Abstract
Endoplasmic reticulum–mediated quality control (ERQC) is a well-studied process in yeast and mammals that retains and disposes misfolded/unassembled polypeptides. By contrast, how plants exert quality control over their secretory proteins is less clear. Here, we report that a mutated brassinosteroid receptor, bri1-5, that carries a Cys69Tyr mutation, is retained in the ER by an overvigilant ERQC system involving three different retention mechanisms. We demonstrate that bri1-5 interacts with two ER chaperones, calnexin and binding protein (BiP), and is degraded by a proteasome-independent endoplasmic reticulum–associated degradation (ERAD). Mutations in components of the calnexin/calreticulin cycle had little effect on the fidelity of the Arabidopsis thaliana ERQC for bri1-5 retention. By contrast, overexpression of bri1-5, treatment with an ERAD inhibitor, RNA interference–mediated BiP silencing, or simultaneous mutations of Cys-69 and its partner Cys-62 can mitigate this quality control, resulting in significant suppression of the bri1-5 phenotype. Thus, bri1-5 is an excellent model protein to investigate plant ERQC/ERAD in a model organism.
INTRODUCTION
In eukaryotic cells, the vast majority of secretory and membrane proteins complete their folding processes in the endoplasmic reticulum (ER). This folding compartment harbors a sophisticated endoplasmic reticulum quality control (ERQC) system that ensures the export of only correctly folded and properly assembled polypeptides to their final destinations (Ellgaard and Helenius, 2003). Incompletely folded and improperly assembled proteins are retained in the ER for additional attempts of chaperone-assisted folding/assembly or destroyed via a process known as endoplasmic reticulum–associated degradation (ERAD) (Romisch, 2005). Such an ERQC system is capable of discriminating native and nonnative proteins by recognizing several common features of improperly/incompletely folded proteins, such as immature Asn-linked glycans, exposed hydrophobic amino acids, and unpaired Cys residues (Sitia and Braakman, 2003).
There are at least three different mechanisms for detecting these structural features and retaining misfolded proteins (Sitia and Braakman, 2003). The best studied is the so-called calnexin/calreticulin (CNX/CRT) cycle, which is specific for glycoproteins (Caramelo and Parodi, 2007). This system depends on UDP-glucose:glycoprotein glucosyltransferase (UGGT), which is capable of discriminating a misfolded protein from its native counterpart by recognizing exposed hydrophobic residues and catalyzing reglucosylation of the Asn-linked oligosaccharide Man9GlcNAc2. The resulting Glc1Man9GlcNAc2-containing glycoprotein then interacts with CNX and/or CRT, two ER resident lectin-like chaperones (Williams, 2006), leading to its retention in the ER. The second system relies on the luminal binding protein (BiP), an ER-localized member of the HSP70 family, for both recognition and retention (Buck et al., 2007). BiP is composed of an N-terminal ATP binding domain and a C-terminal domain that binds to hydrophobic patches on improperly/incompletely folded proteins in an ATP-dependent manner (Flynn et al., 1991; Blond-Elguindi et al., 1993). The third detection mechanism recognizes free thiol groups and retains nonnative proteins by forming mixed disulfides with protein disulfide isomerases or other ER resident proteins with oxidoreductase activity (Reddy et al., 1996; Anelli et al., 2003, 2007).
Most of our current knowledge on ERQC/ERAD was obtained from studies using yeast and mammalian systems. Despite the facts that ERQC is essential for plant growth and development (Boisson et al., 2001; Burn et al., 2002; Gillmor et al., 2002) and that plants contain many highly conserved ERQC/ERAD components (Sung et al., 2001; Persson et al., 2003; Houston et al., 2005; Kirst et al., 2005), little is known about how the plant ERQC/ERAD system operates to retain and/or dispose of misfolded/unassembled secretory proteins in the ER. Studies using engineered substrates with protoplasts or transgenic plants have so far provided strong evidence for the existence of a similar ERQC system and several distinct ERAD pathways (Di Cola et al., 2001, 2005; Brandizzi et al., 2003; Tamura et al., 2004; Muller et al., 2005; Pimpl et al., 2006).
BRI1 is a leucine-rich-repeat (LRR) receptor–like kinase that functions as a cell surface receptor for brassinosteroids (BRs) (Li and Chory, 1997; Wang et al., 2001; Kinoshita et al., 2005). Loss-of-function mutations in BRI1 resulted in a characteristic set of morphological changes that include dwarfed stature, a prolonged vegetative phase, and altered skotomorphogenesis (Clouse et al., 1996; Li and Chory, 1997). Genetic and biochemical studies in the past decade have discovered a linear signaling pathway that involves BRI1 and its coreceptor BAK1 (Li et al., 2002; Nam and Li, 2002), a BRI1 inhibitor, BKI1 (Wang and Chory, 2006), a GSK3-like kinase, BIN2 (Li and Nam, 2002), a protein phosphatase, BSU1 (Mora-Garcia et al., 2004), and two plant-specific transcriptional factors, BES1 and BZR1 (Wang et al., 2002; Yin et al., 2002; Zhao et al., 2002). It was hypothesized that in the absence of BR, BIN2 is a constitutively active kinase that phosphorylates BES1 and BZR1, promoting their degradation (He et al., 2002; Yin et al., 2002), retaining them in the cytosol via interaction with 14-3-3 proteins (Bai et al., 2007; Gampala et al., 2007; Ryu et al., 2007), and inhibiting their DNA binding activities in the nucleus (Vert and Chory, 2006; Gampala et al., 2007). BR binding to BRI1 triggers dimerization and activation of BRI1 and BAK1 (Wang et al., 2005), leading to inhibition of BIN2 and possibly activation of BSU1 and accumulation of nonphosphorylated BES1 and BZR1 in the nucleus to regulate gene expression.
A recent study revealed that the dwarf phenotype of bri1-9, which carries a Ser-to-Phe mutation in the ligand binding domain of BRI1, is caused by ER retention of a structurally imperfect but functionally competent BR receptor (Jin et al., 2007). A suppressor screen looking for extragenic mutations that revert the cabbage-like dwarf mutant to wild-type-looking plants identified several mutations in the Arabidopsis thaliana homolog of UGGT. It was shown that bri1-9 is monoglucosylated by UGGT and interacts with both CNX and BiP. Loss of UGGT activity significantly compromises the fidelity of the CNX/CRT-mediated glycoprotein quality control machinery. As a result, a considerable amount of bri1-9 exits the ER and is correctly targeted to the cell surface, where it can function like normal BR receptors to initiate a phosphorylation-mediated signaling cascade.
In addition to bri1-9, there are six other bri1 alleles containing missense mutations in the extracellular domain of the Arabidopsis BR receptor (Noguchi et al., 1999; Vert et al., 2005). Little is known about how these mutations affect BRI1 function. It was hypothesized previously that these mutations either directly affect BR binding or interfere with BRI1 homodimerization or heterodimerization (Friedrichsen et al., 2000). To test if ER retention of a mutated BR receptor underlies the dwarf phenotype of these known mutants, we performed a biochemical survey and discovered that bri1-5, which harbors a Cys69Tyr mutation in the N-terminal cap domain of the BR receptor (Noguchi et al., 1999), is also retained in the ER by at least three different mechanisms and is degraded by a proteasome-independent ERAD process. We have shown that inhibition of ERAD, overexpression of bri1-5, or silencing BiP expression can suppress the dwarf phenotype of bri1-5. Our discovery indicated that bri1-5 is an excellent model protein to study ER quality control and ERAD in a genetic model organism.
