Summary
Eukaryotic cytochrome c oxidase (COX), the last enzyme of the mitochondrial respiratory chain, is a multimeric enzyme of dual genetic origin, whose assembly is a complicated and highly regulated process. COX displays a concerted accumulation of its constitutive subunits. Data obtained from studies performed with yeast mutants indicate that most catalytic core unassembled subunits are post-translationally degraded. Recent data obtained in the yeast Saccharomyces cerevisiae have revealed another contribution to the stoichiometric accumulation of subunits during COX biogenesis targeting subunit 1 or Cox1p. Cox1p is a mitochondrially encoded catalytic subunit of COX which acts as a seed around which the full complex is assembled. A regulatory mechanism exists by which Cox1p synthesis is controlled by the availability of its assembly partners. The unique properties of this regulatory mechanism offer a means to catalyze multiple-subunit assembly. New levels of COX biogenesis regulation have been recently proposed. For example, COX assembly and stability of the fully assembled enzyme depend on the presence in the mitochondrial compartments of two partners of the oxidative phosphorylation system, the mobile electron carrier cytochrome c and the mitochondrial ATPase. The different mechanisms of regulation of COX assembly are reviewed and discussed.
Keywords: Mitochondria, cytochrome oxidase, cytochrome c, F1F0-ATPase, Cox1p translational regulation
1. Cytochrome C Oxidase Assembly is a Highly Regulated Process
Cytochrome c oxidase (COX) or complex IV of the mitochondrial respiratory chain plays a fundamental role in energy production of aerobic cells. This multimeric enzyme of the inner mitochondrial membrane catalyzes the last step of respiration, the transfer of electrons from cytochrome c to molecular oxygen. By coupling electron transfer with protons translocation from the mitochondrial matrix to the intermembrane space, COX also contributes to the storage of energy in the form of an electrochemical gradient that will be used by the oxidative phosphorylation system for the synthesis of ATP. COX central role in aerobic metabolism is highlighted by its participation in respiratory control. When phosphorylated in a cAMP-dependent fashion, COX is converted into the rate limiting step of respiration, being inhibited at high intramitochondrial ATP/ADP ratio (reviewed in (1)).
Eukaryotic COX is formed by 11-13 subunits (11 in the yeast Saccharomyces cerevisiae and 13 in Homo sapiens) of dual genetic origin. Subunits 1, 2 and 3 are large, highly hydrophobic, transmembrane proteins encoded in the mitochondrial genome. They form the catalytic core of the enzyme and contain metal prosthetic groups. Copper and heme A, a unique heme compound found exclusively in COX, form three redox centers: a CuA center in subunit 2 and a heme a and a CuB-heme a3 binuclear center in subunit 1. The nomenclature of the subunits varies from yeast to mammals and it is clarified in Table 1. The remaining small subunits that surround the core of the enzyme are encoded in the nuclear genome. They are necessary for the assembly/stability of the holo-enzyme and for its dimerization (Table 1). They are also involved in the modulation of the catalytic activity and in the protection of the core from reactive oxygen species (ROS). Some of the nuclear encoded subunits have evolved isoforms able to confer different kinetic properties to the enzyme. For example, subunit Cox5p in the yeast S. cerevisiae, exists in two isoforms, Cox5ap and Cox5bp, which are expressed according to the oxygen availability. In humans, tissue specific isoforms have been reported for four nuclear encoded subunits, COX4 (homolog of yeast Cox5p), COX6a, COX6b and COX7a (2, 3).
Table 1.
Homologue COX subunits in yeast and mammals.
| YEAST | MAMMALS | REQUIREMENT FOR COX* | |||
|---|---|---|---|---|---|
| GENE | PROTEIN | GENE | PROTEIN | ASSEMBLY | ACTIVITY |
| Catalytic core (mtDNA encoded subunits) | |||||
| COX1 | Cox1p | MTCOXI | COX1 | + | + |
| COX2 | Cox2p | MTCOXII | COX2 | + | + |
| COX3 | Cox3p | MTCOXIII | COX3 | + | + |
| Core protective shield (nDNA encoded subunits) | |||||
| COX4 | Cox4p | COXVb | COX5b | + | + |
| COX5 | Cox5p | COXIV | COX4 | + | + |
| COX6 | Cox6p | COXVa | COX5a | + | + |
| COX7 | Cox7p | COXVIIa | COX7a | + | + |
| COX9 | Cox7ap | COXVIc | COX6c | + | + |
| COX8 | Cox8p | COXVIIc | COX7c | + | + |
| COX12 | Cox9p | COXVIb | COX6b | - | - |
| COX13 | Cox10p | COXVIa | COX6a | - | - |
| / | / | COXVIIb | COX7b | + | + |
| / | / | COXVIII | COX8 | + | + |
Catalytic activity or assembly of the yeast enzyme.
Overall, COX is a ductile enzyme, whose activity can be modulated according to the energetic requirement of the cell through isoform subunits composition and physiologically controlled phosphorylation. Although COX structure was resolved in 1996 (4) and an impressive amount of experimental data have shed light into the organization and catalytic properties of the enzyme, many questions of fundamental bioenergetic interest still remain uncovered (reviewed in (5)). COX assembly is a fascinating process, which can be described as a sequential and ordinate addition of subunits and co-factors to an initial seed consisting of Cox1p. COX assembly represents a multistep progression through discrete short-term intermediates requiring a plethora of more than 30 diverse assistant factors, a complex coordinated event that we have just begun to elucidate. In addition to its biological significance, the increasing interest to disclose the process of COX assembly derives from biomedical statistics showing that COX deficiency is the most frequent cause of mitochondrial encephalomyopathies, a heterogeneous group of human disorders characterized by alteration of aerobic energy production (OXPHOS defects). Notably, all the cases of Mendelian inherited COX deficiencies, for which the genetic cause is already known, are the consequence of COX assembly defects. These deficiencies are associated with mutations in nuclear genes encoding COX assembly factors (SCO1 and SCO2, necessary for copper insertion into COX2; COX10 and COX15, necessary for heme a biosynthesis; SURF1, involved in an early step of assembly and LRP130, homologue of the yeast COX1 translational activator Pet309p) (reviewed in (6, 7)). The non-structural factors accomplish diverse functions at all the levels of the assembly process. These roles include transcription and mRNA maturation, translation of COX mitochondrial genes, as well as import into mitochondria of nuclear encoded subunits and insertion of transmembrane subunits into the inner mitochondrial membrane. Essential additional roles involve heme A biosynthesis, copper homeostasis and insertion into the apoenzyme and formation of assembly intermediates. All these functions are interconnected and co-regulated (reviewed in (8)). Furthermore, an additional level of regulation has begun to emerge in the last few years, which connects COX assembly with other players of the OXPHOS system, like cytochrome c and ATP synthase (Fig. 1).
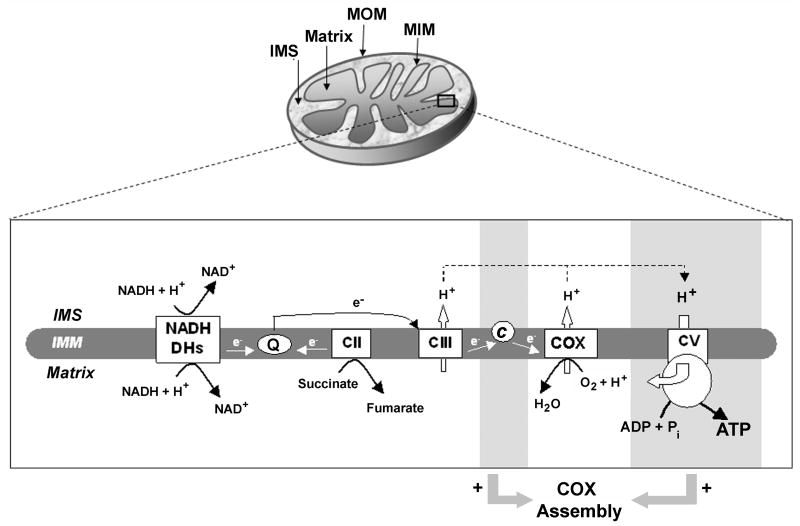
Figure 1.
