). Surprisingly, the null mutant line 3, as well as line 6, which has truncated transcripts, showed normal seed set. These results suggested that the truncated products of line 4 and line 5 had dominant effects but that they were not completely penetrant, as some seed was set. We also crossed line 3 with insertion lines in PDIL2-3 (CS856199) and PDIL1-1 (SALK_015253C). However, neither double mutant showed fertilization-related phenotypes, which might reflect the highly redundant functions of the 22 PDI-like genes in Arabidopsis genome.
Since these two insertion lines showed seed set phenotypes and both sides of the T-DNA insertion in line 5 were recovered, we chose line 5 for further analyses.
Mutant Plants Are Defective in Pollen Tube Guidance Due to Sporophytic Effects
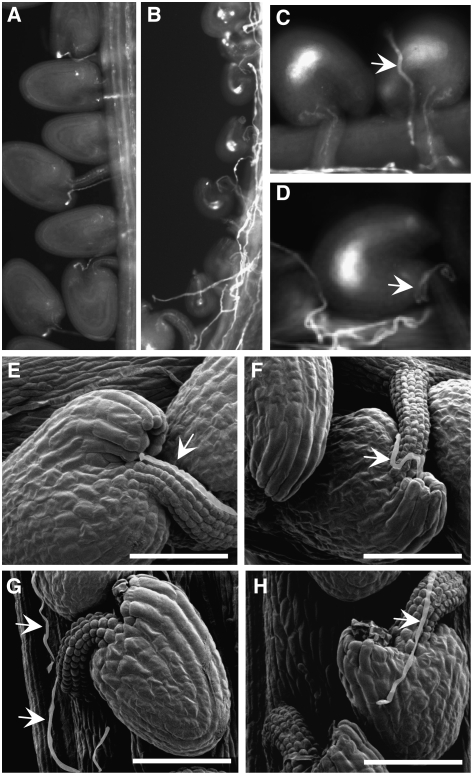
To determine what caused the reduced seed set phenotype, we checked pollen tube growth in pistils of the line 5 homozygous mutant (Figure 3). Approximately 30% of the ovules in the homozygote attracted pollen tubes to the micropyle (Figure 3B), but ∼70% did not, although many pollen tubes were seen growing on the placenta and funicular tissue (Figures 3B and 3C). The pollen tubes appeared to have lost their way after they grew out of the transmitting tract. The undeveloped ovules either received no pollen tubes (58.6%) (Figures 3B and 3C, left) or had pollen tubes randomly growing onto the ovule surface and on the funiculus (10.2%) (Figures 3C, right, and 3D). These observations indicated that funicular and microplylar guidance to undeveloped ovules was disrupted in this line. To further confirm the observed pollen tube guidance defects, we used scanning electron microscopy. Many ovules had no pollen tubes in the vicinity (Figure 3G), or pollen tubes wound around the surface of undeveloped ovules (Figure 3H), indicating funicular and micropylar defects in pollen tube guidance.
Figure 3.
Line 5 Homozygous Plants Show Disrupted Pollen Tube Guidance.
(A) to (D) Decolorized aniline blue staining of 3 d after pollination (DAP) pistils.
(A) Wild-type siliques showing fertilized ovules and pollen tube growth.
(B) Mutant siliques showing randomly growing pollen tubes and undeveloped ovules.
(C) A representative mutant ovule with pollen tube (arrow) growing on the ovule surface.
(D) An ovule with pollen tube (arrow) growing around the funiculus without targeting the micropyle.
(E) and (F) Scanning electron micrographs of 2-DAP pistils. Two ovules from a wild-type plant show normal pollen tube (arrow) growth and correct targeting to the micropyle.
(G) A mutant ovule with a pollen tube (arrow) not targeted to the funiculus or micropyle.
(H) A mutant ovule with a pollen tube (arrow) growing on the surface. Bars = 50 μm in (E) to (H).
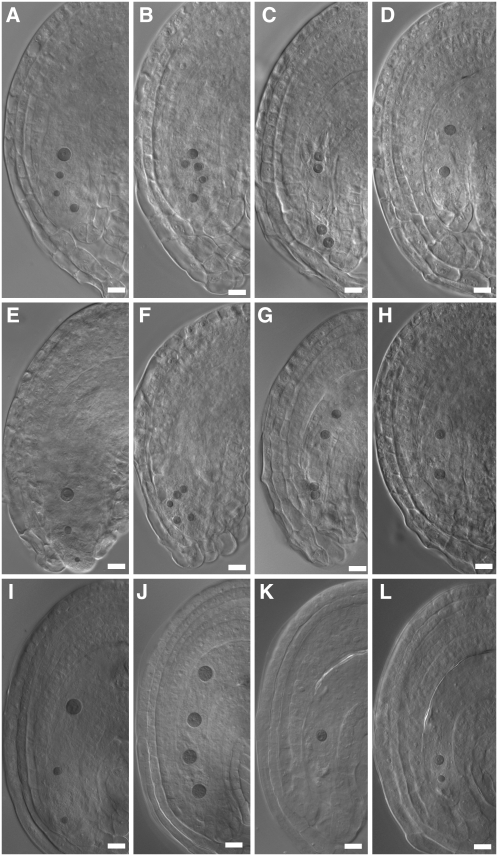
To determine if the reduced seed set was caused by defects of gametophyte origin, we performed reciprocal crosses with wild-type plants using plants that were heterozygous for the T-DNA insertion. The kanamycin resistance marker was silenced in line 5, so we used PCR to determine the genotype of the progeny. Despite the reduced seed set, the transmission of the T-DNA insertion was normal through both parents (Table 1), suggesting that the reduced seed set phenotype was caused by sporophytic effects. Although we had not noticed any abnormalities in ovule structure (Figure 3), we decided to check ovule and embryo sac development in both heterozygous and homozygous plants. In heterozygous plants, ovule development was normal (Figures 4A to 4D), but ovules of line 5 homozygotes often showed a protruding embryo sac phenotype, presumably caused by shorter integuments (Figure 4E). Furthermore, although many line 5 embryo sacs were normal (Figures 4A and 4E), others were delayed at varying stages of maturation (two, four, or eight nuclei), in both heterozygous and homozygous plants (Figures 4B to 4D and 4F to 4H; see Supplemental Table 1 online). Embryo sacs of line 4 homozygotes showed similar phenotypes and thus are not shown.
Table 1.