RESULTS
An Endoglycosidase H–Based Survey Identified bri1-5 as an ER-Localized Protein
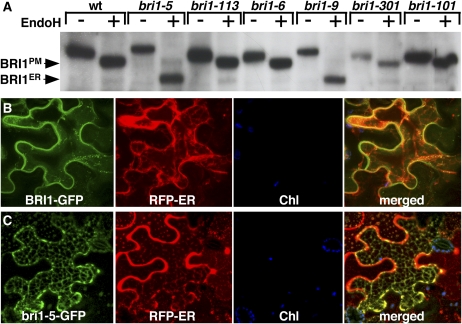
Our previous study revealed that the bri1-9 mutation, which changes the highly conserved Ser-662 residue in the BR binding domain to Phe, results in ER retention of a structurally defective yet biochemically competent BR receptor (Jin et al., 2007). To test if a similar mechanism is responsible for the BR-insensitive dwarf phenotype of other extracellular bri1 mutations, including a dozen bri1 alleles discovered in tilling projects, we conducted a biochemical survey using the Endoglycosidase H (Endo H) assay, which is able to differentiate a plasma membrane–localized glycoprotein from its ER-localized counterpart due to further modification of the Asn-linked glycans in the secretory pathway. Total protein crude extracts of bri1 mutants or transgenic plants expressing various bri1–green fluorescent protein (GFP) fusion proteins were treated with or without Endo H, separated by SDS-PAGE, and analyzed by protein gel blotting with a BRI1 antibody (Mora-Garcia et al., 2004). As shown in Figure 1A and summarized in Supplemental Figure 1 online, out of 16 bri1 extracellular alleles surveyed, only bri1-5 and bri1-9 were completely sensitive to the Endo H digestion, suggesting that bri1-5, similar to bri1-9, is mainly retained in the ER. The difference in mobility between bri1-5 or bri1-9 and the wild-type BRI1 or other mutated bri1 proteins is due to different glycoforms on the BR receptors, bri1-5 and bri1-9 containing only high-mannose-type glycans while the others have a mixture of complex glycans with a few high-mannose-type glycans (explaining a slight reduction in molecular weight upon Endo H treatment). The ER localization of bri1-5 was further confirmed by confocal analysis of transiently expressed bri1-5-GFP in tobacco (Nicotiana benthamiana) leaf epidermal cells. As shown in Figures 1B and 1C, the wild-type BRI1-GFP fusion protein is mainly localized on the plasma membrane, while bri1-5-GFP exhibits a reticulate fluorescence pattern that overlaps nicely with that of red fluorescent protein (RFP)–ER known to be localized in the ER (Chakrabarty et al., 2007).
Figure 1.
bri1-5 Is Retained in the ER.
(A) Endo H assays of several known bri1 mutants. Crude protein extracts of 4-week-old bri1 mutants were denatured and incubated with or without Endo H; the resulting protein samples were separated by SDS-PAGE and analyzed by protein gel blot using an anti-BRI1 antibody. The arrows indicate the positions of bands representing BRI1 in the plasma membrane (BRI1PM) and in the ER (BRI1ER).
(B) and (C) Confocal microscopic analysis of BRI1-GFP (B) and bri1-5-GFP (C). Shown from left to right are fluorescence patterns of GFP fusion proteins (green), the ER-localized RFP (red), autofluorescent chloroplasts (Chl; blue), and the merged images of green, red, and blue fluorescent signals in Agrobacterium-infiltrated tobacco leaf epidermal cells.
bri1-5 Interacts with CNXs in a Monoglucosylation-Dependent Manner
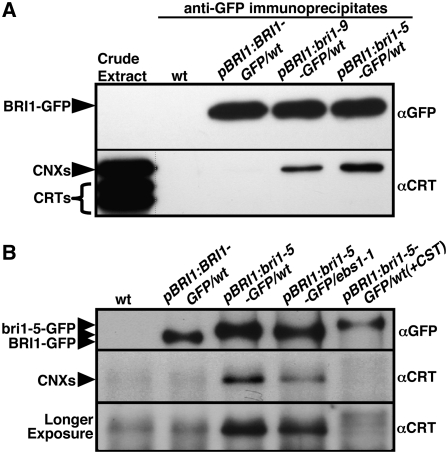
It was known that bri1-9 is retained in the ER via the CNX/CRT cycle (Jin et al., 2007). To test if bri1-5 interacts with any of the Arabidopsis CNXs/CRTs, we expressed bri1-5-GFP, bri1-9-GFP, or BRI1-GFP driven by the native BRI1 promoter in transgenic Arabidopsis plants. A polyclonal anti-GFP antibody was used to immunoprecipitate BRI1-GFP, bri1-9-GFP, or bri1-5-GFP from protein crude extracts of the resulting transgenic plants, and the presence of CNX/CRT in the immunoprecipitates was examined by immunoblotting with an anti-maize (Zea mays) CRT antibody that can detect all Arabidopsis CNXs/CRTs (Persson et al., 2003). As shown in Figure 2A, both bri1-9-GFP and bri1-5-GFP were coimmunoprecipitated with CNXs but not with any of the three CRTs. As a control, neither CNX nor CRT was found to interact with the wild-type BRI1-GFP.
Figure 2.
bri1-5 Interacts with CNXs in a Monoglucosylation-Dependent Manner.
(A) bri1-5 interacts more strongly with CNXs than bri1-9 does.
(B) Introduction of the ebs1-1 mutation or treatment with CST inhibits the bri1-5–CNX interaction.
For both (A) and (B), crude protein extracts of transgenic plants expressing GFP fused to the indicated alleles of BRI1 and driven by the BRI1 promoter were immunoprecipitated using a polyclonal anti-GFP antibody. The presence of BRI1- or bri1-GFP and CNXs in the resulting immunoprecipitates was analyzed by protein gel blot using anti-GFP or anti-maize CRT antibody, respectively. The longer exposure of the protein gel blot filter in (B) was used to show no detectable CNX signal in the anti-GFP immunoprecipitate of the CST-treated sample. Arrowheads show the positions of various protein bands indicated by the labels, and the top two arrowheads in (B) indicate the positions of bri1-5-GFP, with the top one for bri1-5-GFP containing three glucose residues on each glycan due to CST treatment.
To determine if such a bri1-5–CNX interaction depends on the presence of monoglucosylated glycans on bri1-5, we crossed the pBRI1:bri1-5-GFP transgene into the ebs1-1 mutant lacking the Arabidopsis UGGT, which was previously shown to monoglucosylate the ER-localized bri1-9, and used the resulting transgenic mutants for a similar coimmunoprecipitation experiment (Jin et al., 2007). As shown in Figure 2B, lane 4, the ebs1-1 mutation significantly reduced but did not completely eliminate the bri1-5–CNX interaction. The remaining bri1-5–CNX interaction is likely caused by the formation of monoglucosylated glycan on bri1-5 through sequential removal of two terminal glucose residues from Glc3Man9GlcNAc2 and/or glycan-independent bri1-5–CNX interaction (Danilczyk and Williams, 2001). We also treated the pBRI1:bri1-5-GFP transgenic seedlings with castanospermine (CST), which prevents the removal of the first glucose residues from the Glc3Man9GlcNAc2 core glycan (Sasak et al., 1985). This glucosidase inhibitor has been widely used to show monoglucosylated glycan–dependent interaction of CNX/CRT with their clients (Parodi, 2000). As revealed in Figure 2B, lane 5, CST treatment dramatically inhibited the CNX–bri1-5 interaction, and little CNX signal was detected in the anti-GFP immunoprecipitate of the CST-treated sample even after prolonged exposure. These results demonstrated that bri1-5 interacts with CNXs in a monoglucosylation-dependent manner.