Cytochrome c oxidase assembly depends on the presence in mitochondria of cytochrome c and fully assembled ATPase in the yeast Saccharomyces cerevisiae. (A) Schematic representation of mitochondria, the mitochondrial respiratory chain and the oxidative phosphorylation system. Mitochondrial ultrastructural analysis reveals a double membrane, outer (MOM) and inner (MIM), which delimit two compartments, the intermembrane space (IMS) and the matrix. The inner membrane form invaginations called cristae, where the enzymes forming the respiratory chain and OXPHOS system are located. The respiratory chain (MRC) is formed in yeast by a series of NADH dehydrogenases acting as the complex I of higher eukaryotes, complex II or succinate dehydrogenase, complex III or bc1 complex, and complex IV or cytochrome c oxidase (COX), together with the electron carriers ubiquinone (Q) and cytochrome c. All the elements forming the MRC act in concert to transfer electrons (e-) from reducing equivalents (NADH and FADH) to molecular oxygen, in a process coupled to the formation of a proton gradient in the intermembrane space that is used by the ATPase to drive the synthesis of ATP. The assembly of the different elements of the MRC and OXPHOS system is interconnected. For example, as discussed in the text, the presence of both cytochrome c and fully assembled ATPase (both marked with grey boxes), is required (+) for COX assembly. They affect COX assembly through mechanisms that remain to be defined.
The aim of this review is to summarize the new emerging data regarding COX assembly, specifically concerning the several levels of its regulation.
2. Concerted Accumulation of Cox Subunits and Formation of Subassembly Intermediates
2.1 Regulation of Cox1p translation
COX assembly is characterized by a concerted accumulation of its constitutive subunits. Data obtained mainly from studies performed with yeast mutants indicate that most unassembled subunits, particularly the ones forming the catalytic core, are post translationally degraded (9, 10). In particular, non-matured or unassembled newly synthesized subunit 1 and the other catalytic core subunits that do not proceed to COX holoenzyme assembly are targeted to degradation by the AAA ATP-dependent proteases of the inner mitochondrial membrane (reviewed in (11)). Active degradation will avoid the accumulation of unassembled proteins that could have a tendency to aggregate and disturb membrane homeostasis. It will also limit the accumulation of partially matured core subunits that, as recently proposed for yeast subunit 1, could contribute to the production of unstable pro-oxidant intermediates (12).
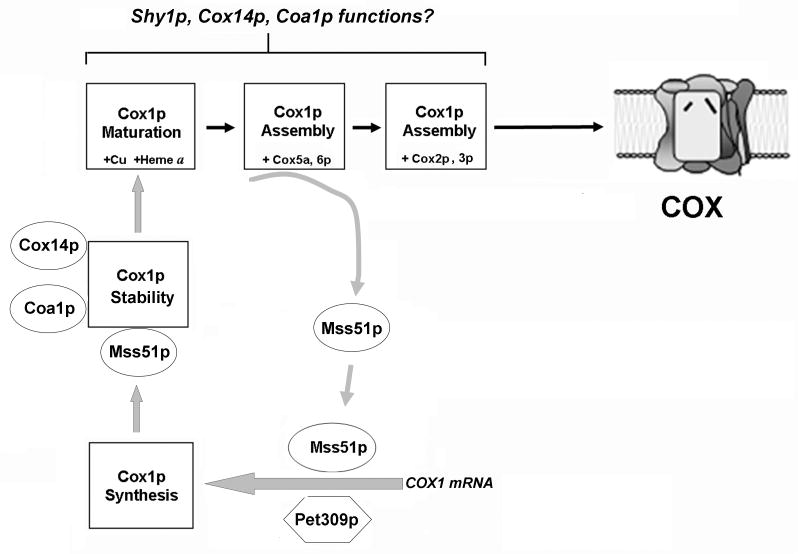
Recently, another contribution to the stoichiometric accumulation of subunits during COX biogenesis in the yeast Saccharomyces cerevisiae has been reported. It consists of a regulatory mechanism by which the synthesis of subunit 1 is regulated by the availability of its assembly partners (13-16). Decreased Cox1p synthesis relative to other mitochondrial translation products had been previously reported as a secondary observation in yeast pet111 mutants that have a lesion in a translational activator of COX2 mRNA (17) and in cox7 mutants, lacking the nuclear-encoded subunit 7 (18). This phenotype could not be accounted for by a defect in the translation system because the mutations were not in proteins related to this mitochondrial activity. Translation of COX1 in S. cerevisiae is under the control of the MSS51 and PET309 gene products (Fig.2) that are also involved in maturation of the COX1 mRNAs (19, 20). A direct connection between Cox1p expression regulation and COX holoenzyme assembly came from studies in which mutations in MSS51 or overexpression of the wild type gene were found able to suppress the COX assembly defect of null mutant of shy1 by enhancing synthesis of Cox1p (13). Shy1p is a COX assembly factor that functions in maturation and/or assembly of Cox1p (13, 15, 21, 22). The precise role of Shy1p, still unknown, is of considerable interest because mutations in its human homologue, SURF1, are responsible for most diagnosed cases of Leigh's syndrome presenting a COX deficiency (23, 24). Cox1p synthesis is decreased in most COX assembly mutants, including shy1 mutants, but it is restored to normal levels by mss51 suppressors of shy1 or by mutations in COX14 (14), which codes for another COX assembly factor (25). Like other translational activators, Mss51p acts on the 5′-UTR of its cognate mRNA molecule, to promote translation (15, 16), binding that could be necessary for optimal initiation of translation by Pet309p (16). However, mss51 mutants, unlike pet309 mutants, can not be suppressed by changes in the 5′-UTR of COX1 mRNA (14, 15). In addition, Mss51p acts on a target in the protein coding sequence of COX1 mRNA that could be necessary to promote elongation (15). Mss51p and newly synthesized Cox1p form a transient complex (14, 15) that is stabilized by Cox14p (14). These interactions have been postulated to down-regulate Cox1p synthesis when COX assembly is impaired (14). According to this model, the release of Mss51p from the ternary complex and its availability for Cox1p synthesis occur at a downstream step in the assembly pathway, most likely catalyzed by Shy1p (14, 26) (Fig. 2).
Figure 2.
Early steps in COX biogenesis in the yeast Saccharomyces cerevisiae connect synthesis, maturation and assembly of Cox1p through a regulatory loop involving the translational factor Mss51p and other proteins required for COX assembly. See explanation in the text.
Despite the already large number of factors known to be involved in COX assembly, new factors continue to be discovered today. Coa1p was recently described as a new player in the subunit 1 synthesis regulatory loop (26, 27) (Fig.2). Coa1p is a protein associated with the inner mitochondrial membrane where it is part of the high molecular weight complex containing Cox14p, Mss51p and newly synthesized Cox1p (26, 27). The interaction Cox14p-Mss51p is unaffected in a Δcoa1 mutant, while, in the absence of Cox14p, the binding of Coa1p-Mss51p is disrupted, suggesting that the interaction of Coa1p with the Cox1p-Cox14p-Mss51p complex is through Cox14p (27). Shy1p is not part of the Cox1p-Cox14p-Mss51p-Coa1p complex (27), but it has been shown to co-precipitate with Coa1p (26, 27), suggesting that these two factors probably interact once Mss51p has disengaged from the complex (27). As for cox14 mutants, mutations in coa1 do not affect the synthesis but the accumulation of Cox1p in the holoenzyme, suggesting that Coa1p also plays a role in the feedback regulation of Cox1p expression (26, 27). The COX assembly defect in a strain carrying a null allele of coa1 can be suppressed by overexpression of Mss51p and Cox10p, the heme a farnesyl transferase required for biosynthesis of heme a (28). Both suppressors can act synergistically when the two proteins are co-expressed, linking Coa1p to both translational regulation and maturation of Cox1p (27). Pierrel and colleagues have speculated that Coa1p could stabilize the Cox1p-Cox14p-Mss51p complex until Shy1p interacts with Coa1p in a step involving heme A insertion into Cox1p and further progression in the assembly process (27). While Cox1p maturation certainly occurs within these initial Cox1p complexes, the precise role of each factor remains to be elucidated.