Segregation Analysis of the Line 5 (SALK_046705) Insertion Mutant
| Parental Genotype | Progeny Genotype | |||
|---|---|---|---|---|
| Male | Female | PDIL2-1/PDIL2-1 | PDIL2-1/pdil2-1 | pdil2-1/pdil2-1 |
| PDIL2-1/pdil2-1 | PDIL2-1/pdil2-1 | 36 | 63 | 31 |
| PDIL2-1/PDIL2-1 | PDIL2-1/pdil2-1 | 47 | 46 | – |
| PDIL2-1/pdil2-1 | PDIL2-1/PDIL2-1 | 83 | 99 | – |
Figure 4.
Embryo Sac Development in Mutant Line 5 and PDIL2-1 Transgenic Plants.
Heterozygous ([A] to [D]) and homozygous ([E] to [H]) ovules of line 5. Normal embryo sac ([A] and [E]), embryo sac with eight nuclei ([B] and [F]), no cellularization, four nuclei ([C] and [G]), and two nuclei ([D] and [H]).
(I) to (L) Representative embryo sac of CDS, truncation 1, and truncation 2 transgenic lines (see Figure 5A), with normal embryo sac (I), four nuclei (J), one nuclei (K), and two nuclei (L), respectively.
Bars = 20 μm.
We also performed reciprocal crosses between the homozygous mutants and wild-type plants. When the mutants were used as female, the number of undeveloped ovules was similar to those in a self cross (Table 2). Similar results were obtained when heterozygous plants were used for the crosses (Table 2), suggesting that the reduced seed set phenotype was of maternal sporophytic origin but affected female gametophyte development.
Table 2.
Seed Set Analyses Following Reciprocal and Self Crosses
| Male | Female | Siliques Counted | Developed Seeds | Undeveloped Ovules | Percentage of Undeveloped Ovules |
|---|---|---|---|---|---|
| Col-0 | Col-0 | 12 | 514 | 25 | 4.6 |
| Line 5 −/− | Col-0 | 8 | 432 | 19 | 4.2 |
| Line 5 −/− | Line 5 −/− | 6 | 58 | 196 | 77.1 |
| Col-0 | Line 5 −/− | 10 | 78 | 223 | 74.1 |
| Line 5 +/− | Col-0 | 5 | 181 | 44 | 19.6 |
| Col-0 | Line 5 +/− | 5 | 78 | 110 | 58.1 |
| Line 4 −/− self | 5 | 102 | 151 | 59.7 | |
| Line 4 +/− self | 10 | 120 | 137 | 53.3 |
Expression of Truncated PDIL2-1 Phenocopies the Reduced Seed Set Phenotype
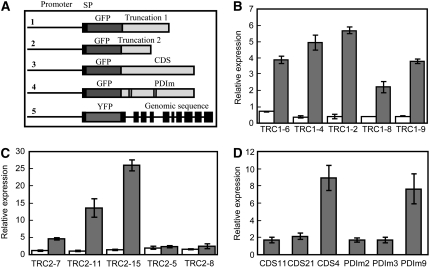
To test the prediction that the reduced seed set phenotypes were caused by the expression of truncated transcripts, we transformed wild-type plants with constructs in which the gene products would correspond to those in the line 4 or line 5 insertion lines, named truncation1 (TRC1) and truncation2 (TRC2), respectively. Figure 5A shows the constructs used for this purpose (#1 and #2) as well as three other constructs used in subsequent experiments. For each construct, we used the endogenous promoter (1.66 kb) and inserted either green fluorescent protein (GFP) or yellow fluorescent protein (YFP) between the signal peptide (SP) and the coding sequence.
Figure 5.
Construct Structures and Real-Time Quantitative PCR Analyses of Transgenic Lines.
(A) Schematic structure of five transformation constructs. SP denotes signal peptide. Truncation 1 and Truncation 2 (TRC1 and TRC2) correspond to insertion sites of line 4 and line 5. CDS denotes the coding sequence of PDIL2-1. PDIm denotes the coding sequence of PDIL2-1 with both active sites mutated.
(B) to (D) Real-time quantitative PCR analyses of the transgenic lines. Endogenous and total PDIL2-1 expression (endogenous and from the transgene) were examined in truncation 1 (B), truncation 2 (C), or CDS and PDIm (D) transgenic plants. White bars in (B) and (C) represent endogenous expression, and gray bars represent total expression. Error bars represent ±sd of four technical replicates.
The resulting T1 transgenic plants were selected by BASTA, and resistant plants were transferred to soil. Siliques were dissected to analyze the seed set. For each truncation construct, some plants had reduced seed set (Table 3). To confirm expression of the transgene, real-time quantitative PCR experiments were used to compare plants showing the phenotype with those that did not. For each construct, plants showing the phenotype (TRC1-2, TRC1-4, TRC1-6, TRC2-7, TRC2-11, and TRC2-15) also had higher levels of the truncated transcripts (Figures 5B and 5C). The endogenous PDIL2-1 expression level was only slightly altered in the truncation 1 and truncation 2 transformants, whether or not the plant showed the reduced seed set phenotype (Figures 5B and 5C). These results indicate that the phenotypes were caused by the transgene, rather than by a cosuppression effect.
Table 3.
Characterization of the PDIL2-1 Transgenic Plants
| Construct | Plants with a Reduced Seed Set Phenotype | Total BASTA-Resistant Plants | Undeveloped Ovules* | Developed Seeds* | Percentage of Undeveloped Ovules |
|---|---|---|---|---|---|
| Truncation 1 | 6 | 18 | 524 | 797 | 39.7 |
| Truncation 2 | 9 | 35 | 1108 | 779 | 58.7 |
| CDS | 6 | 15 | 570 | 597 | 48.8 |
| CDSM | 0 | 20 | – | – | – |
Truncation 1 corresponds to Flag_427G04, which would encode the first 306 amino acids of PDIL2-1, and Truncation 2 corresponds to SALK_046705, which would encode the first 130 amino acids. CDS refers to the coding sequence and CDSM to the mutated coding sequence. Asterisks represent data only from plants with the phenotype. For phenotypic analysis, five siliques were scored for each plant.
We also checked the pollen tube growth in siliques of the transgenic plants and found that undeveloped ovules failed to attract pollen tubes (see Supplemental Figure 1 online). In these transgenic lines and similar to the insertion lines, we found normal embryo sacs (Figure 4I) and arrested embryo sacs (Figures 4J to 4L). Most of the arrested embryo sacs arrested early (one or two nuclei) (Figures 4J to 4L; see Supplemental Table 1 online). The phenotypes were stronger than in lines 4 and 5 and correlated with the reduced seed set (see Supplemental Table 1 online).