bri1-5 Is Degraded by ERAD
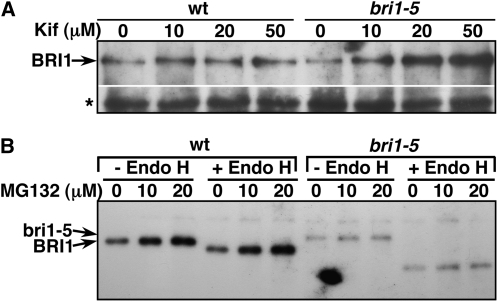
Our repeated protein gel blot analyses revealed that the steady state level of bri1-5 is significantly lower than that of wild-type BRI1 in addition to its slower mobility on SDS-PAGE. Given the fact that bri1-5 is retained in the ER, we suspected that the low abundance of bri1-5 is caused by ERAD, a safeguard mechanism to prevent the overaccumulation of irreparably misfolded proteins in the folding compartment (McCracken and Brodsky, 1996). To test this hypothesis directly, we treated bri1-5 seedlings with kifunensine (Kif), an inhibitor of ER and Golgi α1,2-mannosidases that is known to prevent ERAD of many terminally misfolded proteins (Tokunaga et al., 2000). Figure 3A shows that Kif treatment significantly increased the steady state level of bri1-5 in a dose-dependent manner. Interestingly, a similar treatment with MG132, a widely used inhibitor of the proteasome that prevents degradation of a wide range of ERAD substrates (Schmitz and Herzog, 2004), had little effect on the steady state level of bri1-5 but caused a noticeable increase in the amount of wild-type BRI1 (Figure 3B). Thus, we concluded that bri1-5 is degraded by a proteasome-independent ERAD pathway.
Figure 3.
bri1-5 Is Degraded by a Proteasome-Independent ERAD Process.
(A) The steady state level of bri1-5 is greatly increased by treatment with Kif.
(B) MG132 treatment stabilizes the wild-type BRI1 but has little effect on bri1-5.
For both (A) and (B), leaves of 4-week-old soil-grown plants were removed and incubated for 24 h in liquid half-strength MS medium containing the indicated concentrations of Kif or MG132, and the treated samples were analyzed by Endo H assay followed by protein gel blot for the effect of the inhibitors on the stability of BRI1 and bri1-5. The asterisk in (A) denotes a nonspecific band used as our loading control.
bri1-5 Is a Functional BR Receptor
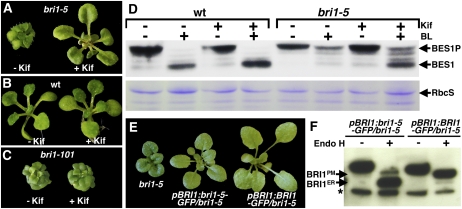
In the course of carrying out the Kif experiment, we noticed that prolonged (5 d) Kif treatment of bri1-5 seedlings led to considerable phenotypic rescue of the bri1-5 mutation under both light and dark growth conditions (Figure 4A; see Supplemental Figure 2 online), while a similar treatment had no noticeable effect on either wild-type seedlings or bri1-101 mutants (Figures 4B and 4C; Supplemental Figure 2 online), suggesting that bri1-5 is a functionally competent BR receptor. We suspected that Kif-induced bri1-5 overaccumulation might saturate its ER retention machinery, thus allowing some bri1-5 proteins to escape the ER and be correctly targeted to the cell surface, where they can function as normal BR receptors. Unfortunately, the Endo H assay was not able to differentiate the plasma membrane–localized bri1-5 from its ER-localized form in Kif-treated seedlings, since Kif completely prevents Golgi-mediated glycan modifications, resulting in high-mannose-type glycans on all glycoproteins. Instead, we tested if Kif treatment led to an increase in BR sensitivity by examining the BR-induced dephosphorylation of BES1, a robust biochemical marker for an activated BR signaling pathway (Vert and Chory, 2006). As shown in Figure 4D, the Kif-treated bri1-5 plants exhibited a detectable increase in their BR sensitivity. The treated bri1-5 mutants accumulated a detectable amount of nonphosphorylated BES1, and the amount of the phosphorylated BES1 was reduced in response to brassinolide (BL) application. By contrast, bri1-5 mutants treated with BL alone still accumulated a detectable amount of phosphorylated BES1. Additional support for bri1-5 being a functional BR receptor came from our discovery that overexpression of bri1-5-GFP in bri1-5 mutants resulted in a significant suppression of the bri1-5 dwarf phenotype, as shown in Figure 4E. Endo H assay of several pBRI1:bri1-5-GFP/bri1-5 transgenic plants revealed the presence of an Endo H–resistant form of bri1-5 that is likely to be present at the cell surface to mediate BR signaling (Figure 4F).
Figure 4.
bri1-5 Is a Functionally Competent BR Receptor.
(A) Five-day Kif treatment suppresses the bri1-5 mutation.
(B) Five-day Kif treatment has no effect on wild-type seedlings.
(C) Five-day Kif treatment has no detectable effect on bri1-101 mutants.
(D) Kif-treated bri1-5 mutants exhibit enhanced BR response. In response to BR treatment, almost all phosphorylated BES1 proteins (BES1P) were dephosphorylated (BES1) in wild-type seedlings treated with (lanes 3 and 4) or without (lanes 1 and 2) Kif, whereas only a small amount of BES1P was dephosphorylated in bri1-5 (lanes 5 and 6) without Kif. By contrast, a large amount of BES1P was dephosphorylated in response to BR in Kif-treated bri1-5 seedlings (lanes 7 and 8).
For (A) to (D), 2-week-old seedlings were transferred to fresh half-strength MS medium containing 10 μM Kif for continued growth and removed 5 d later for photographing ([A] to [C]) or incubation in liquid half-strength MS medium supplemented with or without 1 μM BL (D). Total protein crude extracts were analyzed by immunoblotting using an anti-BES1 antibody, and Coomassie blue staining of the small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (RbcS) was used as the loading control.
(E) Overexpression of pBRI1:bri1-5-GFP suppresses the dwarf phenotype of the weak bri1-5 mutant. Shown are the bri1-5 mutant and two representative transgenic bri1-5 lines expressing the pBRI1:bri1-5-GFP or pBRI1:BRI1-GFP transgene.
(F) Overexpression of bri1-5-GFP leads to export of a detectable pool of defective BR receptor. Anti-GFP immunoprecipitates were treated with or without Endo H, separated by SDS-PAGE, and analyzed by protein gel blot with a monoclonal anti-GFP antibody. The arrows indicate the positions of bands representing BRI1 in the plasma membrane (BRI1PM) and in the ER (BRI1ER), and the asterisk denotes a nonspecific band used as a loading control.
Loss of UGGT Activity Enhances the bri1-5 Mutation
Our previous study with bri1-9 revealed that loss-of-function mutations in UGGT significantly compromise the high fidelity of the CNX/CRT-mediated ER quality control mechanism that retains the mutated BR receptor, allowing export of a significant fraction of bri1-9 to the cell surface, where the mutated BR receptor responds to BR to initiate a phosphorylation-mediated signaling cascade (Jin et al., 2007). Since the bri1-5–CNX interaction also depends on monoglucosylated glycans, we suspected that loss of the UGGT activity might have a similar effect on bri1-5. To directly test our hypothesis, we crossed bri1-5 into ebs1-1, ebs1-2, and ebs1-3 to generate bri1-5 ebs1 double mutants. To our surprise, the ebs1 mutations did not suppress but instead enhanced the bri1-5 phenotype (Figure 5A), suggesting that the CNX/CRT cycle is not a major retention factor for bri1-5 but instead functions redundantly with other ER proteins in keeping bri1-5 in the ER. One or more retention factors unique for bri1-5 are likely overaccumulated in ebs1 mutants due to the so-called unfolded protein response (UPR) (Jin et al., 2007), which upregulates the expression of many ER chaperones and folding catalysts, explaining the stronger dwarf phenotype of the bri1-5 ebs1 double mutants. By contrast, the UGGT-mediated CRT/CNX system plays a major role in retaining bri1-9.
Figure 5.
Mutations of the CNX/CRT Cycle Components Fail to Suppress the bri1-5 Mutation.