At present, it is not clear if Cox1p translation in other organisms is also subject to regulation by downstream events. In mammals, as well as in yeast, mitochondrial DNA encoded subunits are fast degraded when not assembled into the holoenzyme. However, no extensive data are available regarding the rate of Cox1p synthesis in mammalian COX assembly mutants. Among the few cases reported, COX subunit 1 synthesis was shown slightly reduced by 20% of wild type levels in fibroblasts from a cox10 knockout mouse (29). Mitochondrial genes in higher eukaryotes do not have 5′untranslated region and their expression does not seem to depend on gene-specific translational activators. In addition, mammalian homologues of Mss51p, Cox14p and Coa1p have not been identified to date. This does not necessarily exclude the possibility that they may exist in mammalian and other genomes but have not yet been recognized because of their smaller size and/or divergent sequences. For example, the products of yeast PET309 and human LRPPRC (LRP130), responsible for the Canadian form of Leigh syndrome (30), display very weak sequence similarity, even though they both bind to mitochondrial RNAs and are essential for COX expression. It is also possible that translational factors such as Pet309p and Mss51p could have more than one function (i.e. mRNA metabolism, membrane insertion of the newly synthesized protein), only some of which are conserved across different organisms. We expect that understanding the nature and players involved in the mechanism regulating COX subunit 1 biogenesis and assembly in yeast will help to identify their functional homologues in humans.
2.2 Formation of assembly intermediates
COX assembly was modeled a decade ago by Nitjmans and colleagues as a linear process consisting of the sequential incorporation of COX subunits to an initial seed formed by subunit 1. Data supporting COX subunit 1 biogenesis as the first step of COX assembly came from analyses by Blue-native electrophoresis of human cells treated with cycloheximide, a specific inhibitor of cytoplasmic translation. They showed the existence of Cox1p as either unassembled or in subassemblies (31). The same COX assembly intermediates were observed when the cells were treated for short times with doxycycline, a specific inhibitor of mitochondrial translation, which reduces the level of COX subunits available for assembly (31). In these experiments, intermediates containing the other mitochondrial DNA encoded core subunits 2 and 3, but not COX1, were not detected (31). These results suggested that COX1 alone constitutes the first assembly intermediate.
Subunit 1-containing intermediates were also subsequently detected in COX deficient human and mouse cells (29, 32-34) and in yeast cells carrying cox2 mutations (35), suggesting that COX assembly is largely conserved from lower to higher eukaryotes.
According to the model proposed by Nitjmans and coworkers, the addition of subunits COX4 and COX5a (yeast Cox5ap and Cox6p, respectively) to the first assembly intermediate (S1) formed by COX1, results in the progression to the second assembly intermediate (S2). These nuclear DNA-encoded subunits are in direct contact with the core of the mature enzyme. COX5a caps the matrix side of COX1, while COX4 interacts with COX2 through its C-terminal domain and COX1 through its single transmembrane domain (4). Interestingly, three assembly intermediates containing COX1 but not of the other COX subunits were detected in SURF1 and SCO2 as well as in COX10 mutant fibroblasts from human patients (33, 34). These COX1 containing intermediates could correspond to COX1 maturation intermediates. (Fig.2). Insertion of heme A into COX1 occurs before the addition of subunits 4 and 5a (yeast subunits 5 and 6), as suggested by the accumulation of the COX1-COX4-COX5a intermediate in SCO1 and SCO2 mutant fibroblasts (33, 34), but not in COX10 and COX15 deficient cells (33, 36, 37). These findings suggest that the presence of heme A in COX1 might stabilize its binding to COX4 and COX5a. In addition, the heme a and heme a3 – CuB catalytic centers in subunit 1 are buried into the transmembrane portion of the protein. The binuclear center in subunit 1 is coordinated by histidine residues in helices VI, VII and X while heme a is coordinated by histidine residues in helices II and X (4). Because the helices involved must be adjacent to coordinate heme, it is plausible that heme a insertion could play a role in COX subunit 1 folding. Although the precise moment of subunit 1 biogenesis when the metal prosthetic groups are inserted into the complex remains to be determined, this clearly seems to be an early event in COX assembly and probably occurs in either a co-translational or a co-membrane insertion manner. After the formation of the COX1-COX4-COX5a subassembly, the COX assembly process continues with the formation of the third proposed intermediate (S3) by the addition to S2 of most of the remaining subunits with the exception being COX6a and COX7a/b (yeast subunits 10 and 7) that are finally added to complete the holoenzyme (31, 33).
Recently, Stiburek and coworkers used COX deficient mitochondria from different tissues of patients with mutations in the COX assembly factors SCO2 and SURF1 to identify eight different COX assembly intermediates/unassembled subunits, six of which were detectable also in wild type cells (34). The analysis of these intermediates provided novel information to refine the COX assembly model previously proposed. First, it appears that mammalian COX4 and COX5a subunits form a dimer before their incorporation into S1. Noticeably, the equivalent yeast Cox5p-Cox6p dimer was also detected in a COX mutant in which assembly is compromised in the latter stages of the process (38). This result is consistent with the observation that in yeast the presence of subunit 6 is required for subunit 5 stability (39). Second, an additional intermediate seems to exist between S2 and S3, consisting of at least mammalian subunits COX1-COX2-COX4-COX5a (34). It remained unanswered whether COX3 is also present in this intermediate because an antibody against subunit 3 was not used in this study. In a different report, however, COX3 was not detected in the S2 intermediate in human fibroblasts in which copper metallation of COX2 is impaired (33). In yeast, an assembly intermediate formed by Cox1p-Cox3p-Cox5ap-Cox6p, and not Cox4p, Cox6ap, Cox7p, Cox7ap and Cox8p, was detected in two cox2 point mutant strains (35), suggesting that Cox2p is not necessary for the incorporation of Cox3p in the subassembly intermediate in yeast.
COX2 may associate with COX1-COX4-COX5a intermediate upon its copper metallation by the specific SCO1-SCO2 copper chaperones, as suggested by the observation that SCO1 and SCO2 mutant fibroblasts from patients with COX deficiency accumulate the COX1-COX4-COX5a intermediate (33, 34). Significant experimental evidence suggests a possible involvement of the mammalian COX assembly factor SURF1, the homologue of the yeast SHY1, in facilitating the COX1-COX4-COX5a interaction. In both organisms, human and yeast, SURF1/shy1 mutant cells retain some residual COX activity of approximately 15% of wild-type cells, indicating that, though strongly compromised, the COX assembly process is still maintained in these cells. The precise function of SURF1/Shy1p in COX assembly is currently unknown. However, a large body of information has been gathered by studying spontaneous suppressor mutations and high copy suppressors of yeast shy1 mutants, which have indicated that SURF1/Shy1p plays a role in the formation of an early COX assembly intermediate containing subunit 1. As mentioned above, yeast strains carrying mutated and null alleles of shy1 spontaneously revert to a respiratory competent phenotype by mutations in MSS51 which increase the rate of Cox1p synthesis (13). The Mss51p mediated suppression is enhanced by both, co-overexpression of COX10 (27) and co-overexpression of the COX subunits Cox5p and Cox6p, the latter being partners of Cox1p in an early assembly intermediate mentioned above (22). In this context, we have recently reported that overexpression of HAP4 suppresses the respiratory deficient phenotype of yeast shy1 mutant strains. Hap4p is the catalytic subunit of the CCAAT binding site transcriptional activator Hap2p,3p,4p,5p complex, which globally activates transcription of nuclear genes involved in mitochondrial respiration during transition from fermentation to respiration. The shy1 suppression by HAP4 is mediated by a specific increase in the expression of subunits Cox5p and Cox6p (22). Interestingly, overexpression of NF-YA, the catalytic subunit of the human NF-Y transcriptional activator complex homologue of the yeast HAP complex, is able to increase the mitochondrial COX in SURF1 deficient fibroblasts through a mechanism that remains to be characterized (22).