Expression Pattern and Subcellular Localization of PDIL2-1 in Arabidopsis
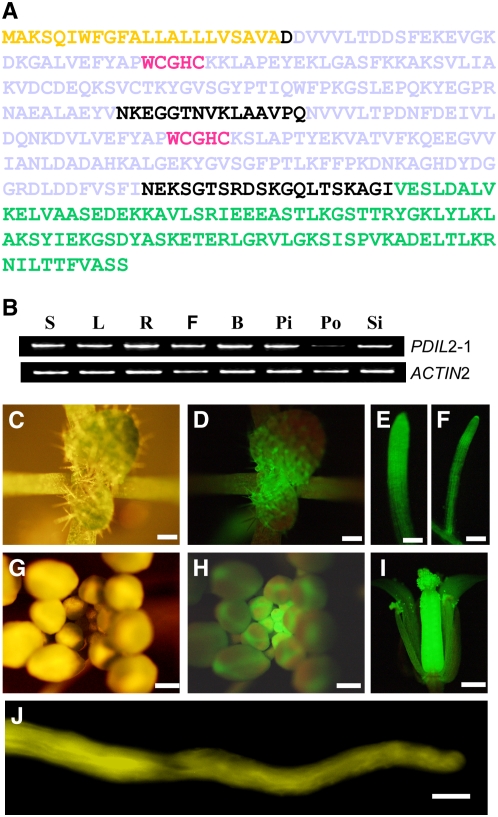
PDIL2-1 has two typical active thioredoxin (CGHC) domains, usually named a and a′, but it lacks the inactive thioredoxin domains b and b′ as well as the acidic c domain and the ER retention signal, KDEL. Because it has a D domain at its C terminus, PDIL2-1 belongs to the PDI-D family (Monnat et al., 1997). There is also a predicted signal peptide at the N terminus. The protein structure of PDIL2-1 is shown in Figure 6A.
Figure 6.
Protein Structure and Expression Pattern of PDIL2-1.
(A) Protein sequence of PDIL2-1. Yellow denotes the signal peptide (SP), gray denotes the two thioredoxin domains (TRX), red denotes the active sites, and green denotes the D domain.
(B) Expression of PDIL2-1 in different organs was examined by RT-PCR using gene-specific and control (ACT2) primers. S, seedlings; L, leaves; R, roots; F, opened flowers; B, vlosed flower buds; Pi, unfertilized pistils; Po, pollen; Si, siliques.
(C) to (J) Expression pattern determined by YFP fusion reporter (ProPDIL2-1:SP-eYFP-PDIL2-1).
(C) and (G) Vegetative and reproductive apical region.
(D) and (H) The same region under UV light (GFP filter).
(E), (F), and (I) Expression of PDIL2-1 in lateral and main roots and in a flower, respectively.
(J) In vitro–germinated pollen tube.
Bars = 500 μm in (C), (D), and (G) to (I), 100 μm in (E) and (F), and 10 μm in (J).
To determine the expression pattern of PDIL2-1, we performed RT-PCR analysis using cDNA samples extracted from different Arabidopsis organs. PDIL2-1 was expressed in all the tissues examined, with a lower expression level in pollen relative to other tissues (Figure 6B).
To determine expression at the protein level, we used a YFP fusion construct (ProPDIL2-1:SP-YFP-PDIL2-1) (Figure 5A, construct #5). At least 100 ovules from six independent T1 transgenic plants were checked and all showed similar YFP expression patterns. As shown in Figures 6C to 6J, ProPDIL2-1:SP-YFP-PDIL2-1 was highly expressed in vegetative and reproductive meristems, in the main and lateral roots, and in flowers. Although the RT-PCR results showed lower expression in pollen (Figure 6B), the YFP fusion protein was easily detected in pollen tubes germinated in vitro (Figure 6J).
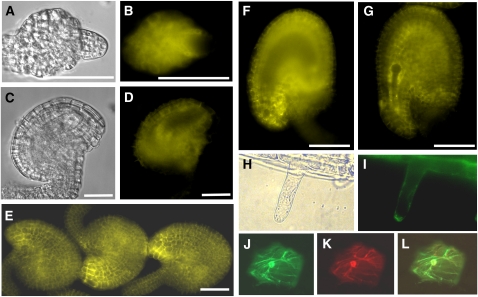
To obtain detailed expression information for flowers, where pollen tube guidance takes place, we checked the YFP fusion pattern in ovules of different developmental stages. In early stage 12 flowers, the funiculus curves backward and the inner and outer integuments do not cover the nucellus. The YFP signal was observed in both inner and outer integuments and also in nucellar cells (Figures 7A and 7B). In late stage 12 and in stage 13 flowers, YFP was also expressed in the integument cells but was not observed in the embryo sac (Figures 7C to 7F). Expression in the mature ovule was striking, with enhanced fluorescence in the micropylar region, as shown in Figures 7E and 7F. After fertilization, YFP was observed in the seed integuments but not in the embryo (Figure 7G).
Figure 7.
PDIL2-1 Expression in Ovules and Subcellular Localization.
(A) and (B) Ovule of early stage 12 flowers under differential interference contrast (DIC) (A) or YFP channel (B). YFP signal is visible in integument cells and nucellus cells.
(C) and (D) Ovule of late stage 12 flowers under DIC (C) or YFP channel (D). YFP is visible in integument cells.
(E) Mature ovules of stage 13 flowers. YFP signal is visible in integument cells and is highly accumulated at the micropyle.
(F) An ovule of stage 13 flowers. Focus is on the plane of the embryo sac to show no signal inside of the embryo sac.
(G) A fertilized ovule with no YFP signal in the developing embryo.
(H) Root hair under transmitted light.
(I) ProPDIL2-1:SP-YFP-PDIL2-1 expression in root hairs.
(J) GFP-KDEL, an ER marker.
(K) PDIL2-1-tdTomato red fusion protein.
(L) Overlay of (J) and (K).
Bars = 20 μm in (A) to (D) and 50 μm in (E) to (G).
To determine the subcellular localization of PDIL2-1, we used two strategies. First, we checked localization in plants expressing ProPDIL2-1:SP-YFP-PDIL2-1. In root hair cells, YFP was observed around the nuclei and in some organelles, consistent with ER localization (Figures 7H and 7I). The YFP signal was also localized in the ER in leaf and hypocotyl cells (see Supplemental Figures 2A and 2B online). Second, we made a construct with a red fluorescent protein fused to the C terminus of PDIL2-1, driven by the cauliflower mosaic virus (CaMV) 35S promoter. This construct was cobombarded into onion epidermal cells with a GFP-KDEL construct to mark the ER. As shown in Figures 7J to 7L, PDIL2-1 colocalized with the ER marker. A construct that lacked the D domain was also ER localized (see Supplemental Figures 2C to 2E online).