(A) Phenotype comparison between bri1-5, three bri1-5 ebs1 double mutants, and the bri1-5 cnx1 cnx2 triple mutant grown in soil for 1 month. Plant images were assembled from different photographs taken at the same magnification.
(B) Endo H analysis of BRI1 and bri1-5 in the wild type and bri1-5, bri1-5 ebs1, and bri1-5 cnx1 cnx2 mutants. The asterisk indicates a nonspecific band used as our loading control.
Eliminating Two CNXs Does Not Lead to Escape of bri1-5 from the ER
Further support for the involvement of other factors in ER retention of bri1-5 came from an analysis of Arabidopsis mutants lacking CNXs. The Arabidopsis genome encodes only one UGGT but two CNXs and three CRTs (Huang et al., 1993; Boyce et al., 1994; Persson et al., 2003). Searching the SIGNAL T-DNA insertion database (Alonso et al., 2003) identified a null mutant for each CNX (see Supplemental Figures 3A, 3B, and 3D online). As predicted, mutation in either gene had little effect on the bri1-5 phenotype (see Supplemental Figure 3E online). Consistent with the ebs1 bri1-5 analysis, simultaneous elimination of the two Arabidopsis CNX genes was not able to suppress the bri1-5 mutation (Figure 5A). Unlike ebs1 mutations, the cnx1 cnx2 double mutation did not enhance the bri1-5 phenotype. This might be simply explained by the inability of the cnx1 cnx2 double mutation to induce UPR (see Supplemental Figure 3C online). Because the CNX/CRT cycle is also involved in facilitating protein folding (Williams, 2006), it is possible that eliminating UGGT or two CNXs can lead to escape of bri1-5 with a more misfolded state that no longer responds to BR. To test this possibility directly, we performed the Endo H assay with bri1-5 ebs1 and bri1-5 cnx1 cnx2 mutants. As shown in Figure 5B, no detectable Endo H–resistant bri1-5 was found in either mutant. Taken together, these data strongly suggest that the UGGT-based CRT/CNX cycle functions redundantly with other retention factors in trapping bri1-5 in the folding compartment.
BiP Is Involved in ER Retention of bri1-5
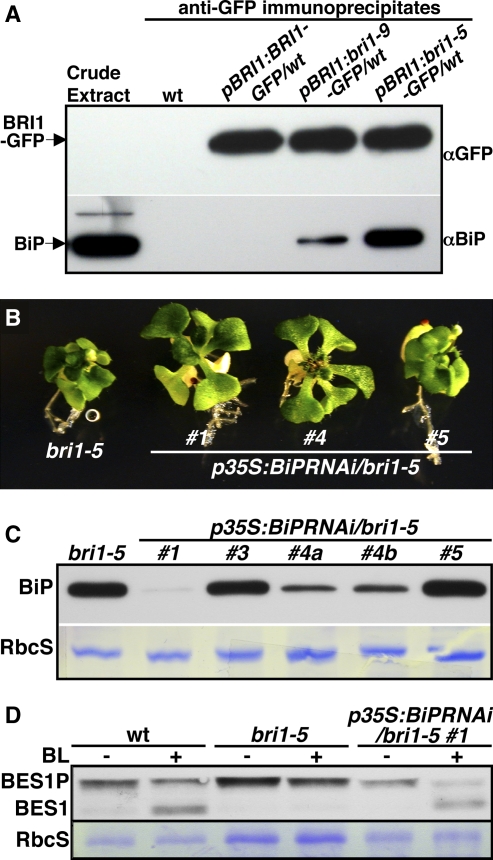
One possible candidate for an additional ER retention factor of bri1-5 is BiP, which was previously shown to interact with bri1-9 (Jin et al., 2007). To determine if BiP is involved in ER retention of bri1-5, we performed a coimmunoprecipitation experiment with pBRI1:bri1-5-GFP, pBRI1:bri1-9-GFP, and pBRI1:BRI1-GFP transgenic plants. As shown in Figure 6A, bri1-5 exhibits a much stronger interaction with BiP than bri1-9 does. To investigate if BiP plays a role in retaining bri1-5 in the ER, we generated p35S:BiPRNAi transgenic plants with a BiP1 cDNA fragment that is highly conserved among all three Arabidopsis BiP genes and the strong, constitutively active 35S promoter of the Cauliflower mosaic virus. Despite numerous attempts, we were not able to obtain any transgenic plants in wild-type or bri1-9 backgrounds, suggesting crucial roles of BiPs in many cellular processes. Out of five p35S:BiPRNAi/bri1-5 transgenic plants, three exhibited a partially suppressed bri1-5 phenotype (Figure 6B), including one that died before reaching maturity, and the other two were morphologically indistinguishable from the bri1-5 mutant. Protein gel blot analysis revealed that these two partially suppressed lines accumulated much less BiP than the corresponding wild-type control, while the two nonsuppressed lines exhibited no change in BiP abundance (Figure 6C). Protein gel blot analysis of BES1 phosphorylation status indicated that the partially suppressed bri1-5 BiPRNAi transgenic line regained partial response to BL (Figure 6D). Taken together, our results suggested that BiP is a major player in retaining bri1-5 in the ER.
Figure 6.
BiPs Are Involved in ER Retention of bri1-5.
(A) Coimmunoprecipitation of bri1-5 and bri1-9 with BiP. Anti-GFP immunoprecipitates (used in Figure 2) were analyzed by immunoblotting with anti-GFP (top strip) and anti-BiP (bottom strip) antibodies.
(B) Phenotypic comparison between bri1-5 and three BiPRNAi transgenic bri1-5 mutants. The compact rosette phenotype of bri1-5 was partially suppressed in lines 1 and 4.
(C) Protein gel blot analysis of total protein crude extracts of bri1-5 and BiPRNAi/bri1-5 transgenic lines with an anti-BiP antibody.
(D) Immunoblot analysis of the BL-induced BES1 dephosphorylation of the wild type, bri1-5, and one BiPRNAi/bri1-5 line.
In both (C) and (D), Coomassie blue staining of the small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (RbcS) was used as the loading control.
A Thiol-Mediated Mechanism Contributes to ER Retention of bri1-5
It is generally thought that in LRR proteins, the N-terminal Cys pair (Cys-62 and Cys-69 in BRI1) forms a disulfide bond that plays a structural role in shielding hydrophobic residues of the first LRR (van der Hoorn et al., 2005). It is thus quite possible that the mutation of Cys-69 to Tyr in bri1-5 creates a free thiol group at Cys-62 that can be recognized by a thiol-mediated ER retention mechanism. Such a system is present in mammalian cells for retaining proteins with exposed thiol groups and involves the formation of reversible disulfide bonds with an ER resident protein, ERp44 (Anelli et al., 2003). BLAST search against the entire Arabidopsis genome sequence failed to detect the presence of an ERp44 homolog.
To test if such a mechanism contributes to ER retention of bri1-5, we created two additional mutant bri1-GFP fusion proteins, bri1C62Y-GFP and bri1-5C62Y-GFP, by site-directed mutagenesis and transformed them individually into bri1-5 mutants to compare their relative biological activities. We reasoned that if a thiol-mediated ER retention is involved in keeping bri1-5 in the ER, eliminating both Cys residues of the N-terminal Cys pair would lead to an increased export of the mutated bri1 protein out of the ER and a strong bri1-5 suppression activity, while mutating Cys-62 to Tyr should have a similar effect on BRI1 as the bri1-5 mutation. Multiple transgenic lines per construct were examined for rosette size of young seedlings and inflorescence height of adult plants. As shown in Figure 7A and Supplemental Figures 4 and 5 online, bri1-5C62Y had a much stronger suppressing activity than the bri1-5-GFP fusion did, while bri1C62Y exhibited similar activity to bri1-5, suggesting the involvement of Cys-62 in ER retention of bri1-5. Consistent with our morphological analysis, the Endo H–resistant form of bri1 accumulated more bri1-5C62Y than either bri1-5 or bri1C62Y (Figure 7B).