The presence of SURF1 orthologues in terminal oxidase operons of several prokaryotes (40) in which COX contains the evolutionary conserved core subunits (Cox1p, Cox2p, Cox3p) but not the core surrounding subunits such as Cox5p and Cox6p, suggests a role of SURF1/Shy1p in forming the catalytic core of the enzyme. Studies in Rhodobacter sphaeroides, suggest that bacterial SURF1 is required for either insertion of heme A at the a3 center or stabilization of the a3-CuB binuclear center in COX1 (21). A function of SURF1/Shy1p in maturation of the a3 center would imply the existence of a second chaperone specific for the heme A of the cytochrome a center. Smith and coworkers proposed that SURF1 could perform this role indirectly by interacting with subunits 1 or 2 to facilitate the formation of the heme a3-CuB center. This would be in agreement with a previous observation suggesting an interaction of SURF1 with COX2 in human cultured cells (41). No interaction of yeast Shy1p was detected with either newly synthesized Cox1p or Cox2p (14). Perhaps such an interaction could occur only after the Cox1p-Cox5p-Cox6p subassembly is formed and Cox2p has been matured by insertion of copper at the CuA site. This possibility is strongly supported by the fact that the COX1-COX4-COX5a (yeast Cox1p-Cox5p-Cox6p) sub-complex accumulates in fibroblasts from SURF1 patients (33) as in fibroblasts from SCO1 and SCO2 patients (33, 34) but not in heme A deficient fibroblasts of patients with lesions in COX10 or COX15 (33) (37). SCO1 and SCO2 are required for the formation of CuA (42) and hence maturation of subunit 2. In turn, the association of metallated COX2 with COX1-COX4-COX5a could affect the stability of the CuB-hem a3 center by capping the heme-insertion channel proposed to be formed in the COX1-COX4-COX5a subassembly (43). As mentioned above, SURF1/Shy1p could play a role in facilitating this interaction. The results showing suppression of yeast shy1 mutants by increasing the amount of Cox1p-Cox5p-Cox6p (22) support this possibility, although a direct role of Shy1p on Cox1p maturation can not be excluded. Further support of the role of Shy1p as an assembly factor was recently reported by showing that Shy1p promotes COX biogenesis through association with different protein modules, potential COX assembly intermediates containing Cox1p and Cox5p among other proteins (26). In addition, Shy1p, together with Cox14p, were also found to interact with partially and fully assembled forms of COX associated with complex III of the mitochondrial respiratory chain. Based on the analysis of several high molecular weight complexes containing Shy1p and Cox14p, it has been proposed that these two COX assembly factors could act beyond the early stages of COX assembly. They could act in the late stages of the process when the COX subunits necessary for the interactions with complex III have been already incorporated (26). Shy1p and Cox14p would accompany partially assembled forms of COX, maintaining their competency for the incorporation of additional subunits, during the process of COX biogenesis (26).
Although most studies on COX assembly focus on the early steps leading to the assembly of the catalytic core subunits, some data are starting to emerge concerning the late steps of assembly in which addition of nuclear encoded subunits lead to complete the formation of the functional holoenzyme. The roles of Shy1p and Cox14p seem to affect these late steps as explained above. In addition, a subassembly intermediate formed by Cox7p-Cox7ap was detected in yeast cox2 mutants (35). Similarly, a high molecular weight complex containing Cox7p-Cox7ap-Cox8p in association with the chaperone Pet100p was observed in yeast wild-type cells (38). However, the complete composition of this complex still remains to be elucidated. The finding of subassembly complexes exclusively containing nuclear DNA-encoded subunits (35, 38) can explain their stability in comparison with the mitochondrial DNA-encoded subunits, which are quickly degraded when the assembly is compromised. This observation also supports the possibility that some nuclear encoded subunits exist in an unassembled pool in the inner mitochondrial membrane ready to enter the assembly process when required.
In summary, COX assembly is a progressive process in which the holoenzyme is built around a seed formed by COX subunit 1, which biogenesis, as part of the formation of the catalytic core, constitutes the principal regulatory step as depicted in Fig. 2.
Cox Assembly Regulation by the Integrity of Cox Functional Partners in the Mitochondrial Membranes
COX assembly has been shown to be sensitive to the protein composition of the mitochondrial membranes. As discussed below, the presence of both the electron carrier cytochrome c and the F1F0-ATPase, are essential for proper COX assembly and stability. Although this double requirement has been observed and studied by several research groups, the mechanisms involved remain to be fully understood.
3.1. Cytochrome c is required for COX assembly
In S. cerevisiae, cytochrome c occurs in two isoforms (iso-1 and iso-2) encoded by nuclear genes CYC1 and CYC7 (44, 45), respectively. CYC1 expression is regulated by oxygen and accounts for 95% of cytochrome c present in mitochondria in normoxic conditions (46). The 5% of iso-2-cytochrome c expressed in null mutants of CYC1 is enough to support respiratory growth albeit showing a reduced rate compared to wild type cells (47). Interestingly, the double null mutant Δcyc1Δcyc7 strain does not only present a complete absence of cytochrome c and as a consequence is unable to respire, but also shows an additional deficiency in COX (48). This phenotype is also observed in mutants of CYC3, a gene encoding for the cytochrome c heme lyase required for the covalent attachment of heme to apocytochrome c (49). Cytochrome c mutants have the characteristics of bona fide COX assembly mutants, including lower levels of heme aa3 and a drastic reduction in the steady state levels of COX mitochondrial subunits Cox1p, Cox2p and Cox3p. In COX assembly mutants, the incorporation of these subunits into the complex is aborted, and the turnover for their degradation increases. In addition, like in other COX mutants, failure of COX assembly in cytochrome c mutants results in down regulation of Cox1p synthesis (50) through the same mechanism involving Mss51p and Cox14p (14) as described above.
The role of cytochrome c in COX assembly is not well understood. Oxidized cytochrome c can accept electrons from cytochrome c1 of the bc1 complex, from cytochrome b2 of lactate dehydrogenase and also from the sulfhydryl oxidase Erv1p (51, 52). The main function of reduced cytochrome c is to donate electrons to COX. Conceivably, cytochrome c could also promote an oxidation or reduction event essential for COX assembly. For example, cytochrome c could be involved in heme A biosynthesis, a possibility that has been already excluded (53). The recent discovery of an interaction of cytochrome c with Erv1p could suggest an interesting possibility involving the import of COX assembly chaperons. The import and folding into the mitochondrial intermembrane space of several copper chaperons required for COX assembly is mediated through the recently described Mia40p pathway (54). These proteins contain a characteristic twin CX9C motif, as in Cox17p, Cox19p and Cox23p, critical for their import. After passage through the mitochondrial outer membrane TOM channel, these proteins are covalently trapped by Mia40p via disulfide bridges. Mia40p also contains cysteine residues, which are oxidized by the Erv1p, functioning as a disulfide relay system that catalyzes the import of proteins into the IMS by an oxidative folding mechanism (54). How Erv1p itself is oxidized to become competent for new rounds of Mia40p oxidation was initially unclear. Recently, it has been reported a connection between the disulfide relay import system and the mitochondrial respiratory chain consisting of electron transfer from Erv1p to molecular oxygen via interaction with cytochrome c (51, 52). Cytochrome c efficiently oxidizes Mia40p in oxygen-limiting conditions (52). Although oxidized cytochrome c facilitates Mia40p oxidation, it was found non-essential in normoxic conditions suggesting the existence of additional electron acceptors for oxidizing reduced Erv1p (52). These results weaken the possibility that cytochrome c prevents COX assembly by blocking the import of its copper chaperones.