PDIL2-1 Can Rescue a PDI Null Mutant of Yeast
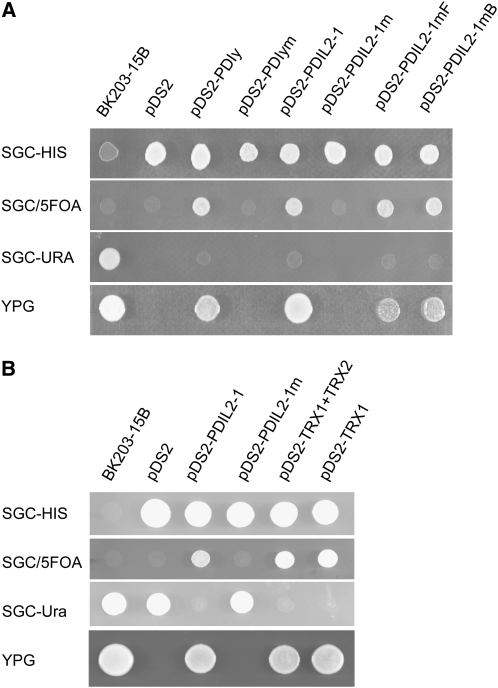
To determine whether PDIL2-1 is a functional enzyme, we performed a yeast complementation experiment. The TRG1/PDI1 gene of yeast encodes an ER-resident protein that is structurally related to PDI and essential for growth. The complementation experiments were performed in BK203-15B cells carrying a deletion of the TRG1 gene, which can be rescued by the pWBK-PDI centromeric plasmid (Kramer et al., 1989). This centromeric vector carries a URA3 gene, which allows positive selection for growth in the absence of uracil and negative selection in the presence of 5-fluoroorotic acid (5-FOA). As shown in Figure 8A, the PDIL2-1 construct complemented the yeast mutant and allowed growth on YPG plates in the absence of pWBK-PDI. Furthermore, constructs in which mutations in either of the two thioredoxin active sites (a and a′) were introduced only inefficiently complemented the yeast mutant. The inactive mutant PDIL2-1m, which has both active sites mutated, was unable to complement under these conditions. None of the colonies picked from the 5-FOA plate grew on plates lacking uracil, demonstrating that growth on YPG plates was not due to the presence of the centromeric plasmid pWBK-PDI (Figure 8A). We therefore conclude that PDIL2-1 has PDI activity.
Figure 8.
PDIL2-1 Has PDI Activity.
(A) Yeast complementation analysis showing that PDIL2-1 is a functional isomerase.
(B) Truncated PDIL2-1 also showed enzyme activity in yeast.
In both panels, the left column lists the medium used for growth. The first column is yeast strain BK203-15B, and the second column is empty pDS2 vector. Transformants with different constructs are marked at the top. The wild-type yeast PDIy and mutated PDIym (with both CGHC boxes mutated to SGHS) were used as positive and negative controls, respectively. PDIL2-1 and PDIL2-1m were constructed similarly to the yeast PDI constructs. PDIL2-1mF and PDIL2-1mB are different mutations of PDIL2-1, with either the first or second CGHC boxes mutated. TRX1 and TRX2 represent the two TRX domains. Note that only constructs with a functional PDI can grow on YPG plates.
Some Truncated PDIL2-1 Versions Have Isomerase Activity
Plants that were heterozygous or homozygous for the T-DNA insertions (line 4 and line 5) and transformants carrying truncated PDIL2-1 constructs (constructs #1 and #2) all showed reduced seed set (Table 3), suggesting that the truncated protein in these lines had a dominant effect. To determine whether a dominant-negative or a gain-of-function mutation could explain the phenotype, we analyzed the isomerase activity of the truncated PDIL2-1 proteins. As shown in Figure 8B, constructs in which the D domain was truncated (TRX1+TRX2) or in which only the first thioredoxin domain was present (TRX1) both complemented the yeast PDI mutation. These results strongly support the gain-of-function explanation.
If the truncated protein of PDIL2-1 caused a gain-of-function effect, overexpression of PDIL2-1 might have the same effect. To test this possibility, we transformed wild-type plants with the PDIL2-1 cDNA driven by its endogenous promoter and separately with a construct in which both active sites were mutated, also driven by its endogenous promoter (Figure 5A, #3 and #4). Of the 15 plants transformed with the wild-type version of the cDNA, six showed reduced seed set, while all 20 transformants with the mutated version showed normal seed set (Table 3). We also observed some developmentally delayed embryo sacs in the transgenic plants with the wild-type version (see Supplemental Table 1 online), supporting the idea that it was the action of PDIL2-1 in the sporophyte that affected embryo sac development. Expression of the transgene was tested by real-time quantitative PCR. As shown in Figure 5D, transformants with different expression levels were obtained for both constructs, but all expressed PDIL2-1 to levels higher than those found in wild-type plants. These results support the idea that the T-DNA insertion lines 4 and 5 are dominant gain-of-function mutants.
DISCUSSION
PDIL2-1 Acts in Maternal Sporophytic Tissues to Affect Embryo Sac Development
Two T-DNA insertion lines in PDIL2-1 (i.e., line 4 and line 5) showed reduced seed set in both heterozygous and homozygous plants. This phenotype was more severe in homozygous plants, which may indicate that the truncated protein interferes or competes for substrate with the normal protein that is also present in the heterozygotes. The null mutant line 3 had full seed set, indicating the dominant nature of the truncated gene products of PDIL2-1. Pollen tube staining and scanning electron microscopy observations showed disrupted pollen tube guidance, and this phenotype was likely caused by the delay in embryo sac maturation.
From reciprocal crosses we found that the reduced seed set phenotype was associated with a maternal sporophytic defect that caused a delay in embryo sac development. However, because both heterozygous and homozygous plants had ovules and embryo sacs that developed normally, the dominant effect cannot be considered fully penetrant. Protein fusion reporter results also support that PDIL2-1 affects sporophytic tissues, since in ovules, PDIL2-1-YFP was expressed only in sporophytic tissues, but not in the embryo sac.