Figure 7.
bri1-5C62Y-GFP Has a Higher bri1-5–Rescuing Activity Than Either bri1-5-GFP or bri1C62Y-GFP.
(A) Phenotypic comparison between 4-week-old wild-type plants and representatives (two per construct) of transgenic bri1-5 mutants expressing pBRI1:bri1-5-GFP, pBRI1:bri1C62Y-GFP, or pBRI1:bri1-5C62Y-GFP. More than 75 T1 transgenic lines per construct were examined for phenotypic analysis. Photographs were taken under the same magnification.
(B) Endo H assay of bri1-GFP of three representative transgenic lines. Transgenic plants expressing GFP-tagged BRI1 and bri1-5 in the wild-type background were also included in the assay to show the positions of the plasma membrane–localized BRI1-GFP and the ER-localized bri1-5-GFP. The top arrow indicates the positions of glycosylated BRI1-GFP and bri1-5-GFP, the middle arrow shows the position of partially deglycosylated plasma membrane–localized bri1-5-GFP (bri1-5-GFPPM), and the bottom arrow indicates the position of fully deglycosylated ER-localized bri1-5-GFP (bri1-5-GFPER).
It is interesting that although the amount of the Endo H–resistant form (likely being localized on the cell surface) is significantly lower than that of wild-type BRI1 (see Supplemental Figure 5B online), the vast majority of the pBRI1:bri1-5C62Y-GFP/bri1-5 transgenic lines are noticeably bigger than the wild-type seedlings of similar age, with longer petioles and elongated leaves when grown on synthetic medium and are much taller than the wild-type controls when grown in soil (Figure 7A; see Supplemental Figures 4 and 5A). This observation suggests that the membrane-localized bri1-5C62Y-GFP might be more active than the wild-type BRI1.
DISCUSSION
An Overvigilant ER Quality Control System in Arabidopsis
In this study, we have demonstrated that the dwarf phenotype of the Arabidopsis bri1-5 mutant is caused by an overvigilant ER quality control system that keeps a structurally defective, yet functionally competent, BR receptor in the folding compartment. We have shown that bri1-5 carries high-mannose-type glycans and interacts with CNX in a monoglucosylation-dependent manner. We have found that bri1-5 can be coimmunoprecipitated with BiP, another ER chaperone known to interact with misfolded/incompletely folded proteins. These experiments allowed us to conclude that bri1-5 is kept in the ER. We have also presented strong evidence for bri1-5 being a functionally competent BR receptor. Overaccumulation of bri1-5 in the ER by treatment with a known ERAD inhibitor, Kif, or overexpression of bri1-5-GFP resulted in an almost complete suppression of the bri1-5 dwarf phenotype. It is interesting that some pBRI1:bri1-5-GFP/bri1-5 transgenic plants grow bigger than the corresponding wild-type plants despite the fact that the level of the Endo H–resistant form of bri1-5-GFP in those transgenic lines was much lower than that of BRI1 in the wild-type control (see Supplemental Figure 5 online), implying that bri1-5 might be even more active than the wild-type BRI1.
This is another example of an overvigilant ERQC retaining/disposing of a structurally imperfect yet functionally competent BR receptor in the folding compartment in Arabidopsis. Our previous study showed that a similar ERQC is responsible for the dwarf phenotype of another well-studied bri1 mutant, bri1-9 (Jin et al., 2007). These results illustrate the importance of keeping a balance between retaining/disposing of potentially toxic proteins and avoiding overvigilance that prevents the export of functionally competent proteins with minimal structural distortion. Export of an aberrant protein could poison its functional partners, while overaccumulation of a terminally misfolded protein in the ER could lead to chronic ER stress and cellular toxicity. On the other hand, an overzealous ER quality control can block the export and promote the destruction of a structurally imperfect yet biochemically active protein, leading to a strong loss-of-function phenotype. An overvigilant ERQC is known to be responsible for the severe clinical phenotype of the most common cystic fibrosis mutation, CFTR(ΔF508), which occurs in >80% cystic fibrosis patients (Sheppard and Welsh, 1999).
The Structural Function of the Cys Pair in the N-Terminal Cap Domain
The biochemical basis for bri1-5 being retained in the ER is in the Cys69Tyr mutation and its effect on the structure of the conserved N-terminal cap domain. This single amino acid change not only puts a bulky aromatic amino acid on the protein surface but also eliminates the conserved Cys-62–Cys-69 disulfide linkage, creating an orphan Cys residue with a free thiol group and destabilizing the N-terminal cap structure.
BRI1 is a member of the plant extracellular LRR proteins containing tandem repeats of a 24–amino acid motif: xLxxL5xLSxNxL(S/T)GxIPxxL20GxLx (where x denotes any amino acid) (Li and Chory, 1997). It is generally believed that most LRR domains exhibit a characteristic curved solenoid structure with one turn corresponding to each LRR that consists of a short parallel β-strand (xxL5x) on the concave inner surface connected by a β-turn or a β-sheet [NxL(S/T)Gx for certain plant extracellular LRR proteins] (Di Matteo et al., 2003) to an antiparallel 310-helical segment (xxL20Gx) on the convex outside surface (Choe et al., 2005). The seven conserved hydrophobic residues of repeating LRRs point inward to form a tightly packed hydrophobic core of the solenoid that is capped at both ends by the N- and C-terminal flanking regions (Choe et al., 2005). Sequence alignment of many known plant extracellular LRR proteins revealed a highly conserved N-terminal capping domain with an x[D/E]xxALLxΦKxxΦx4-10LssWx4-6Cx[W/F]xGVxC consensus sequence (where Φ denotes hydrophobic amino acids) (van der Hoorn et al., 2005).
The recently solved crystal structure of polygalacturonase-inhibiting protein (PGIP) suggests the possible involvement of highly conserved hydrophobic/aromatic amino acids in shielding the hydrophobic interior of the first LRR from solvent (Di Matteo et al., 2003). Biochemical analysis of an Escherichia coli–expressed tomato (Solanum lycopersicum) Leu-rich protein with five LRRs (Kolade et al., 2006) confirmed the formation of a disulfide linkage between the two conserved Cys residues. Based on sequence similarity and molecular modeling (see Supplemental Figure 6 online), we hypothesize that BRI1's N-terminal cap domain adopts a similar structure to that of PGIP, with a long α-helix of 11 amino acids (EIHQLISFKDV) and a short β-sheet (VTC). The disulfide linkage and the stacking of the two aromatic amino acids are thought to stabilize the N-terminal cap structure so that the conserved hydrophobic amino acids of the long α-helix are able to cap the hydrophobic core of the first LRR (van der Hoorn et al., 2005). Elimination of this linkage would significantly affect the packing of the helix with the first LRR, thus exposing its hydrophobic amino acids and those of the first LRR, a structural recognition feature for both UGGT and BiP. This hypothesis might also explain the fact that bri1-5 has stronger interactions with CNXs and BiPs than does bri1-9.
A previous study investigating the structural–functional relationship of the tomato disease resistance protein Cf9 concluded that mutations of the conserved Cys pair significantly reduced Cf9 activity (van der Hoorn et al., 2005). Given what was discovered in this study, it is quite possible that mutations of the two conserved Cys residues might simply lead to ER retention of Cf9, thus reducing the biological activity that requires its cell surface presence.