Cytochrome c could be required for COX assembly in a structural capacity. For example, its interaction with a COX intermediate could be necessary for some step in the assembly pathway. To distinguish between a functional and structural requirement of cytochrome c in COX assembly, Barrientos and colleagues transformed a Δcyc1Δcyc7 double null mutant with a cyc1 mutant gene that was previously reported to express stable but catalytically inactive cytochrome c (50). The mutant allele, cyc1–166, expresses nonfunctional and thermolabile iso-1-cytochrome c and has a serine replacement of the tryptophan at position 65 (W65S), which corresponds to the invariant tryptophan residue found in cytochromes c from all eukaryotic species (55). Analyses of the transformant strains showed a recovery of the COX assembly defect of the double null mutant. The heme a levels of the transformant strain were increased to about 50% of wild type, and the steady state levels of Cox1p, Cox2p and Cox3p were close to normal (50). These results support the hypothesis of cytochrome c playing a structural role in COX assembly because the sole presence of its inactive form was able to rescue the respiratory deficient phenotype of the double null mutant. Interestingly, COX assembly does not seem to depend on stoichiometric concentration of cytochrome c. This possibility is supported by the fact that although the molar concentration of cytochrome c in mitochondria of wild type yeast was estimated to be approximately the same as that of COX, iso-2-cytochrome c, which concentration in a cyc1 mutant is only 12% of the total amount of cytochrome c in wild type yeast, it is able to support the expression of 70% of normal amounts of COX (50). Furthermore, the apo-cytochrome c (heme less protein) cannot substitute for the mature cytochrome, suggesting that the function of cytochrome c probably depends on a properly folded protein (50). In addition to its requirement for assembly, the results obtained with the thermolabile W65S mutant exposed to 37 °C indicated that the main role of cytochrome c is in assembly but that it also contributes toward the stability of the enzyme (50). Although the current literature is consistent with a structural role for cytochrome c in COX assembly, further studies are required to fully understand the mechanism involved.
3.2. COX assembly and stability requires an intact functional ATP synthase
Most of cellular ATP of eukaryote cells is synthesized by the F1F0-ATPase complex of the mitochondrial inner membrane during oxidative phosphorylation. Mitochondrial F1F0-ATP synthase or ATPase is formed by 15–18 distinct subunits. It is composed of two functionally and physically coupled portions, the membrane-embedded F0 sector to which the hydrophilic F1 sector is attached from the matrix side. In S. cerevisiae, the F0 sector is formed by eight subunits (reviewed in (56, 57)). An oligomer of Atp9p forms a ring-like structure that rotates in the membrane bilayer, and together with Atp6p, drives proton translocation across the inner membrane. Six other subunits (Atp8p, Atp4p, the oligomycin sensitivity-conferring protein and subunits d, f, and h) form the stator arm connecting F0 to F1. Two other subunits (e and g) are required for ATPase dimerization. The γ, δ and ε subunits of F1 are part of a central stalk linking it to the Atp9p ring. The rotation of this stalk within the static catalytic F1 hexamer formed by three α and three β subunits is required for ATP synthesis. The remaining seven F1F0-ATPase subunits function in the regulation or oligomerization of the yeast complex (57).
Although the mitochondrial ATPase is functionally coupled to the mitochondrial respiratory chain (MRC) activity through oxidative phosphorylation, no physical interaction of ATPase with MRC enzymes has been reported to date. In tightly coupled mitochondria, the oxygen consumption rate and ATP synthesis activity depend on each other, which results in decreased respiratory activity in many yeast ATPase defective mutants. Short regulatory responses involving COX have been described both in yeast and mammals, including for example allosteric ATP inhibition of COX activity at high ATP/ADP ratios as mentioned above (reviewed in (58)). However, data accumulated over the last twenty years in different organisms suggest the existence of long term regulations in which COX biogenesis and stability are modulated in response to the ATPase activity.
Mutations in several ATPase structural genes in S. cerevisiae, including ATP4 (59), ATP7 (60), ATP8 (61) and ATP9 (62) have been reported to severely affect ATPase assembly and/or function and to produce a pleiotropic effect on COX biogenesis. However, ATPase mutations affect mtDNA stability producing a large amount of “petite” (respiratory deficient) cells, making it difficult to conveniently assess to which degree and how COX assembly is specifically affected in these mutants. In some cases, it was suggested that the COX biogenesis defect was the result of a decrease in Cox1p synthesis (59, 60, 63), while in others protein synthesis was normal and the COX defect was suggested to be caused by a block in heme insertion or synthesis (61). Similarly, ATPase mutants of Schizosaccharomyces pombe, with altered α or β subunits of the F1 portion, also exhibit pleiotropy on COX biogenesis, associated with a reduction on synthesis of the mitochondrial DNA encoded protein subunits of the complex (64). Recently, the role of ATPase on COX assembly has been explored in a S. cerevisiae strain engineered to stably maintain its mtDNA under selection even in the absence of ATPase (63). The authors took advantage of an approach developed by Fox and coworkers (65), based on the insertion of the non-respiratory nuclear gene ARG8 into the mtDNA of a strain carrying a null allele of arg8. In this system, cells growing in the absence of arginine receive selective pressure to maintain their mtDNA. Analysis of the phenotype resulting from an atp6 deletion in this context clearly showed that the absence of this subunit, the last to be incorporated into the ATPase during its assembly, did not compromise the assembly of the rest of the complex (63). As expected, the ATPase activity in this mutant decreased by ∼30% of the wild type, indicating that the F1 portion was expressed and functional in the absence of Atp6p, but was not inhibited by oligomycin, a specific inhibitor of the F1F0 complex proton channel. The lack of Atp6p resulted in a selective decrease in COX accumulation to residual amounts barely detectable. The COX accumulation defect was associated to a decreased synthesis of Cox1p, which probably resulted from a down-regulation of Cox1p translation in the absence of COX assembly through a mechanism involving Mss51p and Cox14p, as previously described for classical COX mutants, as well as for cytochrome c mutants, as mentioned above. We have explored these issues in S. cerevisiae strains developed by our group in which Atp6p assembly into the ATPase complex is prevented by a mutation in its specific chaperone Atp10p. In atp10 mutants, the Cox1p synthesis defect was restored by either deleting COX14 or by integrating a second copy of MSS51 into the nuclear genome (unpublished results), two of the hallmarks of the Cox1p translational regulatory loop previously described (14). Thus, Cox1p synthesis down-regulation is in this context a consequence rather than a cause of COX assembly defects.
The mechanism by which the F1F0-ATPase affects COX biogenesis remains unknown. Rak and colleagues have proposed that the very poor COX synthesis is a vital requirement allowing ATPase mutants to import glycolytic ATP into mitochondria by decreasing the membrane potential. This hypothesis is supported by the fact that COX biogenesis is not altered in uncoupled ATP synthase mutants, missing either subunits δ (66) or ε (67), where maintenance of a membrane potential is severely compromised by significant proton leaks through the F0. While this is a very interesting hypothesis, it remains to be elucidated the COX biogenesis step/s that would be primarily affected by an accumulation of protons in the intermembrane space, e.g. atp6 or atp10 mutants, provoking a failure to assemble COX.