The female gametophyte has been proposed as the source of a guidance signal for the pollen tube, and this is supported by genetic analysis and in vitro laser ablation experiments (Hulskamp et al., 1995; Ray et al., 1997; Higashiyama et al., 2003; Chen et al., 2007). The female gametophyte has no or a trivial effect on the development of surrounding sporophytic tissues, since ovules without embryo sacs had normal sporophyte structures (Bhatt et al., 1999; Yang et al., 1999). However, the sporophyte tissues are crucial for embryo sac development, although how is not completely understood (Yang and Sundaresan, 2000; Yadegari and Drews, 2004). Here, we show that expression of two of its truncated versions or overexpression of PDIL2-1 in sporophytic tissues of the ovule affects embryo sac development.
In addition to the retarded embryo sac development, we also observed some (39.1%) embryo sacs protruding out of ovules in the homozygotes (Figure 4D). This phenotype was also seen in the truncation and CDS overexpression transgenic lines, although in a lesser proportion. A similar phenotype was observed in the mpk3 mpk6 double mutant (Wang et al., 2008), suggesting that PDIL2-1 might be involved in or affect expression of signaling components in this genetic pathway.
It was reported that two Ds transposon insertions in PDIL2-1 (UNE5 and MEE30) caused fertilization-related phenotypes (Pagnussat et al., 2005). However, we found that the PDIL2-1 expression level in UNE5 was equivalent to that in the wild type (see Supplemental Figure 3 online), making it unclear whether the UNE5 phenotype was due to a disruption in PDIL2-1. For UNE5, both sequences flanking the Ds insertion were recovered. The flanking sequences of the left and right borders of the T-DNA in MEE30 are located in different chromosomes (http://www-plb.ucdavis.edu/Labs/sundar/GameDev.htm), suggesting that either multiple insertions, a chromosome rearrangement, or a mutation in the vicinity of the Ds insertion caused the phenotypes observed in these lines and leaving it unresolved if these phenotypes are due to a disruption of PDIL2-1.
Function and Subcellular Localization of PDIL2-1
PDIL2-1 can complement the yeast isomerase mutant, but the enzymatically inactive PDIL2-1m cannot, indicating that PDIL2-1 has PDI activity. The truncated protein, corresponding to the protein that would be produced in T-DNA insertion lines (i.e., line 4 and line 5) also has enzyme activity. These results indicate that the phenotype in the mutant plants might be caused by gain-of-function effects rather than by dominant-negative effects. Transformation experiments using the full-length cDNA driven by the endogenous promoter (Figure 5A, #3) yielded the same phenotype, which also supports the gain-of-function explanation.
Group IV PDIL proteins are regulated by ER stress. The promoter regions of these genes often contain unfolded protein response regulatory cis-acting elements (Urade, 2007), and PDIL2-1 was implicated in the unfolded protein response (Martinez and Chrispeels, 2003; Kamauchi et al., 2005). There is a motif (GGAATTGGACATGGTTTTAATAA) in the first intron of PDIL2-1 that is very similar to the unfolded protein response element, which may be under the regulation of environmental or developmental cues (Martinez and Chrispeels, 2003). The third intron of PDIL2-1 was also proposed to be integrated in the coding sequence under certain conditions (Ner-Gaon and Fluhr, 2006). Such regulatory mechanisms might explain why the construct used for subcellular localization studies (Figure 5A, #5), which is the genomic coding sequence driven by the endogenous promoter, showed normal seed set, while the cDNA driven by the same promoter (Figure 5A, #3) showed reduced seed set.
There are several possibilities for how truncated versions of PDIL2-1, or expression of the cDNA, caused a gain-of-function phenotype. One is altered localization of the truncated proteins. PDI proteins are localized in the ER (an oxidative environment) and can readily oxidize their substrates to form disulfides (Wilkinson and Gilbert, 2004). When localized in a nonoxidative location, such as extracellularly, PDI proteins may exhibit reducing activities (Turano et al., 2002). PDIL2-1 was mainly localized in the ER, as shown in transient assays and in stable transgenic plants (Figure 7), consistent with the localization of another PDI-D protein (Monnat et al., 1997). Truncated PDIL2-1s (i.e., those in line 4 and line 5) lack the D domain, which in Dictyostelium at least, was shown to function as a saturable signal for ER retention (Monnat et al., 1997, 2000). Truncated PDIL2-1 proteins might escape from the ER and be secreted to the cell surface. In our experiments, similar localizations for the truncated and full-length proteins were found in both transient expression and in stable transgenic plants (Figure 7), although we cannot rule out the possibility that some of these PDIL2-1s escaped the ER and were undetectable by our imaging. A strong argument against mislocalization is that the T-DNA insertion lacking only the D domain (line 6 in Figure 2) showed normal seed set. In addition, expression of an extra copy of the full-length protein, with an intact D domain, also caused reduced seed set.
The second possibility is that truncated PDIL2-1 may have an altered substrate binding capability. Proteins homologous to PDIL2-1 have been shown to help the folding of other proteins, suggesting that they may function as chaperones (Wadahama et al., 2007). Whether PDIL2-1 functions as an isomerase or as a chaperone, substrate binding should be essential for its function. In lines 4 and 5, the insertions caused truncations of the protein, with only the first thioredoxin domain remaining intact. For typical PDI proteins, the noncatalytic b and b′ domains are considered to have high affinity for the substrate, while the active thioredoxin domains a and a′ have weak affinity (Darby et al., 1998; Klappa et al., 2000; Pirneskoski et al., 2004). However, PDI-D family proteins do not have b and b′ domains, so it is unknown how PDI-D proteins define their substrates. It is possible that the truncated versions of PDIL2-1 have altered substrate profiles and then gain new functions. The overexpressed PDIL2-1 protein may also bind to an atypical substrate due to its excess and then cause a gain-of-function phenotype.
A third possibility is that truncations might also influence the stability of the protein. If the two truncations make the protein more stable, it might result in excess protein. In the overexpression transgenic plants with seed set phenotypes, the YFP protein level was higher, which is consistent with the idea that the truncated versions of PDIL2-1 are more stable; this might be the most plausible explanation for the phenotype.
METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis thaliana Columbia-0 (Col-0) and T-DNA insertion lines were obtained from the ABRC (Ohio State University). Seeds were surface-sterilized with 20% sodium hypochlorite solution for 10 min, washed three times with sterile water, and plated onto Petri dishes containing basal MS medium (Murashige and Skoog, 1962) supplemented with Arabvitamins (1 mg/L thiamine, 0.5 mg/L pyridoxine, 0.5 mg/L nicotinic acid, and 0.1 mg/L myo-inositol), 0.5 g/L MES (Sigma-Aldrich), and 1% (w/v) sucrose (Fisher Scientific), adjusted to pH 5.8 and solidified with 0.8% agar. Seeds were stratified for 3 d at 4°C in the dark and then grown in long-day conditions (16 h light/8 h dark at 21°C) for 7 d in a Percival growth chamber. After transfer to soil, plants were grown in a greenhouse, with 16 h of daylight, supplemented with lights at an intensity of 75 to 100 μmol m−2 s−1, day/night temperatures of 21°C/18°C, and relative humidity of ∼50%. The insertions in the T-DNA lines were confirmed by PCR. Primers used for PCR are listed in Supplemental Table 2 online.