The Role of BiP in Retaining bri1-5
Despite the fact that bri1-5 interacts with CNX in a monoglucosylation-dependent manner, neither loss-of-function mutation of UGGT nor simultaneous elimination of two Arabidopsis CNX genes was able to suppress the bri1-5 dwarf phenotype or allow some bri1-5 proteins to exit the folding compartment. This is in sharp contrast to bri1-9, which can be rescued by loss-of-function mutations in UGGT (Jin et al., 2007). This is likely due to the fact that there are at least two other mechanisms that retain bri1-5 in the ER: the BiP chaperone system and the thiol-mediated ER retention system. Our results have shown that BiP interacts much more strongly with bri1-5 than with bri1-9, likely caused by a larger hydrophobic surface on bri1-5 than on bri1-9. Interestingly, despite the fact that bri1-5 is retained in the ER by at least three mechanisms, RNA interference–mediated gene silencing of BiP expression was able to partially suppress the bri1-5 phenotype and confer BR responsiveness to the BR receptor mutant. This result suggests that BiP might play a major role in retaining bri1-5 compared with two other mechanisms. This conclusion is consistent with the severe dwarf phenotype of the bri1-5 ebs1 double mutants. The upregulation of BiPs caused by the loss-of-UGGT mutations not only compensates for the UGGT-based CRT/CNX retention system but also traps more bri1-5 in the ER. Alternatively, the partial suppression of the bri1-5 phenotype might be caused by inhibition of ERAD, which is known to involve BiP (Knittler et al., 1995). As a result, the three retention mechanisms are saturated, allowing some bri1-5 proteins to exit the ER.
A Thiol-Mediated Retention System in Arabidopsis
In this study, we also presented strong circumstantial evidence for the involvement of a thiol-mediated ER retention mechanism in keeping a structurally defective yet functionally competent glycoprotein in the ER. Among the three ER retention mechanisms, the thiol-mediated ER retention system is the least studied. Although the phenomenon was first discovered in 1990 when studying the assembly and secretion of the IgM complex (Alberini et al., 1990; Sitia et al., 1990), almost nothing was known about the proteins involved in this process until 2003, when Anelli et al. (2003) identified an ER resident protein, ERp44, as a key retention factor for two ER resident oxidoreductases, Ero1α and Ero1β, as well as several unassembled IgM subunits, such as μ, L, and J chains. A recent study has also provided strong evidence for the involvement of ERp44, along with Ero1α, in regulating the secretion of an adipocyte-specific secretory protein adiponectin (Wang et al., 2007).
The Arabidopsis genome encodes two Ero1 homologs (Dixon et al., 2003) but lacks an ERp44 homolog. Using a complementation assay, we have shown that bri1-5C62Y exhibits a stronger bri1-5–rescuing activity than either bri1-5 or bri1C62Y. Previous experiments showed that treatment with β-mercaptoethanol was able to inhibit the thiol-mediated retention mechanism in mammalian cells and significantly enhanced secretion of its client proteins (Anelli et al., 2003). However, the majority of the bri1-5C62Y proteins are still in the ER. This is consistent with our discovery that bri1-5 is retained in the ER by several independent mechanisms. The thiol-mediated retention system might also be responsible for the enhanced dwarf phenotype of the bri1-5 ebs1 mutant. The loss of UGGT functions in the ebs1 mutants results in the UPR upregulating the expression of many known ER chaperones and folding catalysts that likely include components of the thiol-mediated bri1-5 retention machinery. A biochemical approach, similar to that of ERp44 isolation (Anelli et al., 2003), is needed to identify an Arabidopsis ERp44 functional homolog.
bri1-5 Is Degraded by a Proteasome-Independent ERAD Process
Our results also yielded another unexpected discovery. In contrast with the general belief (Romisch, 2005) and results of a recent study using Arabidopsis protoplasts and the barley (Hordeum vulgare) powdery mildew resistance o protein (Muller et al., 2005) that terminally misfolded proteins are retrotranslocated back into cytosol, where they are degraded by the ubiquitin/proteasome-mediated process, we found that bri1-5 is degraded by a proteasome-independent process. While Kif treatment significantly increased the bri1-5 abundance, little effect on bri1-5 stability was detected when the bri1-5 mutants were treated with MG132, which can block the degradation of the wild-type BRI1 proteins. It remains to be investigated what mechanism is responsible for degrading the ER-accumulated bri1-5. Studies in mammalian and yeast cells have shown that there are several proteasome-independent ERAD processes, such as protease-mediated degradation in the ER/cytosol and lysosomal/autophagic degradation pathway (Schmitz and Herzog, 2004). Previous investigations using artificial substrates suggested that plant cells might also use the lytic vacuoles to degrade ER resident proteins such as BiP and artificial ERAD substrates (Pedrazzini et al., 1997; Brandizzi et al., 2003; Tamura et al., 2004; Pimpl et al., 2006). Since inhibition of bri1-5 ERAD results in a significant phenotypic suppression, screening for second-site mutations or chemical compounds that restore the wild-type morphology to the BR-insensitive dwarf mutant could lead to elucidation of the biochemical mechanism by which the mutated BR receptor is degraded.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana bri1-5 (ecotype Wassilewskija-2) was previously described (Noguchi et al., 1999). ebs1-1, ebs1-2, and ebs1-3 (Jin et al., 2007) were used to generate the bri1-5 ebs1 double mutants. T-DNA insertional mutants (cnx1 [SALK_083600] and cnx2 [SALK_044381]) for both Arabidopsis CNXs were discovered by searching the SIGNAL database (Alonso et al., 2003) and obtained from the ABRC at Ohio State University. The mutants were crossed into bri1-5 to obtain bri1-5 cnx1, bri1-5 cnx2, and bri1-5 cnx1 cnx2 mutants. Seeds of the tilling project–generated bri1 mutants, including CS85498, CS86765, CS86239, CS92187, CS92235, CS92315, CS92322, CS94063, and CS94126, were also obtained from the ABRC. Unless stated otherwise, all transgenic plants generated in this study are in Columbia-0 ecotype background. The seed germination and plant growth conditions were described previously (Li et al., 2001).
Plasmid Constructs and Plant Transformation
The constructs pBRI1:bri1-9-GFP, pBRI1:bri1-5-GFP, pBRI1:bri1C62Y-GFP, and pBRI1:bri1-5C62Y-GFP were generated from pPZP212-BRI1:BRI1-GFP (Friedrichsen et al., 2000) by site-directed mutagenesis using the Stratagene QuickChange II XL site-directed mutagenesis kit. The primers used for site-directed mutagenesis are as follows: bri1-5 (forward, 5′-CTTTCGATGGCGTTACTTACAGAGACGACAAAGTTAC-3′, and reverse, 5′-GTAACTTTGTCGTCTCTGTAAGTAACGCCATCGAAAG-3′); bri1-9 (forward, 5′-CGATGATGTTTCTGGACATGTTTTACAACATGTTGTCTGG-3′, and reverse, 5′-CCAGACAACATGTTGTAAAACATGTCCAGAAACATCATCG-3′), bri1C62Y (forward, 5′-GGTCTTCCAACAAAAACCCGTATACTTTCGATGGCGTTAC-3′, and reverse, 5′-GTAACGCCATCGAAAGTATACGGGTTTTTGTTGGAAGACC-3′). The bri1-5C62Y double mutant was created by introducing the C62Y mutation into the bri1-5–mutated construct with the bri1C62Y primer set. The BRI1-GFP and bri1-5-GFP fusion fragments were removed and cloned into pCHF3 vector (Fankhauser et al., 1999) to create p35S:BRI1-GFP and p35S:bri1-5-GFP constructs, respectively. For the BiPRNAi construct, a 900-bp PCR product from BiP1 (At5g28540) was cloned into a generic vector pHANNIBLE (Wesley et al., 2001) at KpnI and XhoI sites in the antisense direction and at ClaI and XbaI sites in the sense direction using the primer set 5′-gcggtaccagATCGATgagattgtc-3′ and 5′-gcctcgagacaTCTAGAgctcatc-3′ (the underlined sequences are restriction sites for KpnI and XhoI, respectively, while the uppercase sequences are restriction sites for ClaI and XbaI sites, respectively). The resulting BiPRNAi fragment was then cloned into pART27 vector at the NotI site (Gleave, 1992). All transgenes were transformed into wild-type or bri1-5 mutants via the Agrobacterium tumefaciens–mediated floral dipping method (Clough and Bent, 1998).