Recently, several research groups have explored structural constraints to explain the COX deficiency observed in ATPase mutants. In addition to its well established function on ATP generation, the F1F0-ATP synthase has a role in determining the ultra-structure of mitochondria (68-70), which depends on its ability to form dimeric and higher oligomeric super-complexes (68-71). It has been recently reported that mutants of the dimer-specific subunits e and g, which destabilize dimeric and oligomeric F1F0-ATP synthase super complexes, have a decreased mitochondrial membrane potential. The enzymatic activities of F1F0-ATPase as well as the tubular mitochondrial morphology were not affected by the absence of subunits e and g. In these mutants, COX activity was affected to a minor extent (retain ∼80% of wild type COX activity), indicating that ATPase dimerization and oligomerization are not essential for COX assembly (72). To explain the loss of membrane potential, the authors proposed a role for the super-complexes of the F1F0-ATPase in organizing microdomains within the inner membrane, ensuring optimal bioenergetic competence of mitochondria, which when disrupted would affect the overall flux through the respiratory chain and result in a lower membrane potential (72). By contrast, in a more recent study, the mitochondrial membrane potential in yeast mutant lacking either subunit g or e was not found compromised (73). In this study, it was shown that COX activity is actually decreased down to 55% and 75% of wild type levels in strains missing subunits e and g, respectively, while the amount of enzyme appears normal (73). However, the presence of ATPase subunits e and g was reported necessary to maintain the correct organizational state of the super-complex formed by complexes bc1 and COX, which stoichiometry was found altered in the mutant strains (73).
Studies in S. cerevisiae mutants that are unable to form the F1 α3β3 oligomer, either because the α or the β subunit is missing or because the cells are deficient for proteins that mediate F1 assembly (e.g. Atp11p, Atp12p, or Fmc1p), have pleiotropic effects on several MRC enzymes (74). In these mutants, mitochondria were severely deficient not only for COX but also, at a lower extent, for Complex III or bc1 complex (74). These mutants were devoid of mitochondrial cristae, supporting the idea that the F1F0-ATPase is important for biogenesis of the mitochondrial inner membrane, but it remained unclear whether the MRC deficiency resulted from the altered mitochondrial ultra-structure. Finally, the mutants accumulated inclusion bodies containing unassembled F1 α and/or β subunits, an aspect that was considered necessary to prevent the formation of incomplete F1F0 assemblies that could passively transport protons across the inner mitochondrial membrane, thus preventing a total dissipation of the mitochondrial membrane potential, which could be catastrophic for the cell (74). However, the maintenance of a too high membrane potential could also be deleterious by limiting glycolytic ATP import as mentioned above, and the cells would react by blocking the assembly of respiratory enzymes, most significantly, COX.
Although the mechanism underlying the COX biogenesis defect in ATPase mutants is not fully understood, it seems to be conserved from yeast to higher eukaryotes. In human, mtDNA and nDNA mutations affecting ATPase function result in rare devastating encephalomyopathies. In several cases reported, the ATPase deficiency was accompanied by a pleiotropic decrease in COX activity specifically or as part of a more general MRC alteration. NARP (neuropathy, ataxia and retinitis pigmentosa) and MILS (maternally inherited Leigh's syndrome) are mitochondrial disorders associated with mutations in MTATP6. For example, a T9,185C mutation in the mitochondrial gene ATP6 was reported to produce MILS (75). Biochemical analyses in the patients showed that they presented a significant COX activity decline in skeletal muscle (75). Analysis of the NARP mutation T8,993G in cybrids cell lines also showed a MRC deficiency, including a COX defect, which interestingly was alleviated by supplementing the growth media with antioxidants (76). The antioxidant treatment was justified because in these cell lines there was an increase of reactive oxygen species that could be responsible for the MRC damage (76). However, the possibility remained that the antioxidants could act by directly introducing reducing equivalents into the mitochondrial matrix, thereby reducing its pH, which would affect the regulation of mitochondrial respiratory chain enzymes (76). Although most ATPase deficiencies have been attributed to mutations in MTATP6, it has been recently reported a case of ATPase deficiency in the human homologue of the nDNA encoded assembly gene ATP12. As in yeast mutants of the homologue gene (74), the patient presented a pleiotropic COX defect in skeletal muscle (77).
4. Concluding Remarks
Biogenesis of a functional cytochrome c oxidase complex depends on the expression of all the structural and more than two dozen COX-specific assembly genes. The latter impinges on all aspects of the biogenesis process. The assembly of COX, made of subunits of dual genetic origin is a complicated and highly regulated process. The multiple levels of regulation described to date involve the availability of subunits and assembly factors regulated at the transcriptional and translational levels, availability of co-factors, protein import into mitochondria and membrane insertion, and coordination of sequential or simultaneous steps of the process (reviewed in (8)).
New levels of regulation have been recently reported and were reviewed here: 1- Translation of subunit 1 in the yeast S. cerevisiae is contingent with the availability of its assembly partners, thereby acting as a negative feedback loop that paces Cox1p translation to its utilization during assembly of the complex. In the case of a membrane complex such as COX, this form of negative regulation may prevent non-productive aggregation of unassembled hydrophobic membrane proteins by restricting their steady-state concentration. This mechanism could also play a crucial role in facilitating multimeric protein sequential assembly. 2- The arrangement of the mitochondrial chain and oxidative phosphorylation system in the inner mitochondrial membrane plays a crucial role on the stability and assembly of COX. The physical presence of cytochrome c is essential for COX assembly. A fully assembled and functional ATP synthase is also required for maximal levels of COX biogenesis. The discovery of the exact mechanisms involved are expected to shed light into new levels of mitochondrial membrane organization depending on physical and functional constraints.
Finally, in addition of the biological and biochemical interest, the full understanding of the regulation behind COX assembly will also enhance our understanding of aerobic energy production regulation, a process essential for cell performance and survival in normal and disease conditions.
Acknowledgments
We thank Darryl Horn and Rajiv Singh for critically reading the manuscript. Our research is supported by National Institutes of Health Research Grant GM071775A (to A.B.) and a Research Grant from the Muscular Dystrophy Association (to A.B.). F.F. is supported by Telethon-Italy, Fellowship no GFP05008.