Yeast Complementation
The yeast strain BK203-15B (Mata, ura3, leu2, his3, trp1, pdi∷leu2), pDS2 plasmid, and positive and negative controls were kindly provided by David M. Ferrari. Strain BK203-15B, which carries a deletion of the TRG1 gene, is rescued by a centromeric plasmid (pWBK-PDI) that contains a copy of the wild-type TRG1 gene and a URA3 selection marker. The yeast vector pDS2 contains the HIS3 selection marker and has the yeast galactose-inducible GAL1 promoter for the introduced genes. The wild-type PDIL2-1 and a mutated version (with both CGHC boxes mutated to SGHS) were cloned using PCR and inserted into the BamHI and SalI sites of pDS2. Primers used for cloning the PDIL2-1 gene were PDIL2-1 BamHI forward 5′-TAGGATCCATGGCGAAATCTCAGATCTGGTTTG-3′ and PDIL2-1 SalI reverse 5′-TATAGTCGACTTAAGAAGAAGCAACGAACGTGGT-3. Primers used for mutating the first TRX motif were MCC1F 5′-ACGCTCCCTGGAGTGGTCACAGCAAGAAACT-3′ and MCC1R 5′-AGTTTCTTGCTGTGACCACTCCAGGGAGCGT-3′. Primers used for mutating the second TRX motif were MCC2F 5′-TGCACCATGGAGTGGCCACAGCAAATCACT-3′ and MCC2R 5′-AGTGATTTGCTGTGGCCACTCCATGGTGCA-3′. The insertions were sequenced, and transformants from each construct were selected on SGC-HIS medium and then transferred to galactose and 5-FOA (SGC/FOA) medium to counterselect for the presence of the pWBK-PDI centromeric plasmid. Colonies that grew on SGC/FOA were transferred to SGC-URA medium to verify the absence of pWBK-PDI and then analyzed on YPG plates for complementation of the TRG1 deletion.
Phenotypic Analysis
To analyze seed set, siliques 8 to 10 d after fertilization were placed on double-sided tape and transversely dissected under a stereoscope, and then undeveloped and normal ovules were counted. Images of siliques were taken with a Nikon SMZ800 stereoscope. For reciprocal crosses with Col-0, flowers were emasculated at night and crosses were performed 18 to 20 h later. Pollen tube growth in pistils was analyzed using aniline blue staining in pistils 1 to 2 DAP as described (Mori et al., 2006). For scanning electron microscopy, pistils 1 to 2 DAP were carefully opened with a sharp needle and then fixed with FAA (50% ethanol, 3.7% formaldehyde, and 5% acetic acid) for 2 h. After a series of dehydration steps using increasing concentrations of ethanol, siliques were stored in absolute ethanol or were directly subjected to critical point drying, and then siliques were mounted for sputter coating with gold palladium for 25 s and observed on a Hitachi S-4700 scanning electron microscope at an accelerating voltage of 5 kV. For ovule clearing, unpollinated pistils, open flowers, and 3-DAP pistils were dissected to expose ovules, fixed in FAA for 20 min, washed in water, and cleared overnight in Hoyer's solution (chloral hydrate:water:glycerol; 3:1:1; v/v/v). Ovules were observed using DIC microscopy.
Subcellular Localization Construct and Onion Bombardment
The ER marker was kindly provided by Robert Blanvillain and Patrick Gallois (unpublished data). In this pKar6 vector, a chitinase signal peptide sequence (5′-ATGGTGAAGACTAATCTTTTTCTCTTTTTCATCTTTTCACTTCTCCTATCATTATCCTCGGCCGAC-3′) was placed in front of the GFP coding sequence, and a sequence encoding an ER retention signal sequence (5′-AAGGACGAGCTG-3′) was inserted just before the stop codon. To construct the PDIL2-1-tdTomato fusion construct used for cobombardment, the cDNA of PDIL2-1 was amplified from pollen cDNA using forward 5′-CACCATGGCGAAATCTCAGATCTGGTTTGGT-3′ and reverse 5′-CGCTTTCGCGGCCGCTTCCGCAGAAGAAGCAACGAACGTGGTTAGGA-3′ primers, using phusion high fidelity DNA polymerase (New England BioLabs) and then blunt-ligated to tdTomato (Shaner et al., 2004), driven by the CaMV 35S promoter in the pUC19 backbone. The reverse primer contains a sequence encoding the linker AEAAAKA. For onion bombardment, gold particles (1 μm in diameter) were coated with equal amounts (5 μg) of each plasmid DNA and bombarded with a BIOLISTIC-PDS-1000/He particle delivery system (Bio-Rad) at a pressure of 1100 p.s.i. Onion epidermal peels were placed on solidified 0.5× MS medium and held at 22°C in the dark for 24 h before observation.
RT-PCR and Real-Time Quantitative PCR Analysis
RNA was extracted from seedlings, leaves, buds, flowers, unfertilized pistils, pollen, and siliques using the RNeasy kit (Qiagen). For expression analysis, 2 μg of total RNA was reverse-transcribed using the SuperScript II system (Invitrogen) according to the manufacturer's instructions, with oligo poly-T primers in a 20-μL reaction. The gene-specific primers were forward 5′-CACCATGGCGAAATCTCAGATCTGGTTTGGT-3′ and reverse 5′-AGAAGAAGCAACGAACGTGGTTAGGATAT-3′. ACTIN2 expression was used as control, and the primers for ACTIN2 were ACT2Fw 5′-GCCATCCAAGCTGTTCTCTCC-3′ and ACT2Re 5′-TTCTCGATGGAAGAGCTGGT-3′. The primers used for detection of PDIL2-1 expression in the T-DNA insertion lines were P1 5′-CACCATGGCGAAATCTCAGATCTGGTTTGGT-3′, P2 5′-TGTTCACGTATTCAGCCAAAGCT-3′, P3 5′-GCACCAACGTAAAATTAGCTGCAG-3′, and P4 5′- CCTCTCCGTTTCTTTGCTAGC-3′. The GFP-specific primer used to check the expression of the transgenic construct was 5′-AGAAGCGCGATCACATGGTC-3′.