Treatment of Arabidopsis Seedlings with Chemicals
Young growing rosette leaves from 4-week-old soil-grown plants were removed and incubated at 22°C for 24 h in liquid half-strength Murashige and Skoog (MS) medium containing different concentrations of BL, MG132 {N-[(phenylmethoxy)carbonyl]-l-leucyl-N-[(1S)-1-formyl-3-methylbutyl]-l-leucinamide; Sigma-Aldrich}, CST, and Kif (Toronto Research Chemicals). To test if certain chemicals were able to suppress the bri1-5 phenotype, 2-week-old bri1-5 mutants or control plants grown on half-strength MS medium were transferred to fresh half-strength MS medium containing different concentrations of Kif for continued growth under the same growth conditions.
Protein Extraction, Protein Gel Blot, Gel Staining, and Coimmunoprecipitation
The seedlings grown on half-strength MS medium for 3 weeks were collected and ground in liquid N2. Preparation of protein crude extracts, immunoprecipitation, and protein gel blot assay were described previously (Jin et al., 2007). The protein gels were stained with 0.05% (w/v) Coomassie Brilliant Blue R 250 (Fisher Scientific) in a staining solution containing 50% (v/v) methanol and 10% (v/v) acetic acid followed by overnight washing with a destaining solution containing 5% (v/v) methanol and 7% (v/v) acetic acid. The seedlings of pBRI1:bri1-5-GFP/ebs1-1 used for the bri1-5-CNX or bri1-5-BiP coimmunoprecipitation experiments were obtained from the cross between the pBRI1:bri1-5-GFP transgenic line and ebs1-1 (Jin et al., 2007).
Endo H Treatment
The leaf tissues from 4-week-old soil-grown adult plants were extracted with 2× SDS sample buffer. After boiling for 5 min, the leaf samples were centrifuged for 10 min at 10,000g. The resulting supernatant was transferred into a new Eppendorf tube for Endo H digestion following the manufacturer's recommended protocol (New England Biolabs). Both nontreated and Endo H–treated samples were then analyzed by protein gel blot with either anti-BRI1 or anti-GFP antibodies.
Transformation of Tobacco Leaves and Confocal Microscopy
Leaves of 6-week-old tobacco (Nicotiana benthamiana) plants were used for transient expression of p35S:BRI1-GFP and p35S:bri1-5-GFP via Agrobacterium-mediated infiltration (Lee and Yang, 2006). Forty-eight hours after infiltration, the leaf tissues were viewed directly with a Leica TCS-SP5 confocal microscope (Leica Microsystems) to examine the localization patterns of BRI1-GFP and bri1-5-GFP. GFP and RFP were excited using 488- and 543-nm laser light, respectively. Images were acquired with a 0.5-μm Z step at a resolution of 512 × 512 pixels using a ×63/1.30 glycerin-immersion objective and analyzed by Leica LAS AF software (version 1.8.2).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BRI1, NM120100; EBS1, NM_105791; CNX1, NM_125573.3; CNX2, NM_120816.2; and BIP1, NM_122737.3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Summary of the Endo H–Based Survey of 16 bri1 Alleles Carrying Extracellular Mutations.
Supplemental Figure 2. Kif Treatment Rescues the Deetiolation Phenotype of bri1-5.
Supplemental Figure 3. The Effects of cnx1 and cnx2 Mutations on Plant Growth, Gene Expression, and the bri1-5 Mutation.
Supplemental Figure 4. Quantitative Analysis of Leaf Shape and Inflorescence Stem of pBRI1:bri1-GFP/bri1-5 Transgenic Lines.
Supplemental Figure 5. bri1-5C62Y Might Be More Active Than the Wild-Type BRI1.
Supplemental Figure 6. Model of the N-Terminal Half of BRI1's Extracellular Domain.
Supplementary Material
Acknowledgments
We thank the ABRC for providing seeds of various Arabidopsis mutants and the BiP1 cDNA clone, F. Tax for the gift of bri1-5 seeds, R. Boston for the anti-CRT antibody, Y. Yin for the anti-BES1 antiserum, Y. Guo and G. Ren for technical help, and members of the Li laboratory for discussion. This work was supported in part by grants from the National Institutes of Health (Grant GM-60519) and the Department of Energy (Grant ER15672) to J.L.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Zhi Hong (hzhi@umich.edu) and Jianming Li (jian@umich.edu).
Online version contains Web-only data.
References
- Alberini, C.M., Bet, P., Milstein, C., and Sitia, R. (1990). Secretion of immunoglobulin M assembly intermediates in the presence of reducing agents. Nature 347 485–487. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anelli, T., Alessio, M., Bachi, A., Bergamelli, L., Bertoli, G., Camerini, S., Mezghrani, A., Ruffato, E., Simmen, T., and Sitia, R. (2003). Thiol-mediated protein retention in the endoplasmic reticulum: The role of ERp44. EMBO J. 22 5015–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli, T., Ceppi, S., Bergamelli, L., Cortini, M., Masciarelli, S., Valetti, C., and Sitia, R. (2007). Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 26 4177–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, M.Y., Zhang, L.Y., Gampala, S.S., Zhu, S.W., Song, W.Y., Chong, K., and Wang, Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond-Elguindi, S., Cwirla, S.E., Dower, W.J., Lipshutz, R.J., Sprang, S.R., Sambrook, J.F., and Gething, M.J. (1993). Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75 717–728. [DOI] [PubMed] [Google Scholar]
- Boisson, M., Gomord, V., Audran, C., Berger, N., Dubreucq, B., Granier, F., Lerouge, P., Faye, L., Caboche, M., and Lepiniec, L. (2001). Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce, J.M., Coates, D., Fricker, M.D., and Evans, D.E. (1994). Genomic sequence of a calnexin homolog from Arabidopsis thaliana. Plant Physiol. 106 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi, F., Hanton, S., DaSilva, L.L., Boevink, P., Evans, D., Oparka, K., Denecke, J., and Hawes, C. (2003). ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 34 269–281. [DOI] [PubMed] [Google Scholar]
- Buck, T.M., Wright, C.M., and Brodsky, J.L. (2007). The activities and function of molecular chaperones in the endoplasmic reticulum. Semin. Cell Dev. Biol. 18 751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn, J.E., Hurley, U.A., Birch, R.J., Arioli, T., Cork, A., and Williamson, R.E. (2002). The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32 949–960. [DOI] [PubMed] [Google Scholar]
- Caramelo, J.J., and Parodi, A.J. (2007). How sugars convey information on protein conformation in the endoplasmic reticulum. Semin. Cell Dev. Biol. 18 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty, R., Banerjee, R., Chung, S.M., Farman, M., Citovsky, V., Hogenhout, S.A., Tzfira, T., and Goodin, M. (2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: Probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20 740–750. [DOI] [PubMed] [Google Scholar]
- Choe, J., Kelker, M.S., and Wilson, I.A. (2005). Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 309 581–585. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., Langford, M., and McMorris, T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilczyk, U.G., and Williams, D.B. (2001). The lectin chaperone calnexin utilizes polypeptide-based interactions to associate with many of its substrates in vivo. J. Biol. Chem. 276 25532–25540. [DOI] [PubMed] [Google Scholar]
- Di Cola, A., Frigerio, L., Lord, J.M., Ceriotti, A., and Roberts, L.M. (2001). Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. USA 98 14726–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cola, A., Frigerio, L., Lord, J.M., Roberts, L.M., and Ceriotti, A. (2005). Endoplasmic reticulum-associated degradation of ricin A chain has unique and plant-specific features. Plant Physiol. 137 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo, A., Federici, L., Mattei, B., Salvi, G., Johnson, K.A., Savino, C., De Lorenzo, G., Tsernoglou, D., and Cervone, F. (2003). The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc. Natl. Acad. Sci. USA 100 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, D.P., Van Lith, M., Edwards, R., and Benham, A. (2003). Cloning and initial characterization of the Arabidopsis thaliana endoplasmic reticulum oxidoreductins. Antioxid. Redox Signal. 5 389–396. [DOI] [PubMed] [Google Scholar]