References
- 1.Ludwig B, Bender E, Arnold S, Huttemann M, Lee I, Kadenbach B. Cytochrome C oxidase and the regulation of oxidative phosphorylation. Chembiochem. 2001;2:392–403. doi: 10.1002/1439-7633(20010601)2:6<392::AID-CBIC392>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 2.Kadenbach B, Stroh A, Becker A, Eckerskorn C, Lottspeich F. Tissue- and species-specific expression of cytochrome c oxidase isozymes in vertebrates. Biochim Biophys Acta. 1990;1015:368–72. doi: 10.1016/0005-2728(90)90042-3. [DOI] [PubMed] [Google Scholar]
- 3.Linder D, Freund R, Kadenbach B. Species-specific expression of cytochrome c oxidase isozymes. Comp Biochem Physiol B Biochem Mol Biol. 1995;112:461–9. doi: 10.1016/0305-0491(95)00093-3. [DOI] [PubMed] [Google Scholar]
- 4.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–44. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 5.Belevich I, Verkhovsky MI. Molecular mechanism of proton translocation by cytochrome c oxidase. Antioxid Redox Signal. 2008;10:1–30. doi: 10.1089/ars.2007.1705. [DOI] [PubMed] [Google Scholar]
- 6.Solans A, Zambrano A, Barrientos A. Cytochrome c oxidase deficiency: from yeast to human. Preclinica. 2004;2:336–348. [Google Scholar]
- 7.Pecina P, Houstkova H, Hansikova H, Zeman J, Houstek J. Genetic defects of cytochrome c oxidase assembly. Physiol Res. 2004;53:S213–23. [PubMed] [Google Scholar]
- 8.Fontanesi F, Soto IC, Horn D, Barrientos A. Assembly of mitochondrial cytochrome c oxidase, a complicated and highly regulated cellular process. Am J Physiol Cell Physiol. 2006;291:C1129–47. doi: 10.1152/ajpcell.00233.2006. Epub 2006 Jun 7. [DOI] [PubMed] [Google Scholar]
- 9.Nakai T, Mera Y, Yasuhara T, Ohashi A. Divalent metal ion-dependent mitochondrial degradation of unassembled subunits 2 and 3 of cytochrome c oxidase. J Biochem. 1994;116:752–8. doi: 10.1093/oxfordjournals.jbchem.a124592. [DOI] [PubMed] [Google Scholar]
- 10.Nijtmans LG, Spelbrink JN, Van Galen MJ, Zwaan M, Klement P, Van den Bogert C. Expression and fate of the nuclearly encoded subunits of cytochrome-c oxidase in cultured human cells depleted of mitochondrial gene products. Biochim Biophys Acta. 1995;1265:117–26. doi: 10.1016/0167-4889(94)00203-q. [DOI] [PubMed] [Google Scholar]
- 11.Arnold I, Langer T. Membrane protein degradation by AAA proteases in mitochondria. Biochim Biophys Acta. 2002;1592:89–96. doi: 10.1016/s0167-4889(02)00267-7. [DOI] [PubMed] [Google Scholar]
- 12.Khalimonchuk O, Bird A, Winge DR. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J Biol Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- 13.Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–82. doi: 10.1038/sj.emboj.7600358. Epub 2004 Aug 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–61. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zambrano A, Fontanesi F, Solans A, de Oliveira RL, Fox TD, Tzagoloff A, Barrientos A. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:523–35. doi: 10.1091/mbc.E06-09-0803. Epub 2006 Nov 2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poutre CG, Fox TD. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–47. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calder KM, McEwen JE. Deletion of the COX7 gene in Saccharomyces cerevisiae reveals a role for cytochrome c oxidase subunit VII in assembly of remaining subunits. Mol Microbiol. 1991;5:1769–77. doi: 10.1111/j.1365-2958.1991.tb01926.x. [DOI] [PubMed] [Google Scholar]
- 19.Decoster E, Simon M, Hatat D, Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol Gen Genet. 1990;224:111–8. doi: 10.1007/BF00259457. [DOI] [PubMed] [Google Scholar]
- 20.Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–43. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith D, Gray J, Mitchell L, Antholine WE, Hosler JP. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J Biol Chem. 2005;280:17652–6. doi: 10.1074/jbc.C500061200. Epub 2005 Mar 11. [DOI] [PubMed] [Google Scholar]
- 22.Fontanesi F, Jin C, Tzagoloff A, Barrientos A. Transcriptional Activators HAP/NF-Y Rescue a Cytochrome c Oxidase Defect in Yeast and Human Cells. Hum Mol Genet. 2007 Nov 27; doi: 10.1093/hmg/ddm349. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet. 1998;20:337–43. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 24.Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am J Hum Genet. 1998;63:1609–21. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glerum DM, Koerner TJ, Tzagoloff A. Cloning and characterization of COX14, whose product is required for assembly of yeast cytochrome oxidase. J Biol Chem. 1995;270:15585–90. doi: 10.1074/jbc.270.26.15585. [DOI] [PubMed] [Google Scholar]
- 26.Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 2007;26:4347–58. doi: 10.1038/sj.emboj.7601862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzagoloff A, Nobrega M, Gorman N, Sinclair P. On the functions of the yeast COX10 and COX11 gene products. Biochem Mol Biol Int. 1993;31:593–8. [PubMed] [Google Scholar]
- 29.Diaz F, Thomas CK, Garcia S, Hernandez D, Moraes CT. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum Mol Genet. 2005;14:2737–48. doi: 10.1093/hmg/ddi307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A. 2003;100:605–10. doi: 10.1073/pnas.242716699. Epub 2003 Jan 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nijtmans LG, Taanman JW, Muijsers AO, Speijer D, Van den Bogert C. Assembly of cytochrome-c oxidase in cultured human cells. Eur J Biochem. 1998;254:389–94. doi: 10.1046/j.1432-1327.1998.2540389.x. [DOI] [PubMed] [Google Scholar]
- 32.Tiranti V, Galimberti C, Nijtmans L, Bovolenta S, Perini MP, Zeviani M. Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum Mol Genet. 1999;8:2533–40. doi: 10.1093/hmg/8.13.2533. [DOI] [PubMed] [Google Scholar]
- 33.Williams SL, Valnot I, Rustin P, Taanman JW. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J Biol Chem. 2004;279:7462–9. doi: 10.1074/jbc.M309232200. Epub 2003 Nov 7. [DOI] [PubMed] [Google Scholar]
- 34.Stiburek L, Vesela K, Hansikova H, Pecina P, Tesarova M, Cerna L, Houstek J, Zeman J. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem J. 2005;392:625–32. doi: 10.1042/BJ20050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horan S, Bourges I, Taanman JW, Meunier B. Analysis of COX2 mutants reveals cytochrome oxidase subassemblies in yeast. Biochem J. 2005;390:703–8. doi: 10.1042/BJ20050598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antonicka H, Leary SC, Guercin GH, Agar JN, Horvath R, Kennaway NG, Harding CO, Jaksch M, Shoubridge EA. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum Mol Genet. 2003;12:2693–702. doi: 10.1093/hmg/ddg284. Epub 2003 Aug 19. [DOI] [PubMed] [Google Scholar]
- 37.Antonicka H, Mattman A, Carlson CG, Glerum DM, Hoffbuhr KC, Leary SC, Kennaway NG, Shoubridge EA. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am J Hum Genet. 2003;72:101–14. doi: 10.1086/345489. Epub 2002 Dec 09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Church C, Goehring B, Forsha D, Wazny P, Poyton RO. A role for Pet100p in the assembly of yeast cytochrome c oxidase: interaction with a subassembly that accumulates in a pet100 mutant. J Biol Chem. 2005;280:1854–63. doi: 10.1074/jbc.M410726200. Epub 2004 Oct 26. [DOI] [PubMed] [Google Scholar]
- 39.Glerum DM, Tzagoloff A. Submitochondrial distributions and stabilities of subunits 4, 5, and 6 of yeast cytochrome oxidase in assembly defective mutants. FEBS Lett. 1997;412:410–4. doi: 10.1016/s0014-5793(97)00799-0. [DOI] [PubMed] [Google Scholar]
- 40.Poyau A, Buchet K, Godinot C. Sequence conservation from human to prokaryotes of Surf1, a protein involved in cytochrome c oxidase assembly, deficient in Leigh syndrome. FEBS Lett. 1999;462:416–20. doi: 10.1016/s0014-5793(99)01571-9. [DOI] [PubMed] [Google Scholar]
- 41.Nijtmans LG, Artal Sanz M, Bucko M, Farhoud MH, Feenstra M, Hakkaart GA, Zeviani M, Grivell LA. Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 2001;498:46–51. doi: 10.1016/s0014-5793(01)02447-4. [DOI] [PubMed] [Google Scholar]