Real-time quantitative PCR and calculation of relative expression were performed as described by Alandete-Saez et al. (2008). In brief, 3 μg of total RNA from inflorescences were used for reverse transcription. A MyIQ Real-Time PCR detection system (Bio-Rad) and EvaGreen Dye (Biotium) were used to detect gene expression. The IPP2 gene (At3g02780) was used as an internal control. Each real-time PCR experiment contained four technical replicates, and five independent transgenic plants from each truncation construct were used to check expression levels. The primers used for endogenous PDIL2-1 were EndoF 5′-CAACAAAGCTGGTCACGATTATG-3′ and EndoR 5′-GGTCCCAGATTTCTCGTTGATG-3′; primers used for both the endogenous gene and the transgene were BothF 5′-AAAGGAGCTCTCGTCGAGTTTTAC-3′ and BothR 5′-CCTAGCTTTTCATACTCTGGAGCAA-3′.
Constructs and Plant Transformation
To obtain the endogenous promoter, a 1.66-kb fragment upstream of the ATG plus the sequence encoding the signal peptide was amplified using the wild type Col-0 genomic DNA as template and forward 5′- GAGCTCTGCATCACATCTGCGAATCATCA-3′ and reverse 5′- ACTAGTAGCTACGGCTGAAACCAGAAGCA-3′ primers. The amplified fragment was ligated to the T-easy vector (Clontech). After sequencing, the fragment was cleaved by SacI and SpeI and then ligated to the destination vector pB7WGY2 for an eYFP fusion or to pB7WGF2 for an eGFP fusion. The destination vectors used were obtained from VIB-Ghent University (Karimi et al., 2002). The genomic region of PDIL2-1 was amplified using forward 5′-CACCGACGATGTGGTTGTTTTGACTGACGA-3′ and reverse 5′-TTAAGAAGAAGCAACGAACGTGGTTAGGA-3′ primers. The same primers were used to obtain a fragment that would encode the full-length protein, but the cDNA was used as template. For the construct that would encode truncated PDIL2-1, to phenocopy the mutant, the forward primer was the same used for the fragment that would encode the full-length protein. The reverse primer used for truncation 1 (corresponding to line 4) was 5′-TTAGGCTTTGTGTGCATCAGCATC-3′, and the reverse primer used for truncation 2 (corresponding to line 5) was 5′-TTATCCTTCCTTGTTCACGTATTCAGC-3′. The purified PCR products were cloned into the pENTR D-TOPO vector (Invitrogen) and sequenced. The resulting vector was used in an LR reaction using LR Clonase II (Invitrogen) to insert the coding sequence into the destination vectors.
Plants were transformed by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). Seeds were sown on soil and seedlings were selected by spraying with BASTA (Farnam Companies) at a 1:400 dilution. Resistant plants were transferred to freshly prepared soil.
Pollen Germination and Microscopy
Arabidopsis in vitro pollen tube growth was conducted at 22°C, as described (Boavida and McCormick, 2007). Microscopy imaging was performed using an inverted Axiophot microscope (Carl Zeiss) with either bright-field or epifluorescence optics. Images were captured using a Spot digital camera (Diagnostic Instruments), exported using AxioVision (Carl Zeiss), and processed using Photoshop 7.0 (Adobe).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PDIL2-1 (At2g47470, NM_130315), PDIL1-1 (At1g21750, NM_102024), PDIL2-3 (At2g32920, NM_128852), PDIL5-3 (At3g20560, NM_112948), ACTIN2 (At3g18780, NM_112764), IPP2 (At3g02780, NM_111146), and yeast TRG1 (M76982). Accession numbers of T-DNA insertion lines are as follows: line 3, FLAG_015D01; line 4, FLAG_427G04; line 5, SALK_046705; line 6, SALK_082179; line 7, SALK_135268c; line 8, SALK_148421; line 9, SALK_097094; line 10, CS859015, PDIL1-1 (SALK_015253C), PDIL2-3 (CS856199), and PDIL5-3 (SALK_003716).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Pollen Tube Growth Defects.
Supplemental Figure 2. Subcellular Localization of PDIL2-1 and of the Truncated PDIL2-1 Missing the D Domain.
Supplemental Figure 3. Expression of PDIL2-1 in the Wild Type and UNE5.
Supplemental Table 1. Embryo Sac Developmental Stages in Lines 4 and 5 and PDIL2-1 Transgenic Plants.
Supplemental Table 2. Primers Used for Identification of T-DNA Insertion Sites.
Supplementary Material
Acknowledgments
We thank David M. Ferrari for yeast strain BK203-15B (Mat a, ura3, leu2, his3, trp1, pdi∷leu2) and control constructs. We thank Roger Tsien (University of California, San Diego) for providing the tdTomato gene. The ER marker construct in the pKar6 vector was generously provided by Robert Blanvillain and Patrick Gallois (unpublished data). We thank Yan Zhang and Monica Alandete-Saez for discussions throughout the course of this work. We thank UC-Berkeley undergraduate research apprentice program students Trong Lee, John Yoon, and Kenny Chung for technical assistance and David Hantz and Julie Calfas for greenhouse care. This work was supported by National Science Foundation Plant Genome Program Grant 0211742 and by USDA Current Research Information System 5335-21000-030-00D. M.R. was partially supported by Vaadia-BARD Postdoctoral Fellowship Award FI-391-2006 from BARD, the U.S.–Israel Binational Agricultural Research and Development Fund.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sheila McCormick (sheilamc@nature.berkeley.edu).
Online version contains Web-only data.