- Ellgaard, L., and Helenius, A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4 181–191. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284 1539–1541. [DOI] [PubMed] [Google Scholar]
- Flynn, G.C., Pohl, J., Flocco, M.T., and Rothman, J.E. (1991). Peptide-binding specificity of the molecular chaperone BiP. Nature 353 726–730. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Joazeiro, C.A., Li, J., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala, S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor, C.S., Poindexter, P., Lorieau, J., Palcic, M.M., and Somerville, C. (2002). Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- He, J.X., Gendron, J.M., Yang, Y., Li, J., and Wang, Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston, N.L., Fan, C., Xiang, J.Q., Schulze, J.M., Jung, R., and Boston, R.S. (2005). Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 137 762–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., Franklin, A.E., and Hoffman, N.E. (1993). Primary structure and characterization of an Arabidopsis thaliana calnexin-like protein. J. Biol. Chem. 268 6560–6566. [PubMed] [Google Scholar]
- Jin, H., Yan, Z., Nam, K.H., and Li, J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Cano-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., and Chory, J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433 167–171. [DOI] [PubMed] [Google Scholar]
- Kirst, M.E., Meyer, D.J., Gibbon, B.C., Jung, R., and Boston, R.S. (2005). Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol. 138 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittler, M.R., Dirks, S., and Haas, I.G. (1995). Molecular chaperones involved in protein degradation in the endoplasmic reticulum: Quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92 1764–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolade, O.O., Bamford, V.A., Ancillo Anton, G., Jones, J.D., Vera, P., and Hemmings, A.M. (2006). In vitro characterization of the cysteine-rich capping domains in a plant leucine rich repeat protein. Biochim. Biophys. Acta 1764 1043–1053. [DOI] [PubMed] [Google Scholar]
- Lee, M.W., and Yang, Y. (2006). Transient expression assay by agroinfiltration of leaves. Methods Mol. Biol. 323 225–229. [DOI] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., and Nam, K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li, J., Nam, K.H., Vafeados, D., and Chory, J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wen, J., Lease, K.A., Doke, J.T., Tax, F.E., and Walker, J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 213–222. [DOI] [PubMed] [Google Scholar]
- McCracken, A.A., and Brodsky, J.L. (1996). Assembly of ER-associated protein degradation in vitro: Dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Garcia, S., Vert, G., Yin, Y., Cano-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, J., Piffanelli, P., Devoto, A., Miklis, M., Elliott, C., Ortmann, B., Schulze-Lefert, P., and Panstruga, R. (2005). Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212. [DOI] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K.A., and Tax, F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi, A.J. (2000). Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem. J. 348 1–13. [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini, E., Giovinazzo, G., Bielli, A., de Virgilio, M., Frigerio, L., Pesca, M., Faoro, F., Bollini, R., Ceriotti, A., and Vitale, A. (1997). Protein quality control along the route to the plant vacuole. Plant Cell 9 1869–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, S., Rosenquist, M., Svensson, K., Galvao, R., Boss, W.F., and Sommarin, M. (2003). Phylogenetic analyses and expression studies reveal two distinct groups of calreticulin isoforms in higher plants. Plant Physiol. 133 1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl, P., Taylor, J.P., Snowden, C., Hillmer, S., Robinson, D.G., and Denecke, J. (2006). Golgi-mediated vacuolar sorting of the endoplasmic reticulum chaperone BiP may play an active role in quality control within the secretory pathway. Plant Cell 18 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, P., Sparvoli, A., Fagioli, C., Fassina, G., and Sitia, R. (1996). Formation of reversible disulfide bonds with the protein matrix of the endoplasmic reticulum correlates with the retention of unassembled Ig light chains. EMBO J. 15 2077–2085. [PMC free article] [PubMed] [Google Scholar]
- Romisch, K. (2005). Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 21 435–456. [DOI] [PubMed] [Google Scholar]
- Ryu, H., Kim, K., Cho, H., Park, J., Choe, S., and Hwang, I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasak, V.W., Ordovas, J.M., Elbein, A.D., and Berninger, R.W. (1985). Castanospermine inhibits glucosidase I and glycoprotein secretion in human hepatoma cells. Biochem. J. 232 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, A., and Herzog, V. (2004). Endoplasmic reticulum-associated degradation: Exceptions to the rule. Eur. J. Cell Biol. 83 501–509. [DOI] [PubMed] [Google Scholar]
- Sheppard, D.N., and Welsh, M.J. (1999). Structure and function of the CFTR chloride channel. Physiol. Rev. 79 S23–S45. [DOI] [PubMed] [Google Scholar]
- Sitia, R., and Braakman, I. (2003). Quality control in the endoplasmic reticulum protein factory. Nature 426 891–894. [DOI] [PubMed] [Google Scholar]
- Sitia, R., Neuberger, M., Alberini, C., Bet, P., Fra, A., Valetti, C., Williams, G., and Milstein, C. (1990). Developmental regulation of IgM secretion: The role of the carboxy-terminal cysteine. Cell 60 781–790. [DOI] [PubMed] [Google Scholar]
- Sung, D.Y., Vierling, E., and Guy, C.L. (2001). Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 126 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Yamada, K., Shimada, T., and Hara-Nishimura, I. (2004). Endoplasmic reticulum-resident proteins are constitutively transported to vacuoles for degradation. Plant J. 39 393–402. [DOI] [PubMed] [Google Scholar]
- Tokunaga, F., Brostrom, C., Koide, T., and Arvan, P. (2000). Endoplasmic reticulum (ER)-associated degradation of misfolded N-linked glycoproteins is suppressed upon inhibition of ER mannosidase I. J. Biol. Chem. 275 40757–40764. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A., Wulff, B.B., Rivas, S., Durrant, M.C., van der Ploeg, A., de Wit, P.J., and Jones, J.D. (2005). Structure-function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17 1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert, G., and Chory, J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441 96–100. [DOI] [PubMed] [Google Scholar]
- Vert, G., Nemhauser, J.L., Geldner, N., Hong, F., and Chory, J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21 177–201. [DOI] [PubMed] [Google Scholar]
- Wang, X., and Chory, J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313 1118–1122. [DOI] [PubMed] [Google Scholar]
- Wang, X., Goshe, M.B., Soderblom, E.J., Phinney, B.S., Kuchar, J.A., Li, J., Asami, T., Yoshida, S., Huber, S.C., and Clouse, S.D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17 1685–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., Yang, Y., Fujioka, S., Yoshida, S., Asami, T., and Chory, J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2 505–513. [DOI] [PubMed] [Google Scholar]
- Wang, Z.V., Schraw, T.D., Kim, J.Y., Khan, T., Rajala, M.W., Follenzi, A., and Scherer, P.E. (2007). Secretion of the adipocyte-specific secretory protein adiponectin critically depends on thiol-mediated protein retention. Mol. Cell. Biol. 27 3716–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410 380–383. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Williams, D.B. (2006). Beyond lectins: The calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci. 119 615–623. [DOI] [PubMed] [Google Scholar]
- Yin, Y., Wang, Z.Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., and Chory, J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109 181–191. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Peng, P., Schmitz, R.J., Decker, A.D., Tax, F.E., and Li, J. (2002). Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 130 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.