- 42.Leary SC, Kaufman BA, Pellecchia G, Guercin GH, Mattman A, Jaksch M, Shoubridge EA. Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum Mol Genet. 2004;13:1839–48. doi: 10.1093/hmg/ddh197. Epub 2004 Jun 30. [DOI] [PubMed] [Google Scholar]
- 43.Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim Biophys Acta. 2006;1763:759–72. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Sherman F, Taber H, Campbell W. Genetic determination of iso-cytochromes c in yeast. J Mol Biol. 1965;13:21–39. doi: 10.1016/s0022-2836(65)80077-8. [DOI] [PubMed] [Google Scholar]
- 45.Downie JA, Stewart JW, Brockman N, Schweingruber AM, Sherman F. Structural gene for yeast iso-2-cytochrome c. J Mol Biol. 1977;113:369–84. doi: 10.1016/0022-2836(77)90147-4. [DOI] [PubMed] [Google Scholar]
- 46.Sherman F, Stewart JW, Margoliash E, Parker J, Campbell W. The structural gene for yeast cytochrome C. Proc Natl Acad Sci U S A. 1966;55:1498–504. doi: 10.1073/pnas.55.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman F, Stewart JW, Jackson M, Gilmore RA, Parker JH. Mutants of yeast defective in iso-1-cytochrome c. Genetics. 1974;77:255–84. doi: 10.1093/genetics/77.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearce DA, Sherman F. Degradation of cytochrome oxidase subunits in mutants of yeast lacking cytochrome c and suppression of the degradation by mutation of yme1. J Biol Chem. 1995;270:20879–82. doi: 10.1074/jbc.270.36.20879. [DOI] [PubMed] [Google Scholar]
- 49.Dumont ME, Ernst JF, Hampsey DM, Sherman F. Identification and sequence of the gene encoding cytochrome c heme lyase in the yeast Saccharomyces cerevisiae. EMBO J. 1987;6:235–41. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrientos A, Pierre D, Lee J, Tzagoloff A. Cytochrome oxidase assembly does not require catalytically active cytochrome C. J Biol Chem. 2003;278:8881–7. doi: 10.1074/jbc.M212427200. Epub 2003 Jan 08. [DOI] [PubMed] [Google Scholar]
- 51.Dabir DV, Leverich EP, Kim SK, Tsai FD, Hirasawa M, Knaff DB, Koehler CM. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–11. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;179:389–95. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barros MH, Tzagoloff A. Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 2002;516:119–23. doi: 10.1016/s0014-5793(02)02514-0. [DOI] [PubMed] [Google Scholar]
- 54.Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–69. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 55.Schweingruber ME, Stewart JW, Sherman F. Primary site and second site revertants of missense mutants of the evolutionarily invariant tryptophan 64 in iso-1-cytochrome c from yeast. J Biol Chem. 1979;254:4132–43. [PubMed] [Google Scholar]
- 56.Velours J, Arselin G. The Saccharomyces cerevisiae ATP synthase. J Bioenerg Biomembr. 2000;32:383–90. doi: 10.1023/a:1005580020547. [DOI] [PubMed] [Google Scholar]
- 57.Ackerman SH, Tzagoloff A. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog Nucleic Acid Res Mol Biol. 2005;80:95–133. doi: 10.1016/S0079-6603(05)80003-0. [DOI] [PubMed] [Google Scholar]
- 58.Kadenbach B. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim Biophys Acta. 2003;1604:77–94. doi: 10.1016/s0005-2728(03)00027-6. [DOI] [PubMed] [Google Scholar]
- 59.Paul MF, Velours J, Arselin de Chateaubodeau G, Aigle M, Guerin B. The role of subunit 4, a nuclear-encoded protein of the F0 sector of yeast mitochondrial ATP synthase, in the assembly of the whole complex. Eur J Biochem. 1989;185:163–71. doi: 10.1111/j.1432-1033.1989.tb15098.x. [DOI] [PubMed] [Google Scholar]
- 60.Spannagel C, Vaillier J, Arselin G, Graves PV, Velours J. The subunit f of mitochondrial yeast ATP synthase--characterization of the protein and disruption of the structural gene ATP17. Eur J Biochem. 1997;247:1111–7. doi: 10.1111/j.1432-1033.1997.01111.x. [DOI] [PubMed] [Google Scholar]
- 61.Marzuki S, Watkins LC, Choo WM. Mitochondrial H+-ATPase in mutants of Saccharomyces cerevisiae with defective subunit 8 of the enzyme complex. Biochim Biophys Acta. 1989;975:222–30. doi: 10.1016/s0005-2728(89)80252-x. [DOI] [PubMed] [Google Scholar]
- 62.Hadikusumo RG, Meltzer S, Choo WM, Jean-Francois MJ, Linnane AW, Marzuki S. The definition of mitochondrial H+ ATPase assembly defects in mit-mutants of Saccharomyces cerevisiae with a monoclonal antibody to the enzyme complex as an assembly probe. Biochim Biophys Acta. 1988;933:212–22. doi: 10.1016/0005-2728(88)90072-2. [DOI] [PubMed] [Google Scholar]
- 63.Rak M, Tetaud E, Godard F, Sagot I, Salin B, Duvezin-Caubet S, Slonimski PP, Rytka J, di Rago JP. Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J Biol Chem. 2007;282:10853–64. doi: 10.1074/jbc.M608692200. [DOI] [PubMed] [Google Scholar]
- 64.Boutry M, Goffeau A. Alterations of the alpha or beta subunits of the mitochondrial ATPase in yeast mutants. Eur J Biochem. 1982;125:471–7. doi: 10.1111/j.1432-1033.1982.tb06707.x. [DOI] [PubMed] [Google Scholar]
- 65.Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci U S A. 1996;93:5253–7. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duvezin-Caubet S, Caron M, Giraud MF, Velours J, di Rago JP. The two rotor components of yeast mitochondrial ATP synthase are mechanically coupled by subunit delta. Proc Natl Acad Sci U S A. 2003;100:13235–40. doi: 10.1073/pnas.2135169100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guelin E, Chevallier J, Rigoulet M, Guerin B, Velours J. ATP synthase of yeast mitochondria. Isolation and disruption of the ATP epsilon gene. J Biol Chem. 1993;268:161–7. [PubMed] [Google Scholar]
- 68.Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–30. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arselin G, Giraud MF, Dautant A, Vaillier J, Brethes D, Coulary-Salin B, Schaeffer J, Velours J. The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur J Biochem. 2003;270:1875–84. doi: 10.1046/j.1432-1033.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- 70.Giraud MF, Paumard P, Soubannier V, Vaillier J, Arselin G, Salin B, Schaeffer J, Brethes D, di Rago JP, Velours J. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim Biophys Acta. 2002;1555:174–80. doi: 10.1016/s0005-2728(02)00274-8. [DOI] [PubMed] [Google Scholar]
- 71.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schagger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–8. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bornhovd C, Vogel F, Neupert W, Reichert AS. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J Biol Chem. 2006;281:13990–8. doi: 10.1074/jbc.M512334200. [DOI] [PubMed] [Google Scholar]
- 73.Saddar S, Dienhart MK, Stuart RA. The F1Fo-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J Biol Chem. 2008 doi: 10.1074/jbc.M708440200. Jan 10, Epublished ahead of print. [DOI] [PubMed] [Google Scholar]
- 74.Lefebvre-Legendre L, Salin B, Schaeffer J, Brethes D, Dautant A, Ackerman SH, di Rago JP. Failure to assemble the alpha 3 beta 3 subcomplex of the ATP synthase leads to accumulation of the alpha and beta subunits within inclusion bodies and the loss of mitochondrial cristae in Saccharomyces cerevisiae. J Biol Chem. 2005;280:18386–92. doi: 10.1074/jbc.M410789200. [DOI] [PubMed] [Google Scholar]
- 75.Castagna AE, Addis J, McInnes RR, Clarke JT, Ashby P, Blaser S, Robinson BH. Late onset Leigh syndrome and ataxia due to a T to C mutation at bp 9,185 of mitochondrial DNA. Am J Med Genet A. 2007;143:808–16. doi: 10.1002/ajmg.a.31637. [DOI] [PubMed] [Google Scholar]
- 76.Mattiazzi M, Vijayvergiya C, Gajewski CD, DeVivo DC, Lenaz G, Wiedmann M, Manfredi G. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet. 2004;13:869–79. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 77.De Meirleir L, Seneca S, Lissens W, De Clercq I, Eyskens F, Gerlo E, Smet J, Van Coster R. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J Med Genet. 2004;41:120–4. doi: 10.1136/jmg.2003.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]