References
- Alandete-Saez, M., Ron, M., and McCormick, S. (2008). GEX3, expressed in the male gametophyte and in the egg cell of Arabidopsis thaliana, is essential for micropylar pollen tube guidance and plays a role during early embryogenesis. Molecular Plant 1 586–598. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Baker, S.C., Robinson-Beers, K., Villanueva, J.M., Gaiser, J.C., and Gasser, C.S. (1997). Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.M., Lister, C., Page, T., Fransz, P., Findlay, K., Jones, G.H., Dickinson, H.G., and Dean, C. (1999). The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 19 463–472. [DOI] [PubMed] [Google Scholar]
- Boavida, L.C., and McCormick, S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52 570–582. [DOI] [PubMed] [Google Scholar]
- Byzova, M.V., Franken, J., Aarts, M.G., de Almeida-Engler, J., Engler, G., Mariani, C., Van Lookeren Campagne, M.M., and Angenent, G.C. (1999). Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev. 13 1002–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.H., Li, H.J., Shi, D.Q., Yuan, L., Liu, J., Sreenivasan, R., Baskar, R., Grossniklaus, U., and Yang, W.C. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell 19 3563–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Darby, N.J., Penka, E., and Vincentelli, R. (1998). The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J. Mol. Biol. 276 239–247. [DOI] [PubMed] [Google Scholar]
- Ellerman, D.A., Myles, D.G., and Primakoff, P. (2006). A role for sperm surface protein disulfide isomerase activity in gamete fusion: Evidence for the participation of ERp57. Dev. Cell 10 831–837. [DOI] [PubMed] [Google Scholar]
- Engel, M.L., Chaboud, A., Dumas, C., and McCormick, S. (2003). Sperm cells of Zea mays have a complex complement of mRNAs. Plant J. 34 697–707. [PubMed] [Google Scholar]
- Gruber, C.W., Cemazar, M., Heras, B., Martin, J.L., and Craik, D.J. (2006). Protein disulfide isomerase: The structure of oxidative folding. Trends Biochem. Sci. 31 455–464. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Kuroiwa, H., Kawano, S., and Kuroiwa, T. (1998). Guidance in vitro of the pollen tube to the naked embryo sac of Torenia fournieri. Plant Cell 10 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama, T., Kuroiwa, H., and Kuroiwa, T. (2003). Pollen-tube guidance: Beacons from the female gametophyte. Curr. Opin. Plant Biol. 6 36–41. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Yabe, S., Sasaki, N., Nishimura, Y., Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001). Pollen tube attraction by the synergid cell. Science 293 1480–1483. [DOI] [PubMed] [Google Scholar]
- Houston, N.L., Fan, C., Xiang, J.Q., Schulze, J.M., Jung, R., and Boston, R.S. (2005). Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 137 762–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp, M., Schneitz, K., and Pruitt, R.E. (1995). Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M.A., and Preuss, D. (2002). Plotting a course: Multiple signals guide pollen tubes to their targets. Dev. Cell 2 273–281. [DOI] [PubMed] [Google Scholar]
- Kamauchi, S., Nakatani, H., Nakano, C., and Urade, R. (2005). Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272 3461–3476. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Klappa, P., Koivunen, P., Pirneskoski, A., Karvonen, P., Ruddock, L.W., Kivirikko, K.I., and Freedman, R.B. (2000). Mutations that destabilize the a' domain of human protein-disulfide isomerase indirectly affect peptide binding. J. Biol. Chem. 275 13213–13218. [DOI] [PubMed] [Google Scholar]
- Klucher, K.M., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, B., Kramer, W., Williamson, M.S., and Fogel, S. (1989). Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol. Cell. Biol. 9 4432–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, I.M., and Chrispeels, M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnat, J., Hacker, U., Geissler, H., Rauchenberger, R., Neuhaus, E.M., Maniak, M., and Soldati, T. (1997). Dictyostelium discoideum protein disulfide isomerase, an endoplasmic reticulum resident enzyme lacking a KDEL-type retrieval signal. FEBS Lett. 418 357–362. [DOI] [PubMed] [Google Scholar]
- Monnat, J., Neuhaus, E.M., Pop, M.S., Ferrari, D.M., Kramer, B., and Soldati, T. (2000). Identification of a novel saturable endoplasmic reticulum localization mechanism mediated by the C-terminus of a Dictyostelium protein disulfide isomerase. Mol. Biol. Cell 11 3469–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8 64–71. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Ner-Gaon, H., and Fluhr, R. (2006). Whole-genome microarray in Arabidopsis facilitates global analysis of retained introns. DNA Res. 13 111–121. [DOI] [PubMed] [Google Scholar]
- Pagnussat, G.C., Yu, H.J., Ngo, Q.A., Rajani, S., Mayalagu, S., Johnson, C.S., Capron, A., Xie, L.F., Ye, D., and Sundaresan, V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614. [DOI] [PubMed] [Google Scholar]
- Palanivelu, R., Brass, L., Edlund, A.F., and Preuss, D. (2003). Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114 47–59. [DOI] [PubMed] [Google Scholar]
- Pirneskoski, A., Klappa, P., Lobell, M., Williamson, R.A., Byrne, L., Alanen, H.I., Salo, K.E., Kivirikko, K.I., Freedman, R.B., and Ruddock, L.W. (2004). Molecular characterization of the principal substrate binding site of the ubiquitous folding catalyst protein disulfide isomerase. J. Biol. Chem. 279 10374–10381. [DOI] [PubMed] [Google Scholar]
- Punwani, J.A., Rabiger, D.S., and Drews, G.N. (2007). MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell 19 2557–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S.M., Park, S.S., and Ray, A. (1997). Pollen tube guidance by the female gametophyte. Development 124 2489–2498. [DOI] [PubMed] [Google Scholar]
- Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G.W., and Fischer, R.L. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83 735–742. [DOI] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, A.M., Bosch, M., Bots, M., Nieuwland, J., Feron, R., and Mariani, C. (2004). Pistil factors controlling pollination. Plant Cell 16(Suppl): S98–S106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N.C., Campbell, R.E., Steinbach, P.A., Giepmans, B.N., Palmer, A.E., and Tsien, R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shimizu, K.K., and Okada, K. (2000). Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127 4511–4518. [DOI] [PubMed] [Google Scholar]
- Turano, C., Coppari, S., Altieri, F., and Ferraro, A. (2002). Proteins of the PDI family: Unpredicted non-ER locations and functions. J. Cell. Physiol. 193 154–163. [DOI] [PubMed] [Google Scholar]
- Urade, R. (2007). Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274 1152–1171. [DOI] [PubMed] [Google Scholar]
- Wadahama, H., Kamauchi, S., Ishimoto, M., Kawada, T., and Urade, R. (2007). Protein disulfide isomerase family proteins involved in soybean protein biogenesis. FEBS J. 274 687–703. [DOI] [PubMed] [Google Scholar]
- Wang, H., Liu, Y., Bruffett, K., Lee, J., Hause, G., Walker, J.C., and Zhang, S. (2008). Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20 602–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings, K., and Russell, S.D. (2004). Experimental analysis of the fertilization process. Plant Cell 16(Suppl): S107–S118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, B., and Gilbert, H.F. (2004). Protein disulfide isomerase. Biochim. Biophys. Acta 1699 35–44. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., and Drews, G.N. (2004). Female gametophyte development. Plant Cell 16(Suppl): S133–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Kaur, N., Kiriakopolos, S., and McCormick, S. (2006). EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta 224 1004–1014. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., and Sundaresan, V. (2000). Genetics of gametophyte biogenesis in Arabidopsis. Curr. Opin. Plant Biol. 3 53–57. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.